Abstract
Malaria parasites belong to an ancient lineage that diverged very early from the main branch of eukaryotes. The approximately 90-member plasmodial kinome includes a majority of eukaryotic protein kinases that clearly cluster within the AGC, CMGC, TKL, CaMK and CK1 groups found in yeast, plants and mammals, testifying to the ancient ancestry of these families. However, several hundred millions years of independent evolution, and the specific pressures brought about by first a photosynthetic and then a parasitic lifestyle, led to the emergence of unique features in the plasmodial kinome. These include taxon-restricted kinase families, and unique peculiarities of individual enzymes even when they have homologues in other eukaryotes. Here, we merge essential aspects of all three malaria-related communications that were presented at the Evolution of Protein Phosphorylation meeting, and propose an integrated discussion of the specific features of the parasite's kinome and phosphoproteome.
Keywords: malaria, kinome, evolution, phosphoproteomics, protein kinase, comparative genomics
1. Introduction
(a). Malaria: disease status, need for new control agents
Malaria has been a scourge of mankind since the emergence of our species, and has contributed to human evolution, being the strongest known selective pressure in the recent history of the human genome [1]. The disease still kills almost a million people each year, mostly young children in sub-Saharan Africa. There was an encouraging trend in the recent decades (the death toll owing to malaria was up to 3 million a year in the last quarter of the twentieth century [2]), to which the introduction of combination therapy (notably artemisinin combination therapy, ACT) contributed significantly [3,4]. There is, however, a very worrying development, with a number of occurrences of decreased sensitivity of malaria parasite field isolates to artemisinin [5]. It is of crucial importance to maintain an active drug discovery/development pipeline of new antimalarials with novel modes of action [3]. In view of the success encountered in targeting protein kinases in the context of cancer chemotherapy [5], the kinome of malaria parasites has been proposed as a possible target [6].
(b). Plasmodium: phylogeny, evolutionary origin and life cycle
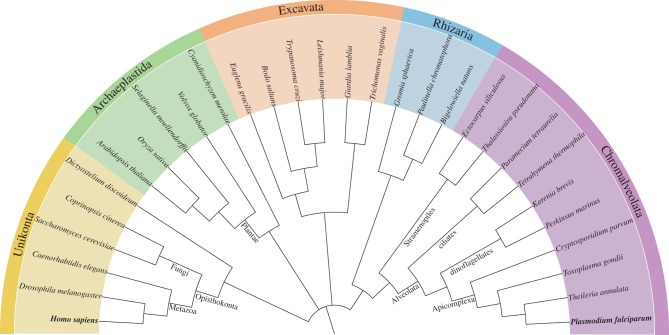
Malaria parasites (genus Plasmodium) constitute a taxon within the Apicomplexa, a group that, together with Stramenopiles, ciliates and dinoflagellates, constitute the Chromalveolata (figure 1). This phylum diverged very early from the main eukaryotic lineage. In line with their phylogenetic relatedness to dinoflagellates, many of which are photosynthetic, malaria parasites and many Apicomplexa contain a remnant plastid and are presumed to descend from photosynthetic ancestors that arose by secondary endosymbiosis; loss of photosynthetic capability is thought to have accompanied the emergence of a parasitic lifestyle [11,12]. The vast phylogenetic distance between malaria parasites and classical eukaryotic model systems such as yeast and metazoans (which are phylogenetically closely related to each other within the Opisthokonta taxon; figure 1) is reflected by profound divergences even in fundamental aspects of biology. These include (to name just a few) an unprecedented mode of transcriptional regulation [13], the coexistence of several different ribosomal RNA sets in the genome [14], and the presence in the nuclear genome of several genes (see Fast et al. [15] for an example) and gene families, such as the calcium-dependent kinases (CDPKs) [16] and the plant-like ApiAP2 transcription factors [17], that most presumably originated in the host cells that were colonized in the two successive rounds of endosymbiosis [12,15].
Figure 1.
Taxonomic arrangement of Plasmodium and model organisms on the eukaryotic tree. Cladogram of representative species in Eukaryota, with Plasmodium and humans indicated in bold. Colours indicate the eukaryotic supergroups defined by Adl et al. [7]. Rhizaria and Chromalveolata are placed together per Hackett et al. [8], and the phylogeny of apicomplexan species is according to Kuo et al. [9]. The tree image was rendered with the Interactive Tree of Life server (iTOL) [10] and edited in Inkscape (http://inkscape.org).
Plasmodium species are obligate parasites with a complex life cycle that requires a vertebrate host and a mosquito vector, of the genus Anopheles in the case of the five species that infect humans. Sporozoites are injected into the bloodstream during a blood meal, and reach the liver where a first (asymptomatic) round of schizogonic replication occurs, generating, in the case of the most virulent species, Plasmodium falciparum, up to 40 000 merozoites. These are geared to invade erythrocytes, where a second round of schizogony occurs; malaria pathogenesis is caused by the synchronous rupture of infected erythrocytes. Transmission to the mosquito requires the differentiated, cell-cycle-arrested sexual forms (male and female gametocytes) that develop in a fraction of the infected erythrocytes. Upon ingestion by the mosquito, asexual parasites are digested, but gametocytes undergo further development into gametes, zygotes and eventually oocysts where sporozoites are generated. These cells accumulate in the insect's salivary gland, where they are primed for infection of a novel human host. Stage transitions during this complex life cycle are likely to require efficient signalling pathways, and evidence for the role of protein phosphorylation in life-cycle progression is emerging [18–20].
2. The plasmodium kinome
(a). In silico characterization
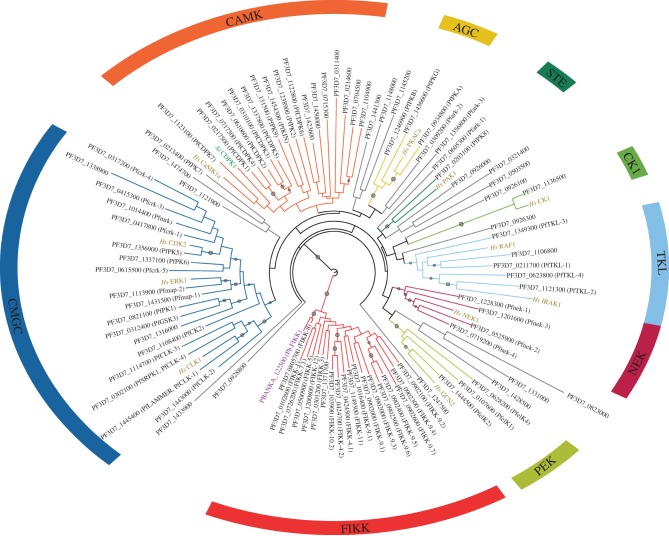
Soon after the Plasmodium falciparum genome sequence became available [21], in silico analyses were performed to identify the parasite's kinome, which was reported to encompass 84 [22] or 99 [23] genes encoding protein kinases, depending on the stringency applied for inclusion of borderline sequences; a more recent study [24] settled for a 91-sequence kinome, excluding atypical kinases (aPKs) such as RIOs and PIKKs (figure 2). Many of the sequences that constitute the P. falciparum kinome cluster within the familiar AGC, CMGC, CK1, TKL and CaMK groups found within the mammalian kinome, and behave as expected, in terms of their biochemical regulation, from their homologues in other organisms. For example, several predicted CDPKs (see below) are indeed stimulated by calcium [29–31], and the activity of at least two enzymes clustering within the cyclin-dependent kinase (CDK) family has been shown to be dependent on the binding of cyclins [32,33], four homologues of which have been identified in the P. falciparum proteome [34]. However, several malarial protein kinases do not cluster within the established eukaryotic protein kinase (ePK) groups that constitute the human kinome. These include the 21 so-called FIKKs, a novel family of atypical protein kinase-like enzymes named after a conserved Phe–Ile–Lys–Lys motif, and a family of CDPKs similar to calcium-regulated protein kinases found in plants but not in metazoans. Conversely, two clusters that are prominent in the human kinome are not represented in the kinome of malaria parasites: the tyrosine kinases and the MAPKK family in the STE group, a group that is itself dramatically reduced in size, with just one member (see below and figure 3 for a distribution of ePK groups among the major eukaryotic taxons).
Figure 2.
Phylogenetic tree of the Plasmodium falciparum kinome. Circular tree of all 91 eukaryotic protein kinases (ePK) in P. falciparum as defined by Talevich et al. [24]. Representative genes from human (Hs), Arabidopsis thaliana (At) and Plasmodium berghei (Pb) are indicated with labels coloured gold, green and purple, respectively. Branch and arc colours indicate kinase classification by ePK major group [25,26], according to Talevich et al. [24], with minor modifications in group assignment according to the gene tree. To construct the tree, the sequences of 91 protein kinases were retrieved from GeneDB (http://genedb.org), P. falciparum 3D7 sequence v. 3. Conserved regions of the kinase domain were aligned with MAPGAPS [27] and unconserved sequence positions, as identified by MAPGAPS, were removed. A gene tree was then inferred from the resulting 245-column alignment using RAxML [28] with the rapid bootstrap analysis and maximum-likelihood tree search algorithm, LG amino acid substitution model, and gamma model of substitution rate heterogeneity. The tree image was rendered with the Interactive Tree of Life server (iTOL) [10] and edited in Inkscape (http://inkscape.org). A grey circle on a branch indicates bootstrap support greater than 50; larger circles indicate greater bootstrap values.
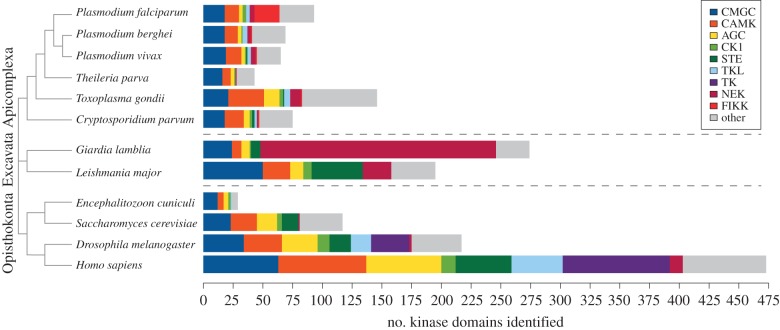
Figure 3.
ePK group distribution across representative of the main eukaryotic taxons. Composition of protein kinase major groups and the apicomplexan-specific FIKK family in the genomes of three Plasmodium species and three other apicomplexans. The kinomes of the model organisms Saccharomyces cerevisiae (Baker's yeast), Drosophila melanogaster (fruitfly) and Homo sapiens (human), as well as the phylogenetically distant parasite Giardia lamblia (figure 1) and the microsporidium Encephalitozoon cuniculi, which has the smallest characterized eukaryotic kinome [35], are included for comparison. The cladogram along the left edge indicates the evolutionary relationships between species. In the stacked bar chart associated with each species, block width indicates number of genes found belonging to each major group of eukaryotic protein kinases; total bar width indicates total kinome size. Data are adapted from Talevich et al. [24] and KinBase (http://kinase.com/kinbase/).
In addition to taxon-restricted kinase families, many ‘orphan’ sequences do not cluster with ePK groups found in other eukaryotes. A good example is that of PfPK7, an orphan kinase that is involved, in line with its absence from the yeast and mammalian kinomes, in a parasite-specific function, namely the regulation of the number of daughter merozoites generated by each schizont [36]. Furthermore, most individual plasmodial enzymes clustering within known ePK groups and families display peculiarities that distinguish them from their homologues in other eukaryotes. These include (sometimes very large) low complexity extensions and insertions in the catalytic domain, usually occurring at loops between secondary structure elements; this feature is not restricted to kinases, but occurs throughout the parasite's proteome, and its evolutionary function is still being debated [37–41]. Many protein kinases, called ‘semi-orphans’ in the initial kinome characterization [22], clearly belong to a specific kinase group or family, but do not have a clear orthologue in other organisms. A good example is the CDK-related kinase Pfcrk-5, which forms a distinct branch with the CDK cluster, and which displays an atypical and apicomplexan-specific motif in the activation loop [24]. Some Plasmodium kinases in a given family appear to contain domains/motifs that are characteristic of other kinase families; a striking example is that of the NIMA-related kinase Pfnek-1: despite overall homology and clear phylogenetic linkage to the NIMA family, Pfnek-1 possesses a MAPKK-like activation motif SMAHS, which is clearly reminiscent of the SMANS motif of mammalian MEK1 and distinct from the FXXT motif usually found at the corresponding position in other members of the NIMA family [42]. Interestingly, Pfnek-1 (and another NIMA-related kinase, Pfnek-3 [43]) has been tentatively implicated in the regulation of an atypical mitogen-activated protein kinase (MAPK), Pfmap-2 [42], which itself has an atypical activation motif [44]. In conjunction with the absence of MEK/MAPKK homologues in the plasmodial kinome [22–24], this might point to an unorthodox MAPK regulation pathway in the parasite (see below).
(b). Functional analysis of the kinome
Protocols for manipulation of the genome of malaria parasites have been available for a decade or so [45]. The roles of individual protein kinases were first investigated following a reverse genetics approach in the rodent malaria model Plasmodium berghei. This revealed essential functions of a MAPK (Pbmap-2) and a CDPK (PbCDPK4) in male gametogenesis [29,46,47], of two NIMA-related kinases in meiosis in the mosquito vector [48,49], and of a CDK in asexual proliferation in erythrocytes [50]. This approach culminated in a kinome-wide study demonstrating that 23 P. berghei ePKs are redundant for asexual erythrocytic parasite development in mice and identifying phenotypes in sexual development for a number of these 23 ePKs [51]. A similar strategy in P. falciparum identified roles for an orphan kinase in proliferation rate linked to the number of progeny merozoites per schizont [36], for an eIF2α kinase in response to starvation stress, similar to GCN2, its closest homologue in yeast [52], and for CDPKs in motility during invasion [53] or egress of merozoites from the erythrocyte [54], to cite a few specific studies; more recently, a kinome-wide approach [19] identified 36 ePKs as refractory to gene disruption, and thus as likely crucial players in asexual proliferation in erythrocytes.
3. plasmodium phosphoproteomics
The determination of the essential protein kinases in the blood stage of P. falciparum [19] not only reveals the protein kinases that might constitute targets for novel antimalarial therapy but also provides a framework to identify key phospho-signalling pathways [55]. Identifying kinase substrates would represent a significant advance in our understanding of such pathways. The huge advances achieved recently in mass spectrometry-based phosphoproteomic approaches allow large portions of the phosphoproteome to be identified in a single experiment [56–58]. Because P. falciparum-infected red blood cells at the schizont stage can be readily isolated from an in vitro culture, this stage in the parasite life cycle was the first to be interrogated by mass spectrometry techniques designed to investigate the global phosphoproteome. Two such studies have recently been published, one revealing 1117 [19] unique phosphor-acceptor sites on over 600 P. falciparum proteins, and another, using lower filter stringency, detecting over 5000 occupied phosphorylation sites [20]. Many of the identified phosphosites overlapped between the two studies.
(a). Tyrosine phosphorylation in Plasmodium falciparum
Among the unexpected findings from these studies was the identification of tyrosine phosphorylation of parasite proteins, because, as mentioned earlier, there are no members of the tyrosine kinase group in the P. falciparum genome. The strongest evidence for tyrosine phosphorylation of P. falciparum proteins was in the activation loop of two protein kinases: PfGSK3 (PFC0525c) and PfCLK3 (PF11_0156) [19]. Phosphorylation of PfGSK3 occurred at Y229, analogous to Y279 and Y216 on mammalian GSK3α/β, the activation loop auto-phosphorylation sites necessary for enzymatic activity [59,60]. The tyrosine phosphorylation of the CDK-like kinase PfCLK3 appears to follow similar lines: this kinase is a serine/threonine kinase related to the pre-mRNA-processing kinase hPRP4 found in higher eukaryotes [22]; both PfCLK3 and hPRP4 cluster within the dual-specificity tyrosine phosphorylated-regulated kinase (DYRK) family [22,61], members of which form an active transient intermediate during translation that is able to cis-auto-phosphorylate on tyrosine [60]. This process is essential for full catalytic activity of the mature enzyme, which then shows only serine/threonine kinase activity. In the case of PfCLK3, phosphorylation occurs on Y526 within a TSY motif in the activation loop [19]. In most DYRKs, however, the phospho-tyrosine is contained within a YXY motif where the second Y is the site of co-translational auto-phosphorylation [60]. Mutation of Y526 in PfCLK3 significantly reduced the serine/threonine kinase activity of the recombinant enzyme [19], indicating that tyrosine phosphorylation of PfCLK3, as for other DYRKs, is essential for a fully active kinase.
The tyrosine phosphorylation of PfCLK3 and PfGSK3 is a somewhat special case. The question remains—are there parasite proteins that are phosphorylated in trans on tyrosine residues? Only a small number of tyrosine-phosphorylated peptides have been identified [19,20], and the confidence levels associated with these peptides are usually very low [19]. This may reflect the low abundance of phosphorylated tyrosines, and additional enrichment techniques such as immunoprecipitation with anti-phosphotyrosine antibodies may be necessary to draw firm conclusions regarding the extent of tyrosine phosphorylation in P. falciparum. It is relevant to mention here that SH2 domains, which are major phosphotyrosine-binding devices in eukaryotic cells [62], appear to be essentially absent from malaria parasites: a query of the plasmodial proteomes with the SH2 HMM profile on InterPro of Pfam (entry PF00017) yielded only one weak hit in P. berghei, in the hypothetical protein PBANKA_114210; although this protein has syntenic orthologues in all other Plasmodium species, the latter do not possess a recognizable SH2 motif. We therefore propose that this is a spurious hit, and that there are no SH2 domains in plasmodia. This is in line with the apparent low occurrence of phosphotyrosine discussed earlier, although we cannot exclude that taxon-specific, cryptic phosphotyrosine-binding domains have evolved in these organisms. It is noteworthy that yeast does not possess SH2 domains, and the only non-metazoan organism in which these domains have been shown to occur is the social free-living amoeba Dictyostelium, whose kinome also lacks typical tyrosine kinases [63].
One exception to the low-confidence phosphotyrosine sites in Plasmodium proteins, however, appears to be the protein PHISTB (PFE1600w). This P. falciparum protein is exported to the cytoplasm of the red blood cell [64], and was unequivocally identified as a tyrosine-phosphorylated protein [20]. What is not clear, however, is whether this tyrosine phosphorylation occurs only once the protein reaches the red blood cell cytoplasm. If this is the case, then it is possible that PHISTB is phosphorylated by a host rather than a parasite tyrosine kinase. The ability of host protein kinases to impact on the parasite erythrocytic life cycle has indeed been previously documented in the case of the parasite-dependent activation of an erythrocyte signalling pathway that is required for parasite survival [65].
(b). Regulation of protein kinases by phosphorylation in Plasmodium falciparum
Among the proteins determined to be phosphorylated in the schizont stage of P. falciparum, 23 kinases were determined to be phosphoproteins [19]. Many of these phosphorylations were on sites within their activation loops and therefore potentially associated with the regulation of catalytic activity [66]. These include, in addition to PfGSK3 and PfCLK3 already discussed, activation loop phosphorylation of the cyclic nucleotide-dependent kinases PfPKA (PFI1685w) and PfPKG (PF14_0346) [19], both of which have crucial functions in the erythrocytic cycle [54,67–70]. How are these kinases regulated? Preliminary studies using phospho-specific antibodies have shown that the activation loop phosphorylation of these protein kinases does not appear to change during the life cycle of the parasite (A. Tobin 2012, unpublished data). This is similar to the constitutively phosphorylated activation loop of mammalian PKA that is mediated by the phosphoinositide-dependent protein kinase PDK1 [71]. That activation loop phosphorylation of parasite protein kinases is dynamic for certain kinases is illustrated by the parasite kinases Pfcrk-1 (PFD0865c) [72] and Pfcrk-3 (PFD0740w) [73]. These kinases are related to the mammalian and fungal CDKs that require both cyclin binding and activation loop threonine phosphorylation for full activity [74]. The threonine activation loop phosphorylation observed for Pfcrk-1 and Pfcrk-3 may therefore be mediated by an upstream kinase in a phospho-signalling cascade analogous to that of the mammalian system. A possible candidate for the kinase acting upstream of Pfcrk-1 and Pfcrk-3 in P. falciparum would be Pfmrk (PF10_0141), as this enzyme displays closest homology to CDK7, a kinase that in mammalian cells acts upstream of the CDKs [75].
Overall, the in silico characterization of the P. falciparum kinome, the assessment of the function of the parasite's protein kinases by reverse genetics, and the global phosphoproteomics approach providing a glimpse into the substrate and effectors of these enzymes, all point to the importance of protein phosphorylation in malaria parasites, and illustrate the phylogenetic divergence between the parasite and its host. In §4, we will consider the specificities of the plasmodial kinome from an evolutionary angle.
4. Evolutionary perspectives on the plasmodium kinome
One of the goals that initially motivated our analysis of ePKs in P. falciparum was to assess the potential of the parasite's kinome as a target for antimalarial drug discovery. In this purpose, we and others have sought to identify distinctive protein features that appear and are conserved in the parasite, but do not appear in the host. Comparative analysis of sequences and protein structures has revealed a number of such features conserved to varying degrees in P. falciparum, related Plasmodium species, and other apicomplexan parasites of medical and veterinary importance. Given the ancient evolutionary divergence between Plasmodium and most model organisms (figure 1), investigation of the cell signalling components in Plasmodium and its relatives provides scientific insights into the early evolution of eukaryotes, as well as a basis for comparison for the study of other protists. Earlier, we pointed out several distinctive features of the P. falciparum kinome and phospho-signalling transduction pathways, and briefly described how reverse genetics and phosphoproteomics can provide information on their involvement in the organism's biological functions. We will now compare the plasmodial kinome with those of other eukaryotes, to address the questions of how parasite-specific kinome features evolved, and whether some of its features are shared by other parasites. This has a direct impact on drug discovery and development: could kinase inhibitors that target parasite-specific features be developed? If so, could the same inhibitors be used to target similar features in related parasites?
(a). Parasitic lifestyle correlates with kinase gene loss
The total number of kinases, i.e. the kinome size, is markedly reduced in P. falciparum and other apicomplexans in comparison with other model eukaryotes. This reduction of the kinome is in line with an overall gene loss observed in the Plasmodium genome. Gene loss and general compaction of the genome (loss of introns, smaller intergenic regions) have been noted as the dominant mode of genomic evolution in obligate intracellular parasites such as the Apicomplexa [76] (see figure 3 for a comparison of ePK group counts across eukaryotes). From a superficial comparison, the kinome of P. falciparum, consisting of roughly 91 ePKs [24] plus at least five aPKs [22], appears to constitute a percentage of the total proteome (1.7% of 5228 protein-coding genes) that is similar to that found in other, non-parasitic eukaryotes: the kinome of the baker's yeast Saccharomyces cerevisiae comprises 117 ePKs (2% of 5770 genes; plus 14 or 10 aPKs, depending on the study) [77,78], the fruitfly Drosophila melanogaster has 223 ePKs (1.4% of approx. 15 000 genes) [79] (plus 15 aPks) and the human kinome 478 (1.9% of approx. 25 000 genes) [80,81] (plus 38 aPKs). However, P. falciparum is the only Plasmodium species in which the FIKK family is vastly expanded; if we consider only typical ePKs, the percentage of the kinases in the kinome decreases from 1.7 to 1.3, towards the lower limit found in eukaryotes (in an astounding comparison, the ciliate Paramecium tetraurelia, related to malaria parasites by the clade Alveolata, possesses 2606 protein kinases, which amounts to 6.6% of its proteome [82]!).
Analysis of the kinomes of several diverse eukaryotic parasitic and free-living species led to a hypothesized ‘core’ set of between 68 and 85 protein kinases found in the common ancestor of all extant eukaryotes [83]. This would suggest that evolutionary necessity has preserved these approximately 85 kinases as the ‘minimal kinome’ of extant free-living eukaryotes. However, obligate parasites may be freed from some of the evolutionary constraints that preserve certain ePK families in other species, and thus show reduction or loss of kinase families that appear essential in other species. An extreme example of such gene loss is the microsporidium Encephalitozoon cuniculi, an obligate parasite that contains only 29 typical ePKs [35]. A similar example in Plasmodium is the absence of key components of MAPK cascades. Typically, this signal transduction pathway is formed by three or four protein kinases associated with distinct ePK families: MAP4K or MEKKK (a tyrosine-like kinase such as Raf, or a member of the STE20 family), MAP3K or MEKK (a member of the STE11 family), MAPKK or MEK (a STE7 member) and MAPK family (best characterized in the ERK1/ERK2, p38 and JNK subfamilies) [84]. However, in Plasmodium and most other apicomplexans, the STE group of kinases is reduced to one or two distantly related members, or even missing entirely [85], although two clear (but atypical) members of the MAPK family are present [86]. One of these, Pfmap-1, has been demonstrated to be phosphorylated on the conserved TXY motif in its activation loop [20], while this otherwise highly conserved activation motif is substituted in the other MAPK (Pfmap-2) by TSH [44]; no evidence for in vivo phosphorylation of this TSH motif is available, but mutating either the Thr or the His (but not the Ser) dramatically affects kinase activity of the recombinant enzyme [44]. Absence of the classical upstream regulators of the MAPK cascade is rare in Eukaryota, but has also been observed in E. cuniculi; in the latter case, not only are the STEs that act as MAPK regulators missing but the microsporidial kinome does not even include MAPK homologues [35]; in view of the association of microsporidia with fungi, which have full MAPK cascades, this is most likely the result of evolutionary gene loss. Similarly, the presence of MAPKK homologues and other STEs in several species of chromalveolates (including the photosynthetic diatom Thalassiosira pseudonana, the parasitic dinoflagellate Perkinsus marinus and the free-living ciliate Tetrahymena thermophila [87]) suggests that the absence of such homologues in Apicomplexa results from gene loss, rather than from evolutionary divergence that preceded appearance of MAPK pathways in the main eukaryotic lineage. In any case, it appears that the parasitic lifestyle of Plasmodium and microsporidia has altered the evolutionary constraints that preserve the complete MAPK cascade found in most other eukaryotes. The functionality of similar cascades is likely to still be necessary in these organisms, but satisfied in other ways. For example, one of the two Plasmodium MAPKs (map-2) is required for the transition from gametocyte to gamete that occurs in response to the new environment met by the parasite upon ingestion by the mosquito; the mode of activation of the enzyme is unclear (as discussed earlier, it has an unusual putative activation motif, and there is no typical MAPKK in the Plasmodium kinome). It has nevertheless been established that in map-2− parasites, the ontogeny of eight flagellated microgametes from a single male gametocyte is blocked [46,47], and hence that MAPK signalling, albeit differing in important aspects, is still exploited by malaria parasites to generate adaptive responses to environmental changes as in higher eukaryotes.
(b). Gene loss is offset by expansions of taxon-specific protein families
The overall trend of gene loss in obligate parasites is occasionally offset by dramatic expansions of specific gene families, including novel kinase families [88].
The kinome of P. falciparum includes a number of ‘orphan’ families that have no orthologues in other eukaryotic lineages [22,23]. The most notable of these is the FIKK family, a novel and rapidly evolving family of exported proteins, including the P. falciparum R45 trophozoite protein, and named after a conserved ‘FIKK’ motif in the second sub-domain of the protein kinase domain (the first ‘K’ corresponding to the conserved lysine in sub-domain II of ePKs) [22]. This family is expanded to 19 copies in P. falciparum, plus one or two pseudogenes, and at least six copies in P. reichenowi, a species closely related to P. falciparum [89]. As discussed earlier, the expansion of the FIKK family accounts for most of the difference in kinome size between P. falciparum and the other Plasmodium species [24]. Most of the FIKK genes in P. falciparum are located in sub-telomeric regions, including a cluster of tandem duplicates on chromosome 9 [89]. Synteny breaks between Plasmodium species frequently appear near FIKK genes. It is hypothesized that the transplantation of the FIKK gene to a sub-telomeric region in the common ancestor of P. falciparum and P. reichenowi placed this gene in a rapidly evolving chromosomal region, enabling rapid gene duplication and diversification along this evolutionary branch [90]. BLAST and HMM profile searches identified only weak similarity of the FIKK C-lobe to kinases in non-apicomplexans, and did not provide a clear signal as to which known kinase family the ancestor of FIKKs may have belonged to (E. Talevich 2012, unpublished data). The maximum-likelihood method we used to infer the P. falciparum kinome gene tree does not indicate the root of the tree, precluding insights into the kinase group at the origin of FIKKs; a more elaborate phylogenetic approach might provide some information in this respect.
The pattern of genome compaction offset by expansion and diversification appears in other obligate parasitic protists. In the coccidians, another sub-clade of Apicomplexa that includes Toxoplasma gondii, there is a novel family of ePKs targeted to the rhoptry (a secretory organelle found in most apicomplexans) [91,92]. This kinase family, dubbed ROPK, is amplified to several dozen copies in all of the coccidians for which the whole-genome annotation is currently available [24]. At least some these kinases are secreted into the host cell and modulate its transcriptional repertoire [91,92].
In Giardia lamblia, a parasitic species belonging to a separate supergroup of protists, Excavata, expansion occurred in another ePK family, that of the NIMA-related kinases (NEKs); again, this amplification is correlated with a lineage-specific feature, in this case, the elaborate cytostructure of the parasite cells [83]. The expansion of the NEK family is seen to a lesser extent in Trichomonas vaginalis [83] and trypanosomes [93], which are also parasitic excavates with elaborate flagellar systems. Three of the four Plasmodium NEK family members are expressed predominantly in sexual stages [48,49,94], which include the only flagellated cell form in the Plasmodium life cycle, the male gamete; however, only one Pfnek has been clearly shown to be male-specific [95,96], whereas two others are associated with female development and appear to be involved in meiosis [48,49].
(c). Kinase gene architecture
Another striking feature of Plasmodium evolution is the remarkably high rate of genomic rearrangement [97]. In protein kinase genes, this phenomenon contributes to variation in protein domain architecture. The 19 copies of the FIKK family in P. falciparum each contain a long amino-terminal extension in addition to the protein kinase domain; however, the sequence in this region, including possible functional accessory domains, is not conserved between gene copies, with the exception of export signals near the very N-terminal end [89]. This suggests rapid diversification of function, a view that is supported by the star-like shape, with branches of similar length, of the FIKK phylogenetic tree [22], and by the following biological observations: (i) different FIKKs are targeted to distinct sub-cellular compartments [90]; (ii) in the case of two FIKKs with the same sub-cellular locations, distinct substrates appear to be targeted by the enzymes [98]; and (iii) the same two FIKKs, the only ones so far to have been the subject of published reverse genetics investigations and for which viable knock-out lines were obtained, displayed phenotypes in the rigidity of the infected erythrocyte, despite phosphorylating distinct proteins [98].
Another multi-member kinase family in malaria parasites is that of the CDPK, which is essentially restricted to Plantae and Chromoalveolata (with a few distantly related members having been reported in trypanosomatids; [93]), and is characterized by a carboxyl-terminal tail containing two to four calcium-binding EF-hand domains. Plasmodium falciparum contains five CDPKs with this architecture (PfCDPK 1–5), but also a sixth sequence with a substantial amino-terminal tail that includes another pair of EF-hand domains [16]. The shuffling and duplication of EF-hand domains in apicomplexan CDPKs has been carefully chronicled with respect to Cryptosporidium parvum [99].
(d). Structural variations within the kinase domain
The kinase domain itself contains 11 conserved structural regions, called subdomains [25]. While these motifs are deeply conserved, as mentioned earlier the loops between subdomains are more variable and can be elaborated through insertions and deletions (indels), as well as amino acid substitutions [26]. This process may sometimes produce new functional features. Indels frequently appear in the catalytic domain of many Plasmodium kinases, and some are conserved across the Plasmodium genus or even more broadly in apicomplexans. Because they usually occur in the loops between secondary structure elements, they are thought not to disrupt the classical kinase fold (see Halbert et al. [73]). In addition to indels, there are significant sequence peculiarities in a number of Plasmodium kinases (especially the so-called orphans, see earlier text) that include small insertions not located in inter-domain loops or atypical activation motifs. It is difficult to predict the effect of such divergent features on the three-dimensional structure. However, the crystal structures made available by the Structural Genomics Consortium provide an opportunity to understand the structural/functional role of such divergent features [100]. There are several Plasmodium protein kinase structures currently available in the Protein Data Bank (PDB), produced by the Structural Genomics Consortium, for which no manuscript has yet been published (to our knowledge, only two PfPK three-dimensional structures had been reported [101,102] prior to the SGC initiative, both of which displayed unusual characteristics). These include three structures of the novel MAPK Pfmap-2 [PDB:3NIE] (see earlier) and its orthologues in P. berghei [PDB:3N9X] and T. gondii [PDB:3RP9]. We look forward to seeing these new structures described in future publications.
The kinome of another apicomplexan, C. parvum, was recently analysed from both evolutionary and structural perspectives, with particular focus on kinase inhibitors that may be developed to treat the disease cryptosporidiosis [99]. In particular, the authors characterized three CDPKs, CpCDPK1 [CryptoDB:cgd3_920; PDB:3DFA, 2WEI], CpCDPK2 [CryptoDB:cgd7_1840; PDB:2QG5, 3F3Z] and CpCDPK4 [CryptoDB:cgd7_40; PDB:3HKO], orthologous to P. falciparum PFB0815w, PFF0520w and PF07_0072, respectively. The authors found a unique insert in CpCDPK4 that remarkably includes a zinc finger between the N and C lobes of the kinase domain; this feature has not been observed in any other protein kinase solved till date, and illustrates the divergent evolutionary path followed by the kinomes of Apicomplexa, and the accompanying emergence of potential functions that are not associated with ePKs in other eukaryotes.
Because the structural coordinates were made available through PDB early on, another team was also able to analyse these structures and identify a possible MAPK-like mode of regulation in CpCDPK2 [24].
(e). Functional roles of pseudokinases in intracellular parasites?
Three highly conserved residues in the protein kinase domain (a lysine and two aspartates located respectively in subdomain II, the catalytic loop and adjacent to the amino-end of the activation segment) are directly involved in catalytic activity in typical ePKs, and the mutation of any of these generally results in loss of kinase activity. A protein that preserves the kinase fold but lacks residues thought to be essential for kinase catalytic activity is predicted to be a pseudokinase (although it must be kept in mind that some predicted pseudokinases have actually been shown to possess phosphotransfer catalytic activity; [103–105]). Pseudokinases can nonetheless still perform other important cellular functions, such as acting as a scaffold to provide substrates or binding partners to other kinases [104]. One putative pseudokinase in the P. falciparum kinome that may play such a role is PFE0170C, which we tentatively classify in the SCY1 family, homologues of the yeast gene Scy1p, a suppressor of Ras mutations [106]. In another lineage of apicomplexans, namely the coccidians such as T. gondii, the diversification of rhoptry kinases (ROPKs) has produced a number of apparent pseudokinases [91]. Interestingly, one of these ROPKs, ROP5, has been shown to play an essential role in the virulence of T. gondii strains [107,108].
5. Concluding remarks
In our endeavour to understand the role of protein phosphorylation in the biology of malaria parasites, the surface of the underlying complexity has barely been scratched. Unlike organisms that are closely related to much better-understood systems such as yeast and mammalian cells [109], and for which cross-reacting tools and reagents can be used to address questions regarding the function of individual kinases, malaria parasites, because of their long independent evolutionary history and ensuing phylogenetic isolation, require de novo investigations. There is a need for ‘reinventing the wheel’, not only because tools must be generated to study the taxon-specific signalling machinery that evolved in Apicomplexa, but also because, to a large extent, sensible inferences cannot be made from knowledge gained in model systems. How could one predict the function of ePKs that do not have even remote homologues in yeast or mammalian cells? This makes research into Plasmodium kinomics more difficult and demanding, but all the more exciting too, as the biology that is and that will be uncovered is likely to shed light from a variant perspective on the biology of protein phosphorylation in other eukaryotes. Another silver lining to this cloud is that the peculiarities that make Plasmodium kinomics research difficult may well be the very features that will enable the discovery and development of new antimalarials based on protein kinase inhibition. The major message we have tried to convey in the preceding sections is that the phylogenetic distance between malaria parasites and their human host translates into important divergences in their respective kinomes, and that most Plasmodium kinases display atypical properties (when compared with mammalian protein kinases) that can be exploited towards selective inhibition.
Acknowledgements
All authors thank Tony Hunter and Tony Pawson for convening an outstanding meeting on the Evolution of protein phosphorylation that stimulated our interest in writing this study. Work in the C.D. laboratory has been supported in the recent years by Inserm, EPFL, the Wellcome Trust and the European Commission (FP6 projects ANTIMAL and SIGMAL and Network of Excellence BioMalPar, and FP7 project MALSIG and Network of Excellence EviMalar), and currently benefits from support from Monash University. Funding for N.K. from the University of Georgia is acknowledged.
References
- 1.Kwiatkowski D. P. 2005. How malaria has affected the human genome and what human genetics can teach us about malaria? Am. J. Hum. Genet. 77, 171–192 10.1086/432519 (doi:10.1086/432519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winstanley P., Ward S. 2006. Malaria chemotherapy. Adv. Parasitol. 61, 47–76 10.1016/S0065-308X(05)61002-0 (doi:10.1016/S0065-308X(05)61002-0) [DOI] [PubMed] [Google Scholar]
- 3.Wells T. N., Poll E. M. 2010. When is enough enough? The need for a robust pipeline of high-quality antimalarials. Discov. Med. 9, 389–398 [PubMed] [Google Scholar]
- 4.Burrows J. N., Chibale K., Wells T. N. 2011. The state of the art in anti-malarial drug discovery and development. Curr. Top. Med. Chem. 11, 1226–1254 10.2174/156802611795429194 (doi:10.2174/156802611795429194) [DOI] [PubMed] [Google Scholar]
- 5.Dondorp A. M., Yeung S., White L., Nguon C., Day N. P., Socheat D., von Seidlein L. 2010. Artemisinin resistance: current status and scenarios for containment. Nat. Rev. Microbiol. 8, 272–280 10.1038/nrmicro2385 (doi:10.1038/nrmicro2385) [DOI] [PubMed] [Google Scholar]
- 6.Doerig C., Meijer L. 2007. Antimalarial drug discovery: targeting protein kinases. Expert Opin. Ther. Targets 11, 279–290 10.1517/14728222.11.3.279 (doi:10.1517/14728222.11.3.279) [DOI] [PubMed] [Google Scholar]
- 7.Adl S. M., et al. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451 10.1111/j.1550-7408.2005.00053.x (doi:10.1111/j.1550-7408.2005.00053.x) [DOI] [PubMed] [Google Scholar]
- 8.Hackett J. D., Yoon H. S., Li S., Reyes-Prieto A., Rummele S. E., Bhattacharya D. 2007. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol. Biol. Evol. 24, 1702–1713 10.1093/molbev/msm089 (doi:10.1093/molbev/msm089) [DOI] [PubMed] [Google Scholar]
- 9.Kuo C. H., Wares J. P., Kissinger J. C. 2008. The Apicomplexan whole-genome phylogeny: an analysis of incongruence among gene trees. Mol. Biol. Evol. 25, 2689–2698 10.1093/molbev/msn213 (doi:10.1093/molbev/msn213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letunic I., Bork P. 2011. Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478 10.1093/nar/gkr201 (doi:10.1093/nar/gkr201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson R. J., Williamson D. H., Preiser P. 1994. Malaria and other Apicomplexans: the ‘plant’ connection. Infect Agents Dis. 3, 29–37 [PubMed] [Google Scholar]
- 12.Kalanon M., McFadden G. I. 2010. Malaria, Plasmodium falciparum and its apicoplast. Biochem. Soc. Trans. 38, 775–782 10.1042/BST0380775 (doi:10.1042/BST0380775) [DOI] [PubMed] [Google Scholar]
- 13.Bozdech Z., Llinas M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1, E5 10.1371/journal.pbio.0000005 (doi:10.1371/journal.pbio.0000005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waters A. P., Syin C., McCutchan T. F. 1989. Developmental regulation of stage-specific ribosome populations in Plasmodium. Nature 342, 438–440 10.1038/342438a0 (doi:10.1038/342438a0) [DOI] [PubMed] [Google Scholar]
- 15.Fast N. M., Kissinger J. C., Roos D. S., Keeling P. J. 2001. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 18, 418–426 10.1093/oxfordjournals.molbev.a003818 (doi:10.1093/oxfordjournals.molbev.a003818) [DOI] [PubMed] [Google Scholar]
- 16.Billker O., Lourido S., Sibley L. D. 2009. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe 5, 612–622 10.1016/j.chom.2009.05.017 (doi:10.1016/j.chom.2009.05.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Painter H. J., Campbell T. L., Llinas M. 2011. The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol. Biochem. Parasitol. 176, 1–7 10.1016/j.molbiopara.2010.11.014 (doi:10.1016/j.molbiopara.2010.11.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doerig C., et al. 2009. Signalling in malaria parasites. The MALSIG consortium. Parasite 16, 169–182 [DOI] [PubMed] [Google Scholar]
- 19.Solyakov L., et al. 2011. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat. Commun. 2, 565 10.1038/ncomms1558 (doi:10.1038/ncomms1558) [DOI] [PubMed] [Google Scholar]
- 20.Treeck M., Sanders J. L., Elias J. E., Boothroyd J. C. 2011. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 10, 410–419 10.1016/j.chom.2011.09.004 (doi:10.1016/j.chom.2011.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner M. J., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 10.1038/nature01097 (doi:10.1038/nature01097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward P., Equinet L., Packer J., Doerig C. 2004. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5, 79 10.1186/1471-2164-5-79 (doi:10.1186/1471-2164-5-79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anamika, Srinivasan N., Krupa A. 2005. A genomic perspective of protein kinases in Plasmodium falciparum. Proteins 58, 180–189 10.1002/prot.20278 (doi:10.1002/prot.20278) [DOI] [PubMed] [Google Scholar]
- 24.Talevich E., Mirza A., Kannan N. 2011. Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol. Biol. 11, 321 10.1186/1471-2148-11-321 (doi:10.1186/1471-2148-11-321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanks S. K., Hunter T. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 [PubMed] [Google Scholar]
- 26.Hanks S. K. 2003. Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 4, 111 10.1186/gb-2003-4-5-111 (doi:10.1186/gb-2003-4-5-111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuwald A. F. 2009. Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics 25, 1869–1875 10.1093/bioinformatics/btp342 (doi:10.1093/bioinformatics/btp342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 29.Billker O., Dechamps S., Tewari R., Wenig G., Franke-Fayard B., Brinkmann V. 2004. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117, 503–514 10.1016/S0092-8674(04)00449-0 (doi:10.1016/S0092-8674(04)00449-0) [DOI] [PubMed] [Google Scholar]
- 30.Green J. L., Rees-Channer R. R., Howell S. A., Martin S. R., Knuepfer E., Taylor H. M., Grainger M., Holder A. A. 2008. The motor complex of Plasmodium falciparum: phosphorylation by a calcium-dependent protein kinase. J. Biol. Chem. 283, 30 980–30 989 10.1074/jbc.M803129200 (doi:10.1074/jbc.M803129200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranjan R., Ahmed A., Gourinath S., Sharma P. 2009. Dissection of mechanisms involved in the regulation of Plasmodium falciparum calcium-dependent protein kinase 4. J. Biol. Chem. 284, 15 267–15 276 10.1074/jbc.M900656200 (doi:10.1074/jbc.M900656200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Roch K., Sestier C., Dorin D., Waters N., Kappes B., Chakrabarti D., Meijer L., Doerig C. 2000. Activation of a Plasmodium falciparum cdc2-related kinase by heterologous p25 and cyclin H. Functional characterization of a P. falciparum cyclin homologue. J. Biol. Chem. 275, 8952–8958 10.1074/jbc.275.12.8952 (doi:10.1074/jbc.275.12.8952) [DOI] [PubMed] [Google Scholar]
- 33.Li Z., Le Roch K., Geyer J. A., Woodard C. L., Prigge S. T., Koh J., Doerig C., Waters N. C. 2001. Influence of human p16(INK4) and p21(CIP1) on the in vitro activity of recombinant Plasmodium falciparum cyclin-dependent protein kinases. Biochem. Biophys. Res. Commun. 288, 1207–1211 10.1006/bbrc.2001.5920 (doi:10.1006/bbrc.2001.5920) [DOI] [PubMed] [Google Scholar]
- 34.Merckx A., et al. 2003. Identification and initial characterization of three novel cyclin-related proteins of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 278, 39 839–39 850 10.1074/jbc.M301625200 (doi:10.1074/jbc.M301625200) [DOI] [PubMed] [Google Scholar]
- 35.Miranda-Saavedra D., Stark M. J., Packer J. C., Vivares C. P., Doerig C., Barton G. J. 2007. The complement of protein kinases of the microsporidium Encephalitozoon cuniculi in relation to those of Saccharomyces cerevisiae and Schizosaccharomyces pombe. BMC Genomics 8, 309 10.1186/1471-2164-8-309 (doi:10.1186/1471-2164-8-309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorin-Semblat D., Sicard A., Doerig C. M., Ranford-Cartwright L., Doerig C. 2008. Disruption of the PfPK7 gene impairs schizogony and sporogony in the human malaria parasite Plasmodium falciparum. Eukaryot. Cell 7, 279–285 10.1128/EC.00245-07 (doi:10.1128/EC.00245-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizzi E., Frontali C. 2001. Low-complexity regions in Plasmodium falciparum proteins. Genome Res. 11, 218–229 10.1101/gr.GR-1522R (doi:10.1101/gr.GR-1522R) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DePristo M. A., Zilversmit M. M., Hartl D. L. 2006. On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene 378, 19–30 10.1016/j.gene.2006.03.023 (doi:10.1016/j.gene.2006.03.023) [DOI] [PubMed] [Google Scholar]
- 39.Zilversmit M. M., Volkman S. K., DePristo M. A., Wirth D. F., Awadalla P., Hartl D. L. 2010. Low-complexity regions in Plasmodium falciparum: missing links in the evolution of an extreme genome. Mol. Biol. Evol. 27, 2198–2209 10.1093/molbev/msq108 (doi:10.1093/molbev/msq108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frugier M., Bour T., Ayach M., Santos M. A., Rudinger-Thirion J., Theobald-Dietrich A., Pizzi E. 2010. Low complexity regions behave as tRNA sponges to help co-translational folding of plasmodial proteins. FEBS Lett. 584, 448–454 10.1016/j.febslet.2009.11.004 (doi:10.1016/j.febslet.2009.11.004) [DOI] [PubMed] [Google Scholar]
- 41.Gardner K. B., Sinha I., Bustamante L. Y., Day N. P., White N. J., Woodrow C. J. 2011. Protein-based signatures of functional evolution in Plasmodium falciparum. BMC Evol. Biol. 11, 257 10.1186/1471-2148-11-257 (doi:10.1186/1471-2148-11-257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorin D., Le Roch K., Sallicandro P., Alano P., Parzy D., Poullet P., Meijer L., Doerig C. 2001. Pfnek-1, a NIMA-related kinase from the human malaria parasite Plasmodium falciparum biochemical properties and possible involvement in MAPK regulation. Eur. J. Biochem. 268, 2600–2608 10.1046/j.1432-1327.2001.02151.x (doi:10.1046/j.1432-1327.2001.02151.x) [DOI] [PubMed] [Google Scholar]
- 43.Low H., Lye Y. M., Sim T. S. 2007. Pfnek3 functions as an atypical MAPKK in Plasmodium falciparum. Biochem. Biophys. Res. Commun. 361, 439–444 10.1016/j.bbrc.2007.07.047 (doi:10.1016/j.bbrc.2007.07.047) [DOI] [PubMed] [Google Scholar]
- 44.Dorin D., Alano P., Boccaccio I., Ciceron L., Doerig C. M., Sulpice R., Parzy D., Doerig C. 1999. An atypical mitogen-activated protein kinase (MAPK) homologue expressed in gametocytes of the human malaria parasite Plasmodium falciparum. Identification of a MAPK signature. J. Biol. Chem. 274, 29 912–29 920 10.1074/jbc.274.42.29912 (doi:10.1074/jbc.274.42.29912) [DOI] [PubMed] [Google Scholar]
- 45.Carvalho T. G., Menard R. 2005. Manipulating the Plasmodium genome. Curr. Issues Mol. Biol. 7, 39–55 [PubMed] [Google Scholar]
- 46.Rangarajan R., Bei A. K., Jethwaney D., Maldonado P., Dorin D., Sultan A. A., Doerig C. 2005. A mitogen-activated protein kinase regulates male gametogenesis and transmission of the malaria parasite Plasmodium berghei. EMBO Rep. 6, 464–469 10.1038/sj.embor.7400404 (doi:10.1038/sj.embor.7400404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tewari R., Dorin D., Moon R., Doerig C., Billker O. 2005. An atypical mitogen-activated protein kinase controls cytokinesis and flagellar motility during male gamete formation in a malaria parasite. Mol. Microbiol. 58, 1253–1263 10.1111/j.1365-2958.2005.04793.x (doi:10.1111/j.1365-2958.2005.04793.x) [DOI] [PubMed] [Google Scholar]
- 48.Reininger L., et al. 2005. A NIMA-related protein kinase is essential for completion of the sexual cycle of malaria parasites. J. Biol. Chem. 280, 31 957–31 964 10.1074/jbc.M504523200 (doi:10.1074/jbc.M504523200) [DOI] [PubMed] [Google Scholar]
- 49.Reininger L., Tewari R., Fennell C., Holland Z., Goldring D., Ranford-Cartwright L., Billker O., Doerig C. 2009. An essential role for the Plasmodium Nek-2 Nima-related protein kinase in the sexual development of malaria parasites. J. Biol. Chem. 284, 20 858–20 868 10.1074/jbc.M109.017988 (doi:10.1074/jbc.M109.017988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rangarajan R., Bei A., Henry N., Madamet M., Parzy D., Nivez M. P., Doerig C., Sultan A. 2006. Pbcrk-1, the Plasmodium berghei orthologue of P. falciparum cdc-2 related kinase-1 (Pfcrk-1), is essential for completion of the intraerythrocytic asexual cycle. Exp. Parasitol. 112, 202–207 10.1016/j.exppara.2005.11.002 (doi:10.1016/j.exppara.2005.11.002) [DOI] [PubMed] [Google Scholar]
- 51.Tewari R., Straschil U., Bateman A., Bohme U., Cherevach I., Gong P., Pain A., Billker O. 2010. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe 8, 377–387 10.1016/j.chom.2010.09.006 (doi:10.1016/j.chom.2010.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fennell C., Babbitt S., Russo I., Wilkes J., Ranford-Cartwright L., Goldberg D. E., Doerig C. 2009. PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress-response to amino-acid starvation. Malar. J. 8, 99 10.1186/1475-2875-8-99 (doi:10.1186/1475-2875-8-99) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato N., et al. 2008. Gene expression signatures and small-molecule compounds link a protein kinase to Plasmodium falciparum motility. Nat. Chem. Biol. 4, 347–356 10.1038/nchembio.87 (doi:10.1038/nchembio.87) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dvorin J. D., et al. 2010. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science 328, 910–912 10.1126/science.1188191 (doi:10.1126/science.1188191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doerig C., et al. 2010. Malaria: targeting parasite and host cell kinomes. Biochim. Biophys. Acta 1804, 604–612 10.1016/j.bbapap.2009.10.009 (doi:10.1016/j.bbapap.2009.10.009) [DOI] [PubMed] [Google Scholar]
- 56.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 10.1016/j.cell.2006.09.026 (doi:10.1016/j.cell.2006.09.026) [DOI] [PubMed] [Google Scholar]
- 57.Cox J., Mann M. 2007. Is proteomics the new genomics? Cell 130, 395–398 10.1016/j.cell.2007.07.032 (doi:10.1016/j.cell.2007.07.032) [DOI] [PubMed] [Google Scholar]
- 58.Lim W. A., Pawson T. 2010. Phosphotyrosine signaling: evolving a new cellular communication system. Cell 142, 661–667 10.1016/j.cell.2010.08.023 (doi:10.1016/j.cell.2010.08.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cole A., Frame S., Cohen P. 2004. Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem. J. 377, 249–255 10.1042/BJ20031259 (doi:10.1042/BJ20031259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lochhead P. A. 2009. Protein kinase activation loop autophosphorylation in cis: overcoming a Catch-22 situation. Sci. Signal. 2, pe4 10.1126/scisignal.254pe4 (doi:10.1126/scisignal.254pe4) [DOI] [PubMed] [Google Scholar]
- 61.Aranda S., Laguna A., de la Luna S. 2011. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 25, 449–462 10.1096/fj.10-165837 (doi:10.1096/fj.10-165837) [DOI] [PubMed] [Google Scholar]
- 62.Huang H., et al. 2008. Defining the specificity space of the human SRC homology 2 domain. Mol. Cell Proteomics 7, 768–784 10.1074/mcp.M700312-MCP200 (doi:10.1074/mcp.M700312-MCP200) [DOI] [PubMed] [Google Scholar]
- 63.Eichinger L., et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 10.1038/nature03481 (doi:10.1038/nature03481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marti M., Good R. T., Rug M., Knuepfer E., Cowman A. F. 2004. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 10.1126/science.1102452 (doi:10.1126/science.1102452) [DOI] [PubMed] [Google Scholar]
- 65.Sicard A., et al. 2011. Activation of a PAK-MEK signalling pathway in malaria parasite-infected erythrocytes. Cell Microbiol. 13, 836–845 10.1111/j.1462-5822.2011.01582.x (doi:10.1111/j.1462-5822.2011.01582.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nolen B., Taylor S., Ghosh G. 2004. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 15, 661–675 10.1016/j.molcel.2004.08.024 (doi:10.1016/j.molcel.2004.08.024) [DOI] [PubMed] [Google Scholar]
- 67.Beraldo F. H., Almeida F. M., da Silva A. M., Garcia C. R. 2005. Cyclic AMP and calcium interplay as second messengers in melatonin-dependent regulation of Plasmodium falciparum cell cycle. J. Cell Biol. 170, 551–557 10.1083/jcb.200505117 (doi:10.1083/jcb.200505117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merckx A., Nivez M. P., Bouyer G., Alano P., Langsley G., Deitsch K., Thomas S., Doerig C., Egee S. 2008. Plasmodium falciparum regulatory subunit of cAMP-dependent PKA and anion channel conductance. PLoS Pathog. 4, e19 10.1371/journal.ppat.0040019 (doi:10.1371/journal.ppat.0040019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leykauf K., Treeck M., Gilson P. R., Nebl T., Braulke T., Cowman A. F., Gilberger T. W., Crabb B. S. 2010. Protein kinase A dependent phosphorylation of apical membrane antigen 1 plays an important role in erythrocyte invasion by the malaria parasite. PLoS Pathog. 6, e1000941 10.1371/journal.ppat.1000941 (doi:10.1371/journal.ppat.1000941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor H. M., McRobert L., Grainger M., Sicard A., Dluzewski A. R., Hopp C. S., Holder A. A., Baker D. A. 2010. The malaria parasite cyclic GMP-dependent protein kinase plays a central role in blood-stage schizogony. Eukaryot. Cell 9, 37–45 10.1128/EC.00186-09 (doi:10.1128/EC.00186-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng X., Ma Y., Moore M., Hemmings B. A., Taylor S. S. 1998. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc. Natl Acad. Sci. USA 95, 9849–9854 10.1073/pnas.95.17.9849 (doi:10.1073/pnas.95.17.9849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doerig C., Doerig C. M., Horrocks P., Coyle J., Carlton J., Sultan A., Arnot D., Carter R. 1995. Pfcrk-1, a developmentally regulated cdc2-related protein kinase of Plasmodium falciparum. Mol. Biochem. Parasitol. 70, 167–174 10.1016/0166-6851(95)00033-W (doi:10.1016/0166-6851(95)00033-W) [DOI] [PubMed] [Google Scholar]
- 73.Halbert J., et al. 2010. A Plasmodium falciparum transcriptional cyclin-dependent kinase-related kinase with a crucial role in parasite proliferation associates with histone deacetylase activity. Eukaryot. Cell 9, 952–959 10.1128/EC.00005-10 (doi:10.1128/EC.00005-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morgan D. O. 1995. Principles of CDK regulation. Nature 374, 131–134 10.1038/374131a0 (doi:10.1038/374131a0) [DOI] [PubMed] [Google Scholar]
- 75.Li J. L., Robson K. J., Chen J. L., Targett G. A., Baker D. A. 1996. Pfmrk, a MO15-related protein kinase from Plasmodium falciparum. Gene cloning, sequence, stage-specific expression and chromosome localization. Eur. J. Biochem. 241, 805–813 10.1111/j.1432-1033.1996.00805.x (doi:10.1111/j.1432-1033.1996.00805.x) [DOI] [PubMed] [Google Scholar]
- 76.Lawrence J. G. 2005. Common themes in the genome strategies of pathogens. Curr. Opin. Genet. Dev. 15, 584–588 10.1016/j.gde.2005.09.007 (doi:10.1016/j.gde.2005.09.007) [DOI] [PubMed] [Google Scholar]
- 77.Hunter T., Plowman G. D. 1997. The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 22, 18–22 10.1016/S0968-0004(96)10068-2 (doi:10.1016/S0968-0004(96)10068-2) [DOI] [PubMed] [Google Scholar]
- 78.Rubenstein E. M., Schmidt M. C. 2007. Mechanisms regulating the protein kinases of Saccharomyces cerevisiae. Eukaryot. Cell 6, 571–583 10.1128/EC.00026-07 (doi:10.1128/EC.00026-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morrison D. K., Murakami M. S., Cleghon V. 2000. Protein kinases and phosphatases in the Drosophila genome. J. Cell Biol. 150, F57–62 10.1083/jcb.150.2.F57 (doi:10.1083/jcb.150.2.F57) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kostich M., English J., Madison V., Gheyas F., Wang L., Qiu P., Greene J., Laz T. M. 2002. Human members of the eukaryotic protein kinase family. Genome Biol. 3, RESEARCH0043 10.1186/gb-2002-3-9-research0043 (doi:10.1186/gb-2002-3-9-research0043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 (doi:10.1126/science.1075762) [DOI] [PubMed] [Google Scholar]
- 82.Arnaiz O., Sperling L. 2011. ParameciumDB in 2011: new tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia. Nucleic Acids Res. 39, D632–D636 10.1093/nar/gkq918 (doi:10.1093/nar/gkq918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manning G., Reiner D. S., Lauwaet T., Dacre M., Smith A., Zhai Y., Svard S., Gillin F. D. 2011. The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol. 12, R66 10.1186/gb-2011-12-7-r66 (doi:10.1186/gb-2011-12-7-r66) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raman M., Chen W., Cobb M. H. 2007. Differential regulation and properties of MAPKs. Oncogene 26, 3100–3112 10.1038/sj.onc.1210392 (doi:10.1038/sj.onc.1210392) [DOI] [PubMed] [Google Scholar]
- 85.Dorin D., Semblat J. P., Poullet P., Alano P., Goldring J. P., Whittle C., Patterson S., Chakrabarti D., Doerig C. 2005. PfPK7, an atypical MEK-related protein kinase, reflects the absence of classical three-component MAPK pathways in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 55, 184–196 10.1111/j.1365-2958.2004.04393.x (doi:10.1111/j.1365-2958.2004.04393.x) [DOI] [PubMed] [Google Scholar]
- 86.Dorin-Semblat D., Quashie N., Halbert J., Sicard A., Doerig C. M., Peat E., Ranford-Cartwright L., Doerig C. 2007. Functional characterization of both MAP kinases of the human malaria parasite Plasmodium falciparum by reverse genetics. Mol. Microbiol. 65, 1170–1180 10.1111/j.1365-2958.2007.05859.x (doi:10.1111/j.1365-2958.2007.05859.x) [DOI] [PubMed] [Google Scholar]
- 87.Stover N. A., Krieger C. J., Binkley G., Dong Q., Fisk D. G., Nash R., Sethuraman A., Weng S., Cherry J. M. 2006. Tetrahymena genome database (TGD): a new genomic resource for Tetrahymena thermophila research. Nucleic Acids Res. 34, D500–D503 10.1093/nar/gkj054 (doi:10.1093/nar/gkj054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuo C. H., Kissinger J. C. 2008. Consistent and contrasting properties of lineage-specific genes in the apicomplexan parasites Plasmodium and Theileria. BMC Evol. Biol. 8, 108 10.1186/1471-2148-8-108 (doi:10.1186/1471-2148-8-108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schneider A. G., Mercereau-Puijalon O. 2005. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics 6, 30 10.1186/1471-2164-6-30 (doi:10.1186/1471-2164-6-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nunes M. C., Goldring J. P., Doerig C., Scherf A. 2007. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Mol. Microbiol. 63, 391–403 10.1111/j.1365-2958.2006.05521.x (doi:10.1111/j.1365-2958.2006.05521.x) [DOI] [PubMed] [Google Scholar]
- 91.Peixoto L., Chen F., Harb O. S., Davis P. H., Beiting D. P., Brownback C. S., Ouloguem D., Roos D. S. 2010. Integrative genomic approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host Microbe 8, 208–218 10.1016/j.chom.2010.07.004 (doi:10.1016/j.chom.2010.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.El Hajj H., Lebrun M., Arold S. T., Vial H., Labesse G., Dubremetz J. F. 2007. ROP18 is a rhoptry kinase controlling the intracellular proliferation of Toxoplasma gondii. PLoS Pathog. 3, e14 10.1371/journal.ppat.0030014 (doi:10.1371/journal.ppat.0030014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parsons M., Worthey E. A., Ward P. N., Mottram J. C. 2005. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6, 127 10.1186/1471-2164-6-127 (doi:10.1186/1471-2164-6-127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lye Y. M., Chan M., Sim T. S. 2006. Pfnek3: an atypical activator of a MAP kinase in Plasmodium falciparum. FEBS Lett. 580, 6083–6092 10.1016/j.febslet.2006.10.003 (doi:10.1016/j.febslet.2006.10.003) [DOI] [PubMed] [Google Scholar]
- 95.Dorin-Semblat D., et al. 2011. Plasmodium falciparum NIMA-related kinase Pfnek-1: sex specificity and assessment of essentiality for the erythrocytic asexual cycle. Microbiology 157, 2785–2794 10.1099/mic.0.049023-0 (doi:10.1099/mic.0.049023-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khan S. M., Franke-Fayard B., Mair G. R., Lasonder E., Janse C. J., Mann M., Waters A. P. 2005. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121, 675–687 10.1016/j.cell.2005.03.027 (doi:10.1016/j.cell.2005.03.027) [DOI] [PubMed] [Google Scholar]
- 97.DeBarry J. D., Kissinger J. C. 2011. Jumbled genomes: missing Apicomplexan synteny. Mol. Biol. Evol. 28, 2855–2871 10.1093/molbev/msr103 (doi:10.1093/molbev/msr103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nunes M. C., Okada M., Scheidig-Benatar C., Cooke B. M., Scherf A. 2010. Plasmodium falciparum FIKK kinase members target distinct components of the erythrocyte membrane. PLoS ONE 5, e11747 10.1371/journal.pone.0011747 (doi:10.1371/journal.pone.0011747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Artz J. D., et al. 2011. The Cryptosporidium parvum kinome. BMC Genomics 12, 478 10.1186/1471-2164-12-478 (doi:10.1186/1471-2164-12-478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gileadi O., et al. 2007. The scientific impact of the structural genomics consortium: a protein family and ligand-centered approach to medically-relevant human proteins. J. Struct. Funct. Genomics 8, 107–119 10.1007/s10969-007-9027-2 (doi:10.1007/s10969-007-9027-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holton S., Merckx A., Burgess D., Doerig C., Noble M., Endicott J. 2003. Structures of P. falciparum PfPK5 test the CDK regulation paradigm and suggest mechanisms of small molecule inhibition. Structure 11, 1329–1337 10.1016/j.str.2003.09.020 (doi:10.1016/j.str.2003.09.020) [DOI] [PubMed] [Google Scholar]
- 102.Merckx A., Echalier A., Langford K., Sicard A., Langsley G., Joore J., Doerig C., Noble M., Endicott J. 2008. Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design. Structure 16, 228–238 10.1016/j.str.2007.11.014 (doi:10.1016/j.str.2007.11.014) [DOI] [PubMed] [Google Scholar]
- 103.Mukherjee K., Sharma M., Urlaub H., Bourenkov G. P., Jahn R., Sudhof T. C., Wahl M. C. 2008. CASK functions as a Mg2+-independent neurexin kinase. Cell 133, 328–339 10.1016/j.cell.2008.02.036 (doi:10.1016/j.cell.2008.02.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kannan N., Taylor S. S. 2008. Rethinking pseudokinases. Cell 133, 204–205 10.1016/j.cell.2008.04.005 (doi:10.1016/j.cell.2008.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taylor S. S., Kornev A. P. 2010. Yet another ‘active’ pseudokinase, Erb3. Proc. Natl Acad. Sci. USA 107, 8047–8048 10.1073/pnas.1003436107 (doi:10.1073/pnas.1003436107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fasolo J., Sboner A., Sun M. G., Yu H., Chen R., Sharon D., Kim P. M., Gerstein M., Snyder M. 2011. Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Dev. 25, 767–778 10.1101/gad.1998811 (doi:10.1101/gad.1998811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Behnke M. S., Khan A., Wootton J. C., Dubey J. P., Tang K., Sibley L. D. 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc. Natl Acad. Sci. USA 108, 9631–9636 10.1073/pnas.1015338108 (doi:10.1073/pnas.1015338108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reese M. L., Zeiner G. M., Saeij J. P. J., Boothroyd J. C., Boyle J. P. 2011. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc. Natl Acad. Sci. USA 108, 9625–9630 10.1073/pnas.1015980108 (doi:10.1073/pnas.1015980108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Manning G., Plowman G. D., Hunter T., Sudarsanam S. 2002. Evolution of protein kinase signaling from yeast to man. Trends. Biochem. Sci. 27, 514–520 10.1016/S0968-0004(02)02179-5 (doi:10.1016/S0968-0004(02)02179-5) [DOI] [PubMed] [Google Scholar]