Abstract
The role of energy deregulation and altered/adapted metabolism in tumor cells is an increasing important issue in understanding cancer. Hereditary leiomyomatosis renal cell carcinoma (HLRCC) is an aggressive form of RCC characterized by germline mutation of fumarate hydratase (FH) followed by somatic loss of the remaining wild type allele, and known to be a highly metastatic and lethal malignancy compared to other RCCs. The intrinsic loss of normal tricarboxylic acid (TCA) cycle presumably aids tumorigenesis due to the necessary metabolic alterations required and the enforced dependence on glycolysis derived energy, mimicking the Warburg effect. Thus, there is considerable utility in establishing a preclinical cell model from these tumors to study energy metabolism deregulation, as well as, developing new targeted therapeutic approaches for TCA cycle enzyme-deficient cancers.
Here we describe a new immortalized cell line, UOK268, derived from a patient’s primary HLRCC-associated kidney cancer. This represents the first primary renal cell line to model TCA cycle gene loss and provides a perfect partner cell line to our previously described metastasis derived HLRCC-associated cell line, UOK262. We identified a novel germline FH missense mutation, p.His192Asp, and the subsequent loss of heterozygosity in UOK268. The UOK268 cell line expressed mutant FH protein, which localized to the mitochondria, but with loss of almost all catalytic activity. The UOK268 cells had severely compromised oxidative phosphorylation and increased glycolytic flux. Ingenuity® pathways analysis of hMitChip3 gene chip data confirmed the altered mRNA expression patterns of genes involved in several important pathways, such as lipid metabolism, apoptosis and energy production/glycolysis. UOK268 provides a unique model of a primary cell line demonstrating an enforced, irreversible Warburg effect and, combined with UOK262, provides a unique in vitro preclinical model for studying the bioenergetics of the Warburg effect in human cancer.
Keywords: Hereditary Leiomyomatosis Renal Cell Carcinoma (HLRCC), FH (Fumarate hydratase) gene, Warburg Effect, Human mitochondrial focused cDNA microarray (hMitChip3)
Introduction
Hereditary leiomyomatosis and renal cell carcinoma (HLRCC) is an inherited autosomal dominant cancer syndrome characterized by predisposition to the development of cutaneous and uterine leiomyomas and a very aggressive form of renal cancer [1;2]. HLRCC-associated renal tumors have distinctive architectural and morphologic features and are particularly aggressive and tend to metastasize early [3]. HLRCC renal tumors differ from other hereditary renal cancers in that they often present as solitary lesions that readily metastasize to regional and distant lymph nodes. The gene causing HLRCC encodes fumarate hydratase (FH), a tumor suppressor and one of the key metabolic enzymes of the mitochondrial tricarboxylic acid (TCA) cycle [4–6]. There has been significant improvement in the pathologic diagnostic accuracy for HLRCC kidney cancer based on its unique morphologic features including the presence of large eosinophilic nucleoli, with clear perinucleolar halos, in papillary pattern [7;8]. Mutations of genes in the tricarboxylic acid (TCA) cycle have been implicated in several aspects of cancer genetics and progression. Yet, the link between the mitochondrial dysfunction and tumorigenesis is still not fully clarified. Despite significant progress in identification of the genetic and molecular factors contributing to the malignant phenotype of HLRCC, profiling of the genetic and genomic status of mitochondrial gene/protein expression for studies of early stage of pre-metastatic HLRCCs are limited.
We have previously characterized the first human FH (−/−) HLRCC kidney cancer metastatic cell line UOK262 [9]. UOK262 was found to have severely compromised mitochondrial oxidative phosphorylation (OXPHOS), exhibited increased aerobic glycolysis, mitochondria dysfunction, high mobility of invasion and high basal reactive oxygen species (ROS level) and represented an example of the “Warburg effect” in human cancer. We have described that the glycolytic switch induced by the loss of FH activated the NADPH oxidase enzyme complex, which may cause DNA and protein damage, resulted in further impaired mitochondrial functions and pathological processes, including tumorigenesis and metastasis [10;11]. Recently, we demonstrated in both UOK268 and UOK262 that inactivation of TCA cycle enzymes resulted in a glycolytic shift and the suppression of the master metabolic regulator, AMPK. Aerobic glycolysis can confer growth advantages by diverting some of the glucose to generate NADPH, acetyl-CoA, and ribose, whereas reduced AMPK signaling activates anabolic factors involved in protein and fatty acid biosyntheses and suppresses the p53 tumor suppressor. Furthermore, reduced AMPK signaling repressed the DMT1 iron transporter, which contributed to elevation of HIF-1α, which promotes aerobic glycolysis in HLRCC [12].
Here we characterize the establishment of a novel HLRCC cell line, UOK268, originated from a patient’s surgically removed primary kidney tumor. UOK268 cells demonstrate compromised oxidative phosphorylation and in vitro dependence on glycolysis consistent with the clinical manifestation of HLRCC. We performed profiling of mitochondrial associated expressed transcripts using third-generation human mitochondria-focused cDNA microarray (hMitChip3). We identified significant differences in the expression on mitochondrial genes related to cell proliferation, glucose metabolism, and nucleic acid and fatty acid biosynthesis in this primary HLRCC cell line. This represents the first human primary renal cell carcinoma tumor cell model with a TCA cycle mutation. The UOK268 cell line should provide a unique in vitro system for the investigation of alterations in the metabolic molecular pathways during early tumorigenesis in HLRCC associated human cancer and possibly those involved in the Warburg phenomenon.
Materials and methods
Patient information and Ethics statement
The patient, who presented with a ½ centimeter, localized type 2 papillary kidney cancer, was evaluated at the U.S. National Cancer Institute (NCI) on a Urologic Oncology Branch protocol approved by the NCI institutional review board and gave written informed consent for participation in this study.
Cell culture and establishment of UOK268 cell line from primary surgical tissue
The UOK268 cell line was established from tumor tissue removed at surgery following the tissue and cell culture protocols and techniques of the Urologic Oncology Branch as previously described with modifications.[13;14] The major modification was the use of high glucose media (25mM D-glucose) and the rapid removal of yellowing, acidified media and replenishment with fresh media due to the cells predicted high glycolysis rate. Shortly after the primary renal tumor was removed and delivered for tissue procurement, a small amount of fresh tumor was minced and placed in a culture flask with a minimal amount of media. The first passage was carried out two to three days after plating by a light treatment with 0.05% trypsin-EDTA, while monitoring the cells growth under an inverted tissue culture microscope. Subsequent passages (over 20) were carried out every 2–3 days by splitting 1 to 2 in the same manner.
Tumor cell lines used in this study include the fumarate hydratase-deficient metastasis derived tumor cell line UOK262 [9] and the 262FH+ cell line created by stable reintroduction of the wild-type fumarate hydrase gene back into UOK262 [12]. The immortalized proximal tubule epithelial cell line HK-2 [15] derived from normal adult human kidney was purchased from American Type Culture Collection (ATCC). The 786-O wt (re-expressing wild-type VHL) cells were a gift from William G. Kaelin, Jr. (Harvard Medical School). All cells were cultivated in DMEM medium containing 25 mM D-glucose and supplemented with 10% fetal calf serum, 100 U/ml PenStrep, and 2 mM L-glutamine. All cell lines were cultured in the same laboratory under the standard cell culture conditions with exceptions where indicated.
Measurements of OXPHOS and glycolysis
The XF96 Extracellular Flux Analyzer (Seahorse Bioscience, MA, USA) was used to detect rapid, real time changes in cellular respiration and glycolysis rate. UOK268 and control cells (786-O wt and UOK262) were cultured in custom XF96 microplates as recommended by the manufacturer. [16] The 786-O wt cell line was used as a control as they had been previously shown in house to be a reliable example of normal levels of OCR and ECAR. Unless otherwise stated, the standard cell seeding concentration was 10,000/well, and the cells were seeded the day before use. Immediately following the addition of fresh medium, basal levels of O2 consumption and proton production were first quantified over approximately 20–25 min. Analysis of extracellular acidification rate (ECAR) reflects lactate excretion and serves as an indirect measure of glycolysis rate, while O2 consumption (OCR) reflects cellular respiration and is directly determined. Experiments were performed by simultaneously measuring three to five replicates of each cell line. All cell lines could be measured in a single experiment. Relative effects were expressed as areas under the curve measurements that were generated by the manufacturer’s software and used to compare the various cell lines, and the observed rates are reported in pMol/min/cell for OCR and mpH/min/cell for ECAR.
Measurement of FH enzyme activity
Whole cell protein quantification in the extract of UOK268 and control cells HK-2 was undertaken by the bicinchoninic acid (BCA) colorimetric assay as previously described.[17] In vitro FH enzyme activity was measured by NADP-malic enzyme coupled assay as previously described.[18] In brief, the increase in absorbance at 340 nM from NADPH formation was measured after adding fumarate (Sigma) at a final concentration of 10 mM into a total volume of 1 ml reaction mixture of cell extract at 30°C for 10 min. All spectrophotometric measurements were conducted using a Beckman DU-530 spectrophotometer.
RNA extraction and purification
When cells reached approximately 90% confluence under microscope, culture medium was removed by aspiration and the cells were rinsed once with 1X phosphate-buffered saline. The cells were lysed directly by adding 17 ml of Trizol Reagent (Invitrogen, Carlsbad, CA, USA) per T175 flask. Total RNA was extracted from freshly cultured cells using the Trizol Reagent protocol, and purified using RNeasy kit (Qiagen, Valencia, CA, USA), following the commercial instructions. Each cell line was cultured in triplicate in parallel with total RNA extracted from each triplicate to reduce random variance.
hMitChip3 microarray
Third-generation human mitochondria-focused cDNA microarray (hMitChip3) containing all the 37 mitochondrial DNA-encoded genes, 1,098 nuclear DNA-encoded and mitochondria-related genes, and 225 controls were printed as described previously [19]. This assessed a total of 1135 mitochondria-related genes include 946 genes associated with 645 molecular functions, 930 genes with 612 biological processes, 476 genes with biological chemistry pathways, 227 genes with 23 reactome events, 237 genes with 320 genetic disorders, and 55 genes with 87 drugs targets [19]. Six μg of total RNA per sample was used for microarray labeling and hybridization as previously described [19]. Slides were scanned using the ScanArray Express Microarray Scanner (PerkinElmer, Boston, MA) as described previously [19]. Each and every gene on the hMitChip3 gene chip was printed in triplicate, and triplicate microarray experiments were performed for each and every RNA samples. Therefore, the RNA level of each and every hMitChip3 gene was measured as many as nine times. The background-subtracted mean values of the multiple measurements were used for microarray data analysis. All microarray experiments were performed in the laboratories of GenProMarkers, Inc. (Maryland, USA).
Microarray database and computational algorithm
Gene expression databases were constructed using FileMaker software (FileMaker Pro, Inc., Santa Clara, CA, USA). Database construction, data filtering and selection were performed as described previously [19]. The quantile normalization method [20] in software R version 2.7.1 (The R Foundation for Statistical Computing) was used to normalize microarray data. The normalized expression data was used to cluster and visualize genes and cell lines by using software Cluster version 3.0 [20] and heatmap was visualized by using software MapleTree (http://rana.lbl.gov/EisenSoftware.htm). All data is MIAME compliant and the raw data has been deposited in a Gene Expression Omnibus (GEO - http://www.ncbi.nlm.nih.gov/geo/) with a series accession number of GSE32439 (provisional).
Gene information analysis
This analysis is essentially the same as described previously. The ID, official symbols and full names of genes on hMitChip3 were updated to human UniGene Build 219 (ftp://ftp.ncbi.nih.gov/repository/UniGene, October 2009) based on IMAGE clone ID (http://image.llnl.gov). Ontology, pathways and phenotypes of genes were compiled from Entrez (ftp://ftp.ncbi.nlm.nih.gov/gene) and DAVID Bioinformatics Resources 2008 (http://david.abcc.ncifcrf.gov). Canonical pathways and molecular and biological function were visualized by using Ingenuity Pathways Analysis software (https://analysisingenuity.com).
Statistics
Statistical calculations were performed on triplicate array experiments using XLSTAT 2006 (XLSTAT, New York, NY, USA). Differentially expressed genes were identified arbitrarily by >1.60-fold change in the average expression of the background-subtracted mean intensity ratios of a gene between comparisons. Using the LIMMA package in software R/Bioconductor (version 2.7.1, the R Foundation for Statistical Computing), we calculated the moderated t-statistic, raw p-values and log2 fold changes in gene expression and FDR (False Discovery Rate) for multiple statistical testing with Benjamini and Hochberg methods [21]. The level of statistical significance was set at a p-value <0.05 and FDR <9%.
Quantitative real time (qRT) PCR validating dysregulated transcripts
Gene expression alterations were analyzed by quantitative real time RT-PCR for 3 up-regulated and 3 down-regulated genes in UOK268 cells as compared to the HK-2 control. Additionally, two genes, PCK2 and HK1, were analyzed by quantitative real time RT-PCR in UOK268, HK-2, UOK262 and 262FH+ (re-expressing wild-type fumarate hydrase) cells. In brief, total RNA was extracted as described above. Twenty nanograms of total RNA was reverse-transcribed to cDNA using SuperScript® III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and random hexamers in a final volume of 20 μl. One microliter of the resulting cDNA was used for PCR amplification in an ABI 7000 real-time PCR system (Applied Biosystems) as recommended by the manufacturer. Primers and fluorogenic probes were designed by Applied Biosystems in the form of Taqman® Gene Expression Assays for the selected genes: RHOT (Hs00373355_m1), PCK2 (Hs00388934_m1), GLUD1 (Hs01632647_g1), MDH2 (Hs00157798_m1), OGDH (Hs01081865_m1), LDHA (Hs00855332_g1) and HK1 (Hs00175976_m1). Expression levels for these genes in UOK268 were normalized to a control housekeeping gene, ACTB (Hs99999903_m1), and CT values were further normalized to those obtained from a normal HK-2 cell line. Samples were run in triplicate and CT values obtained were compared by the Delta CT method. Results are expressed as an average fold-change compared to HK-2 values from three independent experiments.
Antibodies, immunohistochemistry and immunoblotting
Immuno-fluorescence protein in situ and co-localization studies in UOK268 cells (passage 8) were performed by seeding in Lab-TEK II chamber slides (Nalge Nunc International), fixing in 3.7% formaldehyde in PBS, and permeabilizing in 0.2% Triton- X100 for 10 min at room temperature. Cells were then washed three times in PBS and incubated for 1 hour at room temperature with a mouse anti-human FH antibody (1:200 dilution; ab58232, Abcam). Antibodies used for mitochondrial staining analysis included rabbit anti-human SOD2 (1:1000 dilution; ab13533, Abcam). Following three washes in PBS, cells were incubated with either goat anti-rabbit or goat anti-mouse IgG-Alexa fluor 488/Alexa fluro594 (1:500 dilution, Invitrogen) for 30 min at room temperature, washed in PBS, and briefly counterstained with DAPI. After mounting, cells were observed under a Leica DMRxA fluorescent microscope and images were obtained with IPLab v.3.7 software.
Western blotting was performed using standard methods using the following antibodies: FH (ab58232, Abcam), HIF1α (ab12289, Abcam), PCK2 (ab70359, Abcam), HK1 (#2024, Cell Signaling Technologies) and HADHA (ab54477, Abcam) with β-Actin (#A2522, Sigma) used as a loading control.
Results
UOK268 cells demonstrate atypical genetic and pathologic properties consistent with the clinical manifestation of HLRCC
UOK268 cell line was established from a primary HLRCC tumor, which was surgically removed from a 26-year old female patient with early left kidney lesion (Fig. 1A). This lesion was excised from the patient’s posterior renal cyst, with tubulo-papillary fronds inside cystic spaces and solid areas. Cyst walls were lined by atypical, poorly differentiated tumor cells. The tumor cells demonstrated severe nuclear changes, Fuhrman nuclear grade 4, and several demonstrated a pathological feature specific to HLRCC tumor cells of enlarged, heavily stained (eosinophilic) nucleoli surrounded by a noticeable unstained space or halo within normally stained nuclei (Fig. 1B) [7]. The patient possessed a predisposing heterozygous fumarate hydratase mutation at a nucleotide in exon 4, resulting in a His192Asp amino acid change. To confirm whether UOK268 cells contain the identical FH germline mutation, we sequenced genomic DNA from early and late passages of the cell line. As expected, we verified the nucleotide change in the cell line as the patient’s germline missense mutation (c.704 C>G – p.His192Asp) and demonstrated loss of the wild type allele (loss of heterozygousity - LOH), indicating the loss of functional FH protein in the cell line (Fig. 1C).
Figure 1. Morphologic, genetic and functional description of the UOK268 cell line.
(A) Hematoxylin-eosin (H&E) staining of HLRCC kidney cancer removed surgically from the patients left renal tumor tissue (1X) and (B) H&E staining of tissue with 60X magnification to show consistent with atypical HLRCC pathological morphology: large nuclei with predominant nucleoli and atypical perinucleolar clearing.
(C) Genomic DNA mutation analysis revealed only germline mutant allele retained in UOK268 tumor cells.
(D) Western Blot analysis of the levels of HIF1α protein present in the lysates of UOK268, UOK262 and HK-2 cells. The expression levels of β–ACTIN were used as a loading control.
(E) Mutant FH protein is primarily localized to mitochondria of UOK268 cells shown by Alexa-Fluor®-488 labeling with a primary mouse monoclonal antibody to human FH. This overlaps almost completely with the signal from the Alexa-Fluor®-594 labeling with a primary rabbit polyclonal antibody to human mitochondrial protein SOD2.
(F) An In vitro assay of mutant FH enzyme activity from UOK268 tumor cell lysate was conducted by NADP-malic enzyme coupled assay and (G) as shown in Tong, et al,[12] the basal cellular respiration rate (oxygen consumption rate, OCR) of UOK268 and control cells was measured (here we use 786-O wt cell as positive control and UOK262 as negative control). OCR was normalized and shown as fMoles/min/cell.
Abundant mutant FH protein localized on mitochondria with compromised catalytic activity and attenuation of OXPHOS
As the UOK268 cells contained only a mutated version of the FH gene, the expression and localization of the predicted full length mutant protein was investigated. Immunofluorescence was used to detect both FH and superoxide dismutase 2 (SOD2) (the mitochondrial form of the SOD protein family). The FH (labeled with Alexa Fluor® 488) primarily co-localized with mitochondrially expressed SOD2 (labeled with Alexa Fluor® 594), confirming its presence in the mitochondrial matrix (Fig. 1D). Western blot analysis verified the full length mutant FH protein was indeed present. The mutant FH protein (~55kDa) migrated at ~48kDa as previously observed and its expression was more abundant than the HK-2 normal control and similar to that of the UOK262 mutant FH protein and corresponded with high levels of HIF1α, again similar to UOK262 (Fig. 1E, as previously demonstrated in Tong, et al [12]).
To determine whether UOK268 cells have biological and physiological function in TCA cycle, we next performed FH enzyme activity assay and a real-time assessment of the relative contributions of mitochondrial oxidative phosphorylation (OXPHOS). An In vitro assay of mutant FH enzyme activity from UOK268 tumor cell lysate was conducted by NADP-malic enzyme coupled assay as previously described [18]. The UOK268 tumor cell lysate demonstrated a barely detectable level of FH enzyme compared to the HK-2 normal control (Fig. 1F). This suggests that the retention of the germline missense mutated allele in combination with LOH in UOK268 impaired FH catalytic activity.
As shown in Tong, et al, [12] to demonstrate the effect on the mitochondrial OXPHOS in UOK268 cells due to loss of FH activity, an XF96 Extracellular Flux Analyzer was used to obtain real-time metabolic efflux of the parameters pertaining to OXPHOS and glycolysis [16]. OXPHOS was represented by measurement of oxygen consumption rates (OCR) in the process of energy production in UOK268 cells. For comparison, we obtained OCR measurement data from previously used 786-O wt (known to demonstrate normal levels of OCR and ECAR) cells and UOK262 cells. As shown in Fig. 1G, the basal line OCR for UOK268 is displayed as fMoles/min/cell. OCR in UOK268 cells was slightly higher than UOK262, which was described previously as having almost undetectable OCR, while the positive control 786-O wt cells demonstrated a high OCR at 0.012 fMoles/min/cell (Fig. 1G). Since mitochondrial respiration accounts for ~ 90% of cellular oxygen consumption, and cellular protein oxidation accounts for the remaining 10%, the low level of OCR in UOK268 cells probably comes from non-mitochondrial oxygen utilization [22]. This indicates that mitochondrial respiration function in UOK268 cells is diminished to its minimal. The ECAR measurements did not show a significant difference between UOK268 and the control 786-O wt cells, unlike the UOK262 cell line. This test only represents a small amount of time, while observation of the cell lines in culture demonstrated that the UOK262 cell line rapidly turns the acidity marker form pink to yellow due to the secretion of lactate. This phenomenon was observed in the UOK268 cell line as well but over a longer time period, during which the same observation was not seen in 786-O wt cells, which served as a non-HLRCC control.
hMitChip3, mitochondria focused, microarray analysis of UOK268 as a primary FH-deficient HLRCC cell line
To investigate the specific effects of loss of FH activity on the mitochondria and the metabolism of the UOK268 tumor cells, microarray analysis was performed using a mitochondria focused microarray, hMitChip3. This third generation human mitochondria microarray chip contains 1135 mitochondria-related genes (37 mitochondrial DNA-encoded genes, 1,098 nuclear DNA-encoded and mitochondria-related genes and 225 controls) [19]. The microarrays were performed in triplicate from three separate total RNA extracts with three experimental repeats each and produced reproducible data from 997 probes (including control elements), which was subsequently used for unsupervised hierarchical clustering analysis. The 18 microarray were normalized (Suppl. Fig. 1) and the mean values of normalized data were used to calculate the ratios in gene expression between UOK268 and HK-2 cell lines.
The unsupervised hierarchical clustering analysis heat map (Fig. 2) demonstrated clear differences between gene expression patterns, both up-regulation (Fig. 2A) and down-regulation (Fig. 2B), in the UOK268 cell line in comparison to HK-2 cell line.
Figure 2. Heat map and hierarchical clustering of 90 significantly differentially expressed genes between normal renal cell line HK-2 and UOK268.
The clusters show (A) up-regulated genes and (B) down-regulated genes in UOK268 cell line in comparison to HK-2 cell line. Color map corresponds to the fold changes.
To identify the most representative differentially expressed mito-transcripts between FH deficient primary UOK268 and the normal HK-2 line, we employed a statistically supervised method previously described as moderated t test [23]. Based on unsupervised cluster results, we calculated t statistic, p-values, and the false discovery rate (FDR) for each gene between UOK268 and HK-2 cell lines. Statistical calculations were performed on triplicate array experiments using XLSTAT 2006 (XLSTAT, New York, NY, USA). Differentially expressed genes were identified arbitrarily by >1.25-fold change in the average expression of the background-subtracted mean intensity ratios of a gene between comparisons. Using the LIMMA package in software R/Bioconductor (version 2.7.1, the R Foundation for Statistical Computing), and with Benjamini and Hochberg methods [21]. The level of statistical significance was set at a p-value <0.05 and FDR < 9%.
This indentified 182 significantly up-regulated transcripts (Suppl. Table 1) and 127 significantly down-regulated transcripts (Suppl. Table 2) in UOK268, of which 50 of 182 up-regulated transcripts (27.5%) and 55 of 127 down-regulated transcripts (43.3%) had an equal or greater than 2 fold change (Table 1). The combined list of 309 genes was selected for pathway analysis to access the general alterations occurring within the cell line. While from within the 105 genes with greater changes specific genes of interest could be identified for further studies, such as RHOT2, a mitochondrial GTPase involved in mitochondrial trafficking.
Table 1.
Genes significantly Up/Down-regulated greater than 2 fold in UOK268 in comparison to the HK-2 cell line.
| Symbol | Significantly Up-Regulated Genes in UOK268 vs HK-2 | |||
|---|---|---|---|---|
| FC (Raw) | FC (Log2) | p-value | FDR (%) | |
| RHOT2 | 12.00 | 3.58 | 0.000 | 0.65 |
| CPT1C | 7.86 | 2.98 | 0.000 | 0.70 |
| MT1G | 5.48 | 2.45 | 0.003 | 1.93 |
| THAP3 | 4.11 | 2.04 | 0.000 | 0.62 |
| SLC25A44 | 3.75 | 1.91 | 0.011 | 3.58 |
| MDH1 | 3.66 | 1.87 | 0.018 | 4.72 |
| MT1E | 3.60 | 1.85 | 0.011 | 3.52 |
| TRND | 3.58 | 1.84 | 0.000 | 0.56 |
| SLC25A14 | 3.49 | 1.80 | 0.004 | 2.09 |
| HSPA13 | 3.45 | 1.79 | 0.001 | 1.00 |
| MIPEP | 3.40 | 1.77 | 0.002 | 1.25 |
| NKRF | 3.35 | 1.75 | 0.000 | 0.34 |
| CKB | 3.33 | 1.73 | 0.012 | 3.69 |
| UBB | 3.28 | 1.72 | 0.014 | 4.08 |
| DHPS | 3.27 | 1.71 | 0.048 | 8.52 |
| FH | 3.15 | 1.65 | 0.024 | 5.65 |
| P4HB | 3.14 | 1.65 | 0.019 | 4.78 |
| UBE2K | 3.00 | 1.59 | 0.002 | 1.61 |
| ALAS2 | 2.91 | 1.54 | 0.044 | 8.06 |
| YWHAE | 2.88 | 1.52 | 0.006 | 2.63 |
| GPX7 | 2.87 | 1.52 | 0.022 | 5.30 |
| SLC25A29 | 2.85 | 1.51 | 0.017 | 4.59 |
| ACBD3 | 2.83 | 1.50 | 0.014 | 4.09 |
| RNR1 | 2.82 | 1.50 | 0.001 | 1.00 |
| LRRC16B | 2.78 | 1.47 | 0.035 | 6.89 |
| SLC40A1 | 2.76 | 1.46 | 0.004 | 2.20 |
| PLA2G1B | 2.69 | 1.43 | 0.043 | 7.93 |
| CASP5 | 2.64 | 1.40 | 0.000 | 0.70 |
| ACAA1 | 2.58 | 1.37 | 0.000 | 0.58 |
| MTHFD1 | 2.53 | 1.34 | 0.006 | 2.54 |
| RHOT1 | 2.47 | 1.30 | 0.007 | 2.80 |
| ETFA | 2.43 | 1.28 | 0.002 | 1.47 |
| HADHA | 2.39 | 1.26 | 0.002 | 1.47 |
| ATPAF2 | 2.37 | 1.24 | 0.021 | 5.13 |
| RPL5 | 2.30 | 1.20 | 0.024 | 5.65 |
| ETFDH | 2.28 | 1.19 | 0.023 | 5.47 |
| SYNPR | 2.26 | 1.18 | 0.007 | 2.90 |
| LOC100293864 | 2.21 | 1.14 | 0.017 | 4.59 |
| ALDH18A1 | 2.19 | 1.13 | 0.000 | 0.52 |
| HCCS | 2.18 | 1.12 | 0.010 | 3.39 |
| APP | 2.12 | 1.09 | 0.000 | 0.50 |
| MRPS11 | 2.12 | 1.08 | 0.009 | 3.17 |
| GRB10 | 2.11 | 1.08 | 0.001 | 0.88 |
| MTND5 | 2.11 | 1.08 | 0.053 | 9.00 |
| NR4A1 | 2.09 | 1.07 | 0.001 | 0.88 |
| PCK2 | 2.09 | 1.07 | 0.006 | 2.65 |
| BCO2 | 2.09 | 1.06 | 0.042 | 7.87 |
| SLC25A37 | 2.06 | 1.04 | 0.030 | 6.34 |
| MT1B | 2.04 | 1.03 | 0.005 | 2.37 |
| COPA | 2.01 | 1.01 | 0.004 | 2.14 |
| Symbol | Significantly Down-Regulated Genes in UOK268 vs HK-2 | |||
|---|---|---|---|---|
| FC (Raw 1/x) | FC (log2) | p-value | FDR (%) | |
| TIMM8A | 32.02 | −5.00 | 0.000 | 0.29 |
| SLC25A25 | 8.68 | −3.12 | 0.001 | 1.00 |
| ATP5F1 | 6.02 | −2.59 | 0.000 | 0.50 |
| FXYD1 | 5.52 | −2.46 | 0.000 | 0.06 |
| DUOX2 | 4.90 | −2.29 | 0.000 | 0.56 |
| CYP2E1 | 4.67 | −2.22 | 0.000 | 0.52 |
| RAB5B | 4.55 | −2.19 | 0.000 | 0.50 |
| ACAD9 | 4.10 | −2.04 | 0.000 | 0.65 |
| MRPS25 | 4.07 | −2.03 | 0.005 | 2.44 |
| PFN1 | 3.99 | −2.00 | 0.026 | 5.88 |
| GOT2 | 3.94 | −1.98 | 0.000 | 0.65 |
| NDUFV1 | 3.85 | −1.94 | 0.018 | 4.60 |
| MTFMT | 3.59 | −1.84 | 0.000 | 0.57 |
| MRPL21 | 3.53 | −1.82 | 0.000 | 0.70 |
| ACSBG1 | 3.47 | −1.79 | 0.004 | 2.11 |
| TRNL2 (MTTL2) | 3.39 | −1.76 | 0.005 | 2.36 |
| ROPN1B | 3.38 | −1.76 | 0.001 | 0.88 |
| CASP2 | 3.37 | −1.75 | 0.009 | 3.17 |
| IMPDH1 | 3.32 | −1.73 | 0.003 | 1.91 |
| GSTK1 | 3.31 | −1.73 | 0.001 | 0.88 |
| LOC100129794 | 3.25 | −1.70 | 0.001 | 0.88 |
| ACP6 | 3.12 | −1.64 | 0.001 | 0.75 |
| CACNA2D2 | 3.11 | −1.64 | 0.004 | 2.14 |
| LDHA | 3.02 | −1.59 | 0.048 | 8.52 |
| TFB1M | 3.01 | −1.59 | 0.002 | 1.61 |
| CHD3 | 2.95 | −1.56 | 0.001 | 1.11 |
| RPS6KC1 | 2.87 | −1.52 | 0.022 | 5.30 |
| CYP2C18 | 2.86 | −1.51 | 0.002 | 1.36 |
| RAP1B | 2.84 | −1.50 | 0.002 | 1.25 |
| DGUOK | 2.83 | −1.50 | 0.043 | 8.02 |
| TRNL1 (MTTL1) | 2.73 | −1.45 | 0.004 | 2.11 |
| ELOVL1 | 2.65 | −1.40 | 0.032 | 6.64 |
| UVRAG | 2.60 | −1.38 | 0.025 | 5.78 |
| MYH4 | 2.59 | −1.38 | 0.011 | 3.58 |
| GOT1 | 2.57 | −1.36 | 0.015 | 4.20 |
| BCCIP | 2.46 | −1.30 | 0.025 | 5.82 |
| MRPS26 | 2.44 | −1.28 | 0.049 | 8.55 |
| ACSL4 | 2.41 | −1.27 | 0.004 | 1.99 |
| COX7A2 | 2.35 | −1.23 | 0.028 | 6.13 |
| NAT1 | 2.35 | −1.23 | 0.000 | 0.52 |
| DIABLO | 2.30 | −1.20 | 0.000 | 0.29 |
| TOMM20 | 2.29 | −1.19 | 0.028 | 6.16 |
| HNRPC | 2.25 | −1.17 | 0.001 | 0.88 |
| OGDH | 2.25 | −1.17 | 0.003 | 1.73 |
| SOD1 | 2.24 | −1.16 | 0.040 | 7.63 |
| COX7C | 2.23 | −1.16 | 0.010 | 3.25 |
| ALDH6A1 | 2.21 | −1.15 | 0.000 | 0.58 |
| DDX27 | 2.17 | −1.12 | 0.036 | 7.09 |
| MPHOSPH6 | 2.11 | −1.07 | 0.048 | 8.52 |
| ATP5G3 | 2.10 | −1.07 | 0.053 | 9.00 |
| FTMT | 2.06 | −1.04 | 0.003 | 1.94 |
| SLC25A46 | 2.05 | −1.04 | 0.000 | 0.59 |
| NOX5 | 2.04 | −1.03 | 0.047 | 8.52 |
| LONP1 | 2.01 | −1.01 | 0.009 | 3.17 |
| NDUFA9 | 2.01 | −1.01 | 0.016 | 4.42 |
Evaluation of dysregulated mitotranscripts from UOK268 compared with HK-2 with qRT-PCR
To validate the hMitChip3 cDNA microarray results, 6 genes were chosen to be re-evaluated using qRT-PCR. Genes were selected to represent a range of fold changes from within the significance limits, 3 up-regulated (RHOT2 – 12.0, PCK2 – 2.09, GLUD1 – 1.55) and 3 down-regulated (LDHA – 0.33, OGDH – 0.44 and MDH2 – 0.63). All 6 genes demonstrated fold changes in the predicted manner, with the two most up-regulated and the two most down-regulated showing statistically significant changes (Fig. 3A). This demonstrated a good correlation between the microarray results and the qRT-PCR and confirmed that the hMitChip3 cDNA microarray had generated informative data. To demonstrate that these differences between UOK268 and HK-2 were FH mutation specific, rather than differences acquired during cell transformation, western analysis was performed on UOK268, HK-2, UOK262 and 262FH+ (re-expressing wild-type fumarate hydrase) cell lysates for selected up-regulated genes. Western analysis of PCK2 and HK1 proteins demonstrated a specific increase in UOK268 and UOK262 compared to HK-2 that was additionally lost in 262FH+, demonstrating specificity to the absence of working fumarate hydratase (Fig. 3B). This was also observed in the qRT-PCR results for these two genes (Fig. 3C). Interestingly, the HADHA protein was increased in UOK268 and UOK262 compared to HK-2, but this increase was still present in 262FH+ (Fig. 3B). Additionally, the qRT-PCR demonstrated generally lower fold changes and may provide more precise quantification of expression over a wider dynamic range.
Figure 3. Evaluation of dysregulated mitotranscripts from UOK268 compared with HK-2 with qRT-PCR.
(A) Six genes from the hMitChip3 cDNA microarray results were chosen to be re-evaluated using qRT-PCR and the resultant fold changes compared to those from the microarray. Genes were selected to represent a range of fold changes from within the significance limits, 3 up-regulated (RHOT2, PCK2, and GLUD1) and 3 down-regulated (LDHA, OGDH and MDH2). The hMitChip3 cDNA microarray fold change is shown for each gene/transcript (up-regulated in red and down-regulated in green) and the graphs show the average real time RT-PCR fold change (delta CT) of UOK268 relative to HK-2. Student t-tests were used to produce p-values.
(B) Western Blot analysis of the levels of PCK2, HK1, HADHA and FH protein present in the lysates of UOK268, HK-2, UOK262 and 262FH+ (re-expressing wild-type fumarate hydrase) cells. The expression levels of β–ACTIN were used as a loading control.
(C) Quantative RT-PCR results comparing the mRNA levels of PCK2 and HK1 genes between UOK268, HK-2, UOK262 and 262FH+.
Molecular functions, gene expression patterns, biological process and statistics of 309 genes in UOK268
The 309 significantly dysregulated genes (182 up-regulated and 127 down-regulated) were uploaded into the Ingenuity® Pathways Analysis software, annotated and analyzed to identify molecular and biological functions and canonical pathways.
Ingenuity molecular and biological function analysis demonstrated dysregulation of numerous important functions with genes involved in lipid metabolism (57 genes) being the most statistically significant (Table 2). Additionally, the software identified dysregulated genes associated with tumorigenesis (98 genes), apoptosis of the eukaryotic cell (55 genes), cell cycle (14 genes) and energy production (23 genes). The dysregulation of the lipid metabolism and energy production are perhaps logical responses to the loss of an integral citric acid cycle enzyme like fumarate hydratase.
Table 2.
Ingenuity® Pathways Analysis software result tables for the most significant molecular/biological functions and canonical pathways.
| Top Molecular and Biological Functions | ||
|---|---|---|
| Cluster Description | Cluster p-value ranges | Total number of selected genes |
|
| ||
| Lipid Metabolism | 1.87E-11 to 1.78E-02 | 57 |
| Small Molecule Biochemistry | 1.87E-11 to 1.78E-02 | 97 |
| Amino Acid Metabolism | 7.39E-08 to 1.78E-02 | 19 |
| Cellular Function and Maintenance | 7.51E-11 to 1.65E-02 | 33 |
| Cellular Compromise | 8.73E-11 to 1.29E-02 | 29 |
| Top Canonical Pathways | ||
|---|---|---|
| Canonical Pathway | p-value | Ratio of selected genes to pathway genes |
|
| ||
| Mitochondrial Dysfunction | 1.60E-24 | 30/169 (0.178) |
| Oxidative Phosphorylation | 8.17E-17 | 25/165 (0.152) |
| Valine, Leucine and Isoleucine Degradation | 1.54E-13 | 16/111 (0.144) |
| Propanoate Metabolism | 1.93E-11 | 14/130 (0.108) |
| Citrate Cycle | 1.16E-10 | 10/58 (0.172) |
Ingenuity canonical pathway analysis demonstrated mitochondrial dysfunction and oxidative phosphorylation as the pathways with the most significant numbers of genes dysregulated, 30/169 and 25/165 respectively (Table 2). These two pathways contained a large number of overlapping genes involved in oxidation phosphorylation complexes I to V in mitochondrial membrane and this dysregulation is reflected in the low oxygen consumption rate (OCR) in UOK268. Figure 4 represents an adapted version of the mitochondrial dysfunction pathway with the dysregulated genes highlighted (Fig. 4 and Table 3). Other important pathways were also dysregulated including glycosis/gluconeogenesis (6/147 – ACSS1, ADH6, ALDOA, ALDOC, HK1 and LDHA) and apoptosis signaling (6/90 – BID, CASP2, DIABLO, NFKB1, RAF1 and TP53) (Supplementary Fig. 2).
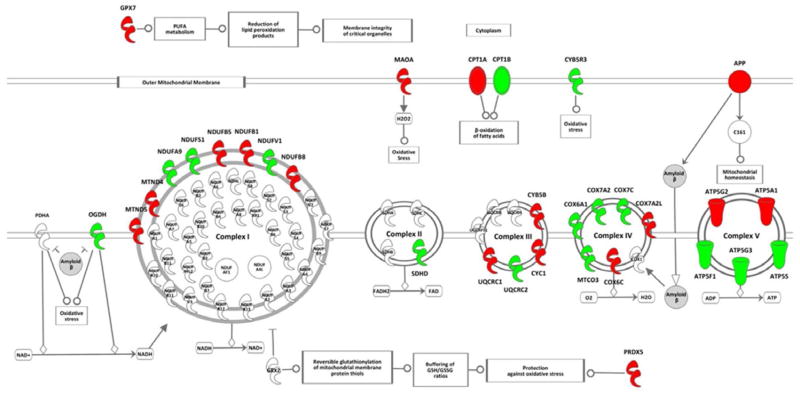
Figure 4. Adapted Ingenuity figure for mitochondrial dysfunction demonstrating the differences in gene transcription in UOK262 and the normal renal cell line HK-2.
The mitochondrial dysfunction pathway was identified by the Ingenuity® Pathways Analysis software (https://analysisingenuity.com) as the most significantly altered biological function in UOK262 compared to the normal renal cell line HK-2. Genes transcripts up-regulated in UOK268 are highlighted in green and down-regulated are highlighted in red. This figure was adapted by removing the aspects of the pathway containing no significantly dysregulated transcripts.
Table 3.
Significantly dysregulated transcripts involved in mitochondrial dysfunction identified by Ingenuity® Pathways Analysis software or known function.
| Gene | Location | Entrez Gene Name | Selection | |
|---|---|---|---|---|
|
| ||||
| ↑ | GPX7 | Cytoplasm | glutathione peroxidase 7 | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | CYB5R3 | Outer Mitochondrial Membrane | cytochrome b5 reductase 3 | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | CPT1B | Outer Mitochondrial Membrane | carnitine palmitoyltransferase 1B (muscle) | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | CPT1A | Outer Mitochondrial Membrane | carnitine palmitoyltransferase 1A (liver) | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | CPT1C | Outer Mitochondrial Membrane | carnitine palmitoyltransferase 1C | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | APP | Outer Mitochondrial Membrane | amyloid beta (A4) precursor protein | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | MAOA | Outer Mitochondrial Membrane | monoamine oxidase A | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | OGDH | Inner Mitochondrial Membrane | oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | GPX4 | Mitochondrial Matrix | glutathione peroxidase 4 (phospholipid hydroperoxidase) | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | PRDX5 | Mitochondrial Matrix | peroxiredoxin 5 | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | NDUFS1 | Complex I | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | NDUFV1 | Complex I | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | NDUFA9 | Complex I | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | MTND4 | Complex I | NADH dehydrogenase, subunit 4 (complex I) | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | MTND5 | Complex I | NADH dehydrogenase, subunit 5 (complex I) | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | NDUFB1 | Complex I | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | NDUFB5 | Complex I | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | NDUFB8 | Complex I | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | SDHD | Complex II | succinate dehydrogenase complex, subunit D, integral membrane protein | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | UQCRC2 | Complex III | ubiquinol-cytochrome c reductase core protein II | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | UQCRC1 | Complex III | ubiquinol-cytochrome c reductase core protein I | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | CYB5B | Complex III | cytochrome b | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | CYC1 | Complex III | cytochrome c-1 | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | COX6A1 | Complex IV | cytochrome c oxidase subunit VIa polypeptide 1 | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | COX7A2 | Complex IV | cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | COX7C | Complex IV | cytochrome c oxidase subunit VIIc | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | MTCO3 | Complex IV | cytochrome c oxidase III | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | COX6C | Complex IV | cytochrome c oxidase subunit VIc | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | COX7A2L | Complex IV | cytochrome c oxidase subunit VIIa polypeptide 2 like | Ingenuity Mitochondrial Dysfunction Pathway |
| ↓ | ATP5F1 | Complex V | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | Known Function |
| ↓ | ATP5G3 | Complex V | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 | Known Function |
| ↓ | ATP5S | Complex V | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s (factor B) | Known Function |
| ↑ | ATP5A1 | Complex V | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | Ingenuity Mitochondrial Dysfunction Pathway |
| ↑ | ATP5G2 | Complex V | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 | Known Function |
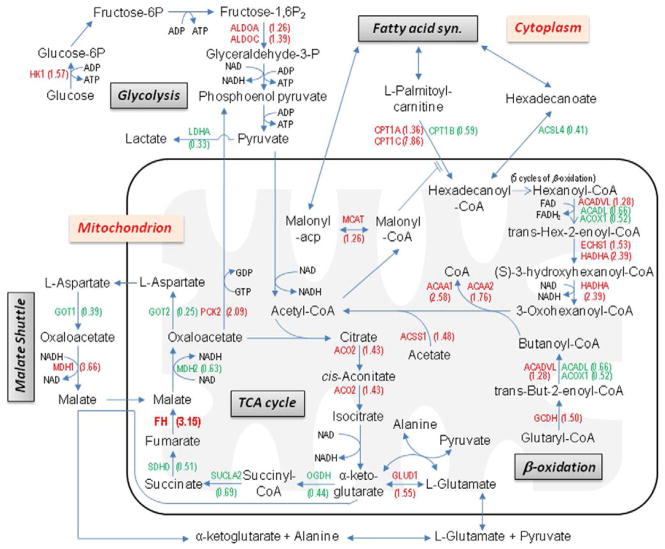
By combining the highlighted functions and pathways from the Ingenuity analysis of the hMitChip3 cDNA microarray results and known pathway data, the effects on the energy metabolism pathways in UOK268 were modeled and illustrated in figure 5. This model provides an overview of the current knowledge of the altered metabolism in UOK268 cells. Dysregulation is seen in all these pathways with genes being both up and down-regulated in response to loss of fumarate hydratase activity and the subsequent tumorigenesis. Additionally, multiple amino acid metabolic pathways are affected by FH loss.
Figure 5. Metabolic pathways with dysregulated genes in renal cancer cell line UOK268 compared to the normal renal epithelial cell line HK-2.
By combining the highlighted functions and pathways from the Ingenuity analysis of the hMitChip3 cDNA microarray results and known pathway data, the effects on the energy metabolism pathways in UOK268 were modeled. The hMitChip3 cDNA microarray fold change is shown for each gene/transcript (up-regulated in red and down-regulated in green).
Discussion
In the 1920s, Otto Warburg made a landmark observation that cancer cells preferentially use glycolysis as opposed to oxidative phosphorylation as the main source of adenosine triphosphate (ATP), even in the presence of normal oxygen levels. He proposed that abnormal energy metabolism was a fundamental aspect of cancer and this observed phenomenon became known as the “Warburg effect” [24–26].
Hereditary leiomyomatosis and renal cell carcinoma (HLRCC) is a form of inherited kidney cancer characterized by affected individuals being at risk of cutaneous and uterine leiomyomas and kidney cancer, normally due to an inactivating mutation of the fumarate hydratase (FH) gene encoding the Krebs cycle enzyme fumarate hydratase [27;28]. HLRCC kidney cancers are notable for their propensity to grow quickly and metastasize early [13]. A high mutation detection rate in HLRCC families [4;5;29] enables the early detection of at-risk individuals and allows for the early initiation of therapy while the tumor is still small. HLRCC kidney tumor cells have lost the ability to completely cycle through the Krebs cycle, due to the loss of fumarate hydratase enzyme activity, and have effectively lost the ability to perform oxidative phosphorylation. Thus, these cancers exist in a state of enforced dependence upon glycolysis and represent a notable example of the Warburg effect.
Cell lines derived from fumarate hydratase-deficient kidney tumors are useful for both the specific investigation of the biology of HLRCC kidney cancers for therapeutic purposes and the general investigation of the mechanism of the Warburg effect in cancer. There exist a number of models of fumarate hydratase deficiency: among them a model consisting of the lung cell line A549 with a stable FH RNAi knockdown [30] and a kidney model consisting of the UOK262 cell line derived from a metastasis of a HLRCC kidney tumor [9]. Additionally, an FH conditional knockout mouse model has been produced from which mouse embryonic fibroblasts (MEFs) have been cultured [31]. The UOK262 cell line retains a missense mutated (p.Gln396Pro) allele of the FH gene and has lost the wild type allele and so produces aberrant mutant fumarate hydratase protein but demonstrates no fumarate hydratase enzyme activity [9]. The UOK262 cell line demonstrates enhanced HIF-1α levels, increased expression of glucose transporter GLUT1 and increased levels of secreted lactate [9;30]. UOK262 does not represent a primary HLRCC kidney tumor, but is derived from a metastasis, therefore the UOK268 cell line described here should more clearly reflect primary/premetastatic HLRCC kidney tumors. The UOK268 cell line, like the UOK262 cell line, retains a missense mutated (p.His192Asp) version of the FH gene. This is a novel mutation of an amino acid which has been shown to be pathogenic when altered to a different amino acid (p.His192Arg) [32]. Like UOK262, UOK268 expresses the mutant allele and the protein product is produced and migrates to the mitochondria, but has no enzymic activity. The presence of this mutated protein may be of additional interest as, although HLRCC can occur with either complete loss of the FH protein or presence of missense mutated protein only, recent studies have suggested a dominant negative effect associated with the missense protein may enhance cancer onset [33]. This may be due to the fact that fumarate hydratase functions as a homotetramer and that a build-up of nonfunctioning/mutated protein containing complexes activates some form of misfolded protein response [25]. This may be particularly relevant to UOK268 as the mutation of p.Arg190His, a commonly occurring HLRCC mutation, investigated in the study occurs only 2 amino acids away from the UOK268 FH mutation [33].
The UOK268 cell line also mimics many of the previously reported features of the UOK262 cell line, with high levels of HIF-1α expression, negligible oxygen consumption, and a dependence upon glucose and glycolysis for energy production. UOK268 is not identical to UOK262 and demonstrates a phenotype that is less aggressive and less extreme and maybe more representative of a primary tumor, as evidenced by the slower growth rate and less elevated degree of glycolytic up-regulation seen here compared to the previously published UOK262 data [12]. This is also evident from recent work showing that the reduced AMPK signaling and repressed DMT1 iron transporter activity was present in both UOK268 and UOK262, but to a lesser extent in UOK268 [12].
Due to the observation of altered mitochondrial morphology in UOK268 and previously in UOK262 [9] and the loss of oxidative phosphorylation seen in both, an initial investigation of mitochondrial function in UOK268 was performed. Additionally, mitochondria dysfunctions have been implicated in a number of diseases and, so far, the genetic basis of 102 hereditable disorders has been attributed to alterations in a quarter of the known nuclear-encoded mitochondrial proteins in humans [34]. To date, focused mitochondrial arrays, such as the hMitChip3 gene chip, have been useful for identifying altered gene expression and variation in the numbers of cancers, inherited mitochondrial diseases and neurological syndromes, including providing assessment of the survival-apoptosis pathways in malignant melanoma [23] and recent reports into patterns of alteration in Alzheimer’s [35] and Parkinson’s disease [36]. Ingenuity molecular and biological function analysis of the hMitChip3 gene chip data demonstrated dysregulation of numerous important functions, with genes involved in lipid metabolism (57 genes) being the most statistically significant. Included within this selection are a group of significantly up-regulated genes (GCDH, ACADVL, ACAA1, ACAA2, HADHA, ECHS1, CPT1A and CPT1C) involved with fatty acid/lipid turn-over, both degradation and synthesis (Figure 5). This is suggestive of the high rate of fatty acid/lipid metabolism preferentially required for membrane production and proliferation. Additionally, dysregulation of genes associated with tumorigenesis (98 genes), apoptosis of the eukaryotic cell (55 genes), cell cycle (14 genes) and energy production (23 genes) was highlighted. The dysregulation of the lipid metabolism and energy production are logical responses to the loss of an integral citric acid cycle enzyme like fumarate hydratase.
Ingenuity canonical pathway analysis demonstrated the most significantly dysregulated pathways were those involving mitochondrial function and oxidative phosphorylation, both pathways contained a large number of overlapping genes involved in oxidation phosphorylation complexes I to V in mitochondrial membrane. This dysregulation is reflected in the low oxygen consumption rate (OCR) in UOK268. Other pathways were also dysregulated including glycolysis/gluconeogenesis and apoptosis signaling, matching the functions of the genes identified above. These data suggest that therapeutic strategies to specifically interfere with glycolytic flux and lipogenesis may provide the foundation for the development of targeted approaches for therapy for fumarate hydratase-deficient cancers. One limitation of using the hMitChip3 gene chip is the obvious concentration upon only those pathways related to the mitochondria. Additional work on other non-mitochondrial related pathways will also be extremely useful and provide other possible avenues for novel therapeutic strategies.
In summary, the fumarate hydratase-deficient kidney cancer cell line, UOK268, represents a unique model for the investigation of primary HLRCC kidney tumors and is a notable example of the Warburg effect in cancer. UOK268 provides unique insights into the metabolic adaption that a primary tumor characterized by impairment of TCA function undergoes, such as the switch to glycolytic dependence and alteration to lipid synthesis, prior becoming an aggressive metastatic tumor. In combination with the UOK262 kidney cell line, this should provide a powerful tool for functional investigation and the evaluation of novel therapeutic drugs for both this specific form of kidney cancer and for other tumors characterized by anaerobic glycolysis.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
We thank Robert A. Worrell for operative tissue procurement and Georgia Shaw for outstanding editorial support.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci U S A. 2001;98:3387–3392. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merino MJ, Torres-Cabala C, Pinto PA, Linehan WM. The morphologic spectrum of kidney tumors in hereditary leiomyomatosis and renal cell carcinoma (HLRCC) syndrome. Am J Surg Pathol. 2007;31:1578–1585. doi: 10.1097/PAS.0b013e31804375b8. [DOI] [PubMed] [Google Scholar]
- 3.Grubb RL, III, Franks ME, Toro J, Middelton L, Choyke L, Fowler S, Torres-Cabala C, Glenn GM, Choyke P, Merino MJ, Zbar B, Pinto PA, Srinivasan R, Coleman JA, Linehan WM. Hereditary leiomyomatosis and renal cell cancer: a syndrome associated with an aggressive form of inherited renal cancer. J Urol. 2007;177:2074–2080. doi: 10.1016/j.juro.2007.01.155. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 5.Alam NA, Rowan AJ, Wortham NC, Pollard PJ, Mitchell M, Tyrer JP, Barclay E, Calonje E, Manek S, Adams SJ, Bowers PW, Burrows NP, Charles-Holmes R, Cook LJ, Daly BM, Ford GP, Fuller LC, Hadfield-Jones SE, Hardwick N, Highet AS, Keefe M, MacDonald-Hull SP, Potts ED, Crone M, Wilkinson S, Camacho-Martinez F, Jablonska S, Ratnavel R, MacDonald A, Mann RJ, Grice K, Guillet G, Lewis-Jones MS, McGrath H, Seukeran DC, Morrison PJ, Fleming S, Rahman S, Kelsell D, Leigh I, Olpin S, Tomlinson IP. Genetic and functional analyses of FH mutations in multiple cutaneous and uterine leiomyomatosis, hereditary leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Hum Mol Genet. 2003;12:1241–1252. doi: 10.1093/hmg/ddg148. [DOI] [PubMed] [Google Scholar]
- 6.Alam NA, Olpin S, Leigh IM. Fumarate hydratase mutations and predisposition to cutaneous leiomyomas, uterine leiomyomas and renal cancer. Br J Dermatol. 2005;153:11–17. doi: 10.1111/j.1365-2133.2005.06678.x. [DOI] [PubMed] [Google Scholar]
- 7.Merino MJ, Torres-Cabala C, Pinto P, Linehan WM. The morphologic spectrum of kidney tumors in hereditary leiomyomatosis and renal cell carcinoma (HLRCC) syndrome. Am J Surg Pathol. 2007;31:1578–1585. doi: 10.1097/PAS.0b013e31804375b8. [DOI] [PubMed] [Google Scholar]
- 8.Zbar B, Glenn G, Lubensky I, Choyke P, Walther MM, Magnusson G, Bergerheim US, Pettersson S, Amin M, Hurley K. Hereditary papillary renal cell carcinoma: clinical studies in 10 families. J Urol. 1995;153:907–912. [PubMed] [Google Scholar]
- 9.Yang Y, Valera VA, Padilla-Nash HM, Sourbier C, Vocke CD, Vira MA, Abu-Asab MS, Bratslavsky G, Tsokos M, Merino MJ, Pinto PA, Srinivasan R, Ried T, Neckers L, Linehan WM. UOK 262 cell line, fumarate hydratase deficient (FH−/FH−) hereditary leiomyomatosis renal cell carcinoma: in vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genet Cytogenet. 2010;196:45–55. doi: 10.1016/j.cancergencyto.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sourbier C, Valera-Romero V, Giubellino A, Yang Y, Sudarshan S, Neckers L, Linehan WM. Increasing reactive oxygen species as a therapeutic approach to treat hereditary leiomyomatosis and renal cell carcinoma. Cell Cycle. 2010;9:4183–4189. doi: 10.4161/cc.9.20.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudarshan S, Sourbier C, Kong HS, Block K, Valera Romero VA, Yang Y, Galindo C, Mollapour M, Scroggins B, Goode N, Lee MJ, Gourlay CW, Trepel J, Linehan WM, Neckers L. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1alpha stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol. 2009;29:4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong WH, Kovtunovych G, Jeong SY, Sougrat R, Yang Y, Sourbier C, Linehan WM, Rouault TA. Reduced AMPK levels and remodeling of iron intracellular iron metabolism in fumarate hydratase-deficient kidney cancer. Cancer Cell. 2011;20:315–327. doi: 10.1016/j.ccr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grubb RL, III, Franks ME, Toro J, Middelton L, Choyke L, Fowler S, Torres-Cabala C, Glenn GM, Choyke P, Merino MJ, Zbar B, Pinto PA, Srinivasan R, Coleman JA, Linehan WM. Hereditary leiomyomatosis and renal cell cancer: a syndrome associated with an aggressive form of inherited renal cancer. J Urol. 2007;177:2074–2079. doi: 10.1016/j.juro.2007.01.155. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Padilla-Nash HM, Vira MA, Abu-Asab MS, Val D, Worrell R, Tsokos M, Merino MJ, Pavlovich CP, Ried T, Linehan WM, Vocke CD. The UOK 257 cell line: a novel model for studies of the human Birt-Hogg-Dube gene pathway. Cancer Genet Cytogenet. 2008;180:100–109. doi: 10.1016/j.cancergencyto.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan MJ, Johnson G, Kirk J, Fuerstenberg SM, Zager RA, Torok-Storb B. HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994;45:48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- 16.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, Chomicz S, Ferrick DA. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 17.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 18.Hatch MD. A simple spectrophotometric assay for fumarate hydratase in crude tissue extracts. Anal Biochem. 1978;85:271–275. doi: 10.1016/0003-2697(78)90299-3. [DOI] [PubMed] [Google Scholar]
- 19.Bai X, Wu J, Zhang Q, Alesci S, Manoli I, Blackman MR, Chrousos GP, Goldstein AL, Rennert OM, Su YA. Third-generation human mitochondria-focused cDNA microarray and its bioinformatic tools for analysis of gene expression. Biotechniques. 2007;42:365–375. doi: 10.2144/000112388. [DOI] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 22.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Su DM, Zhang Q, Wang X, He P, Zhu YJ, Zhao J, Rennert OM, Su YA. Two types of human malignant melanoma cell lines revealed by expression patterns of mitochondrial and survival-apoptosis genes: implications for malignant melanoma therapy. Mol Cancer Ther. 2009 doi: 10.1158/1535-7163.MCT-08-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WARBURG O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 25.Garber K. Energy deregulation: licensing tumors to grow. Science. 2006;312:1158–1159. doi: 10.1126/science.312.5777.1158. [DOI] [PubMed] [Google Scholar]
- 26.Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci U S A. 2001;98:3387–3382. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 29.Toro JR, Nickerson ML, Wei MH, Warren MB, Glenn GM, Turner ML, Stewart L, Duray P, Tourre O, Sharma N, Choyke P, Stratton P, Merino M, Walther MM, Linehan WM, Schmidt LS, Zbar B. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet. 2003;73:95–106. doi: 10.1086/376435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie H, Valera VA, Merino MJ, Amato AM, Signoretti S, Linehan WM, Sukhatme VP, Seth P. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol Cancer Ther. 2009;8:626–635. doi: 10.1158/1535-7163.MCT-08-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Flaherty L, Adam J, Heather LC, Zhdanov AV, Chung YL, Miranda MX, Croft J, Olpin S, Clarke K, Pugh CW, Griffiths J, Papkovsky D, Ashrafian H, Ratcliffe PJ, Pollard PJ. Dysregulation of hypoxia pathways in fumarate hydratase-deficient cells is independent of defective mitochondrial metabolism. Hum Mol Genet. 2010;19:3844–3851. doi: 10.1093/hmg/ddq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardie B, Remenieras A, Kattygnarath D, Bombled J, Lefevre S, Perrier-Trudova V, Rustin P, Barrois M, Slama A, Avril MF, Bessis D, Caron O, Caux F, Collignon P, Coupier I, Cremin C, Dollfus H, Dugast C, Escudier B, Faivre L, Field M, Gilbert-Dussardier B, Janin N, Leport Y, Leroux D, Lipsker D, Malthieu F, McGilliwray B, Maugard C, Mejean A, Mortemousque I, Plessis G, Poppe B, Pruvost-Balland C, Rooker S, Roume J, Soufir N, Steinraths M, Tan MH, Theodore C, Thomas L, Vabres P, Van GE, Meric JB, Verkarre V, Lenoir G, Joulin V, Deveaux S, Cusin V, Feunteun J, Teh BT, Bressac-de PB, Richard S. Novel FH mutations in families with hereditary leiomyomatosis and renal cell cancer (HLRCC) and patients with isolated type 2 papillary renal cell carcinoma. J Med Genet. 2011;48:226–234. doi: 10.1136/jmg.2010.085068. [DOI] [PubMed] [Google Scholar]
- 33.Lorenzato A, Olivero M, Perro M, Briere JJ, Rustin P, Di Renzo MF. A cancer-predisposing “hot spot” mutation of the fumarase gene creates a dominant negative protein. Int J Cancer. 2008;122:947–951. doi: 10.1002/ijc.23209. [DOI] [PubMed] [Google Scholar]
- 34.DiMauro S, Schon EA. Nuclear power and mitochondrial disease. Nat Genet. 1998;19:214–215. doi: 10.1038/883. [DOI] [PubMed] [Google Scholar]
- 35.Reddy PH, McWeeney S, Park BS, Manczak M, Gutala RV, Partovi D, Jung Y, Yau V, Searles R, Mori M, Quinn J. Gene expression profiles of transcripts in amyloid precursor protein transgenic mice: up-regulation of mitochondrial metabolism and apoptotic genes is an early cellular change in Alzheimer’s disease. Hum Mol Genet. 2004;13:1225–1240. doi: 10.1093/hmg/ddh140. [DOI] [PubMed] [Google Scholar]
- 36.Duke DC, Moran LB, Kalaitzakis ME, Deprez M, Dexter DT, Pearce RK, Graeber MB. Transcriptome analysis reveals link between proteasomal and mitochondrial pathways in Parkinson’s disease. Neurogenetics. 2006;7:139–148. doi: 10.1007/s10048-006-0033-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.