Abstract
Eukaryotic protein kinases belong to a large superfamily with hundreds to thousands of copies and are components of essentially all cellular functions. The goals of this study are to classify protein kinases from 25 plant species and to assess their evolutionary history in conjunction with consideration of their molecular functions. The protein kinase superfamily has expanded in the flowering plant lineage, in part through recent duplications. As a result, the flowering plant protein kinase repertoire, or kinome, is in general significantly larger than other eukaryotes, ranging in size from 600 to 2500 members. This large variation in kinome size is mainly due to the expansion and contraction of a few families, particularly the receptor-like kinase/Pelle family. A number of protein kinases reside in highly conserved, low copy number families and often play broadly conserved regulatory roles in metabolism and cell division, although functions of plant homologues have often diverged from their metazoan counterparts. Members of expanded plant kinase families often have roles in plant-specific processes and some may have contributed to adaptive evolution. Nonetheless, non-adaptive explanations, such as kinase duplicate subfunctionalization and insufficient time for pseudogenization, may also contribute to the large number of seemingly functional protein kinases in plants.
Keywords: plant protein kinase, gene family evolution, lineage-specific expansion, comparative genomics
1. Introduction
The eukaryotic protein kinases are defined as enzymes that use the γ-phosphate of adenosine triphosphate (ATP) to phosphorylate serine, threonine or tyrosine residues in protein [1]. Protein kinases are highly similar in having a 250–300 amino acid protein domain that is responsible for the phospho-transfer reaction. Through alignments of protein kinase sequences available at the time, phylogenetic analysis revealed the sequence diversity in this superfamily and provided the first comprehensive classification scheme for protein kinases [2]. When the first plant genome, Arabidopsis thaliana, was sequenced, a surprising number protein kinases, over 1000, were identified [3]. Subsequently, an analysis of human genome sequences indicated the presence of 518 human protein kinases and 106 pseudogenes [4]. Thus, 1–2% of functional genes encode protein kinases, highlighting their importance in many aspects of cellular regulation in both plants and animals.
Plant protein phosphorylation was first detected in Chinese cabbage leaf discs after the application of a plant hormone, cytokinin [5]. Shortly thereafter, studies in common duckweed showed that plant ribosomes were phosphorylated on serine residues [6]. In the same year, the first plant protein kinase was partially purified from pea [7]. However, it was not until the late 1980s and early 1990s that the first few plant protein kinase sequences became available. The first plant protein kinase sequences were identified in pea and in rice through the use of degenerate primers [8]. In 1990, the third plant protein kinase, ZmPK1, was cloned from maize, and was found to have a transmembrane region N-terminal to the kinase catalytic domain and a large putative extracellular domain [9]. This receptor-like kinase (RLK) resembles animal receptor kinases [10,11], but has a kinase domain belonging to a distinct family that is related to the fruitfly Pelle kinase and mammalian interleukin receptor-associated kinases (IRAK) [12], indicating that in plants a different class of kinases was co-opted for functions in transmembrane signal perception and transduction. Another major early finding was the sequencing through Edman degradation of a very abundant protein kinase with resemblance to both calmodulin- and calcium-dependent protein kinases [13], coupling calcium signalling and phosphorylation.
In the ensuing years, many more plant protein kinases homologous to multiple families of animal and fungal protein kinases were identified. However, the biological functions of plant protein kinases were first elucidated for protein kinases that play roles in plant-specific processes. The first published studies demonstrating plant protein kinase function genetically, to our knowledge, are on Pto [14], CTR1 [15] and Tousled [16]. Pto, a protein kinase with a catalytic domain related to those of RLKs, was identified from a tomato cultivar resistant to a bacterial pathogen and confers resistance when introduced into an otherwise susceptible tomato cultivar [14]. CTR1, a kinase in the tyrosine kinase-like (TKL) group, is involved in the negative regulation of plant ethylene signalling [15]. Mutations in the Tousled protein kinase from A. thaliana, the founding member of the Tousled-like kinase group (TLK), were found to result in impaired floral organ development [16]. In the past 20 years, plant protein kinases have been found to be components of signalling networks such as the perception of biotic agents, light quality and quantity, plant hormones, and various adverse environmental conditions. They function in diurnal and circadian regulation, cell cycle regulation, developmental processes, modulating vesicle transport and channel activities, and in regulating cellular metabolism (for reviews, see earlier studies [17–32]).
Recently, a number of plant genome sequences have become available, allowing us to assess the evolutionary history of protein kinases from unicellular algae to land plants with significantly higher resolution than earlier studies that compared only two or four plant species [33,34]. Studies of protein kinases in other eukaryotes have led to detailed classification of this superfamily [35–39]. However, with the exception of studies of individual families, there is no comprehensive classification of plant kinomes using available plant genomic resources and knowledge of evolutionary relationships between kinases in other eukaryotes. Thus, we undertook a comparative study of protein kinases between plants and other eukaryote models. In addition to classifying plant protein kinases into families, we assess the diversity and evolutionary history of plant protein kinases in the context of their functions. The focus of this study is on the eukaryotic protein kinase superfamily. We have also identified other ‘atypical’ protein kinases not similar in sequence to eukaryotic protein kinases [40], but they are not discussed in detail.
2. Data and methods
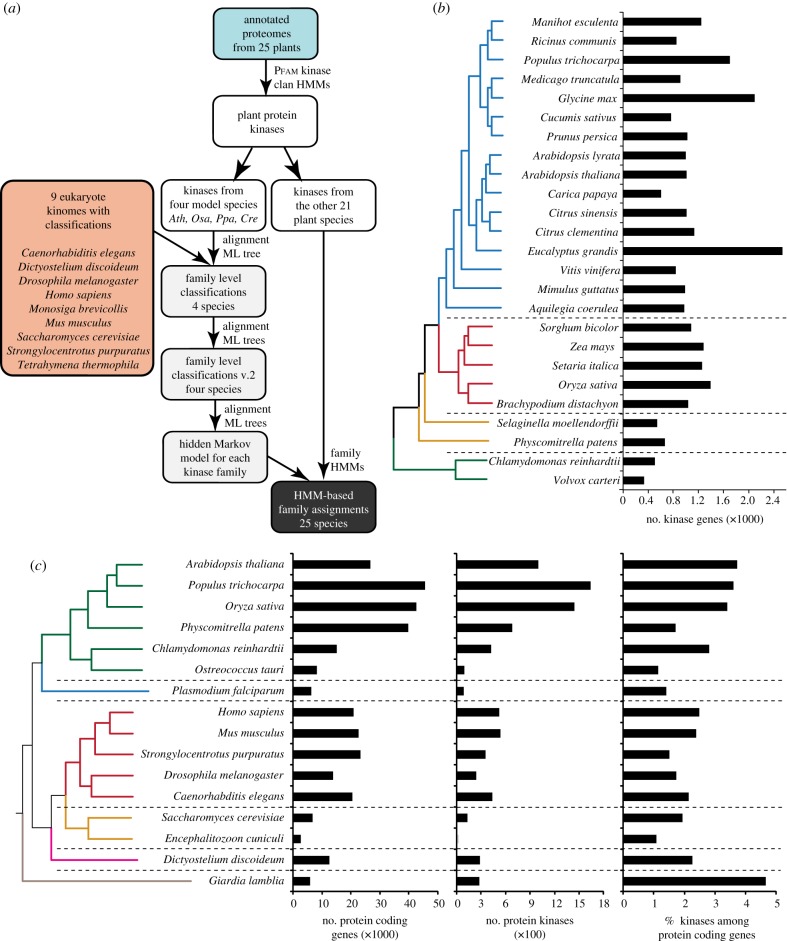
(a). Sequence data and identification of plant protein kinases
To facilitate classification of plant sequences, protein kinase domain sequences and their classification schemes from nine eukaryotes were obtained from KinBase (http://kinase.com/kinbase/FastaFiles/). The nine species included are listed in figure 1a. To identify plant protein kinases, the annotated protein-coding sequences from 25 plant species (figure 1b) were obtained from Phytozome (v. 7, http://www.phytozome.net/). For species with alternatively spliced form annotation, only the longest variant of each gene was analysed further. The putative protein kinases from plants were identified using HMMER v. 3.0 [41] with 16 PKinase clan hidden Markov models (HMM; http://pfam.sanger.ac.uk/clan/pkinase) from Pfam (v. 26; [42]). ‘Trusted cutoff’ values specified by Pfam were used as the thresholds to identify 30 431 putative protein kinase domain sequences. Among these sequences, 29 403 have significant matches to the ‘typical’ protein kinases (Pkinase and Pkinase_Tyr domains) and were analysed further. Both typical and atypical protein kinase information can be found in the electronic supplementary material, appendix S1.
Figure 1.
Protein kinase identification and number of protein kinases in eukaryotes. (a) Input data and analysis pipeline for identifying and classifying plant protein kinases. Model plant species: Ath, Arabidopsis thaliana; Osa, Oryza sativa; Ppa, Physcomitrella patens; Cre, Chlamydomonas reinhardtii. HMM, hidden Markov model; ML, maximum likelihood. (b) Phylogenetic relationships between and numbers of protein kinase genes in 25 plant species. Branch colour: blue, dicotyledon species; red, monocotyledon species; orange, bryophytes; green, green algae. (c) Phylogenetic relationships between selected eukaryotic species and sizes of protein kinase superfamily relative to gene numbers. Branch colour: green, Viridiplantae; blue, Apicomplexa; red, Metazoa; orange, Fungi and Microsporidia; magenta, Amoebozoa; grey, Excavata.
Very few of the plant genomes analysed have been curated manually and there are probably annotation errors. In addition, the A. thaliana and Oryza sativa (rice) genomes have hundreds of pseudogenes that belong to the protein kinase superfamily [43]. Thus, the protein kinase domain sequences were not considered further if the domain alignments covered less than 50 per cent of the Pfam domain models. This resulted in the exclusion of 2841 sequences (see the electronic supplementary material, appendix S1). The same filter was applied to the protein kinase sequences from nine other eukaryotes because there are a few severely truncated or erroneous entries, particularly from Tetrahymena thermophila. The plant protein kinase domain sequences used in subsequent analysis can be found in the electronic supplementary material, appendix S2.
(b). Classification of protein kinases from four model species
To classify plant protein kinase sequences into families, a phylogenetic approach was used. First, the relationships between plant protein kinases and kinases from nine eukaryotes were established (figure 1a). The nine eukaryote protein KinBase dataset contains classification information, including group, family and, in some cases, subfamily levels. Plant kinases were classified based on this scheme. Because it is not feasible to build a phylogeny for all sequences from 25 species, four ‘plant model species’, A. thaliana (dicotyledon), O. sativa (monocotyledon), Physcomitrella patens (moss) and Chlamydomonas reinhardtii (a green alga), were chosen for the first round of classification. These four species were chosen because they are relatively well annotated and are representatives from major lineages in plant evolution. This first pass classification was then used to classify the other 21 plant species as outlined in §2c.
The analysis pipeline for classifying protein kinases in the four model species includes three rounds of phylogenetic analyses (figure 1a). In the first round, a phylogenetic tree was generated for 3656 protein kinases from the four plant models and 218 protein kinase representatives from nine species with kinase classifications. One representative kinase was chosen from each protein kinase family and each of the following five major taxonomic groups: Amoebozoa—Dictyostelium discoideum, Alveolata—T. thermophila, Choanoflagellida—Monosiga brevicollis, Fungi—Saccharomyces cerevisiae, Metazoa—five species. Owing to annotation quality considerations, for metazoan protein kinases, preference was given to human and mouse sequences. The protein kinase domain protein sequences were aligned using Clustal Omega [44] for generating a maximum-likelihood (ML) tree with RAxML-Light (v. 1.0.5, http://sco.h-its.org/exelixis/software.html) using the CAT model (category approximation of GAMMA model of rate heterogeneity) to account for rate heterogeneities [45] and the JTT (Jones, Taylor, Thornton) substitution matrix [46]. On the basis of phylogeny, a plant protein kinase, K, from a model plant was assigned to a protein kinase family, F, if K is in a monophyletic group with all representative sequences from F. Plant sequences that could not be readily assigned were designated as plant-specific.
In the second round, the goal was to verify the family classification from round 1 by building an ML tree for each family. Alignment and tree building procedures were the same as round 1. The RLK/Pelle family has hundreds to more than 1000 members in plants [34]. Thus, instead of building a tree with all putative RLK/Pelles, sequences in this family were first classified into subfamilies based on prior classification schemes [34] and a phylogenetic tree was built for each subfamily. During this step, some sequences were classified in a different family from that assigned in round 1. In the final round, alignments and phylogenetic trees were generated based on round 2 classifications to determine whether the classifications remained consistent. If not, the consistent family designation in at least two rounds of analysis was used. The final round alignments and phylogenetic trees and classification of protein kinases in the four model plants are compiled in the electronic supplementary material, appendix S3.
(c). Classification of protein kinases from 21 other plants
Two approaches were tested for classifying protein kinases from the other 21 plants into kinase groups, families and subfamilies. In the first approach, a protein kinase, K, from the 21 species dataset was first searched against the four model plant protein kinases with BLAST [47]. The classification of K was based on its top matching protein kinases from the model species (expect value < 1e − 5). To evaluate the classification accuracy, protein kinases from one model species were assigned into families based on classification of their top matches from one of the other three models (table 1). The classification accuracy differs widely depending on the evolutionary distances between species. Using rice protein kinases to classify A. thaliana ones, the accuracy is approximately 97 per cent. Using moss classification, more than 10 per cent of the A. thaliana protein kinases are mis-classified. Using green algal classifications, 65–80% of protein kinases in A. thaliana, rice and moss are mis-classified.
Table 1.
Accuracy of protein kinase family assignments.
| subjecta | classificationa | totalb | correctc | % correct |
|---|---|---|---|---|
| BLAST-basedd | ||||
| Ath | Osa | 1015 | 983 | 96.85 |
| Ath | Ppa | 1015 | 904 | 89.06 |
| Ath | Cre | 1015 | 344 | 33.89 |
| Osa | Ath | 1422 | 1358 | 95.50 |
| Osa | Ppa | 1422 | 1053 | 74.05 |
| Osa | Cre | 1422 | 298 | 20.96 |
| Ppa | Ath | 689 | 574 | 83.31 |
| Ppa | Osa | 689 | 554 | 80.41 |
| Ppa | Cre | 689 | 240 | 34.83 |
| Cre | Ath | 530 | 113 | 21.32 |
| Cre | Osa | 530 | 115 | 21.70 |
| Cre | Ppa | 530 | 119 | 22.45 |
| HMM-basedd | ||||
| Ath | HMM | 1015 | 1014 | 99.90 |
| Osa | HMM | 1422 | 1420 | 99.86 |
| Ppa | HMM | 689 | 686 | 99.56 |
| Cre | HMM | 530 | 518 | 97.74 |
aAbbreviations follow species names in figure 1b. Subject refers to the species where the protein kinases were to be classified. Classification refers to species where the family assignments were used to evaluate classification accuracy.
bTotal number of kinase domain sequences examined.
cNumber of cases where query kinase was in the same family as the subject kinase.
dSee §2 for details.
In the second approach, an HMM was built for each family according to the family sequence alignment of model plants (see electronic supplementary material, appendix S4 for HMMs). The HMMs were then used to search against protein kinases from the other 21 plants. The family assignment of a protein kinase sequence was that of the top-scoring HMM (see electronic supplementary material, appendix S5 for classification results and numbers of members in each family). The accuracy of the HMM-based approach is very high, ranging from 98 per cent in classifying C. reinhardtii protein kinases to 99.9 per cent for A. thaliana kinases (table 1). This significant improvement is probably due to the fact that HMM better covers the family sequence space than a single top match sequence. Because of this significant improvement in classification accuracy over the similarity-based approach, in all subsequent analysis, the HMM-based assignments of plant protein kinase families were used.
3. Plant protein kinase superfamily size and diversity
(a). Sizes of plant protein kinase superfamilies when compared with other eukaryotes
In the 10 years since the first plant protein kinase sequence was reported [8], the number of known plant protein kinases rose to over 500, and, because of the rapid progress in sequencing the A. thaliana genome, 175 of these plant kinases came from A. thaliana [18]. In the published annotation of the A. thaliana genome, there are approximately 1000 protein kinases [3], an number five times larger than that in budding yeast, and two to three times larger than various other eukaryotes, including mammals (figure 1c). Later analysis indicated that the protein kinase superfamily is even larger in other flowering plant species, with over 1500 members in rice and poplar [33,34,48]. Why are there more protein kinases in plants than in most other eukaryotes? The mechanisms underlying gene family expansion are similar between eukaryotes and involve tandem duplication in linked regions, retrogene formation, chromosomal duplications and whole genome duplication [49]. The differences in the size of the protein kinase repertoire between flowering plants and other eukaryotes suggest a relatively higher degree of lineage-specific expansion of this superfamily in plants. The main mechanisms contributing to expansion are elevated rates of tandem as well as whole genome duplication in plants relative to those in other eukaryotes [49]. Polyploidization is much more prominent in plants than in any other eukaryote; as many as 70 per cent of extant plant species are polyploids [50]. There have probably been at least three rounds of palaeo-polyploidization in the A. thaliana lineage after its split from monocotyledon species 150 Ma [51–53].
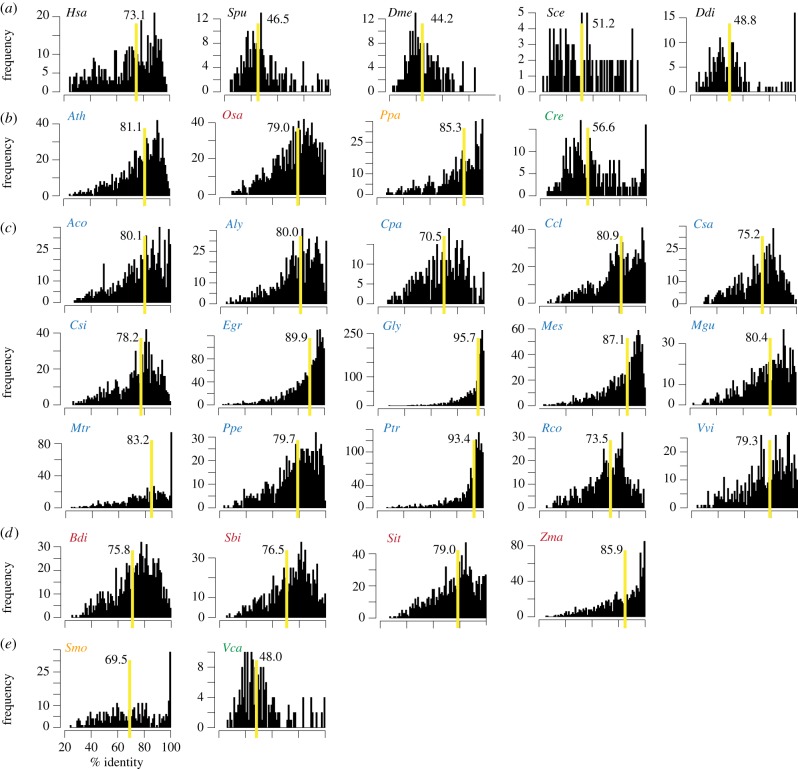
To determine whether there are more protein kinases derived from recent duplication events in plants compared with other eukaryotes, protein sequence identity between all paralogous protein kinases in a species was determined and used as a proxy for timing of duplication. Species that have more recently duplicated and retained protein kinases will have more paralogues with higher identities to each other. The paralogue identity distributions were compared between the four model plant species and five non-plant eukaryotes, including human (Homo sapiens), sea urchin (Strongylocentrotus purpuratus), fruitfly (Drosophila melanogaster), budding yeast (S. cerevisiae) and slime mould (D. discoideum). Among five non-plant eukaryotes, the median identity of protein kinase paralogues is highest in humans (73.1%), significantly higher than the other four species (Wilcoxon rank sum test, all p < 1e − 11; figure 2a). On the other hand, the three model land plants (A. thaliana, Ath; O. sativa, Osa; P. patens, Ppa; figure 2b) have significantly larger median identities compared with that of humans (all p < 1e−10). In C. reinhardtii (Cre; figure 2b), which does not have a history of whole genome duplication, the median identity of kinase paralogues is significantly lower than observed in the model land plants and in humans. This is consistent with the notion that there are more recent protein kinase duplicates in land plants than in other eukaryotes. In addition to elevated duplication rate, another explanation is that many plant protein kinase duplicates tend to be retained. This was shown in a comparative analysis of protein families from A. thaliana, poplar, rice and moss where protein kinases were shown to have significantly higher expansion rates than other protein families [54].
Figure 2.
Distributions of pairwise identities between closest protein kinase paralogues. (a) Non-plant eukaryote representatives. (b) Four model plant species with species abbreviations coloured according to the convention in figure 1b. (c) Additional dicotyledon species. (d) Additional monocotyledon species. (e) Additional bryophyte and green alga. The x-axis indicates paralogue per cent identity, and the y-axis indicates the number of paralogous pairs in an identity bin. The species abbreviations follow the species names shown in figure 1. The yellow line indicates the median paralogue identity value (also shown) for each species.
(b). Expansion of the protein kinase superfamily in Viridiplantae
The expansion of protein kinases appears to be quite rapid, given that there are approximately 600 more protein kinases in poplar than in A. thaliana (figure 1b) and that these two species diverged only approximately 70 Ma. On the other hand, the moss P. patens has 685 protein kinases, substantially less than those of flowering plants (figure 1b). In green algae, the protein kinase superfamily is even smaller with 426 and 93 members in C. reinhardtii and Ostreococcus tauri, respectively [34]. These findings indicate that within Viridiplantae (green plant species, including land plants and green algae), the protein kinase superfamily has steadily grown in size in the lineage leading to flowering plants. In addition, such expansion correlates with an increase in developmental complexity. Both C. reinhardtii and O. tauri are unicellular Chlorophyta green algae hypothesized to have diverged from the Charophyta green algae and land plant lineage approximately 1200 Ma [55]. The divergence time between C. reinhardtii and O. tauri is unclear. Although both of these green algae are free-living unicellular organisms, C. reinhardtii has 333 more protein kinases than O. tauri.
The explanation for this large difference is most likely to due to extensive reduction in genome size that occurred in O. tauri [56]. The sizes of O. tauri intergenic regions are significantly shorter than other eukaryotes with similar genome sizes. In addition, there is substantial reduction of gene family size in general. Compared with other eukaryotes, only 1.1 per cent of annotated protein-coding genes in O. tauri are protein kinases, similar to two obligate intracellular parasites Encephalitozoon cuniculi (1.1%; [39]) and Plasmodium falciparum (1.4%; [38]). Interestingly, the E. cuniculi genome has undergone a significant degree of genome compaction and gene loss [57]. A recent analysis in another intracellular parasite, Giardia lamblia, indicates the presence of 278 protein kinases [37]. However, 198 of these G. lamblia protein kinases belong to the never in mitosis/Aspergillus-related kinase (NEK) family. Thus, excluding the dramatic expansion in NEKs, there remains only 80 protein kinases (1.3% of protein-coding genes) in G. lamblia. Some flowering plants have also undergone gene loss. For example, A. thaliana has probably lost DNA through rearrangement events [58]. However, A. thaliana also has a similar percentage of genes that are protein kinases compared with poplar, rice and moss (figure 1c). Thus, it appears that just as many protein kinases were lost compared with genes in general in flowering plant species with genome reduction.
(c). Protein kinase superfamily size variation among plant species
Although earlier studies have provided important clues on the expansion of the plant protein kinase superfamily, the number of species examined was small. Thus, it was not clear whether the patterns observed would remain consistent if more species were analysed. In the past few years, over 25 Viridiplantae genomes (including 21 flowering plants, one bryophyte, one moss and two algae; figure 1b) have been sequenced with draft annotation available. To further examine how the sizes of plant protein kinase superfamilies differ between plant species, protein kinase sequences were identified from the annotated protein sequences. Among them, approximately 1 per cent are classified as ‘atypical’ protein kinases. Excluding the atypical protein kinases, 26 966 protein kinase domain sequences from 26 775 annotated genes were identified.
There is substantial variation in the protein kinase superfamily size among plant species. In addition to the differences between land plants and algae noted in §3b, the numbers of protein kinases among flowering plants differ by more than fourfold (figure 1b). The analysed species with the largest protein kinase superfamily is rose gum eucalyptus, Eucalyptus grandis, with 2532 kinase genes. By contrast, papaya (Carica papaya) has only 600 kinase genes. It should be noted that the lower numbers of kinases in some plant species are due to incomplete genome sequencing coverage and should be regarded as lower-bound estimates. To circumvent the incomplete genome issue, a comparison of protein kinase superfamily sizes can be conducted in consideration of annotated gene numbers. Interestingly, there exists a significant correlation between the number of protein kinase genes and the total gene number in a genome (Pearson's correlation, r = 0.63, p < 1e − 3). This finding indicates that, despite the large variation in the size of the protein kinase repertoire among flowering plants, a similar proportion of genes encode protein kinases. The implication is that the protein kinase superfamily as a whole grows and shrinks with the rest of the gene families within each genome. Alternatively, this correlation may suggest that the propensity for expanding the protein kinase superfamily is similar among different plant species. In the following sections, we examine the approximate timing of plant protein kinase duplications and assess the degree of differential expansion among plant protein kinase families.
(d). Approximate timing of plant protein kinase duplications
To assess why these species differ so greatly in the number of protein kinase paralogues, an all-against-all similarity search was conducted for protein kinases in each species. The distributions of identities between the most closely related paralogous pairs are shown in figure 2b. In eucalyptus, Glycine max (soybean), and Populus trichocarpa (cottonwood), which have more than 1600 protein kinases, the paralogue identity distributions tend to peak at approximately 95 per cent, significantly higher than other plants. For example, the eucalyptus protein kinase paralogue identities are significantly higher than those of A. thaliana (Wilcoxon rank sum test, p < 1e − 16). Thus, very recent gene duplications contribute significantly to the larger protein kinase repertoire in some plants. Through this analysis, it also becomes apparent that some species, such as Medicago truncatula, Selaginella moellendorffii and C. reinhardtii, have an excess of protein kinases with 100 per cent identity (figure 2b). Some of these identical kinases may be the result of mis-annotation. Thus, an analysis eliminating sequences with 100 per cent similarity was also conducted (see electronic supplementary material, appendix S6), and our conclusion remained unchanged. In addition to identical sequences, there are also a substantial number of nearly identical sequences. In some cases, these may be allelic variants that are identified as distinct loci, as noted in the Selaginella genome study [59].
On the basis of this expanded analysis, it is in general true that plant protein kinase genes experienced much more recent duplication compared with the other five eukaryotic species examined. However, there are some notable exceptions. For example, the protein kinase paralogue identities from castor bean (Ricinus communis), cucumber (Cucumis sativus) and papaya were not significantly different from those of human protein kinases (p > 0.1 in all cases). Interestingly, unlike plant species such as soybean [60] and cottonwood [61] with significantly higher paralogue identities and a recent history of polyploidization, there is no apparent recent whole genome duplication event detected in the cucumber or papaya genomes [62,63]. In addition, castor bean, cucumber and papaya have some of the lowest numbers of protein kinase genes among the plant species analysed (figure 1b). These findings reinforce the idea that whole genome duplication contributes significantly to the expansion of the protein kinase superfamily in plants. We should emphasize that the ancestral lineage leading to castor bean, cucumber and papaya probably experienced at least one round of whole genome duplication 70–100 Ma [51–53]. However, the protein kinase superfamily in papaya, cucumber and castor bean is substantially smaller than other species, particularly those with recent whole genome duplications. Thus, it is likely that many of the protein kinase duplicates derived from whole genome duplication were lost in a timeframe of tens to hundreds of million years. This timeframe for gene loss is consistent with that found in a global study of gene family expansion in four land plant species [54].
(e). Plant protein kinase families and their differential expansion
To evaluate the evolution of plant protein kinases at the family level, plant protein kinases were first classified using an established scheme based on phylogenetic analyses of animal, fungal and protist protein kinases [1,2,35]. The number of kinase sequences is large (more than 26 000), creating computing issues for both the multiple sequence alignment and the phylogenetic analysis phases. Thus, protein kinases were first classified in four model plant species (A. thaliana, rice, moss and C. reinhardtii), based on location in the same monophyletic group as kinases with known, consistent family designations (see electronic supplementary material, appendix S3 for alignments, phylogenies and classification). The model plant classification was then used to classify protein kinases from the other 21 species (see §2). The numbers of plant protein kinases in different families are shown in figure 3a.
Figure 3.
Sizes of protein kinase families in 25 plant species. (a) Major groups and families. (b) RLK/Pelle subfamilies. The species abbreviations follow the species names shown in figure 1b. The number of protein kinases in each family/subfamily is colour coded according to the colour key on the lower right. Kinases that cannot be readily assigned to a group are classified as ‘plant-specific’. Within each group, some plant or algal kinases cannot be assigned to an existing family. They are designated as ‘Pl’ or ‘Cr’ for Plant and C. reinhardtii, respectively. Similarly, some kinases can be assigned to a family but not at the sub-family level. In addition to ‘Pl’ and ‘Cr’ designations, they are designated as ‘Pp’ or ‘Os’ specifying P. patens and O. sativa-specific kinases, respectively.
Through this analysis, the majority of model plant protein kinases can be classified into known groups. Nonetheless, there remains a small number of plant kinases that cannot be assigned at the group level (figure 3a, plant-specific). Some of the plant-specific kinases are found only in green algae (Group-Cr-2), or only in green algae and bryophytes (Group-Pl-1; here ‘Pl’ indicates plant). On the other hand, Group-Pl-3 and -4 are highly conserved across plants and have relatively few members. Most plant kinases that can be classified at the group level can also be readily classified into known families, indicating that many of these families were established prior to the divergence of the plant, animal and fungal lineages or earlier. Nonetheless, there remain unclassified plant-specific families in all major protein kinase groups (figure 3a, Pl families in different groups). There are also families that are shared only between green algae and fungi (CAMK1-Scer), between green algae, bryophyte and T. thermophila (CAMK1-Tthe); between green algae and animals (TK and several TKL families); and between land plants and T. thermophila (CMGC-Pl-Tthe). These ‘mosaic’ patterns suggest potential gene losses in other major lineages or horizontal gene transfers that remain to be further investigated.
Among families that are present in green algae, bryophytes and flowering plants, three major patterns emerge when comparing the variation in numbers of protein kinases across families and species. The first is protein kinase families that consistently have relatively low (1–5) copy numbers (figure 3a, blue series). Given that many of these families have animal or fungal orthologues, low copy numbers suggest that there has been no or limited changes in these families since the divergence of the plant and animal/fungal lineages approximately 1 Ga. Considering that there were repeated whole genome duplications in plants, these low copy number families had the opportunity to expand via duplication, but apparently most duplicates were not retained. For example, the CDK–CCRK, CDK–CDK7, and CDK–CDK8 subfamilies in most species consist of only one gene, except in soybean, where a very recent whole genome duplication occurred (approx. 59 and 14 Ma) [60]. In fact, soybean protein kinase families in general are larger than other species. Thus, the significantly larger protein kinase repertoire in soybean can partly be attributed to the possibility that there has been insufficient time for pseudogenization of duplicates.
The second pattern is families with moderate sizes (6–30 members; figure 3a, green series and yellow), and the third is large families (more than 30 members; figure 3a, red series, magenta series). In general, the sizes of these families are in the order: algae < bryophytes < flowering plants, consistent with the notion that the protein kinase superfamily has expanded over the course of plant evolution. Among plant species, the family with the largest size differences is the RLK/Pelle family with two to three genes in green algae, approximately 300 genes in bryophytes and 374–2205 members in flowering plants. The RLK/Pelle family was established because the kinase domains of various RLKs are more closely related to fly Pelle and human IRAKs than to protein kinases from any other families [12]. This family was later defined as the IRAK family. For historical reasons, we use the ‘RLK/Pelle designation’ throughout. Earlier studies have established that this family has experienced extensive expansion in the land plant lineage [33,34,64]. When the RLK/Pelle family is classified at the subfamily level (figure 3b), a few subfamilies have low copy number (e.g. RLCK-II) but most others have moderate to large sizes. RLK/Pelle subfamilies in soybean are in general larger than in other species, consistent with the patterns observed for other protein kinase families. The most striking expansion of the RLK/Pelle family is seen in eucalyptus where multiple subfamilies have more than 300 members (figure 3b). However, other protein kinase families in eucalyptus are of similar size as other plant species (figure 3a). Thus, the very large number of protein kinases in eucalyptus is mostly a consequence of expansion in the RLK/Pelle gene family.
4. Functional diversity of plant protein kinases
(a). Kinase families that have remained low in copy number in both plants and other eukaryotes
(i). Conserved central roles in metabolic signalling and stress response
As shown in figure 3a, many protein kinase families remain low in copy number, despite ample opportunity to duplicate. Considering that the flowering plant species studied are either palaeo- or recent polyploids, the implication is that many of these protein kinases were duplicated but the duplicates were subsequently lost. Most of these small families are conserved between plants and animals, suggesting that they play roles in conserved processes. Several subfamilies that have undergone limited expansion in most eukaryotic lineages have central roles in metabolic signalling and regulating metabolic changes in response to stress, and include AMPK/SNF1, PDK1, S6K, IRE1 and GCN2. Here, we discuss how the functions of these genes have remained conserved and/or diverged in the plant lineage.
AMPK/SNF1 and LKB1
The AMP-activated protein kinase (AMPK)/SNF1 (sucrose-non-fermenting 1) kinases are conserved across eukaryotes and play central roles in sensing energy status and maintaining energy homeostasis [65]. Like their yeast and animal counterparts, the A. thaliana AMPK homologues SnRK1.1/AtKIN10 and SnRK1.2/AtKIN11 also play roles as central regulators of energy metabolism and homeostasis under stress conditions [66,67]. The involvement of SNF1 kinases in response to starvation also appears to be evolutionarily conserved, and in moss, AMPK/SNF1 kinases enable plants to cope with periods of dark [65,68]. SNF1/AMPK functions and interaction partners are probably broadly conserved across kingdoms. AMPK/SNF1 is activated by LKB1, and PAK1, TOS3 and ELM1 in mammals and yeast, respectively [69,70]. The two A. thaliana LKB subfamily members, GRIK1 and 2, can complement a yeast pak1/tos3/elm1 mutant and can phosphorylate AtSNF1 [71,72].
PDK1
In metazoans and yeast, phosphoinositide-dependent kinase-1 (PDK1) serves as a master regulator of AGC kinase activity, phosphorylating several AGC kinases through interaction with the PDK1 interacting fragment domain in response to 3-phosphoinositide generation [73,74]. A. thaliana PDK1 activates RSK-2 AGC kinases in response to a different lipid signal, phosphatidic acid [75–77]. A. thaliana plants lacking both PDK1-related genes are dwarf and defective in reactive oxygen species-mediated signalling, but still viable [78]. This is in contrast to mice where loss of PDK1 results in embryo lethality [79]. It appears that the activation of AGC kinases by PDK1 is conserved in plants, animals and yeast, but that the details of that regulation as well as downstream effects have diverged.
S6K/AGC-Pl
Because we cannot clearly resolve the relationship between animal ribosomal S6 kinases (S6Ks) and their reported plant homologues, the plant S6K-like genes are designated AGC-Pl in this study. The p70 S6Ks are stimulated in response to nutrients and modulate protein translation by phosphorylating downstream targets such as ribosomal protein S6, EF2 kinase and eIF4B [77]. There are two p70S6K-related proteins in A. thaliana. The S6K2 gene, AT3G08720, can phosphorylate human and plant S6 ribosomal protein, suggesting that targets have been conserved [80]. AtS6K1 is located in the cytoplasm and nucleus, whereas S6K2 is only located in the nucleus and nucleolus; in animals, there are two isoforms of S6K with analogous distinct localization patterns [81]. S6Ks also are phosphorylated by PDK1 in both plants and animals [81]. As in animals, S6K1 is activated by an activation complex called TORC in response to stress, although plant S6Ks do not have the conserved TOR signalling motif found in animals [81]. Arabidopsis thaliana S6K1 interacts with the RBR1–E2F pathway to inhibit cell proliferation. Interestingly, the TSC/Rheb/Tor/S6k pathway in Drosophila was also shown to regulate E2F1 levels and to work with RB1 to regulate cell cycle progression and also cell survival [82]. Plants with reduced levels of AtS6K1 and 2 exhibit chromosome instability and a failure to repress cell proliferation under nutrient limiting conditions [83]. This contrasts with the absence of aneuploidy and effects on cell survival in fruitfly, and indicates that although there are similarities in regulation and targets, the functions of S6K have diverged in plants.
IRE1
One remarkable example of conservation is the mechanism by which IRE1 (inositol-requiring 1) senses endoplasmic reticulum (ER) stress and activates the unfolded protein response. Upon sensing unfolded proteins, IRE1 in yeast and its orthologues in metazoans and (presumably) in plants is activated by autophosphorylation [84,85]. Activated IRE1 then splices the mRNA of a transcription factor, Hac1 in yeast, XBP1 in metazoans [85] and bZIP60 in A. thaliana and rice [86,87]. In all cases, the spliced isoform of the transcription factor regulates the expression of stress response genes, but the mechanism by which splicing activates the transcription factor differs in yeast, animals and plants [85,87].
GCN2
GCN2 (general control nonrepressed 2) regulates translation in response to nutrient stress through its phosphorylation of eIF2α [88]. Yeast GCN2 is also activated by ultraviolet radiation and is required for a checkpoint delaying progression from G1 to S phase [89]. In mammals and Schizosaccharomyces pombe, there are GCN2 orthologues as well as three and two additional eIF2 kinases, respectively, that each respond to different stresses [88,90]. Arabidopsis thaliana contains a single GCN2 gene that can complement amino acid starvation response in gcn2 mutant yeast cells [91] and also EF2α phosphorylation in response to multiple stresses, but not osmotic stress in contrast to yeast GCN2 [92,93].
(ii). Regulation of mitosis and cytokinesis
Another group of protein kinase families conserved between plants and animals that are low copy number includes Aurora and some cyclin-dependent kinase (CDK) subfamilies. Protein kinases in these two families play central roles in the regulation of mitosis, cytokinesis and cell cycle control.
Aurora
In yeast and metazoans, Aurora kinases play important roles in mitosis and cytokinesis (reviewed in Carmena et al. [94]). Plants have α- and β-type Aurora kinases, which are distinct from the A, B and C types in animals and B type found in yeast [95]. Arabidopsis thaliana Aurora kinases co-localize with mitotic structures, centromeres and/or the cell plate and also phosphorylate histone 3, a substrate of animal Aurora kinases, suggesting conservation of mitotic functions [96]. However, as suggested by sequence divergence, plant Aurora kinases have evolved plant-specific functions. A recent study showed that a double knockout of the α Aurora kinases AtAUR1 and AtAUR2 is gametophyte lethal and that the α kinases function in division plane orientation [97]. The β-type Aurora kinase could not complement the mutant, indicating that it has a different function. Interestingly, a distinct group of Aurora-related kinases has dramatically expanded in green algal species (figure 3a). A C. reinhardtii Aurora-like kinase, CALK, is required for flagellar disassembly and its phosphorylation state is used to measure flagellar length [98,99]. In mammals, Aurora kinase A, which regulates entry into mitosis, also has a non-mitotic role promoting disassembly of the primary cilium, a structure evolutionarily related to the motile flagella of Chlamydomonas [100]. In both cases, flagellar/ciliary disassembly requires microtubule destabilization, suggesting that signalling pathways leading to disassembly are conserved in Chlamydomonas and mammals; however, CALK is quite divergent from Aurora kinase A [98,100].
CDKA and CDKB
In plants, some CDK subfamilies are highly conserved with low copy number, such as CDKA, CDKB, while the others have expanded fairly significantly. In some cases, the functions of plant orthologues of small CDK gene families have remained highly conserved. For example, yeast requires one gene, Cdc2/Cdc28, to drive cell cycle progression, and in A. thaliana, this function is supplied by CDKA [101]. In our analysis, CDKA does not form a monophyletic group with Cdc2/Cdc28 and is classified as a plant-specific CDK family (CDK-Pl). In A. thaliana, there are three plant-specific CDKB genes that work with CDKA to regulate the G2/M transition [102].
CDK7
Similar to CDKA, the plant CDK7 subfamily members also seem to have conserved functions with their yeast and animal counterparts, the cyclin-activating kinases (CAKs). Animal CDK7, the major CAK, regulates transcription by phosphorylating the C-terminal domain (CTD) of RNA Pol II [103,104]. The CAK and transcriptional regulation functions are carried out by two separate CDK7 genes in yeast. The sole rice CDK7 orthologue appears to have both CAK and transcriptional regulation functions [105,106]. Arabidopsis thaliana has three CDK7-related genes, of which two (CDKD2 and 3) phosphorylate both the RNA Pol II CTD and human CDK2 (reviewed in Inagaki & Umeda [107]).
CDK20
In contrast to CDKA and CDK7, the functions of the small CDK subfamily CDK20 (originally named CCrK) have diverged in plants. In A. thaliana, a cell-cycle-related kinase (CRRK) subfamily member, CDKF1, functions as a CAK [108]. The mammalian CRRK, while required for cell proliferation, does not have CAK activity [109]. Interestingly, CCRK-related genes are involved in the regulation of cilia assembly in vertebrates [110] and flagellar length in green algae [111]. These examples indicate that even though CCRK remains largely a single copy gene in all lineages examined (figure 3a), it has been recruited for different functions in different organisms.
Other cyclin-dependent kinases
Other CDK members with non-mitotic roles have also remained low in copy number in the plant lineage. CDK8 subfamily members regulate transcription in metazoans and in yeast, functioning as part of a complex called Mediator, to modulate the activity of RNA Pol II [112,113]. The A. thaliana CDK8 orthologue, HEN3, is required for floral cell differentiation [114] and also functions as part of Mediator complex to regulate transcription in A. thaliana [115], indicating that this function is also conserved in plants.
(iii). Low copy number families with divergent or unknown functions
Similar to CDK20 discussed earlier, some subfamilies with low copy numbers in plants and animals can also diverge significantly in their functions. WEE1 and ULK are clear examples as they regulate plant-specific processes. On the other hand, functional similarities of RCK and TLK homologues between eukaryotes are not entirely clear.
WEE1
In fission yeast and metazoans, WEE1 plays a central role in the cell cycle, regulating cell cycle progression by phosphorylating and inactivating CDK1, which is in turn activated by the phosphatase Cdc25 (reviewed in Doonan & Kitsios [104]). In budding yeast, where there is no gap between S-phase and mitosis, the WEE1 homologue, Swe1p, functions in a morphogenesis checkpoint instead, monitoring the actin cytoskeleton [116,117]. On the basis of presence of homologous genes, the same CDK1–WEE1–CDC25 regulatory loop was thought to operate in plants. However, recent evidence suggests that entry into mitosis is regulated differently in plants than in fission yeast and metazoans [118]. Supporting this, plant WEE1 functions in endo-reduplication and as a regulator of a DNA integrity checkpoint during the S phase rather than as a central regulator of cell cycle progression [119–121].
ULK
ULK (uncoordinated 51-like kinase) subfamily members regulate autophagy in metazoans and yeast, and have also been shown to regulate vesicle transport in neurons (reviewed in Chan [122]). The only ULK-ULK4 kinase in A. thaliana, RUNKEL (RUK), is an essential gene, but is involved in cell division rather than in autophagy [123], even though autophagy pathways appear to be conserved in plants and metazoans [124]. The ULK-fused subfamily members are components of the sonic hedgehog signalling pathway and regulate Gli transcription factors involved in cell proliferation and cell fate [125]. In A. thaliana, which lacks sonic hedgehog signalling components, the only ULK-fused homologue is located at the phragmoplast and regulates cytokinesis [126].
RCK
One interesting example of conservation of function is the role of the ros cross-hybridizing kinase (RCK) male germ-cell-associated kinase (MAK) homologues in flagella/cilia morphology [30]. Note that the plant MAK homologues were classified as mitogen-activated protein kinase (MAPK) in Rodriguez et al. [30], which is inconsistent with our analysis results. The C. reinhardtii MAK homologue [127], CrLF4, regulates flagellar length [128]. CrLF4 homologues in C. elegans and mouse regulate the morphology and length of cilia [129,130]. The three A. thaliana MAK-related genes are expressed in the male gametophyte and pollen tube [131], but their functions are not known.
TLK
The TLK was first identified in A. thaliana as a regulator of flower initiation and development [16] and transcriptional silencing [132]. TLKs are conserved throughout eukaryotes, but are absent from yeast. Metazoan and plant TLK orthologues appear to share functions in chromatin dynamics, indicated by their ability to phosphorylate the same substrates: Histone 3B and ASF1, a histone chaperone protein that functions in chromatin assembly [133–137].
CMGC_DYRK
Dual specificity yak-related kinases comprise three subfamilies: (i) DYRK, (ii) homeodomain interacting kinases (HIPKs), and (iii) pre-mRNA processing protein 4 kinases (PRP4). The first member of the DYRK family, YAK1, was identified in yeast, and since then members have been found in all eukaryotes (reviewed in Aranda et al. [138]). In yeast, Yak1 regulates stress and nutrient response transcription factors [139,140]. Interestingly, YAK1 genes are not found in animals, which have the related DYRK1 and DYRK2 genes [138]. Nearly all plants have one to three YAK1-related genes, but none of the DYRK genes in plants have been characterized. Plants do not have apparent HIPKs, which in animals regulate transcription by interacting with homeodomain proteins [141], but do have expanded PRP4 kinase subfamilies. The functions of these genes also remain uncharacterized. Given the importance of PRP4 kinases in splicing in yeast [142] and mammals [143], it would be interesting to investigate their function in splicing in plants.
(b). Protein kinase families with moderate degrees of expansion
(i). Families with divergent functions but conserved signalling network components
A number of families are found in most eukaryotes but have experienced moderate degrees of expansion in plants. Members in several of these families, including MAPK, MAPK kinase (MAP2K), MAPK kinase kinase (MAP3K) and RSK-2 have been shown to interact with homologous signalling network partners in plants, fungi and animals. However, they play apparently different roles in plants, presumably because of the morphological, developmental and physiological divergence and differences in life histories between plants and other eukaryotes.
MAPK
MAPKs in plants, metazoans and yeast link extracellular and intracellular signals to downstream responses. In plants, MAPKs play diverse roles in development, and response to abiotic and biotic stresses (reviewed in Rodriguez et al. [30]). Several signalling modules involving MAPKs in both plants and animals have been defined. Although there is a high degree of conservation of structure of these modules, MAPK signalling pathways in plants are different from those in other eukaryotes [30]. Furthermore, the MAPK family has expanded more in land plants (7–31 members) relative to yeast (6) and animals (6–14), suggesting more opportunities for diversification of functions in the plant lineage. In rice, poplar and A. thaliana, there is a high degree of conservation of MAPKs, and most orthologous relationships are clear [30].
STE7 and STE11
These two related families of protein kinases, STE7 (Sterility 7) and STE11, function as MAP2K and MAP3K, respectively. Note that Raf kinases also function as MAP3K but belong to the TKL group. Like MAPKs, the STE7 and STE11 subfamilies have expanded in the land plant lineage (3–19 and 14–55 members, respectively) relative to yeast (four) and metazoans analysed (4–10). The expansion of the STE11 lineage is particularly pronounced: there are three times as many STE11 kinases in A. thaliana than STE7. These two subfamilies have been reviewed extensively elsewhere [30]. The first complete MAPK signalling pathway in plants was determined for the innate immunity pathway activated by the flagellin peptide, flg22. Flg22 binds to the FLS2 receptor, an RLK, activating the MAP3K, MEKK, which in turn activates the MAP2Ks MKK4 and MKK5, which activate MPK3 and MPK6 leading to downstream transcription of defence response genes [144]. Strikingly, this signalling framework is the same used in animals to signal innate immunity with the exception that the pathogen-associated molecular pattern (PAMP) is perceived by a Toll-like receptor that then acts through a cytosolic IRAK kinase [144]. Note that IRAK belongs to the RLK/Pelle family, which has undergone dramatic expansion in plants [34] but remains a small family in metazoans (1–4 members). Since this first example was published, MAPK pathways regulating cell division [145,146], stomata development (reviewed in Liu et al. [147]) and abiotic stress have been elucidated (reviewed in Sinha et al. [148]).
RSK-2
The most extensively studied AGC kinases in plants belong to the RSK-2 subfamily that is related to but evolutionarily distinct from RSK (p90 ribosomal S6 kinase). Members are involved in many aspects of cell growth and proliferation, and are activated by MAPK signalling cascades as well as by PDK1 (reviewed in Anjum & Blenis [149]). In land plants, the RSK kinases have undergone moderate expansion and are involved mainly in plant-specific roles, including blue light perception [150], polar auxin transport [151] and stress response [152,153]. Despite this divergence in function between plants and other eukaryotes, the regulation of RSK and RSK-2 activity by PDK1 is conserved (see §4a(i)).
(ii). Divergent functions but conserved regulatory mechanisms
The regulation of kinase activity of some moderately expanded families, OSTL1, CHK1 and CK1, is achieved by conserved mechanisms, even though the functions of and/or the signalling pathways regulating these kinases have diverged.
OSTL
The AMPK/SNF-related kinase family, OSTL (open stomata-like; SnRK2), which is related to but more divergent from the AMPK/SNF1 family discussed in §4a(i), has undergone pronounced expansion and divergence in plants [28]. Studies in rice, wheat and A. thaliana have revealed roles for SnRK2s in osmotic stress, the plant hormone abscisic acid (ABA) and/or sugar metabolism signalling (reviewed in Coello et al. [154]). There is evidence that SnRK2s are regulated by protein phosphatase 2C (PP2C), a mechanism used to regulate AMPK/SNF1 in animals and yeast. For example, the A. thaliana ABA-activated OST1/SnRK2.6 kinase is deactivated by the PP2C, HAB1, upon ABA binding to an ABA receptor [155]. The crystal structure of the SnRK2.6–HAB1 complex revealed that PP2C binds to both SnRK2s and ABA receptors through a common domain. Algae SnRKs lack this domain, indicating that SnRK2 ABA signalling is a land-plant-specific adaptation [156].
CHK1
In animals, CHK1 and the closely related CHK2 play roles in DNA damage response. Earlier studies indicated that there are no obvious plant CHK1 orthologues [157], but in our analysis, some plant kinases appear to be more closely related to CHK1 than to other CAMK families. CHK1, or the SNF1-related SnRK3, subfamily members are calcineurin B-like protein (CBL) interacting protein kinases (CIPKs). Plant CBLs are related to calcineurin B proteins in animals and yeast. However, CBL–CIPK signalling pathways are specific to plants; in yeast and animals calcineurin B binds to phosphatases, not kinases [158]. The plant CAMKL–CHK1 subfamily includes salt overly sensitive2 (SOS2), which interacts with the calcium-binding protein, SOS3, to regulate ion homeostasis and confer salt tolerance [159,160]. CIPKs have also been implicated in osmotic stress responses, ABA signalling, nitrate sensing and K+ transport, with the CBL-binding partner determining pathway specificity [161,162]. Like SnRK2s, CIPKs interact with PP2Cs, although whether CIPKs are dephosphorylated by PP2Cs has not yet been determined [156].
CK1
Casein kinase 1 (CK1) is evolutionarily conserved across eukaryotes and regulates a wide variety of cellular processes [163]. Many plant lineages have more than twice the number of CK1 paralogues compared with humans. Most plant CK1s have not been functionally characterized; however, there is evidence that A. thaliana CK1 proteins are involved in microtubule organization [164]. Not much is known about the regulation of plant CK1 activity, but achieving target specificity through the differential subcellular targeting of different isoforms appears to be a mechanism for controlling CK1 activity across eukaryotes [165,166]. Another group of CK1-related genes (CK1_CK1-Pl) is found only in the plant lineage. Rice EARLY FLOWERING1 encodes a plant-specific CK1 that negatively regulates gibberellic acid (GA) signalling by phosphorylating a DELLA domain protein, also a member of a plant-specific family of GA regulators [167], indicating that these family members participate in plant-specific processes.
(iii). Expansion and involvement in plant-specific processes
The plant-specific expansion of kinase families such as GSK3, NEK, CTR1, plant-specific TKLs and WNK/NRBP may take on functions in plant-specific processes that are probably adaptive. The plant-specific TKLs have clearly evolved to regulate processes that are specific to plants.
GSK3
Glycogen synthase kinase 3 (GSK3) is a key regulator of several developmental processes, including cell migration [168], metabolism [169] and cell proliferation. CMGC_GSK3 kinases are conserved in animals, fungi and plants, but while mammals have two copies of GSK3, this subfamily has expanded (up to more than 20 members) in land plants. In C. reinhardtii, the only GSK3 homologue is required for flagellar assembly [170]. In plants, GSK3 kinases function in brassinosteroid hormone signalling, abiotic and biotic stress pathways, and flower development (reviewed in Saidi et al. [32]). The diversification of GSK3 genes several times during land plant evolution suggests that they have been important for adaptation [32].
NEK
NimA was first isolated from Aspergillus niger, and NimA-related kinases (NEK) are found in all eukaryotes, ranging from one family member in yeast to 14 in mammals. Although NEKs have divergent functions, a common feature is their regulation of microtubules (reviewed in Moniz et al. [171]). NEK genes have roles in cell cycle regulation as well as ciliagenesis, and it is thought that expansion in organisms with cilia/flagella might be due to the need to coordinate the cell cycle with cilia development [171,172]. In G. lamblia, the NEK family has expanded to an astounding 198 of the 278 protein kinases, 70 per cent of which are likely to be catalytically inactive [37]. The dramatic expansion of the NEK family may be due to the fact that G. lambila has eight flagella and two nuclei, requiring more regulatory kinases; however, the expanded NEK genes are not orthologous to NEKs in expanded subfamilies in ciliates, which are also binucleate [37]. Notably, green algae C. reinhardtii and Volvox carteri have twice as many NEKs as land plants, suggesting that expansion may also be related to the presence of flagella in these algae. In plants, A. thaliana NEK6 has been the most extensively characterized. It interacts with microtubules to regulate epidermal cell morphogenesis [173,174] and is also involved in stress responses [175,176].
DRK-1 and DRK-2
In A. thaliana, expansion of TKLs has occurred in families whose members are involved in plant-specific processes. Because of high sequence divergence, the classification of TKLs is challenging. Particularly, in our analysis, many plant kinases that are regarded as Raf homologues (or referred to as MAP3Ks on some occasions) belong to the TKL group but do not appear to be in a monophyletic group with metazoan Raf kinases. One such example are the DRKs (downstream of receptor kinases) originally found in the slime mould D. discoideum [177] that resolved into two clades: DRK-1 and DRK-2. The DRK-2 family includes EDR1, which regulates SA-inducible defence signalling [178] and CTR1, which regulates ethylene responses [15]. Originally, on the basis of sequence similarity with Raf kinase, which has MAP3K activity, CTR1 was proposed to activate MAPK signalling cascades in response to ethylene (reviewed in Colcombet & Hirt [25]). However, a biochemical function as a MAP3K has not been demonstrated. EDR1 is recruited to the ER via its association with the plant-specific E3 ligase/kinase KEEP ON GOING, a member of the small TKL_Pl-1 subfamily; this complex may regulate signalling complexes located at the ER [179]. Rice DSM1, another DRK-2 member, mediates drought stress response [180]. The function of other DRK-2 and all DRK-1 members remains unclear.
TKL: plant-specific subfamilies
The TKL_Pl-4 kinases belong to multiple expanded, little studied subfamilies. HIGH TEMPERATURE1 is expressed in guard cells and regulates stomata in response to CO2 [181]. Three additional members (STY 8, STY17 and STY46) phosphorylate the transit peptides on chloroplast-targeted preproteins and are required for chloroplast differentiation [182]. The TKL_Pl-5 subfamily member, VH1-interacting kinase (VIK), interacts with VH1/BRL2, and mutations lead to defects in leaf venation and auxin and BR response [183]. VIK also phosphorylates the A. thaliana tonoplast monosaccharide transporter to regulate vacuolar sugar accumulation [184].
WNK/NRBP
WNK (with no lysine) and nuclear receptor binding protein (NRBP) families are considered together because they consistently form a monophyletic group. In addition, the duplication event that led to these two families appears to have taken place in the animal/fungal lineage after its divergence from plants (see electronic supplementary material, appendix S3). WNK kinases are distinct from other kinases because a conserved lysine in the catalytic cleft is found in subdomain I instead of subdomain II. They are found in most eukaryotes, but were lost in yeast [185]. In mammals, WNKs have been extensively studied owing to their effects on renal ion transport (reviewed in Huang et al. [186]). The C. elegans WNK1 gene functions in cell volume recovery after hypertonic stress [187]. Not much is known about the functions of WNKs in plants. Plant WNK genes in plants are transcriptionally regulated by the circadian clock and abiotic stress [188,189]. Soybean GmWNK1 regulates root architecture in response to ABA and osmotic signals [190]. On the basis of common functions of WNKs in osmotic stress responses [190], members in this family may be retained owing to their adaptive roles in protecting cells against water loss.
(c). Families with significant degree of expansion
(i). Significantly expanded families tend to play roles in stress response
The CDPKs and RLK/Pelle have undergone the most significant degree of expansion among protein kinase families in all land plant lineages. On the basis of our knowledge of their functions, their expansion and subsequent functional divergence may have allowed plants to perceive and/or respond to various environmental signals.
CDPK
The calcium-dependent protein kinases (CDPKs) comprise a large family of Ca2+ regulated kinases found in plants [13] and alveolates [38] (see Talevich et al. [191], in this issue) that have a domain similar to calmodulin and bind Ca2+ directly. Different CDPKs bind to Ca2+ with differing affinities, potentially allowing different CDPKs to respond to different Ca2+ signals [20]. CDPKs play roles in development, for example regulating transcription in response to hormone levels [192]. Several CDPKs are known to function in abiotic stress response and ABA signalling pathways through phosphorylation of targets, including ion channels and transporters (reviewed in Das & Pandey [193]). CDPK activity in guard cells regulates the opening and closing of stomata in response to ABA [194,195] and methyl jasmonate [196]. Recently, CDPKs have also been found to be key components of innate immunity signalling pathways [197,198].
RLK/Pelle
Plant RLK/Pelle family members play diverse roles in development, regulating cell-type specificity and organ identity (reviewed in De Smet et al. [199], as well as in defence response [200] and, to a lesser extent, in abiotic stress response [201–203]. It has been shown that RLK/Pelle subfamilies with developmental regulatory functions tend to have lower degrees of expansion than those involved in defence response [33]. However, subsequent study has shown that this distinction is not as clear-cut as was once believed [34]. Involvement in basal immunity is a function shared by both animal and plant RLK/Pelle genes, and the functions of RLK/Pelles in innate immunity pathways as pattern recognition receptors have been particularly well-studied [204]. Several lines of evidence suggest that RLK/Pelle expansion is tied to the need for plants to adapt to changing biotic conditions [34]. Although little domain gain within RLK extracellular domains has occurred since the divergence of the vascular plant lineage from moss, the domains that have been acquired have been implicated in sensing biotic signals. In addition, there is evidence that RLK/Pelle genes have been co-opted for roles in sensing biotic signals. For example, LRR-I subfamily members have evolved legume-specific roles in symbiosis. Finally, RLK/Pelle subfamilies show substantial differential expansion in different plant lineages, and subfamilies showing differential expansion, such as DLSV (DUF26, SD-1, LRR-VIII and VWA, a moss-specific new RLK subfamily), L-LEC, LRR-XII, SD2b, WAK and WAK/LRK10L-1 also tend to be enriched in genes that have been implicated in biotic stress response based on either function or expression evidence [34].
5. Why there are so many plant protein kinases?
In this study, plant protein kinases were identified and classified based on assignments from other eukaryotes. Many of these families and even subfamilies have clear plant homologues, suggesting their early establishment during the course of eukaryote evolution. The plant protein kinase superfamily is in general larger than that in other eukaryotes. Our findings indicate that the larger protein kinase repertoire is the consequence of recent duplications. Species such as soybean and cottonwood, where very recent genome duplications have taken place, have more kinases than most other plant species. Together with frequent tandem duplication found in this gene family [12,33,34], these duplication mechanisms are the main proximal cause for the higher degree of expansion of the protein kinase superfamily in plants.
The propensity to expand, however, differs greatly among protein kinase families and subfamilies because clearly not all duplicates were retained. Despite frequent duplication, many plant protein kinases remain low in copy number. In some cases, such as WEE1 and TLK, there are only single members within the families established prior to the divergence of eukaryotic species. Considering their functions, many are involved in ‘house-keeping’ functions such as regulation of metabolism, cell cycle and mitosis. Their low copy number and house-keeping role is consistent with the finding that duplicability of house-keeping genes tends to be low [205]. However, the plant lineage diverged from the animal and fungi lineages over a billion years ago. Extensive developmental and physiological adaptation and life-history differences have led to significant differences in how these highly conserved kinases function. As a result, many of them now play roles in plant-specific processes.
Aside from families with low copy number, some families have been moderately or highly expanded. What was the evolutionary force(s) that drove such differential expansion in protein kinase families? Given what we understand now, it appears that the selection pressure on the ability to properly respond to changing environment is a major contributor [54]. In the case of RLK/Pelle family members, which play roles in recognition of non-self biotic factors, continuous selective pressure imposed by pathogens and symbionts potentially drove the rather drastic expansion in this family [34]. The expansion of the CDPK family, whose members are involved in calcium-modulated signalling, could conceivably be a consequence of adaptive evolution where a new CDPK duplicate allowed perception of a different calcium signal through variation in affinity for Ca2+ and/or interacting protein substrates [20]. For the other families (such as multiple TKL families, the WNK/NRBP and the NEK families) that have undergone moderate expansion, the reason for expansion remains unclear. One possibility is that many of these protein kinases were duplicated and retained owing to their roles in plant-specific processes. This notion is consistent with the plant-specific roles played by many plant protein kinases with animal/fungal orthologues. To test this hypothesis, comparative functional studies are required between early diverging plant species such as algae and bryophytes and flowering plants.
In addition to an adaptive explanation for the observation of significantly more kinases in plants, it is possible that some protein kinases have been retained owing to at least two non-adaptive reasons. First, some duplicate protein kinases may be retained simply owing to subfunctionalization where the functions of the ancestral kinase were partitioned among the duplicates [206]. In this situation, neither copy can be lost without clear phenotypic consequences. This possibility, at first glance, may run counter to the well-known finding in the plant research community that there is a substantial functional overlap between duplicate genes. There are a large number of plant studies demonstrating that loss-of-function in multiple paralogues is necessary to reveal mutant phenotypes in laboratory conditions. However, it is apparent that laboratory conditions are not as diverse as the natural environment. Thus, the absence of a phenotype with the loss-of-function of one paralogue cannot be regarded as evidence against subfunctionalization.
The second non-adaptive explanation is that some protein kinases may very well be on their way to becoming pseudogenes. There are several lines of evidence suggesting this may be the case. Note that species with recent whole genome duplications tend to have larger protein kinase repertoires. In a comparative study of A. thaliana and rice genomes, hundreds of protein kinase pseudogenes were found, although protein kinases as a whole tend to have proportionally less pseudogenes than other families [43]. Population genomic studies of A. thaliana have also shown that many members of the RLK/Pelle family, which experienced the most extensive expansion, have alleles with non-sense and/or frame-shift mutations expected to disrupt gene function [207]. Therefore, there may have been insufficient time after duplication to accumulate mutations that render some protein kinases completely non-functional.
Another potential indication that some duplicate kinases are becoming pseudogenes is the prevalence of pseudokinases in plants. Around 13 per cent of A. thaliana protein kinases were hypothesized to be pseudokinases with modifications in some residues critical for catalytic activity [208]. However, some predicted pseudokinases have been shown to have catalytic activity, and the numbers of pseudokinases may be overestimated [209]. Among RLK/Pelle family members, the proportion of pseudokinases is even higher at approximately 20 per cent. In some cases, such pseudokinases have clear biological functions. One of the first examples is the STRUBBELIG kinase, which lacks enzymatic phospho-transfer activity but is essential for proper A. thaliana development [210]. Examples such as STRUBBELIG clearly demonstrate the importance of phosphorylation-independent mechanisms in plant signal transduction. However, a clear demonstration of loss-of-function phenotype is required to argue against the equally likely explanation that loss of kinase activity is due to relaxed or no selection on duplicates.
The relative importance of adaptive and non-adaptive explanations for protein kinase family expansion is unclear. But these discussions do highlight the challenge in elucidating plant protein kinase functions, given their exceptional numbers and similarities in all plant species. Comparative genomic studies examining molecular patterns of protein kinase evolution within and between species may provide some hints as to which protein kinases continue to experience strong purifying selection and are likely to remain functional.
Acknowledgements
We thank Raymond Chollet, Alice Harmon, Douglas Randall, Michael Sussman, John Walker and two anonymous reviewers for discussions and insights. This work was partially funded by NSF MCB-0929100, MCB-1119778 and IOS-1126998 to SHS.
References
- 1.Hanks S. K., Hunter T. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 [PubMed] [Google Scholar]
- 2.Hanks S. K., Quinn A. M., Hunter T. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241, 42–52 10.1126/science.3291115 (doi:10.1126/science.3291115) [DOI] [PubMed] [Google Scholar]
- 3.The Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 10.1038/35048692 (doi:10.1038/35048692) [DOI] [PubMed] [Google Scholar]
- 4.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 (doi:10.1126/science.1075762) [DOI] [PubMed] [Google Scholar]
- 5.Ralph R. K., McCombs P. J., Tener G., Wojcik S. J. 1972. Evidence for modification of protein phosphorylation by cytokinins. Biochem. J. 130, 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trewavas A. 1973. The phosphorylation of ribosomal protein in Lemna minor. Plant Physiol. 51, 760–767 10.1104/pp.51.4.760 (doi:10.1104/pp.51.4.760) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keates R. A. 1973. Cyclic nucleotide-independent protein kinase from pea shoots. Biochem. Biophys. Res. Commun. 54, 655–661 10.1016/0006-291X(73)91473-3 (doi:10.1016/0006-291X(73)91473-3) [DOI] [PubMed] [Google Scholar]
- 8.Lawton M. A., Yamamoto R. T., Hanks S. K., Lamb C. J. 1989. Molecular cloning of plant transcripts encoding protein kinase homologs. Proc. Natl Acad. Sci. USA 86, 3140–3144 10.1073/pnas.86.9.3140 (doi:10.1073/pnas.86.9.3140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker J. C., Zhang R. 1990. Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature 345, 743–746 10.1038/345743a0 (doi:10.1038/345743a0) [DOI] [PubMed] [Google Scholar]
- 10.Hunter T., Lindberg R. A., Middlemas D. S., Tracy S., van der Geer P. 1992. Receptor protein tyrosine kinases and phosphatases. Cold Spring Harb. Symp. Quant. Biol. 57, 25–41 10.1101/SQB.1992.057.01.005 (doi:10.1101/SQB.1992.057.01.005) [DOI] [PubMed] [Google Scholar]
- 11.van der Geer P., Hunter T., Lindberg R. A. 1994. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu. Rev. Cell Biol. 10, 251–337 10.1146/annurev.cb.10.110194.001343 (doi:10.1146/annurev.cb.10.110194.001343) [DOI] [PubMed] [Google Scholar]
- 12.Shiu S. H., Bleecker A. B. 2001. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. USA 98, 10 763–10 768 10.1073/pnas.181141598 (doi:10.1073/pnas.181141598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper J. F., Sussman M. R., Schaller G. E., Putnam-Evans C., Charbonneau H., Harmon A. C. 1991. A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science 252, 951–954 10.1126/science.1852075 (doi:10.1126/science.1852075) [DOI] [PubMed] [Google Scholar]
- 14.Martin G. B., Brommonschenkel S., Chunwongse J., Frary A., Ganal M. W., Spivey R., Wu T., Earle E. D., Tanksley S. D. 1993. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262, 1432–1436 10.1126/science.7902614 (doi:10.1126/science.7902614) [DOI] [PubMed] [Google Scholar]
- 15.Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., Ecker J. R. 1993. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441 10.1016/0092-8674(93)90119-B (doi:10.1016/0092-8674(93)90119-B) [DOI] [PubMed] [Google Scholar]
- 16.Roe J. L., Rivin C. J., Sessions R. A., Feldmann K. A., Zambryski P. C. 1993. The Tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 75, 939–950 10.1016/0092-8674(93)90537-Z (doi:10.1016/0092-8674(93)90537-Z) [DOI] [PubMed] [Google Scholar]
- 17.Stone J. M., Walker J. C. 1995. Plant protein kinase families and signal transduction. Plant Physiol. 108, 451–457 10.1104/pp.108.2.451 (doi:10.1104/pp.108.2.451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardie D. G. 1999. Plant protein serine/threonine kinases: classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 97–131 10.1146/annurev.arplant.50.1.97 (doi:10.1146/annurev.arplant.50.1.97) [DOI] [PubMed] [Google Scholar]
- 19.Shiu S. H., Bleecker A. B. 2001. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001, RE22 10.1126/stke.2001.113.re22 (doi:10.1126/stke.2001.113.re22) [DOI] [PubMed] [Google Scholar]
- 20.Hrabak E. M., et al. 2003. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680 10.1104/pp.102.011999 (doi:10.1104/pp.102.011999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Champion A., Kreis M., Mockaitis K., Picaud A., Henry Y. 2004. Arabidopsis kinome: after the casting. Funct. Integr. Genomics 4, 163–187 10.1007/s10142-003-0096-4 (doi:10.1007/s10142-003-0096-4) [DOI] [PubMed] [Google Scholar]
- 22.Chen M., Chory J., Fankhauser C. 2004. Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87–117 10.1146/annurev.genet.38.072902.092259 (doi:10.1146/annurev.genet.38.072902.092259) [DOI] [PubMed] [Google Scholar]
- 23.Harper J. F., Harmon A. 2005. Plants, symbiosis and parasites: a calcium signalling connection. Nat. Rev. Mol. Cell Biol. 6, 555–566 10.1038/nrm1679 (doi:10.1038/nrm1679) [DOI] [PubMed] [Google Scholar]
- 24.Francis D. 2007. The plant cell cycle: 15 years on. New Phytol. 174, 261–278 10.1111/j.1469-8137.2007.02038.x (doi:10.1111/j.1469-8137.2007.02038.x) [DOI] [PubMed] [Google Scholar]
- 25.Colcombet J., Hirt H. 2008. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413, 217–226 10.1042/BJ20080625 (doi:10.1042/BJ20080625) [DOI] [PubMed] [Google Scholar]
- 26.Demarsy E., Fankhauser C. 2009. Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 12, 69–74 10.1016/j.pbi.2008.09.002 (doi:10.1016/j.pbi.2008.09.002) [DOI] [PubMed] [Google Scholar]
- 27.Dahan J., Wendehenne D., Ranjeva R., Pugin A., Bourque S. 2010. Nuclear protein kinases: still enigmatic components in plant cell signalling. New Phytol. 185, 355–368 10.1111/j.1469-8137.2009.03085.x (doi:10.1111/j.1469-8137.2009.03085.x) [DOI] [PubMed] [Google Scholar]
- 28.Halford N. G., Hey S. J. 2009. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 419, 247–259 10.1042/BJ20082408 (doi:10.1042/BJ20082408) [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., McCormick S. 2009. AGCVIII kinases: at the crossroads of cellular signaling. Trends Plant Sci. 14, 689–695 10.1016/j.tplants.2009.09.006 (doi:10.1016/j.tplants.2009.09.006) [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez M. C., Petersen M., Mundy J. 2010. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649 10.1146/annurev-arplant-042809-112252 (doi:10.1146/annurev-arplant-042809-112252) [DOI] [PubMed] [Google Scholar]
- 31.Gish L. A., Clark S. E. 2011. The RLK/Pelle family of kinases. Plant J. 66, 117–127 10.1111/j.1365-313X.2011.04518.x (doi:10.1111/j.1365-313X.2011.04518.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saidi Y., Hearn T. J., Coates J. C. 2011. Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci. 17, 39–46 10.1016/j.tplants.2011.10.002 (doi:10.1016/j.tplants.2011.10.002) [DOI] [PubMed] [Google Scholar]
- 33.Shiu S. H., Karlowski W. M., Pan R., Tzeng Y. H., Mayer K. F., Li W. H. 2004. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234 10.1105/tpc.020834 (doi:10.1105/tpc.020834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehti-Shiu M. D., Zou C., Hanada K., Shiu S. H. 2009. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26 10.1104/pp.108.134353 (doi:10.1104/pp.108.134353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning G., Plowman G. D., Hunter T., Sudarsanam S. 2002. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514–520 10.1016/S0968-0004(02)02179-5 (doi:10.1016/S0968-0004(02)02179-5) [DOI] [PubMed] [Google Scholar]
- 36.King N., et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788 10.1038/nature06617 (doi:10.1038/nature06617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning G., Reiner D. S., Lauwaet T., Dacre M., Smith A., Zhai Y., Svard S., Gillin F. D. 2011. The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol 12, R66 10.1186/gb-2011-12-7-r66 (doi:10.1186/gb-2011-12-7-r66) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward P., Equinet L., Packer J., Doerig C. 2004. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5, 79 10.1186/1471-2164-5-79 (doi:10.1186/1471-2164-5-79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miranda-Saavedra D., Stark M. J., Packer J. C., Vivares C. P., Doerig C., Barton G. J. 2007. The complement of protein kinases of the microsporidium Encephalitozoon cuniculi in relation to those of Saccharomyces cerevisiae and Schizosaccharomyces pombe. BMC Genomics 8, 309 10.1186/1471-2164-8-309 (doi:10.1186/1471-2164-8-309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miranda-Saavedra D., Barton G. J. 2007. Classification and functional annotation of eukaryotic protein kinases. Proteins 68, 893–914 10.1002/prot.21444 (doi:10.1002/prot.21444) [DOI] [PubMed] [Google Scholar]
- 41.Finn R. D., Clements J., Eddy S. R. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–37 10.1093/nar/gkr367 (doi:10.1093/nar/gkr367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 10.1093/nar/gkp985 (doi:10.1093/nar/gkp985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou C., Lehti-Shiu M. D., Thibaud-Nissen F., Prakash T., Buell C. R., Shiu S. H. 2009. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 151, 3–15 10.1104/pp.109.140632 (doi:10.1104/pp.109.140632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sievers F., et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 (doi:10.1038/msb.2011.75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lartillot N., Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 10.1093/molbev/msh112 (doi:10.1093/molbev/msh112) [DOI] [PubMed] [Google Scholar]
- 46.Jones D. T., Taylor W. R., Thornton J. M. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 47.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 (doi:10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dardick C., Chen J., Richter T., Ouyang S., Ronald P. 2007. The rice kinase database. A phylogenomic database for the rice kinome. Plant Physiol. 143, 579–586 10.1104/pp.106.087270 (doi:10.1104/pp.106.087270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298 10.1016/S0169-5347(03)00033-8 (doi:10.1016/S0169-5347(03)00033-8) [DOI] [Google Scholar]
- 50.Otto S. P., Whitton J. 2000. Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437 10.1146/annurev.genet.34.1.401 (doi:10.1146/annurev.genet.34.1.401) [DOI] [PubMed] [Google Scholar]
- 51.Blanc G., Hokamp K., Wolfe K. H. 2003. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144 10.1101/gr.751803 (doi:10.1101/gr.751803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blanc G., Wolfe K. H. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678 10.1105/tpc.021345 (doi:10.1105/tpc.021345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowers J. E., Chapman B. A., Rong J., Paterson A. H. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438 10.1038/nature01521 (doi:10.1038/nature01521) [DOI] [PubMed] [Google Scholar]
- 54.Hanada K., Zou C., Lehti-Shiu M. D., Shinozaki K., Shiu S. H. 2008. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148, 993–1003 10.1104/pp.108.122457 (doi:10.1104/pp.108.122457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon H. S., Hackett J. D., Ciniglia C., Pinto G., Bhattacharya D. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818 10.1093/molbev/msh075 (doi:10.1093/molbev/msh075) [DOI] [PubMed] [Google Scholar]
- 56.Derelle E., et al. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl Acad. Sci. USA 103, 11 647–11 652 10.1073/pnas.0604795103 (doi:10.1073/pnas.0604795103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katinka M. D., et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450–453 10.1038/35106579 (doi:10.1038/35106579) [DOI] [PubMed] [Google Scholar]
- 58.Hu T. T., et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481 10.1038/ng.807 (doi:10.1038/ng.807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banks J. A., et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 10.1126/science.1203810 (doi:10.1126/science.1203810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmutz J., et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 10.1038/nature08670 (doi:10.1038/nature08670) [DOI] [PubMed] [Google Scholar]
- 61.Tuskan G. A., et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 10.1126/science.1128691 (doi:10.1126/science.1128691) [DOI] [PubMed] [Google Scholar]
- 62.Huang S., et al. 2009. The genome of the cucumber, Cucumis sativus, L. Nat. Genet. 41, 1275–1281 10.1038/ng.475 (doi:10.1038/ng.475) [DOI] [PubMed] [Google Scholar]
- 63.Ming R., et al. 2008. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996 10.1038/nature06856 (doi:10.1038/nature06856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shiu S. H., Bleecker A. B. 2003. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543 10.1104/pp.103.021964 (doi:10.1104/pp.103.021964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hardie D. G. 2011. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 10.1101/gad.17420111 (doi:10.1101/gad.17420111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baena-Gonzalez E., Rolland F., Thevelein J. M., Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942 10.1038/nature06069 (doi:10.1038/nature06069) [DOI] [PubMed] [Google Scholar]
- 67.Jossier M., Bouly J. P., Meimoun P., Arjmand A., Lessard P., Hawley S., Grahame Hardie D., Thomas M. 2009. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 59, 316–328 10.1111/j.1365-313X.2009.03871.x (doi:10.1111/j.1365-313X.2009.03871.x) [DOI] [PubMed] [Google Scholar]
- 68.Thelander M., Olsson T., Ronne H. 2004. Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J. 23, 1900–1910 10.1038/sj.emboj.7600182 (doi:10.1038/sj.emboj.7600182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woods A., et al. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13, 2004–2008 10.1016/j.cub.2003.10.031 (doi:10.1016/j.cub.2003.10.031) [DOI] [PubMed] [Google Scholar]
- 70.Hong S. P., Leiper F. C., Woods A., Carling D., Carlson M. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl Acad. Sci. USA 100, 8839–8843 10.1073/pnas.1533136100 (doi:10.1073/pnas.1533136100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen W., Hanley-Bowdoin L. 2006. Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol. 142, 1642–1655 10.1104/pp.106.088476 (doi:10.1104/pp.106.088476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen W., Reyes M. I., Hanley-Bowdoin L. 2009. Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol. 150, 996–1005 10.1104/pp.108.132787 (doi:10.1104/pp.108.132787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9, 186–197 10.1016/S0960-9822(99)80088-8 (doi:10.1016/S0960-9822(99)80088-8) [DOI] [PubMed] [Google Scholar]
- 74.Bayascas J. R. 2010. PDK1: the major transducer of PI 3-kinase actions. Curr. Top. Microbiol. Immunol. 346, 9–29 10.1007/82_2010_43 (doi:10.1007/82_2010_43) [DOI] [PubMed] [Google Scholar]
- 75.Anthony R. G., et al. 2004. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23, 572–581 10.1038/sj.emboj.7600068 (doi:10.1038/sj.emboj.7600068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zegzouti H., Anthony R. G., Jahchan N., Bogre L., Christensen S. K. 2006. Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl Acad. Sci. USA 103, 6404–6409 10.1073/pnas.0510283103 (doi:10.1073/pnas.0510283103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pearce L. R., Komander D., Alessi D. R. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 10.1038/nrm2822 (doi:10.1038/nrm2822) [DOI] [PubMed] [Google Scholar]
- 78.Camehl I., Drzewiecki C., Vadassery J., Shahollari B., Sherameti I., Forzani C., Munnik T., Hirt H., Oelmuller R. 2011. The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog. 7, e1002051 10.1371/journal.ppat.1002051 (doi:10.1371/journal.ppat.1002051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lawlor M. A., Mora A., Ashby P. R., Williams M. R., Murray-Tait V., Malone L., Prescott A. R., Lucocq J. M., Alessi D. R. 2002. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 21, 3728–3738 10.1093/emboj/cdf387 (doi:10.1093/emboj/cdf387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turck F., Kozma S. C., Thomas G., Nagy F. 1998. A heat-sensitive Arabidopsis thaliana kinase substitutes for human p70s6k function in vivo. Mol. Cell Biol. 18, 2038–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mahfouz M. M., Kim S., Delauney A. J., Verma D. P. 2006. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18, 477–490 10.1105/tpc.105.035931 (doi:10.1105/tpc.105.035931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hsieh T. C., Nicolay B. N., Frolov M. V., Moon N. S. 2010. Tuberous sclerosis complex 1 regulates dE2F1 expression during development and cooperates with RBF1 to control proliferation and survival. PLoS Genet. 6, e1001071 10.1371/journal.pgen.1001071 (doi:10.1371/journal.pgen.1001071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henriques R., et al. 2010. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J. 29, 2979–2993 10.1038/emboj.2010.164 (doi:10.1038/emboj.2010.164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koizumi N., Martinez I. M., Kimata Y., Kohno K., Sano H., Chrispeels M. J. 2001. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant. Physiol. 127, 949–962 10.1104/pp.010636 (doi:10.1104/pp.010636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 10.1038/nrm2199 (doi:10.1038/nrm2199) [DOI] [PubMed] [Google Scholar]
- 86.Deng Y., Humbert S., Liu J. X., Srivastava R., Rothstein S. J., Howell S. H. 2011. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 7247–7252 10.1073/pnas.1102117108 (doi:10.1073/pnas.1102117108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hayashi S., Wakasa Y., Takahashi H., Kawakatsu T., Takaiwa F. 2011. Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 69, 946–956 10.1111/j.1365-313X.2011.04844.x (doi:10.1111/j.1365-313X.2011.04844.x) [DOI] [PubMed] [Google Scholar]
- 88.Wek R. C., Jiang H. Y., Anthony T. G. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 10.1042/BST20060007 (doi:10.1042/BST20060007) [DOI] [PubMed] [Google Scholar]
- 89.Grallert B., Boye E. 2007. The Gcn2 kinase as a cell cycle regulator. Cell Cycle 6, 2768–2772 10.4161/cc.6.22.4933 (doi:10.4161/cc.6.22.4933) [DOI] [PubMed] [Google Scholar]
- 90.Zhan K., Narasimhan J., Wek R. C. 2004. Differential activation of eIF2 kinases in response to cellular stresses in Schizosaccharomyces pombe. Genetics 168, 1867–1875 10.1534/genetics.104.031443 (doi:10.1534/genetics.104.031443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y., Dickinson J. R., Paul M. J., Halford N. G. 2003. Molecular cloning of an Arabidopsis homologue of GCN2, a protein kinase involved in co-ordinated response to amino acid starvation. Planta 217, 668–675 10.1007/s00425-003-1025-4 (doi:10.1007/s00425-003-1025-4) [DOI] [PubMed] [Google Scholar]
- 92.Zhang Y., Wang Y., Kanyuka K., Parry M. A., Powers S. J., Halford N. G. 2008. GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2alpha in Arabidopsis. J. Exp. Bot. 59, 3131–3141 10.1093/jxb/ern169 (doi:10.1093/jxb/ern169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lageix S., Lanet E., Pouch-Pelissier M. N., Espagnol M. C., Robaglia C., Deragon J. M., Pelissier T. 2008. Arabidopsis eIF2alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 8, 134 10.1186/1471-2229-8-134 (doi:10.1186/1471-2229-8-134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carmena M., Ruchaud S., Earnshaw W. C. 2009. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21, 796–805 10.1016/j.ceb.2009.09.008 (doi:10.1016/j.ceb.2009.09.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Demidov D., Van Damme D., Geelen D., Blattner F. R., Houben A. 2005. Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17, 836–848 10.1105/tpc.104.029710 (doi:10.1105/tpc.104.029710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kawabe A., Matsunaga S., Nakagawa K., Kurihara D., Yoneda A., Hasezawa S., Uchiyama S., Fukui K. 2005. Characterization of plant Aurora kinases during mitosis. Plant Mol. Biol. 58, 1–13 10.1007/s11103-005-3454-x (doi:10.1007/s11103-005-3454-x) [DOI] [PubMed] [Google Scholar]
- 97.Van Damme D., De Rybel B., Gudesblat G., Demidov D., Grunewald W., De Smet I., Houben A., Beeckman T., Russinova E. 2011. Arabidopsis alpha Aurora kinases function in formative cell division plane orientation. Plant Cell 23, 4013–4024 10.1105/tpc.111.089565 (doi:10.1105/tpc.111.089565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pan J., Wang Q., Snell W. J. 2004. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev. Cell 6, 445–451 10.1016/S1534-5807(04)00064-4 (doi:10.1016/S1534-5807(04)00064-4) [DOI] [PubMed] [Google Scholar]
- 99.Luo M., Cao M., Kan Y., Li G., Snell W., Pan J. 2011. The phosphorylation state of an aurora-like kinase marks the length of growing flagella in Chlamydomonas. Curr. Biol. 21, 586–591 10.1016/j.cub.2011.02.046 (doi:10.1016/j.cub.2011.02.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pugacheva E. N., Jablonski S. A., Hartman T. R., Henske E. P., Golemis E. A. 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129, 1351–1363 10.1016/j.cell.2007.04.035 (doi:10.1016/j.cell.2007.04.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Joubes J., Chevalier C., Dudits D., Heberle-Bors E., Inze D., Umeda M., Renaudin J. P. 2000. CDK-related protein kinases in plants. Plant Mol. Biol. 43, 607–620 10.1023/A:1006470301554 (doi:10.1023/A:1006470301554) [DOI] [PubMed] [Google Scholar]
- 102.Guitierrez C. 2009. The Arabidopsis cell division cycle The Arabidopsis Book 7, e0120 10.1199/tab.0120 (doi:10.1199/tab.0120). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaldis P. 1999. The cdk-activating kinase (CAK): from yeast to mammals. Cell Mol. Life Sci. 55, 284–296 10.1007/s000180050290 (doi:10.1007/s000180050290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Doonan J. H., Kitsios G. 2009. Functional evolution of cyclin-dependent kinases. Mol. Biotechnol. 42, 14–29 10.1007/s12033-008-9126-8 (doi:10.1007/s12033-008-9126-8) [DOI] [PubMed] [Google Scholar]
- 105.Yamaguchi M., Umeda M., Uchimiya H. 1998. A rice homolog of Cdk7/MO15 phosphorylates both cyclin-dependent protein kinases and the carboxy-terminal domain of RNA polymerase II. Plant J. 16, 613–619 10.1046/j.1365-313x.1998.00338.x (doi:10.1046/j.1365-313x.1998.00338.x) [DOI] [PubMed] [Google Scholar]
- 106.Yamaguchi M., Fabian T., Sauter M., Bhalerao R. P., Schrader J., Sandberg G., Umeda M., Uchimiya H. 2000. Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants. Plant J. 24, 11–20 10.1046/j.1365-313x.2000.00846.x (doi:10.1046/j.1365-313x.2000.00846.x) [DOI] [PubMed] [Google Scholar]
- 107.Inagaki S., Umeda M. 2011. Cell-cycle control and plant development. Int. Rev. Cell Mol. Biol. 291, 227–261 10.1016/B978-0-12-386035-4.00007-0 (doi:10.1016/B978-0-12-386035-4.00007-0) [DOI] [PubMed] [Google Scholar]
- 108.Takatsuka H., Ohno R., Umeda M. 2009. The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. Plant J. 59, 475–487 10.1111/j.1365-313X.2009.03884.x (doi:10.1111/j.1365-313X.2009.03884.x) [DOI] [PubMed] [Google Scholar]
- 109.Wohlbold L., Larochelle S., Liao J. C., Livshits G., Singer J., Shokat K. M., Fisher R. P. 2006. The cyclin-dependent kinase (CDK) family member PNQALRE/CCRK supports cell proliferation but has no intrinsic CDK-activating kinase (CAK) activity. Cell Cycle 5, 546–554 10.4161/cc.5.5.2541 (doi:10.4161/cc.5.5.2541) [DOI] [PubMed] [Google Scholar]
- 110.Ko H. W., Norman R. X., Tran J., Fuller K. P., Fukuda M., Eggenschwiler J. T. 2010. Broad-minded links cell cycle-related kinase to cilia assembly and hedgehog signal transduction. Dev. Cell 18, 237–247 10.1016/j.devcel.2009.12.014 (doi:10.1016/j.devcel.2009.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tam L. W., Wilson N. F., Lefebvre P. A. 2007. A CDK-related kinase regulates the length and assembly of flagella in Chlamydomonas. J. Cell Biol. 176, 819–829 10.1083/jcb.200610022 (doi:10.1083/jcb.200610022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Leclerc V., Leopold P. 1996. The cyclin C/Cdk8 kinase. Prog. Cell Cycle Res. 2, 197–204 10.1007/978-1-4615-5873-6_19 (doi:10.1007/978-1-4615-5873-6_19) [DOI] [PubMed] [Google Scholar]
- 113.Conaway R. C., Conaway J. W. 2011. Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 21, 225–230 10.1016/j.gde.2011.01.013 (doi:10.1016/j.gde.2011.01.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang W., Chen X. 2004. HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131, 3147–3156 10.1242/dev.01187 (doi:10.1242/dev.01187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gonzalez D., Bowen A. J., Carroll T. S., Conlan R. S. 2007. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and Mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell Biol. 27, 5306–5315 10.1128/MCB.01912-06 (doi:10.1128/MCB.01912-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sia R. A., Herald H. A., Lew D. J. 1996. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell 7, 1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McMillan J. N., Sia R. A., Lew D. J. 1998. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 142, 1487–1499 10.1083/jcb.142.6.1487 (doi:10.1083/jcb.142.6.1487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dissmeyer N., Weimer A. K., De Veylder L., Novak B., Schnittger A. 2010. The regulatory network of cell-cycle progression is fundamentally different in plants versus yeast or metazoans. Plant Signal Behav. 5, 1613–1618 10.4161/psb.5.12.13969 (doi:10.4161/psb.5.12.13969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gonzalez N., Gevaudant F., Hernould M., Chevalier C., Mouras A. 2007. The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J. 51, 642–655 10.1111/j.1365-313X.2007.03167.x (doi:10.1111/j.1365-313X.2007.03167.x) [DOI] [PubMed] [Google Scholar]
- 120.De Schutter K., et al. 2007. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19, 211–225 10.1105/tpc.106.045047 (doi:10.1105/tpc.106.045047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cools T., et al. 2011. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23, 1435–1448 10.1105/tpc.110.082768 (doi:10.1105/tpc.110.082768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chan E. Y. 2012. Regulation and function of uncoordinated-51 like kinase proteins. Antioxid. Redox Signal 17, 775–785 10.1089/ars.2011.4396 (doi:10.1089/ars.2011.4396) [DOI] [PubMed] [Google Scholar]
- 123.Krupnova T., et al. 2009. Microtubule-associated kinase-like protein RUNKEL needed for cell plate expansion in Arabidopsis cytokinesis. Curr. Biol. 19, 518–523 10.1016/j.cub.2009.02.021 (doi:10.1016/j.cub.2009.02.021) [DOI] [PubMed] [Google Scholar]
- 124.Hofius D., Munch D., Bressendorff S., Mundy J., Petersen M. 2011. Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 18, 1257–1262 10.1038/cdd.2011.43 (doi:10.1038/cdd.2011.43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Murone M., Luoh S. M., Stone D., Li W., Gurney A., Armanini M., Grey C., Rosenthal A., de Sauvage F. J. 2000. Gli regulation by the opposing activities of fused and suppressor of fused. Nat. Cell Biol. 2, 310–312 10.1038/35010610 (doi:10.1038/35010610) [DOI] [PubMed] [Google Scholar]
- 126.Oh S. A., Johnson A., Smertenko A., Rahman D., Park S. K., Hussey P. J., Twell D. 2005. A divergent cellular role for the FUSED kinase family in the plant-specific cytokinetic phragmoplast. Curr. Biol. 15, 2107–2111 10.1016/j.cub.2005.10.044 (doi:10.1016/j.cub.2005.10.044) [DOI] [PubMed] [Google Scholar]
- 127.Moran T. V., Walker J. C. 1993. Molecular cloning of two novel protein kinase genes from Arabidopsis thaliana. Biochim. Biophys. Acta 1216, 9–14 10.1016/0167-4781(93)90031-8 (doi:10.1016/0167-4781(93)90031-8) [DOI] [PubMed] [Google Scholar]
- 128.Berman S. A., Wilson N. F., Haas N. A., Lefebvre P. A. 2003. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 13, 1145–1149 10.1016/S0960-9822(03)00415-9 (doi:10.1016/S0960-9822(03)00415-9) [DOI] [PubMed] [Google Scholar]
- 129.Burghoorn J., Dekkers M. P., Rademakers S., de Jong T., Willemsen R., Jansen G. 2007. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 104, 7157–7162 10.1073/pnas.0606974104 (doi:10.1073/pnas.0606974104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Omori Y., et al. 2010. Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc. Natl Acad. Sci. USA 107, 22 671–22 676 10.1073/pnas.1009437108 (doi:10.1073/pnas.1009437108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang Y., Zhang W. Z., Song L. F., Zou J. J., Su Z., Wu W. H. 2008. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148, 1201–1211 10.1104/pp.108.126375 (doi:10.1104/pp.108.126375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Y., Liu J., Xia R., Wang J., Shen J., Cao R., Hong X., Zhu J. K., Gong Z. 2007. The protein kinase TOUSLED is required for maintenance of transcriptional gene silencing in Arabidopsis. EMBO Rep. 8, 77–83 10.1038/sj.embor.7400852 (doi:10.1038/sj.embor.7400852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Y., DeFatta R., Anthony C., Sunavala G., De Benedetti A. 2001. A translationally regulated Tousled kinase phosphorylates histone H3 and confers radioresistance when overexpressed. Oncogene 20, 726–738 10.1038/sj.onc.1204147 (doi:10.1038/sj.onc.1204147) [DOI] [PubMed] [Google Scholar]
- 134.Sillje H. H., Nigg E. A. 2001. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11, 1068–1073 10.1016/S0960-9822(01)00298-6 (doi:10.1016/S0960-9822(01)00298-6) [DOI] [PubMed] [Google Scholar]
- 135.Carrera P., Moshkin Y. M., Gronke S., Sillje H. H., Nigg E. A., Jackle H., Karch F. 2003. Tousled-like kinase functions with the chromatin assembly pathway regulating nuclear divisions. Genes Dev. 17, 2578–2590 10.1101/gad.276703 (doi:10.1101/gad.276703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ehsan H., Reichheld J. P., Durfee T., Roe J. L. 2004. TOUSLED kinase activity oscillates during the cell cycle and interacts with chromatin regulators. Plant Physiol. 134, 1488–1499 10.1104/pp.103.038117 (doi:10.1104/pp.103.038117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pilyugin M., Demmers J., Verrijzer C. P., Karch F., Moshkin Y. M. 2009. Phosphorylation-mediated control of histone chaperone ASF1 levels by Tousled-like kinases. PLoS ONE 4, e8328 10.1371/journal.pone.0008328 (doi:10.1371/journal.pone.0008328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Aranda S., Laguna A., de la Luna S. 2011. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 25, 449–462 10.1096/fj.10-165837 (doi:10.1096/fj.10-165837) [DOI] [PubMed] [Google Scholar]
- 139.Lee P., Cho B. R., Joo H. S., Hahn J. S. 2008. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 70, 882–895 10.1111/j.1365-2958.2008.06450.x (doi:10.1111/j.1365-2958.2008.06450.x) [DOI] [PubMed] [Google Scholar]
- 140.Malcher M., Schladebeck S., Mosch H. U. 2011. The Yak1 protein kinase lies at the center of a regulatory cascade affecting adhesive growth and stress resistance in Saccharomyces cerevisiae. Genetics 187, 717–730 10.1534/genetics.110.125708 (doi:10.1534/genetics.110.125708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim Y. H., Choi C. Y., Lee S. J., Conti M. A., Kim Y. 1998. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem. 273, 25 875–25 879 10.1074/jbc.273.40.25875 (doi:10.1074/jbc.273.40.25875) [DOI] [PubMed] [Google Scholar]
- 142.Rosenberg G. H., Alahari S. K., Kaufer N. F. 1991. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol. Gen. Genet. 226, 305–309 10.1007/BF00273617 (doi:10.1007/BF00273617) [DOI] [PubMed] [Google Scholar]
- 143.Schneider M., Hsiao H. H., Will C. L., Giet R., Urlaub H., Luhrmann R. 2010. Human PRP4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat. Struct. Mol. Biol. 17, 216–221 10.1038/nsmb.1718 (doi:10.1038/nsmb.1718) [DOI] [PubMed] [Google Scholar]
- 144.Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., Boller T., Ausubel F. M., Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 10.1038/415977a (doi:10.1038/415977a) [DOI] [PubMed] [Google Scholar]
- 145.Takahashi Y., Soyano T., Kosetsu K., Sasabe M., Machida Y. 2010. HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51, 1766–1776 10.1093/pcp/pcq135 (doi:10.1093/pcp/pcq135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Soyano T., Nishihama R., Morikiyo K., Ishikawa M., Machida Y. 2003. NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 17, 1055–1067 10.1101/gad.1071103 (doi:10.1101/gad.1071103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu Y. K., Liu Y. B., Zhang M. Y., Li D. Q. 2010. Stomatal development and movement: the roles of MAPK signaling. Plant Signal Behav. 5, 1176–1180 10.4161/psb.5.10.12757 (doi:10.4161/psb.5.10.12757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sinha A. K., Jaggi M., Raghuram B., Tuteja N. 2011. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav. 6, 196–203 10.4161/psb.6.2.14701 (doi:10.4161/psb.6.2.14701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Anjum R., Blenis J. 2008. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 9, 747–758 10.1038/nrm2509 (doi:10.1038/nrm2509) [DOI] [PubMed] [Google Scholar]
- 150.Briggs W. R., Christie J. M. 2002. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210 10.1016/S1360-1385(02)02245-8 (doi:10.1016/S1360-1385(02)02245-8) [DOI] [PubMed] [Google Scholar]
- 151.Zourelidou M., Muller I., Willige B. C., Nill C., Jikumaru Y., Li H., Schwechheimer C. 2009. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136, 627–636 10.1242/dev.028365 (doi:10.1242/dev.028365) [DOI] [PubMed] [Google Scholar]
- 152.Rentel M. C., et al. 2004. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427, 858–861 10.1038/nature02353 (doi:10.1038/nature02353) [DOI] [PubMed] [Google Scholar]
- 153.Matsui H., Yamazaki M., Kishi-Kaboshi M., Takahashi A., Hirochika H. 2010. AGC kinase OsOxi1 positively regulates basal resistance through suppression of OsPti1a-mediated negative regulation. Plant Cell Physiol. 51, 1731–1744 10.1093/pcp/pcq132 (doi:10.1093/pcp/pcq132) [DOI] [PubMed] [Google Scholar]
- 154.Coello P., Hey S. J., Halford N. G. 2011. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 62, 883–893 10.1093/jxb/erq331 (doi:10.1093/jxb/erq331) [DOI] [PubMed] [Google Scholar]
- 155.Vlad F., et al. 2009. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21, 3170–3184 10.1105/tpc.109.069179 (doi:10.1105/tpc.109.069179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hauser F., Waadt R., Schroeder J. I. 2011. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–355 10.1016/j.cub.2011.03.015 (doi:10.1016/j.cub.2011.03.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Adachi S., et al. 2011. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 10 004–10 009 10.1073/pnas.1103584108 (doi:10.1073/pnas.1103584108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Luan S., Lan W., Chul Lee S. 2009. Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL-CIPK network. Curr. Opin. Plant Biol. 12, 339–346 10.1016/j.pbi.2009.05.003 (doi:10.1016/j.pbi.2009.05.003) [DOI] [PubMed] [Google Scholar]
- 159.Liu J., Ishitani M., Halfter U., Kim C. S., Zhu J. K. 2000. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl Acad. Sci. USA 97, 3730–3734 10.1073/pnas.060034197 (doi:10.1073/pnas.060034197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Halfter U., Ishitani M., Zhu J. K. 2000. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl Acad. Sci. USA 97, 3735–3740 10.1073/pnas.040577697 (doi:10.1073/pnas.040577697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.D'Angelo C., et al. 2006. Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 48, 857–872 10.1111/j.1365-313X.2006.02921.x (doi:10.1111/j.1365-313X.2006.02921.x) [DOI] [PubMed] [Google Scholar]
- 162.Pandey G. K., Grant J. J., Cheong Y. H., Kim B. G., Li le G., Luan S. 2008. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 1, 238–248 10.1093/mp/ssn003 (doi:10.1093/mp/ssn003) [DOI] [PubMed] [Google Scholar]
- 163.Cheong J. K., Virshup D. M. 2011. Casein kinase 1: complexity in the family. Int. J. Biochem. Cell Biol. 43, 465–469 10.1016/j.biocel.2010.12.004 (doi:10.1016/j.biocel.2010.12.004) [DOI] [PubMed] [Google Scholar]
- 164.Ben-Nissan G., Cui W., Kim D. J., Yang Y., Yoo B. C., Lee J. Y. 2008. Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol. 148, 1897–1907 10.1104/pp.108.129346 (doi:10.1104/pp.108.129346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gross S. D., Anderson R. A. 1998. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal 10, 699–711 10.1016/S0898-6568(98)00042-4 (doi:10.1016/S0898-6568(98)00042-4) [DOI] [PubMed] [Google Scholar]
- 166.Lee J. Y. 2009. Versatile casein kinase 1: multiple locations and functions. Plant Signal Behav. 4, 652–654 10.4161/psb.4.7.8991 (doi:10.4161/psb.4.7.8991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dai C., Xue H. W. 2010. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29, 1916–1927 10.1038/emboj.2010.75 (doi:10.1038/emboj.2010.75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sun T., Rodriguez M., Kim L. 2009. Glycogen synthase kinase 3 in the world of cell migration. Dev. Growth Differ. 51, 735–742 10.1111/j.1440-169X.2009.01141.x (doi:10.1111/j.1440-169X.2009.01141.x) [DOI] [PubMed] [Google Scholar]
- 169.Kim Y. M., Seo Y. H., Park C. B., Yoon S. H., Yoon G. 2010. Roles of GSK3 in metabolic shift toward abnormal anabolism in cell senescence. Ann. NY Acad. Sci. 1201, 65–71 10.1111/j.1749-6632.2010.05617.x (doi:10.1111/j.1749-6632.2010.05617.x) [DOI] [PubMed] [Google Scholar]
- 170.Wilson N. F., Lefebvre P. A. 2004. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot. Cell 3, 1307–1319 10.1128/EC.3.5.1307-1319.2004 (doi:10.1128/EC.3.5.1307-1319.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Moniz L., Dutt P., Haider N., Stambolic V. 2011. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 6, 18 10.1186/1747-1028-6-18 (doi:10.1186/1747-1028-6-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Quarmby L. M., Mahjoub M. R. 2005. Caught Nek-ing: cilia and centrioles. J. Cell Sci. 118(Pt 22), 5161–5169 10.1242/jcs.02681 (doi:10.1242/jcs.02681) [DOI] [PubMed] [Google Scholar]
- 173.Motose H., Tominaga R., Wada T., Sugiyama M., Watanabe Y. 2008. A NIMA-related protein kinase suppresses ectopic outgrowth of epidermal cells through its kinase activity and the association with microtubules. Plant J. 54, 829–844 10.1111/j.1365-313X.2008.03445.x (doi:10.1111/j.1365-313X.2008.03445.x) [DOI] [PubMed] [Google Scholar]
- 174.Sakai T., et al. 2008. Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 53, 157–171 10.1111/j.1365-313X.2007.03327.x (doi:10.1111/j.1365-313X.2007.03327.x) [DOI] [PubMed] [Google Scholar]
- 175.Lee S. J., Cho D. I., Kang J. Y., Kim M. D., Kim S. Y. 2010. AtNEK6 interacts with ARIA and is involved in ABA response during seed germination. Mol. Cells 29, 559–566 10.1007/s10059-010-0070-7 (doi:10.1007/s10059-010-0070-7) [DOI] [PubMed] [Google Scholar]
- 176.Zhang B., et al. 2011. NIMA-related kinase NEK6 affects plant growth and stress response in Arabidopsis. Plant J. 68, 830–843 10.1111/j.1365-313X.2011.04733.x (doi:10.1111/j.1365-313X.2011.04733.x) [DOI] [PubMed] [Google Scholar]
- 177.Goldberg J. M., Manning G., Liu A., Fey P., Pilcher K. E., Xu Y., Smith J. L. 2006. The Dictyostelium kinome: analysis of the protein kinases from a simple model organism. PLoS Genet. 2, e38 10.1371/journal.pgen.0020038 (doi:10.1371/journal.pgen.0020038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Frye C. A., Tang D., Innes R. W. 2001. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl Acad. Sci. USA 98, 373–378 10.1073/pnas.011405198 (doi:10.1073/pnas.011405198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gu Y., Innes R. W. 2011. The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol. 155, 1827–1838 10.1104/pp.110.171785 (doi:10.1104/pp.110.171785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ning J., Li X., Hicks L. M., Xiong L. 2010. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152, 876–890 10.1104/pp.109.149856 (doi:10.1104/pp.109.149856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hashimoto M., Negi J., Young J., Israelsson M., Schroeder J. I., Iba K. 2006. Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat. Cell Biol. 8, 391–397 10.1038/ncb1387 (doi:10.1038/ncb1387) [DOI] [PubMed] [Google Scholar]
- 182.Lamberti G., Gugel I. L., Meurer J., Soll J., Schwenkert S. 2011. The cytosolic kinases STY8, STY17, and STY46 are involved in chloroplast differentiation in Arabidopsis. Plant Physiol. 157, 70–85 10.1104/pp.111.182774 (doi:10.1104/pp.111.182774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ceserani T., Trofka A., Gandotra N., Nelson T. 2009. VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J. 57, 1000–1014 10.1111/j.1365-313X.2008.03742.x (doi:10.1111/j.1365-313X.2008.03742.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wingenter K., et al. 2011. A member of the mitogen-activated protein 3-kinase family is involved in the regulation of plant vacuolar glucose uptake. Plant J. 68, 890–900 10.1111/j.1365-313X.2011.04739.x (doi:10.1111/j.1365-313X.2011.04739.x) [DOI] [PubMed] [Google Scholar]
- 185.McCormick J. A., Ellison D. H. 2011. The WNKs: atypical protein kinases with pleiotropic actions. Physiol. Rev. 91, 177–219 10.1152/physrev.00017.2010 (doi:10.1152/physrev.00017.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Huang C. L., Cha S. K., Wang H. R., Xie J., Cobb M. H. 2007. WNKs: protein kinases with a unique kinase domain. Exp. Mol. Med. 39, 565–573 [DOI] [PubMed] [Google Scholar]
- 187.Choe K. P., Strange K. 2007. Evolutionarily conserved WNK and Ste20 kinases are essential for acute volume recovery and survival after hypertonic shrinkage in Caenorhabditis elegans. Am. J. Physiol. Cell Physiol. 293, C915–C927 10.1152/ajpcell.00126.2007 (doi:10.1152/ajpcell.00126.2007) [DOI] [PubMed] [Google Scholar]
- 188.Nakamichi N., Murakami-Kojima M., Sato E., Kishi Y., Yamashino T., Mizuno T. 2002. Compilation and characterization of a novel WNK family of protein kinases in Arabiodpsis thaliana with reference to circadian rhythms. Biosci. Biotechnol. Biochem. 66, 2429–2436 10.1271/bbb.66.2429 (doi:10.1271/bbb.66.2429) [DOI] [PubMed] [Google Scholar]
- 189.Kumar K., Rao K. P., Biswas D. K., Sinha A. K. 2011. Rice WNK1 is regulated by abiotic stress and involved in internal circadian rhythm. Plant Signal. Behav. 6, 316–320 10.4161/psb.6.3.13063 (doi:10.4161/psb.6.3.13063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang Y., Suo H., Zheng Y., Liu K., Zhuang C., Kahle K. T., Ma H., Yan X. 2010. The soybean root-specific protein kinase GmWNK1 regulates stress-responsive ABA signaling on the root system architecture. Plant J. 64, 230–242 10.1111/j.1365-313X.2010.04320.x (doi:10.1111/j.1365-313X.2010.04320.x) [DOI] [PubMed] [Google Scholar]
- 191.Talevich E., Tobin A. B., Kannnan N., Doerig C. 2012. An evolutionary perspective on the kinome of malaria parasites. Phil. Trans. R. Soc. B 367, 2607–2618 10.1098/rstb.2012.0014 (doi:10.1098/rstb.2012.0014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ishida S., Yuasa T., Nakata M., Takahashi Y. 2008. A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20, 3273–3288 10.1105/tpc.107.057489 (doi:10.1105/tpc.107.057489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Das R., Pandey G. K. 2009. Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr. Genomics 11, 2–13 10.2174/138920210790217981 (doi:10.2174/138920210790217981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mori I. C., et al. 2006. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 4, e327 10.1371/journal.pbio.0040327 (doi:10.1371/journal.pbio.0040327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zou J. J., Wei F. J., Wang C., Wu J. J., Ratnasekera D., Liu W. X., Wu W. H. 2010. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 154, 1232–1243 10.1104/pp.110.157545 (doi:10.1104/pp.110.157545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Munemasa S., Hossain M. A., Nakamura Y., Mori I. C., Murata Y. 2011. The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol. 155, 553–561 10.1104/pp.110.162750 (doi:10.1104/pp.110.162750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Boudsocq M., Willmann M. R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S. H., Sheen J. 2010. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464, 418–422 10.1038/nature08794 (doi:10.1038/nature08794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Coca M., San Segundo B. 2010. AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 63, 526–540 10.1111/j.1365-313X.2010.04255.x (doi:10.1111/j.1365-313X.2010.04255.x) [DOI] [PubMed] [Google Scholar]
- 199.De Smet I., Voss U., Jurgens G., Beeckman T. 2009. Receptor-like kinases shape the plant. Nat. Cell Biol. 11, 1166–1173 10.1038/ncb1009-1166 (doi:10.1038/ncb1009-1166) [DOI] [PubMed] [Google Scholar]
- 200.Dodds P. N., Rathjen J. P. 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548 10.1038/nrg2812 (doi:10.1038/nrg2812) [DOI] [PubMed] [Google Scholar]
- 201.Sivaguru M., Ezaki B., He Z. H., Tong H., Osawa H., Baluska F., Volkmann D., Matsumoto H. 2003. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 132, 2256–2266 10.1104/pp.103.022129 (doi:10.1104/pp.103.022129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Osakabe Y., Maruyama K., Seki M., Satou M., Shinozaki K., Yamaguchi-Shinozaki K. 2005. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17, 1105–1119 10.1105/tpc.104.027474 (doi:10.1105/tpc.104.027474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yang T., Chaudhuri S., Yang L., Du L., Poovaiah B. W. 2010. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285, 7119–7126 10.1074/jbc.M109.035659 (doi:10.1074/jbc.M109.035659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Albert M., Jehle A. K., Mueller K., Eisele C., Lipschis M., Felix G. 2010. Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J. Biol. Chem. 285, 19 035–19 042 10.1074/jbc.M110.124800 (doi:10.1074/jbc.M110.124800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gu Z., Steinmetz L. M., Gu X., Scharfe C., Davis R. W., Li W. H. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66 10.1038/nature01198 (doi:10.1038/nature01198) [DOI] [PubMed] [Google Scholar]
- 206.Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Clark R. M., et al. 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317, 338–342 10.1126/science.1138632 (doi:10.1126/science.1138632) [DOI] [PubMed] [Google Scholar]
- 208.Castells E., Casacuberta J. M. 2007. Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J. Exp. Bot. 58, 3503–3511 10.1093/jxb/erm226 (doi:10.1093/jxb/erm226) [DOI] [PubMed] [Google Scholar]
- 209.Kannan N., Taylor S. S. 2008. Rethinking pseudokinases. Cell 133, 204–205 10.1016/j.cell.2008.04.005 (doi:10.1016/j.cell.2008.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Chevalier D., Batoux M., Fulton L., Pfister K., Yadav R. K., Schellenberg M., Schneitz K. 2005. STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc. Natl Acad. Sci. USA 102, 9074–9079 10.1073/pnas.0503526102 (doi:10.1073/pnas.0503526102) [DOI] [PMC free article] [PubMed] [Google Scholar]