Abstract
Background
Mu opioid receptor (OPRM1) ligands may alter expression of chemokines and chemokine receptors involved in penetration of human immunodeficiency virus (HIV) type 1 into the cell. We suggest that OPRM1 variants may affect the pathophysiology of HIV infection.
Methods
DNA samples from 1031 eligible African Americans, Hispanics, and whites from the Women's Interagency HIV Study (WIHS) who were alive as of April 2006 were analyzed. We performed regression analysis of association of 18 OPRM1 variants with a change of viral load and CD4 cell count during 2 periods: between admission to WIHS and the start of highly active antiretroviral therapy (HAART) (interval X) and between the start of HAART and the most recent WIHS visit (interval Y), and examined the association of these variants with HIV status.
Results
Regardless of genotype, a significant decrease in viral load during interval X was found for each ethnicity. Whites with allele G of the functional polymorphism 118A > G (reference sequence rs1799971) showed a smaller decrease in viral load; those bearing minor alleles IVS1 + 1050A, IVS1 + 14123A, and IVS2 + 31A showed a larger decrease in viral load over interval X (0.01 < P < .05). Hispanics with the same alleles showed a greater increase in CD4 cell count over interval Y (0.01 < P < .05). We found an association between OPRM1 variants and HIV status in African Americans and whites.
Conclusions
OPRM1 polymorphisms may alter the severity of HIV infection before and after HAART.
After our original discovery [1] of the functionality of the 118A > G polymorphism in the gene (OPRM1) encoding the mu opioid receptor, variants of OPRM1 have been extensively studied in large cohorts for their association with stress responsivity, response to pain management, affective disorders, and specific addictive disorders, including cocaine and opioid addictions and alcoholism [2–5, 6]. However, OPRM1 variants have not been studied in a cohort of individuals with human immunodeficiency virus (HIV) infection. OPRM1 is involved in many physiological functions, including stress responsivity, by tonic inhibition of the hypothalamic-pituitary-adrenal (HPA) axis [7]; eating behavior; endocrine function; and cognition [2–5]. Beta-endorphin, an endogenous opioid that activates OPRM1, binds the 118G variant (reference sequence rs1799971) of OPRM1 3 times as tightly as it binds the prototype receptor [1]. Basal levels of cortisol are higher in healthy humans with 1 or 2 copies of 118G [8]. This polymorphism was shown to be functional in the treatment of alcoholism [9].
The OPRM1 is also involved in immune response and HIV and simian immunodeficiency virus expansion [10–18]. We have studied HIV since the early 1980s [19–21]. Opioid ligands may regulate the expression of chemokines and chemokine receptors [12]. Many studies have found cross-desensitization interactions between opioid and chemokine receptors both in vivo and in vitro [13–15]. Both OPRM1 and delta opioid receptor (OPRD1) selective ligands can induce desensitization of CCR1, CCR2, CCR5, CXCR1, and CXCR2, but not CXCR4. Similarly, selective ligands of CCR1, CCR2, CCR5, CCR7, and CX3CR1 can desensitize both OPRM1 and OPRD1 [13–15]. Opioids selectively promote induction of proinflammatory or anti-inflammatory effects, depending on the involvement of OPRM1 or, alternatively, on the kappa opioid receptor (OPRK1), which frequently act in opposition to each other [16, 17].
Specific variants of CCR5 and CCR2 suppress HIV-1 transmission and cause delay in HIV-1 disease progression [22–24]. Genetic differences in major histocompatibility complex (MHC) genes (HLA in humans) have been associated with different rates of progression from HIV infection to AIDS [25–28]. We suggest that the genetic variants that contribute to depression, affective disorders, and stress responsivity may also influence the pathophysiology of HIV infection.
In this study, we looked for an association between selected OPRM1 polymorphisms and HIV status, using HIV-positive individuals (cases) and HIV-negative individuals (controls) recruited at Women's Interagency HIV Study (WIHS) sites [29–32]. We also tested for the association between specific genotypes of OPRM1 and the response to HIV treatment, including changes in the viral load and CD4 cell count, across the period from admission to WIHS to the start of HAART and the period from the start of HAART to the most recent WIHS visit for which data were available. Our findings indicate the potential involvement of the OPRM1 in the pathophysiology of HIV infection.
METHODS
Study Subjects: Recruitment and Diagnostic Procedures
Characteristics of subjects included and excluded in the analysis are shown in Table 1. Unrelated African American, white, and Hispanic women were recruited by WIHS. As of 2002, 3766 subjects were enrolled in 2 cohorts, with the first recruited during 1994–2000 (most subjects were recruited during 1994–1995) and the second recruited during 2001–2002 (available at: https://statepiaps.jhsph.edu/wihs/invest-info/dossier.pdf; accessed 15 June 2011). Only subjects who signed an extended, updated informed consent form between 1 April 2006 and 30 September 2007 (n = 1506) were included, thus excluding those who died between WIHS entry and 2006. Subjects completed our Family Origin Questionnaire on 3 generations. Limited clinical information about subjects was provided by WIHS, including HIV status, CD4 cell count, and viral load. Physical examination, specimen collection, and interview about the history of illness, substance abuse, current medications, and medication adherence were done every 6 months.
Table 1.
Characteristics of Subjects Included or Excluded From the Study, by Human Immunodeficiency Virus (HIV) Infection Status
| No. of Subjects |
|||
|---|---|---|---|
| Characteristic | HIV– | HIV+ | Total |
| Total number of subjects | … | … | 1506 |
| Subjects included in this study | |||
| Ethnicity | |||
| African American | 213 | 478 | 691 |
| White | 46 | 115 | 161 |
| Hispanic | 68 | 111 | 179 |
| Total | 327 | 704 | 1031 |
| Had clinical data for 2 of 3 study points availablea | … | 621 | … |
| Subjects excluded from this study | |||
| Mixed ethnicity | 134 | 287 | 421 |
| Low DNA concentration | … | … | 45 |
| Seroconverted after admission to WIHS study | … | … | 6 |
| Start HAART before admission to WIHS | 0 | 1 | 1 |
| Start HAART after the most recent visit | 0 | 1 | 1 |
| Missing longitudinal data | 0 | 1 | 1 |
| Total | … | … | 475 |
Abbreviations: HIV+, HIV positive; HIV–, HIV negative; WIHS, Women's Interagency HIV Study.
a Study points were defined as admission to WIHS, initiation of highly active antiretroviral therapy, and most recent visit to WIHS for which clinical data were made available. A total number of samples that was either included or excluded from the study is shown in bold.
A total of 475 subjects were excluded because of mixed ethnicity (421 subjects; Table 1), seroconversion after admission to WIHS (6 subjects), and insufficient amount of DNA samples (45 subjects), leaving 1031 subjects for association studies involving HIV status. A subset of HIV-positive subjects with complete data across 3 study points—entrance to WIHS (hereafter, “R”), initiation of HAART (hereafter, “S”), and the most recent WIHS visit for which clinical data were made available (hereafter, “T”)—were used to compare changes in viral load and CD4 cell count during 2 intervals (Table 2). The first interval was defined as the period from R to S (hereafter, “X”), and the second interval was defined as the period from S to T (hereafter, “Y”). A different subset of HIV-positive subjects with complete data across 2 of 3 study points (R and S, or S and T) was used for analysis of the influence of genotype on changes in viral load or CD4 cell count across intervals X and Y (Table 3). Subjects who did not start HAART were excluded from viral load and CD4 cell count analyses. Study point S occurred during the period between 6 months before the start of HAART and 1 month after the initiation of HAART.
Table 2.
Ethnicity of Subjects With Human Immunodeficiency Virus Infection for Whom Viral Loads or CD4 Cell Counts From All 3 Study Points Were Available
| No. of Subjects |
||
|---|---|---|
| Ethnicity | Viral Load | CD4 Cell Count |
| African American | 388 | 412 |
| White | 96 | 105 |
| Hispanic | 94 | 104 |
| Total | 578 | 621 |
Study points were defined as admission to the Women's Interagency HIV Study (WIHS), initiation of highly active antiretroviral therapy, and most recent visit to WIHS for which clinical data were made available.
Table 3.
Ethnicity of Subjects With Human Immunodeficiency Virus Infection for Whom Viral Loads or CD4 Cell Counts From 2 of 3 Study Points Were Available for Use in Statistical Genetics (Logistic Regression) Analysis of the Association Between OPRM1 Polymorphisms and Change in Viral Load and CD4 Cell Count
| No. of Subjects |
||||
|---|---|---|---|---|
| Viral Load |
CD4 Cell Count |
|||
| Ethnicity | Interval X | Interval Y | Interval X | Interval Y |
| African Americans | 390 | 393 | 412 | 412 |
| Whites | 96 | 98 | 105 | 105 |
| Hispanics | 94 | 95 | 104 | 104 |
| Total | 580 | 586 | 621 | 621 |
Study points were defined as admission to the WIHS, Women's Interagency HIV Study (WIHS; R), initiation of highly active antiretroviral therapy (S), and most recent visit to WIHS for which clinical data were made available (T). Interval X was defined as the time from R to S, and interval Y was defined as the time from S to T.
OPRM1 Genotyping
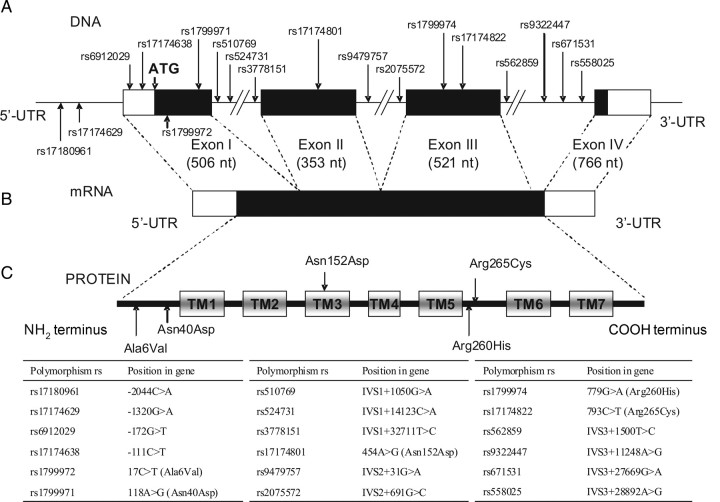
The structure of OPRM1 is shown in Figure 1. DNA from peripheral blood mononuclear cell pellets obtained from the WIHS depository was isolated using the PureGene DNA purification kit (Gentra Systems, Minneapolis, MN) and used for genotyping on an ABI Prism 7900 sequence detection system according to manufacturer's protocol, with custom-synthesized oligonucleotide primers and probes specific for the polymorphism chosen (Supplementary Table 1). For each polymorphism, the results of TaqMan genotyping were confirmed by Sanger sequencing of 22 randomly selected samples, using the same primers.
Figure 1.
Structure of the OPRM1 gene. A, Locations of polymorphisms. B, Structure of messenger RNA (mRNA). C, Structure of the mu opioid receptor peptide. Abbreviations: nt, nucleotides; rs, Single Nucleotide Polymorphism Database (dbSNP) reference sequence; TM, transmembrane domains; UTR, untranslated regions.
Statistical Analysis
Analysis of variance (ANOVA) with repeated measures was used to assess the change in viral load and CD4 cell counts for each ethnicity in the subset of HIV-positive subjects who had data from study points R, S, and T, without stratification by genotype with data. Hardy-Weinberg equilibrium (HWE) for each marker was determined in controls, using the likelihood ratio χ2 test, separately for each ethnicity (54 tests). For a Bonferroni-corrected value of 0.01, individual χ2 tests are significant when P < .0002. Likelihood ratio χ2 tests were used to determine differences in genotype frequencies among ethnicities in the group of controls only. In this test, genotypes were collapsed when the expected number in each cell was <1.
Multiple regression analyses of viral load and CD4 cell count were carried out with cases only, using 2 approaches. In the first approach, multiple regressions were carried out, using the difference in the natural log of the viral load over interval X as the dependent variable, with the length of interval X and each genotype as independent (predictor) variables. One such analysis was carried out for each variant and each ethnicity. As viral load and CD4 cell count measurements were strongly skewed (with a long right tail), they were transformed to natural logs (1 was added to CD4 cell counts). We tested whether genotypes were associated with changes in viral load and CD4 cell counts over intervals X and Y.
In the second approach, analyses were carried out twice for each variant, once in the dominant model (genotypes AA combined with genotypes AB) and once in the recessive model (genotypes AB combined with genotypes BB). Resulting groups were used as predictor variables.
Logistic regression was performed for all cases and controls, including HIV-positive subjects who had not start HAART since admission to WIHS, to assess the association between each variant and HIV status, separately for each ethnicity and with each variant as a predictor variable, using 2 approaches. In the first approach, logistic regression was carried out for each variant, with case/control as the dependent variable and genotype code (0, 1, and 2 for genotypes AA, AB, and BB, respectively) as the independent (predictor) variable. In the second approach, the analysis was carried out twice for each variant in a fashion similar to that described above, using dominant and recessive models.
Analysis of the linkage disequilibrium (LD) of the variants studied was performed separately for each ethnicity, using Haploview 4.2 (available at: http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview; accessed 12 March 2011).
RESULTS
Analysis of Clinical Data: Overall Change of Viral Load and CD4 Cell Counts at Different Study Points, Without Stratification by OPRM1 Genotype
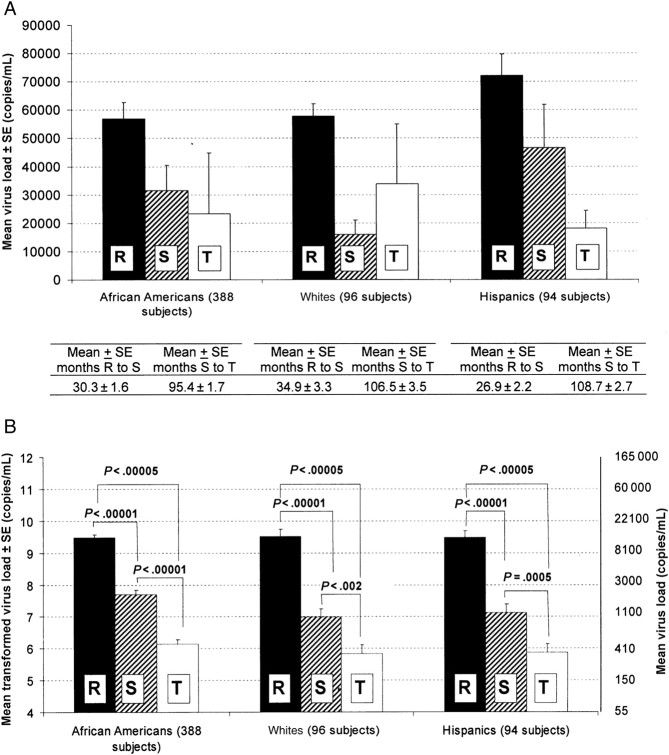
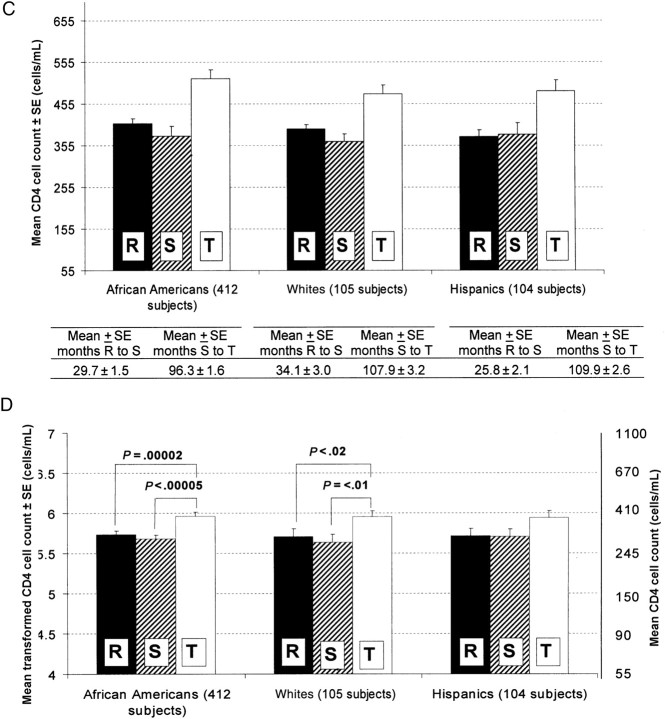
In each ethnicity, there was a significant decrease in viral load between study points R and S and between study points R and T (Figure 2B). ANOVA with repeated measures across study points in the subset of subjects with complete data at all 3 study points showed a significant main effect of interval in African Americans (F[2,274] = 28.10; P < .00001), whites (F[2,394] = 78.52; P < .000001), and Hispanics (F(2,186) = 59.48; P < .000001). Newman-Keuls post hoc tests showed that viral load fell significantly in each ethnicity during intervals X and Y (Figure 2B). The improvement (ie, reduction) of the viral load may have been due to general medical care; to monotherapy, such as azidothymidine (AZT); or to combination therapy. AZT has been shown to be effective in reducing viral load [33, 34].
Figure 2.
Equivalent and natural log–transformed viral loads and CD4 cell counts, at different study points, for subjects of different ethnicities. Study points were defined as admission to the Women's Interagency HIV Study (WIHS; R), initiation of highly active antiretroviral therapy (HAART; S), and most recent visit to WIHS for which clinical data were made available (T). SE, standard error. A, Viral loads in different ethnic groups, by study point and interval between study points. B, Transformed virus loads for statistical analysis. C, CD4 cell counts in different ethnic groups, by study point and interval between study points. D, Transformed CD4 cell counts (+1) for statistical analysis.
At WIHS entry, the majority of the HIV-positive subjects (70.6% of African Americans, 74.3% of whites, and 83.7% of Hispanics) had CD4 cell counts <500 cells/mL (Supplementary Table 1B). There was improvement in CD4 cell count after HAART. Significant increases in CD4 cell counts were found in African Americans (F[2,288] = 17.61; P < .00005) and whites (F[2,208] = 5.09; P < .01) but not in Hispanics (F[2,186]= 1.33; P = .267). Newman-Keuls post hoc tests showed an increase in CD4 cell counts in both ethnic groups only after the start of HAART (Figure 2D). Among Hispanics, there was an apparent change in CD4 cell count that had the same direction as that for African Americans and whites, but this did not reach statistical significance.
Ethnic Differences in Allele Frequencies
In the control group, overall allele frequencies were significantly different among the 3 ethnicities in 8 variants (Supplementary Table 1C). None of the polymorphisms tested in this study showed significant deviation from HWE in controls after correction for multiple testing (P < .0002; Supplementary Table 1D). Polymorphisms with a frequency of <0.05 in both controls and cases were excluded from analysis (Supplementary Table 1C).
Multiple Regression Analysis of Association of Genotypes of OPRM1 With Viral Load
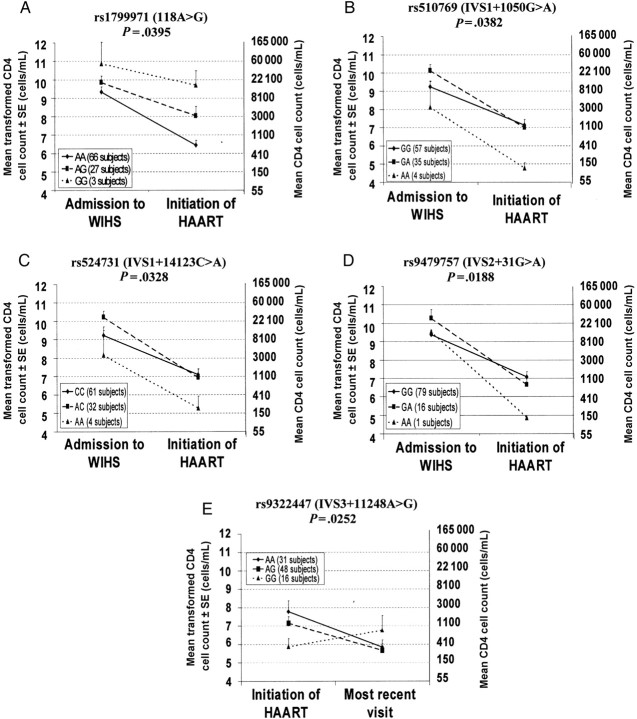
By use of multiple regression analysis in approach 1 in the group of HIV-positive subjects, we found that individuals bearing the minor alleles IVS1 + 1050A, IVS1 + 14123A, and IVS2 + 31A had a greater reduction in viral load over interval X as compared to individuals bearing major alleles of these variants (Figure 3B–D and Table 4). Interestingly, individuals bearing 1 or 2 copies of variant G of polymorphism 118A > G had a smaller reduction in viral load over interval X (Figure 3A and Table 4). Hispanic subjects bearing genotype GG in IVS3 + 11248A > G (rs9322447) showed no decrease in viral load over interval Y as compared to those bearing genotypes AA and AG (Figure 3E and Table 4). Similar results were found using the dominant-recessive model of analysis (approach 2; Table 4).
Figure 3.
Association between genotypes of OPRM1 variants and changes in natural log–transformed viral loads between admission to Women's Interagency HIV Study (WIHS) and the start of highly active antiretroviral therapy (HAART), among whites (A–D) and Hispanics (E). In all panels, genotypes of the homozygous major alleles are denoted by the solid line. Abbreviations: rs, Single Nucleotide Polymorphism Database (dbSNP) reference sequence; SE, standard error.
Table 4.
Significant Findings From Multiple Regression Analysis of the Association Between OPRM1 Genotypes and Changes in Viral Load and CD4 Cell Count During Interval X Only and/or During Interval Y Only Among Subjects With Human Immunodeficiency Virus (HIV) Infection
|
P of Change (Approach 1)a |
||||
|---|---|---|---|---|
| Variant, by HIV marker and interval | African Americans | Whites | Hispanics | P of Change (Approach 2) |
| Viral load, interval X | n = 390 | n = 96 | n = 94 | |
| 118A>G | LFb | .0395c | .8788 | White rec, .0432 |
| IVS1+1050G>A | .9597 | .0382d | .8767 | White rec, .0304 |
| IVS1+14123C>A | .1743 | .0328d | .8610 | White rec, .0213 |
| IVS2+31G>A | .6165 | .0188d | .6597 | White rec, .0206 |
| Viral load, interval Y | n = 393 | n = 98 | n = 95 | |
| IVS3+27669G>A | .5724 | .2761 | .0252e | Hispanic dom, .0143 |
| CD4 cell count, interval Y | n = 412 | n = 105 | n = 104 | |
| IVS1+1050G>A | .9545 | .1124 | .0183f | Hispanic dom, .0292 |
| IVS1+14123C>A | .5666 | .0556 | .0150f | White rec, .0214; Hispanic dom, .0170 |
| IVS1+32711T>C | .2506 | .0575 | .1095 | White rec. .0395 |
| IVS2+31G>A | .6394 | .0693 | .0493f | Hispanic rec. .0493 |
Values in bold italics indicate point-wise significant associations (P < .05). Interval X was defined as the period between admission to WIHS and initiation of HAART, and interval Y was defined as the period between initiation of HAART and the most recent visit to WIHS for which clinical data were made available.
Abbreviations: dom, dominant model; HAART, highly active antiretroviral therapy; rec, recessive model; WIHS, Women's Interagency HIV Study.
a The number of subjects in each analysis differed because of missing data (Table 1).
b LF, Low Frequency, Ethnic groups in which the genotype frequency of specific single-nucleotide polymorphisms was <0.05 were eliminated.
c Subjects with minor allele G showed a smaller decrease.
d Subjects with minor allele A showed a larger decrease.
e Subjects with minor allele G showed a smaller decrease.
f Subjects with minor allele A showed a larger increase.
Multiple Regression Analysis of Association of Genotypes of OPRM1 With CD4 Cell Count
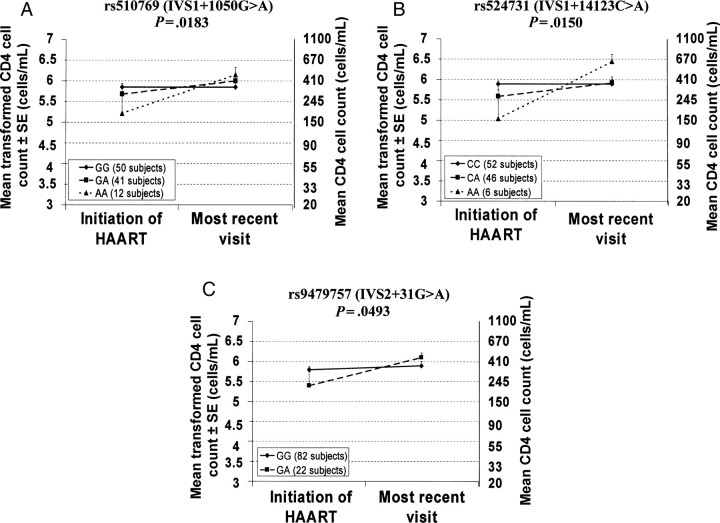
Multiple regression analysis in approach 1 in the HIV-positive group revealed no significant effect of the genotype on the change of CD4 cell count over interval X in any ethnic group studied. We found that Hispanic subjects bearing 1 or 2 copies of the minor alleles IVS1 + 1050A, IVS1 + 14123A, and IVS2 + 31A had a greater increase (ie, improvement) of CD4 cell count over interval Y (Figure 4A–C and Table 4).
Figure 4.
Association between genotypes of OPRM1 variants and changes in natural log–transformed CD4 cell counts (+1) between the start of highly active antiretroviral therapy (HAART) and the most recent visit to the Women's Interagency HIV Study for which clinical data were made available, among Hispanics. In all panels, genotypes of the homozygous major alleles are denoted by the solid line. Abbreviations: rs, Single Nucleotide Polymorphism Database (dbSNP) reference sequence; SE, standard error.
In approach 2, no significant effect of any polymorphism studied on the change of the CD4 cell count over interval X was found in any ethnic group. In Hispanics, a significant effect of the genotypes of IVS1 + 1050G > A and IVS1 + 14123C > A on the change of the CD4 cell count over interval Y was found using the dominant model (P = .0292 and P = .0170, respectively; Table 4) and an effect of IVS2 + 31G > A on the change of the CD4 cell count over the same period in the recessive model (P = .0493), similar to findings from approach 1. In whites, a significant effect of the genotypes of IVS1 + 14123C > A and IVS1 + 32711T > C on the change of the CD4 cell count over interval Y was found using the recessive model (P = .0214 and P = .0395, respectively, Table 4). This effect was not found using approach 1.
Logistic Regression Analysis of OPRM1 Polymorphisms and HIV Status
By use of logistic regression in controls and cases (approach 1; Table 5), we found that, in whites, the minor alleles 2044A, IVS1 + 1050A, IVS1 + 14123A, IVS1 + 32711C, and IVS2 + 31A had lower frequencies in the HIV-positive group as compared to controls. In African Americans, the minor allele IVS2 + 691C was found in greater frequency in the HIV-positive group, and the minor allele 11189G was found in lower frequency in the HIV-positive group.
Table 5.
Significant Findings From Logistic Regression Analysis of OPRM1 Polymorphisms and Human Immunodeficiency Virus (HIV) Infection Status
|
P (Approach 1) |
||||
|---|---|---|---|---|
| Variant | African American (n = 213 HIV– and 478 HIV+) | White (n = 46 HIV– and 115 HIV+) | Hispanic (n = 68 HIV– and 111 HIV+) | P (Approach 2) |
| -2044C>A | LFa | .0078b | LFa | White rec, .0078d |
| IVS1+1050G>A | .1322 | .0278b | .7352 | NS |
| IVS1+14123C>A | .4837 | .0083b | .7866 | White rec, .0149d |
| IVS1+32711T>C | .1894 | .0038b | .1983 | White rec, .0145e; white dom, .0352f |
| IVS2+31G>A | .3837 | .0171b | .6479 | White rec, .0092g |
| IVS2+691G>C | .0491c | .3268 | .1357 | NS |
| IVS3+27669G>A | .0262b | .6006 | .2545 | African American rec, .0304h |
Values in bold italics indicate point-wise significant associations (P < .05).
Abbreviations: dom, dominant model; HIV+, HIV positive; HIV–, HIV negative; NS, not significant; rec, recessive model.
a LF,Low Frequency, Ethnic groups in which the genotype frequency of specific single-nucleotide polymorphisms was <0.05 were eliminated.
b Minor allele frequency was decreased in the HIV+ group.
c Minor allele frequency was increased in the HIV+ group.
d Decreased frequency of genotypes (CA+AA) was found in the HIV+ group.
e Decreased frequency of genotypes (TC+CC) was found in the HIV+ group.
f Decreased frequency of genotype CC was found in the HIV+ group.
g Decreased frequency of genotypes (GA+AA) was found in the HIV+ group.
h Decreased frequency of genotypes (AG+GG) was found in the HIV+ group.
By use of approach 2 (Table 5), we found a significantly lower frequency of genotypes with the minor alleles -2044 (CA + AA), IVS1 + 14123(CA + AA), IVS1 + 32711(TC + CC), and IVS2 + 31(GA + AA) in the HIV-positive group of whites, using the recessive model. Also in whites, a lower frequency of genotype IVS1 + 32711CC was found in the HIV-positive group, using the dominant model. A lower frequency of genotypes 11189(AG + GG) was found in the HIV-positive group of African Americans, using the recessive model.
All findings in multiple regression analyses or logistic regression analyses were only point-wise significant.
Analysis of LD
LD analysis was performed for cases and controls combined, separately for each ethnic group. Complete LD (r2 = 1) was not detected for any combination of variants. However, several variants were in high LD (r2 > 0.7). Variants IVS1 + 1050G > A (rs510769) and IVS1 + 14123C > A were in high LD (r2 > 0.7) in whites and Hispanics. Variants IVS1 + 14123C > A and IVS1 + 32711T > C (rs3778151) were in high LD (r2 > 0.7) in whites only. Variants IVS2 + 691G > C (rs2075572) and IVS3 + 11248A > G were in high LD in all ethnicities (r2 > 0.66). In African Americans, high LD was observed between variants −1320G > A and −111C > T (r2 = 0.90).
DISCUSSION
We selected 3 study points that identify crucial and well-defined points in the clinical history of HIV-infected patients. Date of infection was not verified for most patients. We found high variability in viral loads in the original data (Figure 2A), especially at study point T, which results in different patterns of viral load change in African Americans as compared to that in whites; when data were natural log transformed, we observed the same patterns of viral load change in all 3 ethnicities (Figure 2B). The analyses of viral load and CD4 cell count changes during intervals X and Y indicate strong decline of viral load over both intervals in all 3 ethnic groups (Figure 2B). However, no improvement in CD4 cell count was found in any ethnic group before the start of HAART (Figure 2D). Before the start of HAART, some subjects received monotherapy, including AZT, or 2-drug combination therapy. Administration of AZT alone may result in reduction of viral load by 50% in antiretroviral-naive patients; administration of AZT with other modified nucleotides, including 2′,3′-dideoxyinosine, 2′,3′-dideoxycytidine, and 2′,3′-dideoxy-3′-thiacytidine, reduces the viral load by 80%–90% [33]. AZT treatment was also shown to be effective in reducing maternal-infant HIV transmission [35].
Whites with minor allele G of the OPRM1 variant 118A > G (Asn40Asp), a functional stress responsivity variant, had a smaller decrease in viral load over interval X. In vitro studies by our group showed that the variant Asp40 of OPRM1 binds beta-endorphin, an endogenous ligand of OPRM1, with 3 times the affinity of wild-type OPRM1 [1], suggesting that subjects bearing this polymorphism might have different response to opioid medications. Cell studies by our group showed that the Asp40 variant of OPRM1 had lower forskolin-induced cAMP accumulation and lower receptor-binding-site availability [36]. Studies of postmortem human brain specimens showed that allele 118G is expressed at a lower level than 118A [37]. This polymorphism was shown to be associated with a number of neurobiological conditions, including heroin addiction [38], alcoholism [39], and pharmacological response to naltrexone treatment for alcoholism [9].
The 118G allele has been associated with proinflammatory cytokine levels in a study involving healthy Japanese subjects, among whom the allele frequency of the 118G was 59.3% [40]. Cross-desensitization interactions between OPRM1 and chemokine receptors that are directly involved in HIV proliferation have been shown [13, 14]. Therefore, alteration of OPRM1 activity in subjects bearing the 118G variant may result in altered activity of chemokine receptors (coreceptors for HIV replication), producing a worse clinical outcome in HIV-positive patients. Indeed, a smaller decrease in viral load over interval X in HIV-positive whites bearing the 118G allele was found in this study. Alteration in viral load change in subjects bearing the 118G allele might also be directly related to altered stress responsivity previously found in healthy people bearing this allele [1].
We did not find association between any polymorphism and change in CD4 cell count during interval X in any ethnic group. However, we found an association of the intronic IVS1 + 1050G > A, IVS1 + 14123C > A, and IVS2 + 31G > A variants with the change in CD4 cell count over interval Y in Hispanics. The same polymorphisms were found to be associated with a change in viral load over interval X in whites. In both cases, these polymorphisms showed similar effect. In whites, subjects with minor alleles IVS1 + 1050A, IVS1 + 14123A, and IVS2 + 31A had a larger decrease (ie, improvement) in viral load before the start of HAART. In Hispanics, they had a greater increase (ie, improvement) in CD4 cell count since the start of HAART.
In whites, the smallest group in our study, we found a number of polymorphisms, including a possible promoter variant, -2044C > A, and a number of intronic variants, IVS1 + 1050G > A, IVS1 + 14123C > A, IVS1 + 32711T > C, and IVS2 + 31G > A, to be associated with HIV status. The frequency of the minor allele of these variants was lower in the HIV-positive group, indicating a possible protective effect from infection. Both IVS1 + 1050G > A and IVS1 + 32711T > C were found by our group to be associated with vulnerability to opioid addiction in whites [41]. Allele T of IVS1 + 32711T > C was found to be associated with positive subjective response to the first episode of heroin use in Chinese subjects [42]. Haplotype block (A-A), consisting of 118A > G and IVS1 + 1050G > A, was found to be associated with feelings of amphetamine-induced euphoria, energy, and stimulation in whites [43]. Several polymorphisms that we found to be associated with HIV status in whites (IVS1 + 1050G > A, IVS1 + 14123C > A and IVS2 + 31G > A; Table 5) were also found to be associated with a decrease in viral load over interval X in whites and with an increase in CD4 cell count in Hispanics over interval Y (Table 4). Further study in a white group with a larger number of subjects is needed.
We found in African Americans, the largest ethnic group in our study, associations between HIV status and both the intronic IVS2 + 691G > C and the polymorphism from the 3′ untranslated region IVS3 + 11248A > G. Previously, a minor haplotype GCG, consisting of variants IVS2 + 31G > A, IVS2 + 691G > C, and IVS3 + 183A > G (rs10485057), was found to be associated with smoking initiation and nicotine dependence [44]. Two of these variants, IVS2 + 31G > A and IVS2 + 691G > C, in our study were found to be associated with HIV status in whites and African Americans, respectively.
Studies by our group and others have shown that OPRM1 is involved in modulation of the HPA axis, one of the main systems involved in stress regulation [1, 7]. Stress, depressed mood, and dysthymic disorders have been reported to be associated with unsafe sex practices [45]. Numerous studies indicate that impulsivity is a statistically significant predictor of sexual risk behavior [46] and, therefore, HIV infection. Abuse of drugs as the result of self-medication for depression, in turn, may lead to infection with HIV and/or other infectious diseases, through sharing of needles or by engaging in unsafe sex while under the influence of drugs [19–21]. Many studies have reported an association of OPRM1 variants with vulnerability to heroin addiction [31, 38, 41]. Also, OPRM1 variants have been found to be associated with polysubstance abuse, including cocaine use [47] and alcoholism [39]. Cocaine use or injection drug use, reported by 27% of HIV-positive subjects and 35% of HIV-negative subjects, was associated with inconsistent condom use [48]. Many earlier studies, including studies by our group, linked drug abuse, specifically, use of psychostimulants, including cocaine, with HIV-related risk behavior [49].
We found clinical improvement in subjects of different ethnicities bearing alleles IVS1 + 1959A, IVS1 + 14123A, and IVS2 + 31A (in whites, decline of viral load over interval X; in Hispanics, increase of CD4 cell count over interval Y) and association of the same variants with HIV status. This provides evidence of the involvement of OPRM1 variants and the OPRM1 system in the response to HIV treatment. A number of studies showed that intronic variants located in miRNA binding sites may influence gene expression [50]. Testing the influence of these polymorphisms on expression of OPRM1 in cell cultures could be performed in future studies. We are now testing polymorphisms that we found to have an effect on CD4 cell count or viral load change in the current study for their association with psychiatric conditions, using the same WIHS cohort.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Dr Vadim Yuferov, for comments and a critical review of the manuscript, and Susan Russo, for editorial assistance.
We acknowledge Susan Holman from SUNY Downstate Medical Center (Brooklyn, NY) for facilitation of acquisition of specimen and related clinical data. We also acknowledge Dr Judith Cook for providing additional data for our studies. We are very grateful to the women participating in WIHS, for their time, cooperation, and support.
Disclaimer. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by the NIH (grants P60 DA05130 and MH79880 to M. J. K. and grant MH076537 to H. C.) and the National Natural Science Foundation of China (grant NSFC 30730057 to J. O.).
The WIHS project is supported in part by the National Institute of Allergy and Infectious Diseases (NIAID; grant U01 318345). Data in this manuscript were collected by the WIHS Collaborative Study Group, located at the New York City/Bronx Consortium (principal investigator, Kathryn Anastos); in Brooklyn, NY (Howard Minkoff); at the Washington, DC, Metropolitan Consortium (Mary Young); at the Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt); at the Los Angeles County/Southern California Consortium (Alexandra Levine); at the Chicago Consortium (Mardge Cohen); and at the Data Coordinating Center (Stephen Gange). The WIHS is funded by the NIAID (grants UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590) and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant UO1-HD-32632). The study is cofunded by the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Deafness and Other Communication Disorders. Funding is also provided by the National Center for Research Resources (UCSF-CTSI grant UL1 RR024131).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bond C, LaForge KS, Tian M, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–13. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev. 2005;57:1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Kreek MJ, LaForge KS. Stress responsivity, addiction, and a functional variant of the human mu-opioid receptor gene. Mol Interv. 2007;7:74–8. doi: 10.1124/mi.7.2.7. [DOI] [PubMed] [Google Scholar]
- 4.Kreek MJ. Role of a functional human gene polymorphism in stress responsivity and addictions. Clin Pharmacol Ther. 2008;83:615–8. doi: 10.1038/clpt.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y, Proudnikov D, Yuferov V, Kreek MJ. Drug-induced and genetic alterations in stress-responsive systems: implications for specific addictive diseases. Brain Res. 2010;1314:235–52. doi: 10.1016/j.brainres.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xuei X, Flury-Wetherill L, Bierut L, et al. The opioid system in alcohol and drug dependence: family-based association study. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:877–84. doi: 10.1002/ajmg.b.30531. [DOI] [PubMed] [Google Scholar]
- 7.Ducat E, Ray B, Bart G, et al. Mu-opioid receptor A118G polymorphism in healthy volunteers affects hypothalamic-pituitary-adrenal axis adrenocorticotropic hormone stress response to metyrapone. Addict Biol. 2011 doi: 10.1111/j.1369-1600.2011.00313.x. doi:10.1111/j.1369-1600.2011.00313.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bart G, LaForge KS, Borg L, Lilly C, Ho A, Kreek MJ. Altered levels of basal cortisol in healthy subjects with a 118G allele in exon 1 of the Mu opioid receptor gene. Neuropsychopharmacology. 2006;31:2313–7. doi: 10.1038/sj.npp.1301128. [DOI] [PubMed] [Google Scholar]
- 9.Oslin DW, Berrettini W, Kranzler HR, et al. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28:1546–52. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Li H, Guo L, et al. The mechanism involved in the repression of the mu opioid receptor gene expression in CEM x174 cells infected by simian immunodeficiency virus. J Leukoc Biol. 2009;85:684–91. doi: 10.1189/jlb.0908543. [DOI] [PubMed] [Google Scholar]
- 11.Finley MJ, Happel CM, Kaminsky DE, Rogers TJ. Opioid and nociceptin receptors regulate cytokine and cytokine receptor expression. Cell Immunol. 2008;252:146–54. doi: 10.1016/j.cellimm.2007.09.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steele AD, Henderson EE, Rogers TJ. Mu-opioid modulation of HIV-1 coreceptor expression and HIV-1 replication. Virology. 2003;309:99–107. doi: 10.1016/s0042-6822(03)00015-1. [DOI] [PubMed] [Google Scholar]
- 13.Grimm MC, Ben-Baruch A, Taub DD, et al. Opiates transdeactivate chemokine receptors: delta and mu opiate receptor-mediated heterologous desensitization. J Exp Med. 1998;188:317–25. doi: 10.1084/jem.188.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers TJ, Steele AD, Howard OM, Oppenheim JJ. Bidirectional heterologous desensitization of opioid and chemokine receptors. Ann N Y Acad Sci. 2000;917:19–28. doi: 10.1111/j.1749-6632.2000.tb05369.x. [DOI] [PubMed] [Google Scholar]
- 15.Szabo I, Chen XH, Xin L, et al. Heterologous desensitization of opioid receptors by chemokines inhibits chemotaxis and enhances the perception of pain. Proc Natl Acad Sci U S A. 2002;99:10276–81. doi: 10.1073/pnas.102327699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers TJ, Peterson PK. Opioid G protein-coupled receptors: signals at the crossroads of inflammation. Trends Immunol. 2003;24:116–21. doi: 10.1016/s1471-4906(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 17.Sacerdote P. Opioid-induced immunosuppression. Curr Opin Support Palliat Care. 2008;2:14–8. doi: 10.1097/SPC.0b013e3282f5272e. [DOI] [PubMed] [Google Scholar]
- 18.Donahoe RM, O'eil SP, Marsteller FA, et al. Probable deceleration of progression of Simian AIDS affected by opiate dependency: studies with a rhesus macaque/SIVsmm9 model. J Acquir Immune Defic Syndr. 2009;50:241–9. doi: 10.1097/QAI.0b013e3181967354. [DOI] [PubMed] [Google Scholar]
- 19.Novick DM, Kreek MJ, Des Jarlais DC, et al. Antibody to LAV, the putative agent of AIDS, in parenteral drug abusers and methadone-maintained patients: therapeutic, historical, and ethical aspects. NIDA Res Monogr. 1986;67:318–20. [PubMed] [Google Scholar]
- 20.Des Jarlais DC, Friedman SR, Novick DM, et al. HIV-1 infection among intravenous drug users in Manhattan, New York City, from 1977 through 1987. JAMA. 1989;261:1008–12. doi: 10.1001/jama.261.7.1008. [DOI] [PubMed] [Google Scholar]
- 21.Kreek MJ, Des Jarlais DC, Trepo CL, Novick DM, Abdul-Quader A, Raghunath J. Contrasting prevalence of delta hepatitis markers in parenteral drug abusers with and without AIDS. J Infect Dis. 1990;162:538–41. doi: 10.1093/infdis/162.2.538. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Paxton WA, Wolinsky SM, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–3. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 23.Mummidi S, Ahuja SS, Gonzalez E, et al. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–93. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 24.Smith MW, Dean M, Carrington M, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–65. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 25.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 26.Keet IP, Tang J, Klein MR, et al. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J Infect Dis. 1999;180:299–309. doi: 10.1086/314862. [DOI] [PubMed] [Google Scholar]
- 27.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelak K, Goldstein DB, Walley NM, et al. Infectious Disease Clinical Research Program HIV Working Group; National Institute of Allergy and Infectious Diseases Center for HIV/AIDS Vaccine Immunology (CHAVI). Host determinants of HIV-1 control in African Americans. J Infect Dis. 2010;201:1141–9. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook JA, Burke-Miller JK, Cohen MH, et al. Crack cocaine, disease progression, and mortality in a multicenter cohort of HIV-1 positive women. AIDS. 2008;22:1355–63. doi: 10.1097/QAD.0b013e32830507f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman DN, Feldman JG, Greenblatt R, et al. CYP1A1 genotype modifies the impact of smoking on effectiveness of HAART among women. AIDS Educ Prev. 2009;21:81–93. doi: 10.1521/aeap.2009.21.3_supp.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crystal HA, Hamon S, Randesi M, et al. A C17T polymorphism in the mu opiate receptor is associated with quantitative measures of drug use in African American women. Addict Biol. 2012;17:181–191. doi: 10.1111/j.1369-1600.2010.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapadia F, Cook JA, Cohen MH, et al. The relationship between non-injection drug use behaviors on progression to AIDS and death in a cohort of HIV seropositive women in the era of highly active antiretroviral therapy use. Addiction. 2005;100:990–1002. doi: 10.1111/j.1360-0443.2005.01098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrigan R. Measuring viral load in the clinical setting. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10(Suppl 1)):S34–40. [PubMed] [Google Scholar]
- 34.Schooley RT, Ramirez-Ronda C, Lange JM, et al. Virologic and immunologic benefits of initial combination therapy with zidovudine and zalcitabine or didanosine compared with zidovudine monotherapy. Wellcome Resistance Study Collaborative Group. J Infect Dis. 1996;173:1354–66. doi: 10.1093/infdis/173.6.1354. [DOI] [PubMed] [Google Scholar]
- 35.Connor EM, Sperling RS, Gelber R, et al. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. N Engl J Med. 1994;331:1173–80. doi: 10.1056/NEJM199411033311801. [DOI] [PubMed] [Google Scholar]
- 36.Kroslak T, Laforge KS, Gianotti RJ, Ho A, Nielsen DA, Kreek MJ. The single nucleotide polymorphism A118G alters functional properties of the human mu opioid receptor. J Neurochem. 2007;103:77–87. doi: 10.1111/j.1471-4159.2007.04738.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–24. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 38.Bart G, Heilig M, LaForge KS, et al. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry. 2004;9:547–9. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bart G, Kreek MJ, Ott J, et al. Increased attributable risk related to a functional mu-opioid receptor gene polymorphism in association with alcohol dependence in central Sweden. Neuropsychopharmacology. 2005;30:417–22. doi: 10.1038/sj.npp.1300598. [DOI] [PubMed] [Google Scholar]
- 40.Matsunaga M, Isowa T, Murakami H, et al. Association of polymorphism in the human mu-opioid receptor OPRM1 gene with proinflammatory cytokine levels and health perception. Brain Behav Immun. 2009;23:931–5. doi: 10.1016/j.bbi.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Levran O, Londono D, O'Hara K, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7:720–9. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang D, Shao C, Shao M, et al. Effect of mu-opioid receptor gene polymorphisms on heroin-induced subjective responses in a Chinese population. Biol Psychiatry. 2007;61:1244–51. doi: 10.1016/j.biopsych.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Dlugos AM, Hamidovic A, Hodgkinson C, et al. OPRM1 gene variants modulate amphetamine-induced euphoria in humans. Genes Brain Behav. 2011;10:199–209. doi: 10.1111/j.1601-183X.2010.00655.x. doi:10.1111/j.1601-183X.2010.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Kendler KS, Chen X. The mu-opioid receptor gene and smoking initiation and nicotine dependence. Behav Brain Funct. 2006;2:28. doi: 10.1186/1744-9081-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers G, Curry M, Oddy J, Pratt N, Beilby J, Wilkinson D. Depressive disorders and unprotected casual anal sex among Australian homosexually active men in primary care. HIV Med. 2003;4:271–5. doi: 10.1046/j.1468-1293.2003.00155.x. [DOI] [PubMed] [Google Scholar]
- 46.Hayaki J, Anderson B, Stein M. Sexual risk behaviors among substance users: relationship to impulsivity. Psychol Addict Behav. 2006;20:328–32. doi: 10.1037/0893-164X.20.3.328. [DOI] [PubMed] [Google Scholar]
- 47.Luo X, Kranzler HR, Zhao H, Gelernter J. Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-Americans. Am J Med Genet B Neuropsychiatr Genet. 2003;120B:97–108. doi: 10.1002/ajmg.b.20034. [DOI] [PubMed] [Google Scholar]
- 48.Wilson TE, Massad LS, Riester KA, et al. Sexual, contraceptive, and drug use behaviors of women with HIV and those at high risk for infection: results from the Women's Interagency HIV Study. AIDS. 1999;13:591–8. doi: 10.1097/00002030-199904010-00008. [DOI] [PubMed] [Google Scholar]
- 49.Novick DM, Trigg HL, Des Jarlais DC, Friedman SR, Vlahov D, Kreek MJ. Cocaine injection and ethnicity in parenteral drug users during the early years of the human immunodeficiency virus (HIV) epidemic in New York City. J Med Virol. 1989;29:181–5. doi: 10.1002/jmv.1890290307. [DOI] [PubMed] [Google Scholar]
- 50.Ying SY, Chang CP, Lin SL. Intron-mediated RNA interference, intronic microRNAs, and applications. Methods Mol Biol. 2010;629:205–37. doi: 10.1007/978-1-60761-657-3_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.