Abstract
The zebrafish larva is an optically-transparent vertebrate model with complex organs that is widely used to study genetics, developmental biology, and to model various human diseases. In this article, we present a set of novel technologies that significantly increase the throughput and capabilities of previously described vertebrate automated screening technology (VAST). We developed a robust multi-thread system that can simultaneously process multiple animals. System throughput is limited only by the image acquisition speed rather than by the fluidic or mechanical processes. We developed image recognition algorithms that fully automate manipulation of animals, including orienting and positioning regions of interest within the microscope’s field of view. We also identified the optimal capillary materials for high-resolution, distortion-free, low-background imaging of zebrafish larvae.
Introduction
The zebrafish (Danio rerio) is a widely used vertebrate model for in vivo chemical and genetic screens. Several desirable attributes contribute to the popularity of zebrafish larvae, including their small size, optical transparency, and rapid, ex vivo growth in aqueous medium. Zebrafish models of many important human diseases have been developed1–3. Lead compounds discovered in zebrafish assays have been useful for drug development because of the high level of conservation of drug activity between mammals and zebrafish4. There are also large numbers of zebrafish mutants available, facilitating the study of gene functions and the identification of cellular targets of new compounds. Due to these and other compelling advantages, zebrafish studies have grown at an exponential rate during the last two decades.
The most fundamental challenge limiting the scale of zebrafish studies and screens has been the absence of methodologies for rapid manipulation and imaging. High-resolution imaging requires researchers to immobilize and manually orient each individual larva. Typically this is done by mounting specimens in methylcellulose or low-melting-point agarose, a cumbersome process that takes 5–15 minutes per animal depending on the complexity of the experiment. Although some published screens have involved large numbers of animals5–9, these have been limited to highly specific or low-content assays.
We have previously described the vertebrate automated screening technology (VAST) that allows, for the first time, high-speed screening of zebrafish larvae at cellular resolution10. VAST can process each larva within 20 seconds and an entire multiwell plate within 35 minutes. However, there are considerable advantages to further accelerating zebrafish assays and fully automating the screening process11. Here, we demonstrate novel technologies that significantly increase the throughput of VAST by simultaneously processing multiple fish. Under these conditions, the system throughput becomes limited only by the image acquisition speed rather than by the fluidic and mechanical processes. We also demonstrate image recognition algorithms that fully automate zebrafish manipulation steps such as positioning and orienting animals. In addition, we have analyzed several capillary materials in order to identify the one with the best optical properties for distortion-free, low-background, cellular-resolution imaging of zebrafish.
Materials and methods
Zebrafish
Wild-type and transgenic Gt(T2KSAG)j1282b zebrafish were used in all experiments. Zebrafish larvae were incubated at 28°C in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgSO4) and were anesthetized with 0.2 mg/ml Tricaine during the experiments. All experiments were approved by MIT’s Committee on Animal Care under protocol #0309–025–12.
Multi-thread Vertebrate Automated Screening Technology
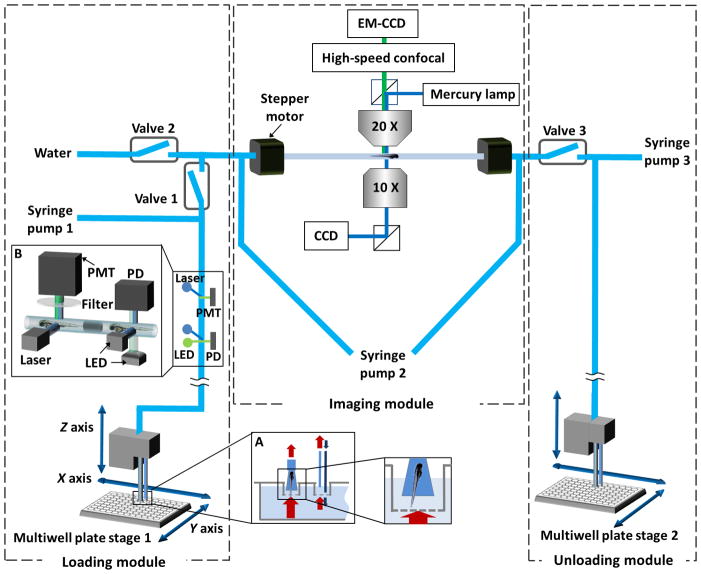
The multi-thread VAST system loads larvae automatically from a 96-well nylon mesh plate that fits into a matching 96-well tray (MultiScreen-Mesh Plate, Millipore Inc.). The multiwell plate is brought into position by a three-axis motor stage (RCA2, Intelligent Actuators). The mesh-filter insert allows easy transfer of larvae. In order to keep the water level constant, two tubes separately aspirate and dispense water (Figure 1 insert A) via the syringe pump. Larvae are detected as they pass through the high-speed zebrafish discriminator, which distinguishes larvae from air bubbles and debris and also differentiates fluorescent from non-fluorescent larvae. The design of the zebrafish discriminator is discussed in detail below. Two high-precision step motors (KP35FM2-044, Japan Servo Co.) hold an ultra-thin glass capillary along its axis of rotation. The capillary has an outer diameter of 700 μm and a wall thickness of 10 μm (BGCT 0.7, Capillary Tube Supplies) and is immersed in a water bath to minimize the index of refraction mismatch. The assembly (i.e. motors, capillary and water bath) is mounted on a three-axis position stage (MPC-200, Sutter Instrument) and held between an upright microscope for epi-fluorescence (and confocal) imaging and an inverted microscope for bright-field imaging. The inverted microscope is connected to a high-speed CCD (GX-1050, Prosilica) and is primarily used to detect, position, and rotate the larvae. A high-speed confocal head (Infinity 3, VisiTech Inc.) with a cooled EM-CCD (iXon+885, Andor Technology) is used for confocal imaging. The fluidic elements are controlled by three pinch valves (075P2NC12, BioChem Fluidics) and three computerized syringe pumps (XLP 6000, Tecan). Fluidic components are connected through silicone tubing (0.8 mm inner diameter, Bio-Chem Fluidics). The entire system is controlled via a code written in MATLAB.
Fig. 1.
Multi-thread Vertebrate Automated Screening Technology (VAST). The platform consists of three subsystems that operate simultaneously: loading, imaging, and unloading. Larvae are automatically loaded to the platform from individual wells of a mesh-filter multiwell plate positioned by a motorized x y stage. The mesh-filter insert allows easy transfer of larvae into the system. In order to avoid a drop in the fluid level within multiwell plate (while larva is being aspirated by the loading nozzle), a circulator is set nearby the loading nozzle (inset A) that consists of a fluid source and an aspirator where the tip of the aspirator is slightly elevated with respect to the tip of the fluid source. A zebrafish discriminator with a brightfield and a fluorescence photodetection system (inset B) discriminates the passage of fluorescent larvae from non-fluorescent ones, air bubbles and debris. Two step motors hold a capillary immersed in a water bath along its axis of rotation; this assembly is mounted on a three-axis position stage (not shown) and held between an upright microscope and an inverted microscope. A multifocal confocal head with a cooled electron-multiplying charge-coupled device (EM-CCD) camera and a second large-area charge-coupled device (CCD) are used for high-speed confocal and wide-field fluorescence imaging, respectively. A high-speed CCD camera connected to the inverted microscope allows rapid bright-field imaging for positioning and orienting the larvae.
Multi-thread Operation
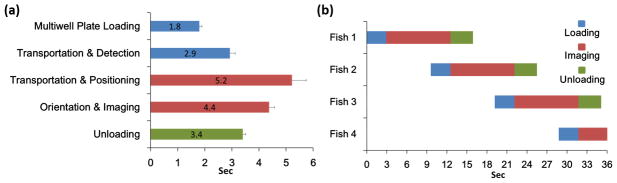
The multi-thread VAST system simultaneously performs three independent operations to enable parallel processing of multiple fish; loading, imaging, and unloading. At the beginning of each cycle, a larva is acquired from a multiwell plate by syringe pump 1 and the zebrafish discriminator is activated to distinguish the contents of the flow (i.e. larvae versus debris or bubbles). After the discriminator detects a larva, pinch valve 1 is opened and syringe pump 1 is stopped, allowing the larva to transit from the loading module to the imaging module. The larva is transported into the imaging module via syringe pump 2 after the flow direction is changed by switching pinch valves 2 and 3. The loading module continuously repeats these steps and each loading cycle takes 4 seconds to complete. In the imaging module, the larva is automatically positioned, rotated, and imaged inside the capillary, which takes 9.6 seconds on average. The timing for imaging was calculated for a single wide-field fluorescence acquisition with a 300 ms exposure time. The larva is then moved to the unloading module and a new larva is transported into the capillary from the loading module. The larva in the unloading module, (held between valve 3 and the imaging module), is then transferred into its corresponding well of the output multiwell plate. Each unloading cycle takes 3.4 seconds and dispenses 500 μL of medium with a flow rate of 165 μL/sec. Timing analysis is based on trials of n=192 animals. As reported previously, over 98% of larvae imaged by VAST survive and develop without noticeable morphological abnormalities10. By multi-threading the loading, imaging and unloading operations, throughput becomes limited only by the slowest operation (i.e. the imaging step; 9.6 seconds), rather than by the total time required to carry out all processes (Figure 2).
Fig. 2.
Timings of multi-thread processing of zebrafish. Blue, red, and green bars indicate the processes comprising loading, imaging, and unloading, respectively. (a) The time required for each handling step (n = 192). Multiwell plate loading process includes positioning the multiwell plate and the loading nozzle, and aspirating a larva into the nozzle from the multiwell plate. Transportation time is the duration it takes to move a larva within the fluidic tube from one point to another. (b) Multi-thread operation: The system simultaneously performs loading, imaging, and unloading operations with three different larvae. As a result, the overall processing time is dictated solely by the duration of the slowest process (i.e. imaging; 9.6 sec), not by the total duration of all processes.
Fluorescence/bright-field automated zebrafish discriminator
A low-cost, high-performance discriminator (Figure 3a) is used to distinguish larvae from debris and air bubbles and to separate fluorescent and non-fluorescent larvae. The discriminator is formed by a bright-field photodetection system and a fluorescence-activated larva sorter. The fluorescence-activated sorter consists of an epi-fluorescence detection system with a GFP filter set, a 10X lens, a photo-multiplier tube (PMT), and a mercury light source.
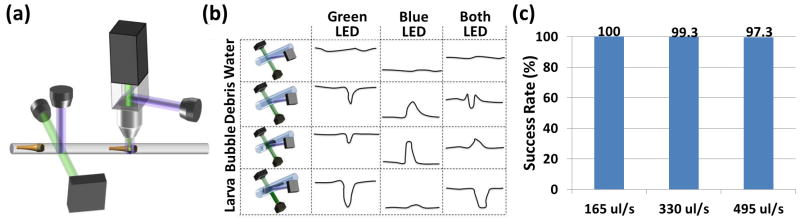
Fig. 3.
Zebrafish discriminator. (a) Schematic representation of the zebrafish discriminator. The system is composed of a bright-field discrimination system and a fluorescence-activated zebrafish sorter. (b) Schematic representation of the mechanism of action of the bright-field discrimination system. By combining both the scattered and transmitted signals the system distinguishes a zebrafish larva from air bubbles and debris. (c) Detection and discrimination reliabilities at increasing flow rates. The reliability is near 100 % at normal operating speeds of 330 μL/s. (n=150).
The bright-field photodetection system is comprised of an integrated photodiode and an amplifier chip (OPT301, Texas Instruments) and two LEDs aligned in transmission and scattering configurations. By simultaneously measuring transmitted light and scattered light at different wavelengths, this device reliably discriminates between larvae, debris, and air bubbles based on the transmission/scattering ratio of different objects (Figure 3b): The transmitted light largely passes through the fluidic tube when the tube is filled with solution. When an object or bubble crosses the light path, the light is absorbed/scattered and therefore the transmitted light intensity received by the photodiode decreases. A live larva tends to absorb light more than scattering it, while a dead larva, debris, or bubble tend to scatter light more than transmitting it. To balance the scattered and transmitted light intensities on the detector, we use two LEDs where one is in the transmission path and the other is in the orthogonal scattering path. LEDs with different emission colors are used to aid the alignment of the transmission/scattering ratio. Although the scattered light intensity is significantly lower than the transmitted light intensity, the emission power of the two LEDs did not require any tuning because of the significant difference in the spectral responsivity of the photodiode to the LEDs’ colors.
Such discrimination of multicellular organisms has traditionally been performed with more complicated setups and image processing algorithms. The reliability of our discriminator was tested under a variety of flow rates, ranging from 165 μL/sec to 495 μL/sec (Figure 3c). For the flow rates that we use in screening (330 μL/sec), the reliability is near 100 %.
Automated identification of zebrafish orientation and position
A key step in the design of VAST is the automation of the fine positioning of the region of interest (ROI) within the relatively small field-of-view (FOV) of the high power objective lenses used for cellular-resolution imaging. To accurately and consistently image the same ROI across many larvae, it is necessary to precisely control both position along the length of the capillary and orientation/rotation around the fixed axis of the capillary. Due to the phenotypic variations that can occur in large-scale genetic and chemical screens, a reliable and flexible algorithm is crucial.
Our algorithm works through a four-step process; coarse positioning of the larva under the FOV, calculation of the direction of its entry into the capillary, identification of its orientation and rotation, and fine positioning of the ROI within the FOV of the high power objective lens.
The coarse positioning of the larvae in the center of the FOV is done through a closed loop feedback between the CCD imaging the FOV and the computer-controlled syringe pump as previously described 10.
Larvae can enter the capillary in either a head- or tail-first orientation. For this reason, the longitudinal (head vs. tail) orientation of the larvae has to be identified before any image processing steps are performed. A 1D pixel intensity profile f(x) for the fish can be computed by summing the pixel intensities across the axis orthogonal to the main tube axis. The center of mass (of pixel intensities), CM, of the larvae is computed by summing the position values weighted by the normalized 1D intensity profile.
The largest intensity variation between the zebrafish and surrounding water/tubing occurs at the very tip of the head, x0, which is identified by finding the position of maximum value of f′, the derivative of the 1D intensity profile. The center of mass falls in the upper part of the abdomen, and therefore the orientation of the larvae can be found by comparing the geometrical center to the center of mass of the fish.
Where D = 1 (D = −1) means that the head is towards left (right).
We next identify the lateral orientation of the larva. Due to the anatomical symmetry and optical transparency of the larvae, it is unreliable to identify the lateral orientation based only on the ventral and dorsal views (Figure 4a). Instead, we acquire a series of 2D images of the larva f(x,y) at a constant frame rate of 180 frames per second (fps) while rotating the capillary 360 degrees at a constant rotation speed (Figure 4b). Using the frame acquisition rate and rotation speed, a rotation angle is assigned for each acquired frame. We then compute the cross correlation12, CC, between each frame and predefined template intensity distributions t(x,y) of age-matched larvae imaged at dorsal and lateral views using the following relationship:
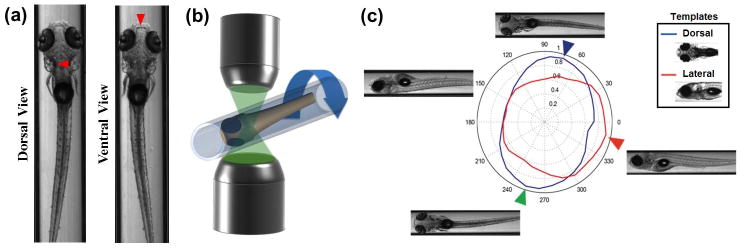
Fig. 4.
Identification of the angle and position of larvae. (a) Dorsally and ventrally oriented larvae. Red arrows indicate the dominant morphological features for distinguishing dorsally vs. ventrally views. It is too difficult to reliably determine the larva’s orientation based solely on comparisons with dorsal and ventral reference images. (b) The larva is rotated along its longitudinal axis through a full 360° and snapshots are acquired at two degree increments at 180 frames per second. (c) The system then correlates the images with a library of prerecorded dorsal and lateral images from stage-matched control larvae (inset). The blue and red curves in the radar chart are results of the image correlations with the dorsal and lateral templates. The blue, green, and red arrows indicate the orientations of maximum correlations with the dorsally, ventrally and laterally orientated templates, respectively. The reliability of the algorithm is 99 % (n=100 larvae)
Where f̄u,v and t̄u,v are the average pixel values for the image and template, respectively. The cross-correlation with the dorsal template yields two narrow peaks at the dorsal and ventral orientations (Figure 4c). However, these peaks alone are insufficient to distinguish between dorsal and ventral sides. The cross-correlation with the lateral template yields a single wider peak that identifies the right versus left side of the fish. This knowledge of left/right orientation and the direction of rotation are then used to differentiate which peaks from the dorsal template corresponds to the ventral vs. dorsal orientations. To handle phenotypic variation across larvae, we have constructed a collection of templates for larvae under the typical morphological abnormalities (e.g. pericardial edema). When the maximum cross correlation between the templates and the larva does not exceed a user defined minimum, our algorithm looks through this collection of templates and recalculates the orientation using the template with the highest correlation. If the cross correlation does not reach a user-defined threshold for any template, the larva is discarded. The reliability of the orientation algorithm is 99 % (n=100 larvae).
Once the dorsal, ventral, and lateral orientations have been identified, the larva is automatically rotated for imaging at the predefined angle specified by the user (e.g. dorsally to image the brain, ventrally to image the heart and brachial arches, obliquely to image the pancreas). At this orientation, the ROI is defined by matching the larva to a third template of aged-matched larvae at the desired angle which contains the location of the ROI.
Analysis of capillary materials for high- quality imaging
Confocal imaging necessitates both low autofluorescence and low optical aberrations. Since larvae are imaged through a capillary in VAST, the choice of capillary material is a critical factor in achieving distortion-free, low-background, high-resolution imaging. A previous study reported that Teflon tubes offer optimal quality for bright-field imaging of larvae within capillaries13, due to the matched refractive index with water. However, limited quantitative data has been reported for fluorescence imaging, which is crucial for most experiments. Polymers such as PTFE (polytetrafluoroethylene) and FEP (fluorinated ethylene propylene), two types of Teflon, cause significant autofluorescence. This dramatically decreases the signal-to-noise ratio (SNR) and lowers the quality of fluorescent images. On the other hand, glass capillaries with ultra-low autofluorescence and extinction coefficients are not refractive index-matched to the surrounding water, which can create distortions and lead to decreased confocality and axial resolutions.
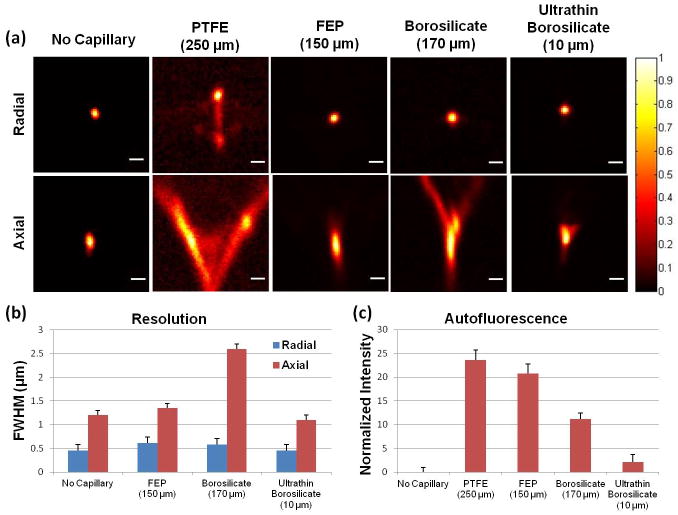
We have tested four capillaries of different materials and thicknesses and quantitatively evaluated the point-spread functions (PSF) of the resultant confocal images. GFP-tagged fluorescent micro beads (0.2 μm in diameter) were mixed in 2 % Type IV optical agarose (Sigma, A3643) to avoid Brownian motion. The beads were then imaged with a multifocal confocal system with pinhole size of 30 μm, through a 100x 1.1 NA water dipping objective (Nikon Instruments). The beads trapped within the agar mold (index matched to water) were imaged either without any intervening glass, inside a PTFE capillary with a wall thickness of 250 μm (Zeus Inc.), inside an FEP capillary with a wall thickness of 150 μm (Zeus Inc.), inside a borosilicate capillary with a wall thickness of 170 μm (Wale Apparatus Co.), or within an ultrathin borosilicate capillary tube with a wall thickness of 10 μm (Capillary Tube Supplies Ltd.). Figure 5a shows a cross-section in the radial and axial planes through the center of a bead for each of the conditions.
Fig. 5.
Analysis of capillary materials for distortion-free low-background high-resolution imaging. (a) From left to right, the point-spread functions (PSF) with different materials/conditions; no capillary, PTFE capillary, FEP capillary, borosilicate glass capillary, and ultra-thin borosilicate glass capillary. The wall thicknesses of capillaries are 250 μm, 150 μm, 170 μm and 10 μm, respectively (shown in brackets). The ultra-thin glass capillary produces the least image distortion. Scale bar is 1 μm. (b) Radial (blue) and axial (red) resolutions of candidate materials. PTFE was not included due to the significant non-Gaussian PSF it produces. (c) Autofluorescence analysis of candidate materials. PTFE and FEP capillaries cause significant autofluorescence.
We computed the axial full-width half-maximum (FWHM) of the PSFs without a capillary and when using the FEP, 170 μm thick borosilicate, and the 10 μm thick borosilicate capillaries. The FWHMs were 1.2 μm, 1.4 μm, 2.6 μm and 1.1 μm, respectively (Figure 5b). Due to the optical aberrations that cause non-Gaussian like PSFs on the PTFE capillary, we were unable to compute a FWHM. Importantly, our analysis shows that the 10 μm thick borosilicate capillary produces significantly less image distortion than the Teflon capillaries with refractive indexes closer to water. Furthermore, our autofluorescence analysis demonstrates that, unlike borosilicate, both types of Teflon (i.e. PTFE and FEP) cause significant autofluorescence (Figure 5c). Thus, we conclude the ultrathin (10 μm thick) borosilicate capillary is the most appropriate choice for high-resolution confocal imaging.
Conclusions
Here, we have demonstrated a fully automated multi-threaded vertebrate screening platform. To significantly increase throughput, we modularized the system into three independent sections that can process multiple zebrafish in parallel. In this configuration, system throughput is limited only by the image acquisition speed rather than by the speeds of fluidic and mechanical processes. A discriminator differentiates the entry of a fluorescent larva from non-fluorescent larva, air bubbles, and small debris. A simple and highly reliable algorithm is implemented for automated identification of the position and rotational angle of the larva. Furthermore, to identify the best capillary materials for confocal and fluorescence imaging, we analyzed various capillaries measuring their PSFs and autofluorescence. We showed that ultra-thin borosilicate capillaries (with wall thickness of 10 μm) provide the highest-resolution distortion-free low-background images.
Supplementary Material
Acknowledgments
All zebrafish used in these studies were raised and maintained in the Koch Institute Zebrafish Core Facility. We thank Dr. Adam Amsterdam for his oversight of the fish facility, and specifically of the transgenic lines used here, and we thank Tim Angellini for technical assistance in raising and maintaining the fish. We also thank Dr. Peter Eimon for his comments and help during the writing of the manuscript. This work was supported by NIH Director’s New Innovator Award (DP2 OD002989), NIH Transformative Research Award (R01 NS073127), Packard Award in Science and Engineering, MIT-Harvard Broad Institute SPARC Award, Foxconn Sponsorship, the Martino’s Centre for Biomedical Imaging Training Grant, and the LaCaixa Scholarship.
Footnotes
Author contributions.
T.Y.C., C.P., A.A., C.W., and M.F.Y. designed experiments; T.Y.C., C.P., and M.F.Y. wrote the manuscript; T.Y.C., C.P., and A.A. performed the experiments.
Competing interest statement.
The authors declare that they have no competing financial interest.
References
- 1.Zon LI, Peterson RT. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 2.Lieschke GJ, Currie PD. Nature Reviews Genetics. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 3.Meeker ND, Trede NS. Dev Comp Immunol. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Kari G, Rodeck U, Dicker AP. Clin Pharmacol Ther. 2007;82:70–80. doi: 10.1038/sj.clpt.6100223. [DOI] [PubMed] [Google Scholar]
- 5.Gehrig J, Reischl M, Kalmar E, Ferg M, Hadzhiev Y, Zaucker A, Song C, Schindler S, Liebel U, Muller F. Nat Methods. 2009;6:911–916. doi: 10.1038/nmeth.1396. [DOI] [PubMed] [Google Scholar]
- 6.Kokel D, Bryan J, Laggner C, White R, Cheung CY, Mateus R, Healey D, Kim S, Werdich AA, Haggarty SJ, Macrae CA, Shoichet B, Peterson RT. Nat Chem Biol. 2010;6:231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, Schier AF. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran TC, Sneed B, Haider J, Blavo D, White A, Aiyejorun T, Baranowski TC, Rubinstein AL, Doan TN, Dingledine R, Sandberg EM. Cancer Res. 2007;67:11386–11392. doi: 10.1158/0008-5472.CAN-07-3126. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho R, de Sonneville J, Stockhammer OW, Savage ND, Veneman WJ, Ottenhoff TH, Dirks RP, Meijer AH, Spaink HP. PLoS One. 2011;6:e16779. doi: 10.1371/journal.pone.0016779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pardo-Martin C, Chang TY, Koo BK, Gilleland CL, Wasserman SC, Yanik MF. Nat Methods. 2010;7:634–636. doi: 10.1038/nmeth.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamplin OJ, Zon LI. Nature Methods. 2010;7:600–601. doi: 10.1038/nmeth0810-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis JP. Vision Interface. 1995:120–123. [Google Scholar]
- 13.Ekker SC, Petzold AM, Bedell VM, Boczek NJ, Essner JJ, Balciunas D, Clark KJ. Zebrafish. 2010;7:149–154. doi: 10.1089/zeb.2010.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.