Abstract
Chronic infections with the human malaria parasite Plasmodium falciparum depend on antigenic variation. P. falciparum erythrocyte membrane protein 1 (PfEMP1), the major erythrocyte surface antigen mediating parasite sequestration in the microvasculature, is encoded in parasites by a highly diverse family of var genes. Antigenic switching is mediated by clonal variation in var expression, and recent in vitro studies have demonstrated a role for epigenetic processes in var regulation. Expression of particular PfEMP1 variants may result in parasite enrichment in different tissues, a factor in the development of severe disease. Here, we study in vivo human infections and provide evidence that infection-induced stress responses in the host can modify PfEMP1 expression via the perturbation of epigenetic mechanisms. Our work suggests that severe disease may not be the direct result of an adaptive virulence strategy to maximize parasite survival but that it may indicate a loss of control of the carefully regulated process of antigenic switching that maintains chronic infections.
The vast majority of infections with the human malaria parasite Plasmodium falciparum result in mild disease or asymptomatic carriage that can last for several months. Many infections do, however, lead to acute, potentially fatal malaria, and the complex interactions between host and parasite that generate severe disease are poorly understood. Chronic infections persist in part because of antigenic variation by the parasite, which evades host immunity through the sequential expression of antigenic variants of P. falciparum erythrocyte membrane protein 1 (PfEMP1) on the surface of infected erythrocytes. PfEMP1 also mediates sequestration in the microvasculature and prevents splenic clearance.
PfEMP1 is encoded by a highly diverse family of var genes [1–3]. These genes are generally silenced: antigenic switching is mediated by clonal variation in their expression [4], and this is associated with recurring waves of high parasitemia observed in nonimmune human volunteers infected with P. falciparum for malariotherapy [5]. Recent in vitro studies have demonstrated that epigenetic processes are involved in the control of antigenic variation by the parasite: expressed or silenced var genes are marked with activating or silencing histone modifications, and var silencing is relaxed in the absence of either of the parasite's NAD+-dependent histone deacetylase enzymes, or sirtuins [6–9].
Var genes are categorized into 5 subgroups, upsA–upsE, according to their genomic locations and conserved upstream promoter sequences [10, 11]. Previous studies have reported that genes in the upsA and/or upsB subgroups tend to be expressed in children with symptomatic or severe malaria [12–15], while upsE genes are associated with symptomatic malaria in pregnancy [16, 17]. The upsA, upsB, and upsE gene subgroups are those predominantly affected by in vitro sirtuin deletions [6, 7]. Furthermore, both sirtuins also affect telomere maintenance [6], a possible factor in the silencing of subtelomeric var genes via the telomere position effect. Together, this evidence suggests that there may be links between sirtuins, var expression, telomere maintenance, and disease severity in patients. We now show for the first time that the expression of epigenetic modifiers during in vivo infections correlates with patterns of var expression, severe malaria, and host stress factors in Gambian children.
MATERIALS AND METHODS
Patient Population and Sampling
P. falciparum genomic DNA and RNA were prepared from venous blood samples from 125 children with symptomatic malarial infections (65 had severe infection, and 60 had mild infection). Patient recruitment, sample collection, and inclusion criteria for this study are described by Walther et al. [18], and data were obtained from samples collected with these criteria between 2005 and 2009 (2 samples were collected in 2005, 5 in 2006, 32 in 2007, 17 in 2008, and 69 in 2009). No systemic differences between the various years were observed. The study was reviewed and approved by the Gambian Government/MRC Ethics Committee, the Ethics Committee of the London School of Hygiene and Tropical Medicine, and the Office of Human Research Administration at the Harvard School of Public Health.
RNA and DNA Preparation
Blood samples were processed within 2 hours of collection. Cells were washed in Roswell Park Memorial Institute (RPMI) 1640 medium, peripheral blood mononuclear cells were removed by density centrifugation (800 g, 30 min) over a Nycoprep 1.077 gradient (Nycomed, Sweden) or over a 50:50 Nycoprep/Histopac gradient (Sigma), and the erythrocyte fraction was then washed 3 times in RPMI 1640 medium. Approximately 100 μL of infected erythrocytes were stored at −80°C for genomic DNA preparation, and approximately 200 μL were stored in Trizol (Invitrogen) at −80°C for RNA preparation. RNA was prepared using the Purelink RNA mini kit (Invitrogen), and DNA was prepared using the DNA blood mini kit (Qiagen). In addition to ring-stage samples obtained directly from patient blood, late trophozoite/schizont stage RNA was obtained by maturing an aliquot of infected erythrocytes in in vitro culture for approximately 36–48 hours until late stage parasites were visible in Giemsa-stained blood smears (an equivalent of about 50 μL erythrocyte material was extracted for this analysis).
Gene Expression Analysis
Reverse-transcription polymerase chain reaction (PCR) and quantitative PCR to measure var, PfSir2a, PfSir2b, and PfTERT transcript abundance was performed using ring-stage RNA as described by Rottmann et al. [13]. Primers for the conserved acidic terminal segment (ATS) region of all var genes (kindly provided by I. Felger, STI, Basel) were added to the set of previously published ups-specific primers. The unpublished primers are as follows: ATS, cccatacacaaccaaytgga (forward) and ttcgcacatatctctatgtctatct (reverse); SBP1, cacttgcaactaccgaatta (forward) and gtaaagcttcttgagccatt (reverse); MAHRP, gatgttccatttggattttt (forward) and gttttccttcaccatcagtt (reverse); seryl-tRNA synthetase, aagtagcaggtcatcgtggtt(forward) and ttcggcacattcttccataa (reverse); myosin, ggttcagaggatggcaacat (forward) and agacctgcgccagaaaacta (reverse); Sir2a, gggaatgtatttgaagcagt (forward) and cgatgtgccaattactaaaa (reverse); Sir2b, gtcccggctagctcttatcc (forward) and aattgggcaccta (reverse); PfTERT, aatcgaagccttccgttatg (forward) and ttgaacatgtaaggccatgaa (reverse); and PF10_0051, catttttaacacatggagggtct (forward) and tgaagtgtatccttgatttgtgc (reverse). ΔΔCt analysis yielded relative copy numbers of each transcript, compared with the average of 2 conserved ring-stage-expressed genes, one encoding spectrin-binding protein (SBP1) and the other encoding membrane-associated histidine-rich protein (MAHRP). PfTERT was additionally measured in mature-stage RNA, controlled to the average transcript levels of 2 housekeeping genes, one encoding myosin and the other encoding seryl-tRNA synthetase.
To measure var expression in the 3D7 laboratory strain, a validated set of primers recognizing almost every var gene in the genome was used [17, 19]. RNA preparation and quantitative PCR were performed as described by Dzikowski et al. [19], with modifications as reported by Merrick et al. [20]. Three biological replicates were performed for each line, and relative copy numbers were calculated by comparison to the average level of transcripts from 5 housekeeping genes.
Parasite Culture and Transfection
The 3D7 strain of P. falciparum was obtained from the Walter and Eliza Hall Institute (Melbourne, Australia). Parasites were maintained in vitro in human O+ erythrocytes at 4% hematocrit in RPMI 1640 medium (Sigma) supplemented with 25 mM HEPES (EMD Biosciences), sodium bicarbonate (Sigma), 50 mg/L hypoxanthine (Sigma), and 0.5% Albumax (Invitrogen) [21]. Sorbitol-synchronized ring-stage parasites at 5% parasitemia were transfected with approximately 100 mg DNA by electroporation and then maintained with 2.5 mg/mL Blasticidin HCl (Sigma) until drug-resistant parasites were visible in blood smears, around 4 weeks after transfection. The presence of the transfected plasmid was confirmed by Southern blotting.
Plasmid Construction
A plasmid containing the coding sequence for PfSir2a (PF13_0152) and ∼1.75kb of its own upstream promoter sequence from the 3D7 strain was constructed in the plasmid vector pLNENRGFP [22]. The PfSir2a coding sequence was amplified from 3D7 genomic DNA, using the primers gatccctaggatgggtaatttaatgatt (forward) and gatccttaagctacattattttcttattttt (reverse), and cloned into pLNENRGFP as an AvrII/AflII fragment. The construct was verified by sequencing. The PfSir2a upstream sequence was then amplified (Sir2proF/Sir2proR) using the primers atgggcccacttaataacgatag (forward) and gatccctaggaatatataattattttaatcttaac (reverse) and cloned into this plasmid as an Apa1/AvrII fragment, replacing the calmodulin promoter sequence.
Telomere Length Analysis
Median telomere length was measured by telomere restriction fragment Southern blotting as previously described [23]. Blots were analyzed with ImageJ software to obtain the median length for each telomere smear. There was sufficient DNA to obtain data from 102 of the 125 parasite samples.
Statistical Analysis
Assay data (transcript levels and phenotypic metadata are available in the datasheet [Supplementary Materials]) were included from 116 cases (56 with severe infection and 60 with mild infection) from 2007 to 2009. A total of 73 initial features were collapsed by single linkage clustering using mutual information into 46 groups, using a threshold of 40% shared information (available in the datasheet [Supplementary Materials]). Two extreme outliers representing mislabeled data were removed, and all pairs were tested using Spearman rank correlation for continuously valued features and the Mann–Whitney U test (for 2-valued factors) or the Kruskal-Wallis rank sum test (for multivalued factors) for discrete factors. Significance was assessed using an empirical bootstrap P value over 5000 randomizations (nonparametric tests and bootstrapping were chosen to avoid effects from remaining experimental outliers). P values were adjusted for multiple hypothesis testing, using false discovery rate (FDR) q values.
RESULTS
Expression of var Subsets and Sirtuins Are Linked in Parasites Causing Severe Malaria
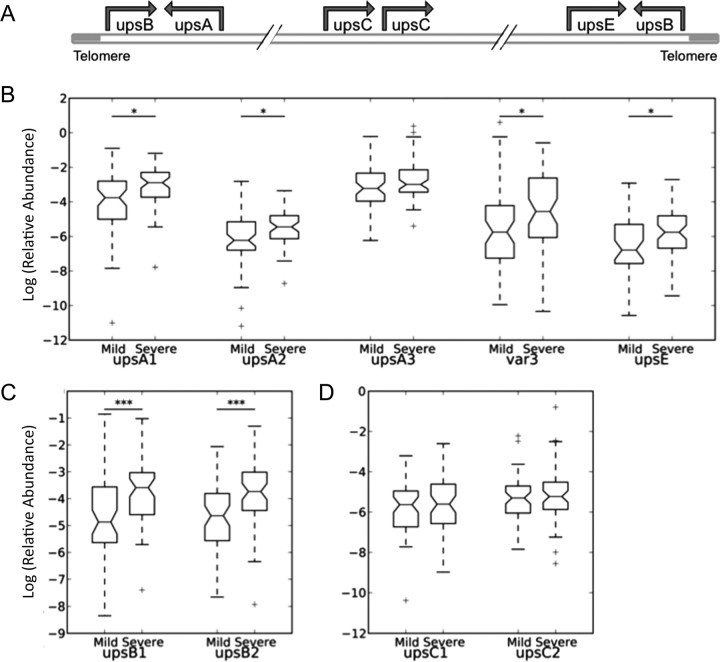
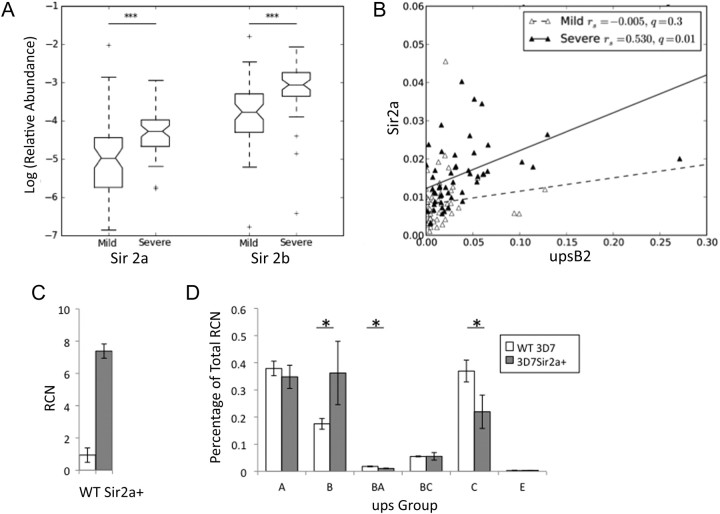
We analyzed isolates of P. falciparum from 116 Gambian children with severe or mild malaria by quantitative PCR to determine patterns of var expression and levels of expression of the sirtuins PfSir2a and PfSir2b. Parasites in patients with severe disease were the highest expressers of upsB genes, the most subtelomeric of the var subgroups (Spearman 2-tailed FDR, q = 0.008 for upsB1 and q = 0.008 for upsB2) (Figure 1A), and there were also nonsignificant trends for higher expression of subtelomeric upsA and upsE genes (q = 0.07 for upsA1, q = 0.06 for upsA2, and q = 0.014 for upsE) (Figure 1B and 1C). By contrast, expression levels of chromosome-internal var genes—the upsC subgroup—were not associated with disease severity (q = 0.8 for upsC1 and q = 0.8 for upsC2) (Figure 1D). Both sirtuins were more highly expressed in parasites causing severe disease than in those causing mild disease (q = 0.009 for PfSir2a and q = 0.009 for PfSir2b) (Figure 2A). Two other genes, which encode the catalytic subunit of telomerase and a putative adenosine diphosphate–adenosine triphosphate translocator, were not correlated with disease severity. Furthermore, high PfSir2a expression correlated strongly with high expression of upsB genes (as expected, since severe disease and upsB var expression are correlated) (Figure 2B, Supplementary Figure 1). This correlation was seen only in parasites from patients with severe cases of malaria (q = 0.01 for upsB2/Sir2a), possibly indicating a threshold effect whereby the sirtuin and virulence genes are only highly expressed during severe disease. A similar strong correlation was not observed between PfSir2b transcript levels and upsB var expression levels (Supplementary Figure 2), suggesting a greater impact in vivo of PfSir2a on var silencing.
Figure 1.
Transcript abundance of each group of var genes in parasites causing severe or mild malaria. Transcript abundance is relative to 2 conserved ring-stage transcripts. Data are for isolates from 56 patients with mild malaria and 60 with severe malaria. Boxes indicate medians and interquartile ranges (IQRs), and whiskers indicate 1.5 times the IQRs. *Nominal statistical significance (false discovery rate [FDR] q < 0.1) and ***statistical significance (FDR q < 0.01), by the nonparametric Mann–Whitney U test. A, Chromosomal arrangement of var gene groups. B, Transcript abundance of upsA and E var genes. Three degenerate primer pairs recognize conserved regions within upsA genes; one recognizes the conserved upsA subgroup, termed var3; one recognizes the unique upsE upstream sequence. C, Transcript abundance of upsB var genes (2 degenerate primer pairs recognizing upsB upstream sequences). D, Transcript abundance of upsC var genes (2 degenerate primer pairs recognizing upsC upstream sequences).
Figure 2.
Sirtuin expression is elevated in parasites causing severe malaria. A, Abundance of sirtuin transcripts in parasites causing severe or mild malaria. Transcript abundance and box plots are defined as described in Figure 1. B, Correlation between upsB var and PfSir2a transcript abundance. Stratification by disease severity demonstrates significant coregulation only in severe disease (Spearman q = 0.01; linear fit shown). Both transcripts are also significantly upregulated in severe malaria (Figures 1B and 2A). C, Quantitative polymerase chain reaction shows that PfSir2a expression increases several-fold in 3D7PfSir2a+, compared with the parent. Transcript abundance was measured in the schizont stage, at which PfSir2a expression peaks in 3D7 (wild type [WT]). Relative copy number (RCN) for PfSir2a was calculated by comparison to the average abundance of transcripts for seryl-tRNA synthetase and myosin. D, var expression profiles were measured in 3D7 (white bars) and 3D7PfSir2a+ (black bars). RCNs for every individual var gene were assembled by ups group, and the proportion of the total var transcripts for each group was calculated. “upsBA” and “upsBC” refer to groups of vars with hybrid upstream sequences. Three biological replicates were performed, and differences between 3D7 and PfSir2a+ were assessed by 2-tailed t test. *P < .05.
Overexpression of PfSir2a in vitro Affects var Expression
Paradoxically, in vitro data suggests that subtelomeric var genes are generally upregulated rather than downregulated in genetic knockouts of the sirtuins [6, 7]. To test the effect of PfSir2a upregulation on var expression in vitro, a strain overexpressing PfSir2a was generated (Figure 2C). The modified strain, 3D7PfSir2a+, showed deregulated var expression with a bias toward subtelomeric upsB var genes, whereas the parental strain predominantly expressed chromosome-internal var genes (Figure 2D).
Telomere Length in Field Isolates Is Highly Variable but Independent of Sirtuin and var Expression
The control of gene expression among upsA, upsB, and upsE var genes, which lie in subtelomeric regions of the parasite's genome, may be influenced by the telomere position effect (a phenomenon in which distance from the telomere can influence patterns of gene expression and silencing due to proximity to the nuclear periphery). Telomeres in the 3D7PfSir2a+ line were shortened, compared with those of the parental strain (Figure 3A), while the genetic knockout of PfSir2a showed dramatically elongated telomeres [7]. PfSir2a therefore had an influence on telomere maintenance, providing a possible mechanism for the alterations in var expression observed. Median telomere length was determined for 102 of the Gambian isolates, and significant variation was seen (mean, 1.23 kb [range, 0.78–1.78 kb]; SD, 0.21 kb) (Figure 3B). The diversity of telomere lengths among these local isolates was almost as great as that measured among 56 laboratory isolates with diverse worldwide origins (mean, 1.34 kb [range, 0.80–2.30 kb]; SD, 0.31 kb). Variable telomerase levels may contribute to this, since telomere lengths correlated significantly with expression of the telomerase catalytic subunit, PfTERT, in mature, replicative-stage parasites (q = 0.04) (Figure 3C). However, no significant relationships were observed between telomere length and disease severity nor between telomere length and sirtuin or var expression.
Figure 3.
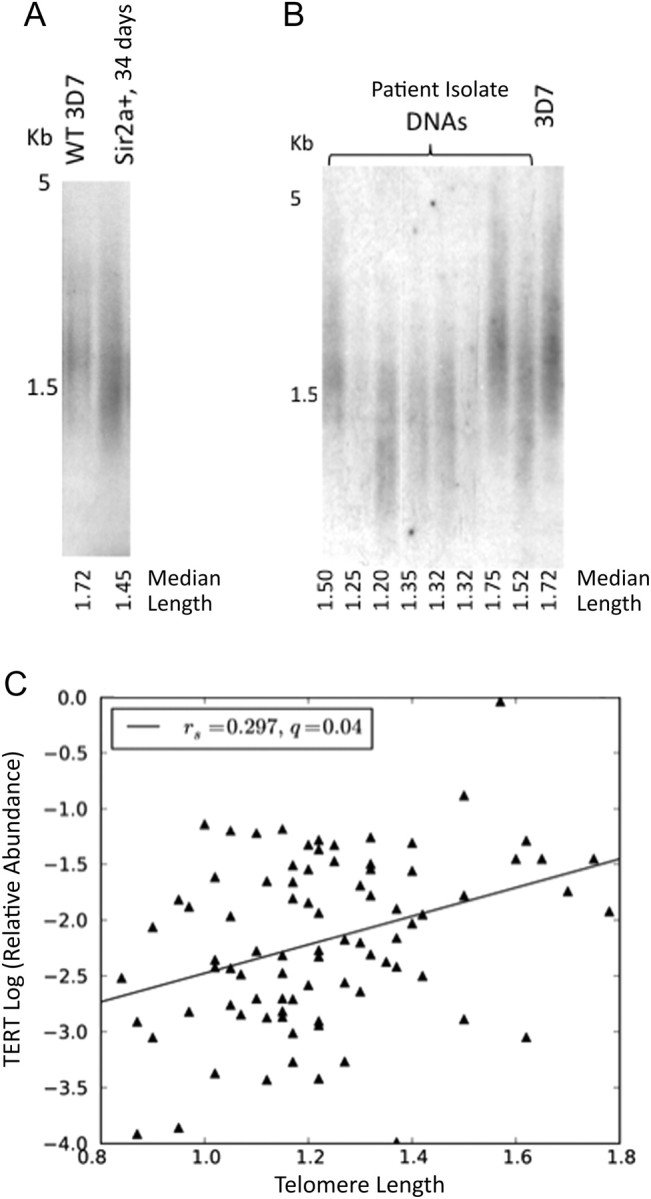
Telomere length is highly variable in field isolates. A, Telomere restriction fragment (TRF) Southern blot showing telomere lengths in genomic DNA from 3D7 (wild type [WT]), compared with 3D7Sir2a+ (34 days after transfection). B, TRF Southern blot showing telomere lengths in parasite DNA from Gambian field isolates. C, Telomere lengths in field isolates plotted against relative transcript abundance of PfTERT in mature-stage parasites. Transcript abundance was measured as described in Figure 2C (Spearman q = 0.04; linear fit shown).
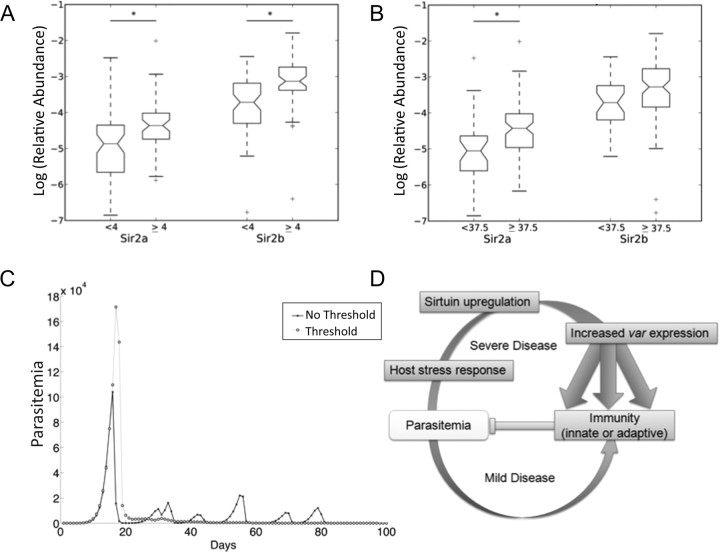
High Temperature and High Blood Lactate Level in Patients Are Linked to High Sirtuin Expression
To identify potential stimuli for the upregulation of a sirtuin-dependent epigenetic program, patient data were interrogated for host factors associated with sirtuin or var expression. Transcriptional responses in both sirtuins and var genes to certain physiological stresses have been demonstrated in vitro [24–27], and microarray analysis of patient-derived strains previously revealed elevated transcription of PfSir2a in a group of isolates apparently responding to starvation [28]. This is of particular interest because sirtuins in model organisms respond to nutritional and metabolic status via the NAD+ cofactor. Here, high fever and high blood lactate level both correlated with high expression of sirtuins (q = 0.01 for temperature/Sir2a, and q = 0.02 for Sir2b; q = 0.03 for lactate/Sir2a, and q = 0.02 for Sir2b) (Figure 4A and 4B). Lactate level also correlated with PfSir2a and PfSir2b expression when analyzed as a continuous variable (Supplementary Figure 3).
Figure 4.
High temperature and high blood lactate level in patients are linked to high sirtuin expression. A, PfSir2a and PfSir2b transcript abundance in patients with blood lactate level <4 mmol/L or ≥4 mmol/L. Box plots and significance are defined as described in Figure 1. Spearman Sir2a q = 0.03, Sir2b q = 0.02; discretization at severe malaria concentration threshold shown for visualization, Kruskal-Wallis Sir2a q = 0.05, Sir2b q = 0.03, n = (57, 52). B, PfSir2a and PfSir2b transcript abundance in patients with temperatures <37.5°C or ≥37.5°C. Spearman Sir2a q = 0.03, Sir2b q = 0.02; for blood lactate as a continuous variable; discretization of blood lactate at severe malaria concentration threshold shown for visualization, with Kruskal–Wallis values Sir2a q = 0.05, Sir2b q = 0.03, n = (57, 52). B, PfSir2a and PfSir2b transcript abundance in patients with temperatures above or below 37.5°C. Spearman Sir2a q = 0.01, Sir2b q = 0.02; for temperature as a continuous variable; discretization of temperature at standard severe malaria temperature threshold shown for visualization, with Kruskal–Wallis Sir2a q = 0.02, Sir2b q = 0.14, n = (30, 85). C, A representative pair of model simulations, illustrating infection dynamics with and without the threshold effect. All parameters are kept equivalent except the distribution of switch probabilities (see Methods), governed in this case by an exponential distribution with mean μ of 2. Other parameter values: Pc* = 30, Pm* = 8, threshold = log (parasitized RBCs per ml) = 6. D, Possible relationships between patient stress factors and sirtuin expression, var gene expression, and, hence, disease severity.
Modification of Parasite Gene Expression by Host Stressors Determines Infection Dynamics in a Within-Host Model
To explore how host stress factors might influence parasites to negatively affect disease outcome, a simple extension of a previously published mathematical model for the progression of malaria infections [29] was developed, incorporating a stressor threshold effect of parasite density. The model assumed that above this threshold, var expression was perturbed, broadening the distribution of switching between variants and raising the probability of expressing antigenic variants with high growth rates (more information on the model is available in the Supplementary Materials; Supplementary Figure 4 and Supplementary Table 1). Simulated infections with the threshold effect were characterized by much higher initial peak parasitemias and by more heterogenous var expression than in the absence of such a threshold (Figure 4C). These peaks stimulated a strong immune response, suppressing further distinct peaks as the infection progressed and reducing parasite clearance time. The effect of the stressor threshold was therefore to generate an acute rather than a chronic infection, with a loss of tight control of antigenic switching.
DISCUSSION
This study shows that severe malaria in Gambian children is linked to high expression of both the P. falciparum sirtuin genes and the subtelomeric var genes, particularly of the upsB type. We provide evidence that parasite sirtuins may respond transcriptionally to host stress factors such as fever and elevated blood lactate level, altering var expression by affecting the subtelomeric regions of the parasite genome and ultimately contributing to the risk of developing severe malaria. Thus, although virulence is generally thought to be an outcome of underlying host and parasite genetics, the epigenetic environment may also play an important role in disease severity.
The link between increased expression of specific var gene subsets and disease severity in this Gambian population is consistent with results of previous studies from other geographical regions [12–14]. Although hemoglobinopathies, such as HbS, could affect disease severity, these were at a very low prevalence in the study population. This study represents the first evidence that expression of histone-modifying enzymes in P. falciparum is linked to disease severity and to the expression of particular virulence genes. We support our in vivo findings with in vitro genetic evidence showing that overexpression of PfSir2a can result in alterations in the expression of var gene subsets. Perturbations in var expression are also seen in the complete absence of PfSir2a [6, 7]. In model systems, both the knockout and the overexpression of certain silencing factors can likewise cause similar phenotypes [30]. The PfSir2a-overexpressing line may be more relevant than the genetic knockout to the in vivo situation, since no natural knockout of either sirtuin has been reported and the in vivo relationships between epigenetic factors and virulence genes are likely to be quantitative and dynamic rather than absolute and irreversible.
The other phenotype observed following overexpression of PfSir2a was a shortening in telomere length [6, 7]. Previously a lengthening in telomere length was observed following disruption of PfSir2a. Telomere length could affect var expression by altering the influence of telomere position effect at these loci. However, telomere length was not linked to var expression or disease severity in the field isolates examined in this study. Significant changes in telomere length probably require many replicative cycles, in contrast with acute epigenetic changes that could alter var transcription patterns directly via changes in chromatin structure.
In this study, we found that the level of sirtuin expression was linked to the environmental signals of temperature and lactate levels. This result suggests that some regulatory circuits in P. falciparum may diverge from a prior assertion that gene transcription in P. falciparum follows a hard-wired cascade unresponsive to stresses such as drug treatment [31]. Indeed Rosenberg et al. have observed that var expression in vitro can be affected by the biological stresses of oxidative damage and glucose starvation [26]. The in vivo parasite may in fact possess sophisticated responses to natural physiological stresses, including fever and variation in bloodstream metabolites, to control expression of specific virulence genes. Indeed, the human sirtuin SirT1 responds to a variety of stresses and upregulates a heat-inducible transcription factor in response to heat shock [32], while in cultured malaria parasites, heat shock can elevate transcription of both PfSir2a and var genes [25, 27]. Of note, sirtuins can also be regulated at the protein level, via posttranslational modification or availability of the NAD+ cofactor [33], which could additionally influence parasite phenotypes in ways not addressed here.
Here, we present a simple within-host model to simulate the impact of a stressor threshold above which there is an increase in the expression of var genes and PfEMP1 on the surface of infected erythrocytes. We find that this would indeed result in a switch from a chronic to an acute infection. Within this model framework, semi-immune individuals may therefore be represented as having initially well-developed adaptive, nonspecific immune responses, as well as innate responses, which suppress initial peaks of parasitemia below the threshold and prevent the escalation of parasite growth and var expression. By contrast, individuals with less adaptive immunity are less capable of suppressing parasite growth below the threshold and are more likely to develop high host stress levels, inflammatory innate immune responses, and severe disease (Figure 4D). These distinct patterns of infection dynamics raise the question of whether deregulation of var gene expression could be an adaptive response by the parasite to promote rapid growth in the face of an activated immune system (indicated by fever) or whether it represents an aberrant loss of control over antigenic variation.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the author. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We are grateful to Ebako Takem, Augustine Ebonyi, Idrissa Sambou, Simon Correa, Demba Jammeh, and other colleagues at the MRC Laboratories in The Gambia for their support.
Financial support. This work was supported by a Burroughs Welcome Fund New Investigator in the Pathogenesis of Infectious Diseases Award (to M. T. D.), the National Institutes of Health (R01AI057919), the Medical Research Council of the United Kingdom for research at MRC Gambia, and a Charles H. Hood Foundation fellowship (to C. J. M.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Baruch DI, Pasloske BL, Singh HB, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 2.Smith JD, Chitnis CE, Craig AG, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–10. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su XZ, Heatwole VM, Wertheimer SP, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 4.Scherf A, Hernandez-Rivas R, Buffet P, et al. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–26. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins WE, Jeffery GM. A retrospective examination of the patterns of recrudescence in patients infected with Plasmodium falciparum. Am J Trop Med Hyg. 1999;61:44–8. doi: 10.4269/tropmed.1999.61-044. [DOI] [PubMed] [Google Scholar]
- 6.Duraisingh MT, Voss TS, Marty AJ, et al. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Tonkin CJ, Carret CK, Duraisingh MT, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66:1296–305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 10.Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar J. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraemer SM, Smith JD. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol Microbiol. 2003;50:1527–38. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 12.Kyriacou HM, Stone GN, Challis RJ, et al. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol. 2006;150:211–8. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rottmann M, Lavstsen T, Mugasa JP, et al. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun. 2006;74:3904–11. doi: 10.1128/IAI.02073-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaestli M, Cockburn IA, Cortes A, Baea K, Rowe JA, Beck HP. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J Infect Dis. 2006;193:1567–74. doi: 10.1086/503776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen AT, Magistrado P, Sharp S, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med. 2004;199:1179–90. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viebig NK, Gamain B, Scheidig C, et al. A single member of the Plasmodium falciparum var multigene family determines cytoadhesion to the placental receptor chondroitin sulphate A. EMBO Rep. 2005;6:775–81. doi: 10.1038/sj.embor.7400466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salanti A, Staalsoe T, Lavstsen T, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–91. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 18.Walther M, Jeffries D, Finney OC, et al. Distinct roles for FOXP3 and FOXP3 CD4 T cells in regulating cellular immunity to uncomplicated and severe Plasmodium falciparum malaria. PLoS Pathog. 2009;5:e1000364. doi: 10.1371/journal.ppat.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merrick CJ, Dzikowski R, Imamura H, Chuang J, Deitsch K, Duraisingh MT. The effect of Plasmodium falciparum Sir2a histone deacetylase on clonal and longitudinal variation in expression of the var family of virulence genes. Int J Parasitol. 2010;40:35–43. doi: 10.1016/j.ijpara.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 21.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–5. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 22.Nkrumah LJ, Muhle RA, Moura PA, et al. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods. 2006;3:615–21. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figueiredo LM, Freitas-Junior LH, Bottius E, Olivo-Marin JC, Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21:815–24. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang J, Zhou H, Rathore D, Sullivan M, Su XZ, McCutchan TF. Ambient glucose concentration and gene expression in Plasmodium falciparum. Mol Biochem Parasitol. 2004;133:125–9. doi: 10.1016/j.molbiopara.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Oakley MS, Kumar S, Anantharaman V, et al. Molecular factors and biochemical pathways induced by febrile temperature in intraerythrocytic Plasmodium falciparum parasites. Infect Immun. 2007;75:2012–25. doi: 10.1128/IAI.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg E, Ben-Shmuel A, Shalev O, Sinay R, Cowman A, Pollack Y. Differential, positional-dependent transcriptional response of antigenic variation (var) genes to biological stress in Plasmodium falciparum. PLoS One. 2009;4:e6991. doi: 10.1371/journal.pone.0006991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Udomsangpetch R, Pipitaporn B, Silamut K, et al. Febrile temperatures induce cytoadherence of ring-stage Plasmodium falciparum-infected erythrocytes. Proc Natl Acad Sci U S A. 2002;99:11825–9. doi: 10.1073/pnas.172398999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daily JP, Scanfeld D, Pochet N, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450:1091–5. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 29.Molineaux L, Diebner HH, Eichner M, Collins WE, Jeffery GM, Dietz K. Plasmodium falciparum parasitaemia described by a new mathematical model. Parasitology. 2001;122:379–91. doi: 10.1017/s0031182001007533. [DOI] [PubMed] [Google Scholar]
- 30.Campbell RB, Sinclair DA, Couling M, Brock HW. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol Gen Genet. 1995;246:291–300. doi: 10.1007/BF00288601. [DOI] [PubMed] [Google Scholar]
- 31.Ganesan K, Ponmee N, Jiang L, et al. A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates. PLoS Pathog. 2008;4:e1000214. doi: 10.1371/journal.ppat.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–6. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu SP, Lin SJ. Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim Biophys Acta. 2010;1804:1567–75. doi: 10.1016/j.bbapap.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]