Abstract
Background
Kaposi sarcoma–associated herpesvirus (KSHV) encodes 12 pre-microRNAs that yield 25 mature microRNAs. We previously reported phylogenetic analysis of the microRNA-coding region of KSHV from Kaposi sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman disease (MCD) patients. We observed a high level of conservation for most sequences but also a divergent cluster of 5 KSHV sequences, including 2 from MCD patients.
Methods
KSHV microRNA sequences from 23 MCD patients and 7 patients with a newly described KSHV-associated inflammatory cytokine syndrome (KICS) were examined by amplification, cloning, and sequencing of a 646-bp fragment of K12/T0.7 encoding microRNA-K12-10 and microRNA-K12-12 and a 2.8-kbp fragment containing the remaining 10 pre-microRNAs.
Results
Phylogenetic analysis showed a distinct variant cluster consisting exclusively of MCD and KICS patients in all trees. Pearson χ2 analysis revealed that 40 single-nucleotide polymorphisms (SNPs) at various loci were significantly associated with MCD and KICS risk. Cluster analysis of these SNPs generated several combinations of 3 SNPs as putative indicators of MCD and KICS risk.
Conclusions
These findings show that MCD and KICS patients frequently have unusual KSHV microRNA sequences and suggest an association between the observed sequence variation and risk of MCD and KICS.
Kaposi sarcoma–associated herpesvirus (KSHV), also known as human herpesvirus 8, is the etiologic agent of Kaposi sarcoma (KS), primary effusion lymphoma (PEL), and a subtype of multicentric Castleman disease (MCD), most commonly seen in people with human immunodeficiency virus (HIV) [1–3]. KSHV-MCD is a rare B-cell lymphoproliferative disorder that mainly affects lymph nodes and other lymphoid tissue. Signs and clinical laboratory abnormalities in KSHV-MCD include fever, wasting, malaise, adenopathy, splenomegaly, cytopenia, hypoalbuminemia, hyponatremia, and elevated inflammatory markers [4, 5]. The overproduction of the inflammatory cytokine human interleukin 6 (hIL-6) and a KSHV-encoded homologue, viral interleukin 6 (vIL-6), is implicated with disease pathogenesis [6, 7]. Elevated circulating levels of these cytokines are noted with symptomatic KSHV-MCD. Other factors such as higher circulating KSHV load, as well as elevated serum interleukin 10 (IL-10) and C-reactive protein, are also associated with worsening of symptoms [8–10]. Recently, a newly characterized IL-6 associated syndrome was reported in patients with HIV and KSHV coinfection that shares many characteristics of KSHV-MCD. Patients with this syndrome, which we refer to as KSHV-associated inflammatory cytokine syndrome (KICS), display MCD-like symptoms and have elevated levels of IL-6 and KSHV but do not have pathological findings of MCD [11].
The KSHV genome is highly conserved overall, displaying about 99% sequence identity across viral strains with the exception of 2 hypervariable genes encoding tyrosine kinase-signaling proteins, K1/VIP and K15/LAMP. K1/VIP is known to show up to 35% variability in encoded amino acid sequence across viral strains and is generally used for viral subtyping [12–15]. Six KSHV subtypes have been identified (subtypes A through F) through K1/VIP sequencing, and these subtypes have been shown to display a geographical distribution [15, 16].
KSHV has 2 transcriptional programs: latency and lytic replication. During latency, viral gene expression is tightly regulated and the viral genome expresses only the minimal number of genes required to maintain latency. These genes are clustered together in a locus known as the latency transcription unit. Conversely, lytic replication involves the expression of most viral genes and results in the production of progeny virions. In KS tumors and PELs, the predominant proportion of cells is latently infected with KSHV; however, up to 5% of cells undergo lytic replication at any time [17]. In KSHV-MCD, however, a high percentage of the B-cell plasmablasts express vIL-6, and other viral lytic gene expression is also relatively widespread [17]. This observation suggests that KSHV lytic genes, and especially vIL-6, may play a greater role in KSHV-MCD than in KS and PEL.
MicroRNAs are short RNA molecules about 19–24 nucleotides in length that have been shown to play a regulatory role in posttranscriptional gene expression. MicroRNAs are initially transcribed from the human genome, as well as from some viral genomes, resulting in primary microRNAs (pri-microRNAs), which are subsequently processed by Drosha resulting in approximately 60-nt hairpin pre-microRNAs. These are exported out of the nucleus and the hairpin is removed by Dicer, leaving a double-stranded RNA duplex. One of the 2 strands is incorporated into the RNA-induced silencing complex, which translocates to its respective messenger RNA (mRNA) target.
The KSHV genome encodes 12 pre-microRNA sequences that have been shown to yield 25 mature microRNAs [18–21]. In total, 10 of the 12 pre-microRNAs are encoded between the v-FLIP (ORF71) and K12/T0.7 genes in the latency transcriptional unit. The remaining 2 microRNAs are present in the coding region of the K12/T0.7 gene. MicroRNAs have been shown to play important roles in diverse cellular processes, including apoptosis, cellular differentiation, cell growth, and many others; however, the targets and functions of KSHV microRNAs are not currently fully known [22, 23]. Targets that have been identified indicate that KSHV microRNAs play a role in maintaining viral latency, evading the host immune system, and controlling lytic replication [24–30].
MicroRNA polymorphisms (miR-polymorphisms) and microRNA single-nucleotide polymorphisms (miR-SNPs) have been shown to affect microRNA processing and function. The miR-polymorphisms and miR-SNPs have been associated with disease risk in heart disease and cancer, including breast cancer and chronic lymphocytic leukemia [31–33].
Our previous study examining the KSHV microRNA sequences in clinical samples from subjects from diverse geographical locations, as well as from different disease groups, showed a high degree of conservation [34]. However, phylogenetic analysis of the microRNA cluster region and T0.7 gene showed a distinct clustering of sequences in patients with KSHV-associated malignancies including 2 sequences from MCD patients. In our subsequent case-control study of AIDS-KS we showed an association between SNPs in the microRNA cluster region and T0.7 gene with KS disease risk [35]. Given the proposed role of KSHV microRNA in maintaining viral latency and evading the immune response and the role of other microRNAs in regulating the levels of cytokines including IL-6, we hypothesized that polymorphisms in KSHV-encoded microRNAs may differentiate patients with KSHV-associated inflammatory syndromes (MCD or KICS) from HIV/KSHV-coinfected patients with no known history of inflammatory symptoms [36]. To expand on our previous observations, we sought to phylogenetically examine the KSHV microRNA-encoding regions in samples from additional KSHV-MCD patients, as well as KICS patients, to investigate a possible association between KSHV-encoded sequence variation and KSHV-MCD/KICS risk.
MATERIALS AND METHODS
Patient Population
MCD and KICS clinical samples were collected from patients enrolled in the National Cancer Institute Institutional Review Board–approved HIV and AIDS Malignancy Branch protocols that allowed for genetic evaluation of KSHV. All patients provided written informed consent. MCD patients all had pathologic diagnosis of KSHV-MCD, and KICS patients had MCD-like symptoms and laboratory abnormalities—including elevated hIL-6, vIL-6, KSHV load, and IL-10—but no evidence of KSHV-MCD [11]. Patients with concurrent KS and/or PEL were nonetheless categorized into these 2 primary groups. In total, 5 of the 7 KICS patients included were those from the original report of this syndrome [11]. All MCD patients and KICS patients were HIV coinfected with the exception of 1 KICS patient. Biospecimens from patients with HIV and KSHV coinfection, but no known history of KSHV-MCD or KICS, were obtained as previously described, and patients were classified as AIDS-KS cases if they had clinical KS or as KSHV control patients if they had no clinical KS [35].
Biospecimen Collection
For MCD and KICS patients, saliva and whole blood specimens were obtained. Saliva was collected by having patients rinse their mouth with mouthwash and subsequently discharge this fluid into a 50-mL conical tube. Cell pellets were separated by centrifugation at 17 000g for 5 minutes. The resulting cell pellets were separated from supernatants and stored at −80°C until use. Lymphocytes were separated from whole blood using standard Ficoll-Paque procedures (MP Biomedicals). Cell pellets were made (1–2 million cells per pellet) and frozen at −80°C until use. DNA extractions were performed using the Qiagen DNA Blood mini kit according to the manufacturer's protocol. Viral load determinations were performed using a quantitative polymerase chain reaction (PCR) assay to identify samples with detectable KSHV DNA for sequencing [37, 38].
Amplification, Cloning, and Sequencing of KSHV MicroRNA Cluster Region, T0.7, and K1
Sequences were amplified via a nested PCR amplification strategy for K1, T0.7, and the microRNA cluster regions as described elsewhere [34]. Direct sequencing was performed by using the inner nested PCR primers for each respective region/fragment. If amplification yields were inadequate, cloning was performed by using P-GEM T-Easy vector system (Promega) to provide amplified templates for sequencing. Overlapping forward and reverse sequencing reactions were performed for each region/fragment sequenced, and the sequence reads were collected using an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). All forward and reverse sequencing reads for K1 and T0.7, as well as the entire microRNA cluster region, were assembled using DNA Baser (version 2.91.5, Heracle Software). Each forward and reverse sequence collected from each patient and the identity of each gene region were confirmed using BLAST.
Phylogenetic Analysis
Sequence alignments for the K1, T0.7, and microRNA cluster region were performed using ClustalX (version 1.83). Nucleic acid sequences were used for T0.7 and the microRNA cluster region. K1 sequences were translated to amino acid sequences prior to alignment. Relevant sequences were obtained from the GenBank database as well as sequences from our case-control study of AIDS-KS [35]. Each sequence region was imported into GeneDoc (version 2.7.000) and trimmed to equal lengths. Bootstrap neighbor-joining analysis was performed on the alignments with MEGA (version 4.0.2) for K1, T0.7, the entire microRNA cluster region, and each of the 3 microRNA cluster region segments.
Statistical Analysis of SNPs
The association of each individual SNP with clinical status (KSHV-MCD/KICS vs AIDS-KS vs control) was ascertained using exact nonparametric χ2 tests [39]. As multiple comparisons were performed, a Bonferroni adjustment was done, and P < .05 was determined to be of marginal significance, P < .005 to be approaching significance, and P < .0005 to be significant. Classification tree analysis was performed employing all 77 SNPs to identify combinations of genetic loci that could be indicators of KSHV-MCD and KICS [40]. Classification trees were constructed using a recursive partitioning algorithm that identified the splits in the data, based on wild-type or SNP categorizations at certain genetic loci to form subsets that would best predict KSHV-MCD and KICS. Not every sample was successfully sequenced at all loci, so multiple imputations were performed to minimize potential bias due to missing values. Ten sets of multiple random imputations were generated using Markov chain Monte Carlo (MCMC) simulation to estimate the parameters of a log-linear model using the S-PLUS missing library software [41–43]. Logit models were then fitted to each of the 10 complete data sets that were created using multiple imputations. The parameters of the models were estimated using Bayesian statistical methods and the WinBUGS 14 software [44]. Vague, but proper, prior distributions were used to fit the logit models and the log-linear imputation models. These prior distributions had very large variances so that they were essentially uninformative but were actual proper distributions and hence ensured successful convergence of the MCMC simulation algorithm. Conditional probabilities of disease status were then combined from the 10 complete data set models for each SNP configuration [41–43].
RESULTS
Patients
The primary analysis of KSHV microRNA phylogenetic diversity in patients with KSHV-MCD or KICS is based on 23 MCD samples, including 2 patients whose sequences had been previously reported and 7 KICS samples. AIDS-KS cases (21) and KSHV controls (14) included 35 cases described elsewhere [34, 35]. Patient characteristics are listed in Table 1.
Table 1.
Summary of Multicentric Castleman Disease and KSHV-Associated Inflammatory Cytokine Syndrome Patient Characteristics and Phylogenetic Data
| Sample ID | Diagnosis | Ethnicity | Birthplace | Sex | PEL Diagnosis | History of Clinical KS | K1 | T0.7 | miRNAs 1–5 | miRNAs 6, 11, 7 | miRNAs 7–9 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MCD-01 | MCD | White | US | M | N | N | A | C | N/D | N/D | N/D |
| MCD-02 | MCD | White | US | M | N | Y | N/D | F | F | F | F |
| MCD-03 | MCD | Hispanic | Nicaragua | M | N | N | B | B | B | B | B |
| MCD-04 | MCD | Hispanic | Puerto Rico | M | N | Y | C | C | C | C | C |
| MCD-05 | MCD | Black | US | M | N | N | A | C | C | C | C |
| MCD-06 | MCD | Black | US | M | N | Y | N/D | B | B | B | N/D |
| MCD-07 | MCD | White | US | M | N | Y | F | F | F | F | F |
| MCD-08 | MCD | White | US | M | N | N | C | C | C | C | C |
| MCD-09 | MCD | White | US | M | N | Y | A | C | C | C | C |
| MCD-10 | MCD | Black | US | M | N | N | A5 | A | A | A | N/D |
| MCD-11 | MCD | African | Sierra Leone | M | N | Y | B | B | B | B | B |
| MCD-12 | MCD | African | Cameroon | M | N | N | N/D | B | B | B | B |
| MCD-13 | MCD | White | US | M | Y, germinotropic lymph | Y | A | F | F | F | F |
| MCD-14 | MCD | Black | US | M | N | Y | A | C | C | C | C |
| MCD-15 | MCD | Black | Liberia | F | N | Y | B | B | B | B | B |
| MCD-16 | MCD | White | US | M | N | N | A | C | C | C | N/D |
| MCD-17 | MCD | Black | Malawi | F | N | Y | N/D | B | F | B | N/D |
| MCD-18 | MCD | White | US | M | N | Y | A | C | C | C | N/D |
| MCD-19 | MCD | White | US | M | N | N | A | C | C | C | C |
| MCD-20 | MCD | Black | Nigeria | M | N | N | A5 | A | A | A | N/D |
| MCD-21 | MCD | White | US | M | N | Y | A | N/D | C | N/D | N/D |
| MCD-22 | MCD | White | US | M | Y | Y | A | C | C | C | C |
| MCD-23 | MCD | Hispanic | Colombia | M | N | N | C | F | F | F | F |
| KICS-01 | KICS | Black | Kenya | M | N | Y | B | F | F | F | F |
| KICS-02 | KICS | Hispanic | Puerto Rico | M | N | Y | N/D | C | C | C | C |
| KICS-03 | KICS | Black | US | M | N | N | N/D | A | A | C | A |
| KICS-04 | KICS | Black | US | M | N | Y | A | A | A | C | A |
| KICS-05 | KICS | Black | Cameroon | M | N | Y | A5 | B | B | B | B |
| KICS-06 | KICS | White | US | M | Y | Y | A | C | N/D | N/D | N/D |
| KICS-07 | KICS | White | US | F | N | Y | C | C | C | C | C |
Subtype designation for K1 and T0.7 are based on traditional subtype designations and previously described precedents for the miRNA cluster region segments [34].
Abbreviations: KICS, KSHV-associated inflammatory cytokine syndrome; KS, Kaposi sarcoma; KSHV, Kaposi sarcoma–associated herpesvirus; MCD, multicentric Castleman disease; miRNA, microRNA; N/D, not determined; PEL, primary effusion lymphoma.
Subtype Determination of Viral Strains
Four K1 subtypes were represented among the MCD and KICS patients for whom K1 sequences could be obtained (Figure 1). Of the 24 K1 amino acid sequences obtained from MCD and KICS patients, 4 (16.67%) were subtype C; 12 (50%) were subtype A, including 3 (12.5%) subtype A5; 4 (16.67%) were subtype B, and 1 (4.16%) was subtype F. The distribution of subtypes was similar between the MCD and KICS patients. KS cases and KSHV controls were subtype A (31.2%) or subtype C (68.8%).
Figure 1.
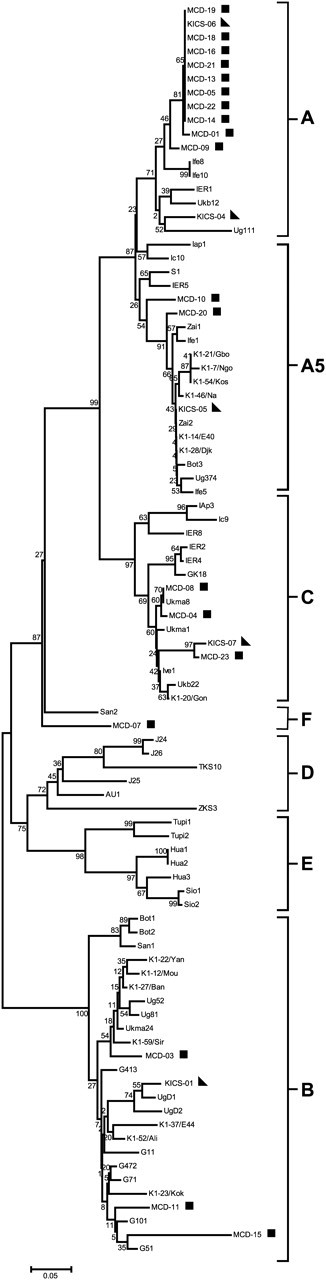
K1 subtype analysis of Kaposi sarcoma–associated herpesvirus (KSHV)–multicentric Castleman disease (MCD) and KSHV-associated inflammatory cytokine syndrome (KICS) patients. All K1 sequences collected were translated prior to analysis and were analyzed by bootstrap neighbor-joining analysis with relevant sequences from GenBank. In total, 100 bootstrap replicates were conducted to determine the topology of the trees using MEGA (version 4.0.2). KSHV-MCD patients are indicated by a square and KICS patients are indicated by a triangle. AIDS–Kaposi sarcoma cases and controls are unmarked.
Phylogenetic Analysis of T0.7 and MicroRNA Cluster Region
Phylogenetic analysis included sequences from KSHV-MCD, KICS, and AIDS-KS patients and KSHV controls from whom sequences could be obtained. Figure 2 shows the phylogenetic analysis of the microRNA cluster region showing a conserved branch (subtypes A and C) and a variant cluster (subtypes B and F) consisting exclusively of sequences from KSHV-MCD/KICS patients. Similar trees were obtained with sequences for which only partial sequences were obtained (Supplementary Figure 1) and for T0.7 (Figure 3). Subtype designation was based on previously reported precedents [34]. In addition to the sequences making up the variant cluster, some miRNA sequences from KSHV-MCD and KICS patients were also present in the conserved branch. Representative microRNA cluster sequences of determined subtypes are available from GenBank (accession numbers JQ029277–JQ029280).
Figure 2.
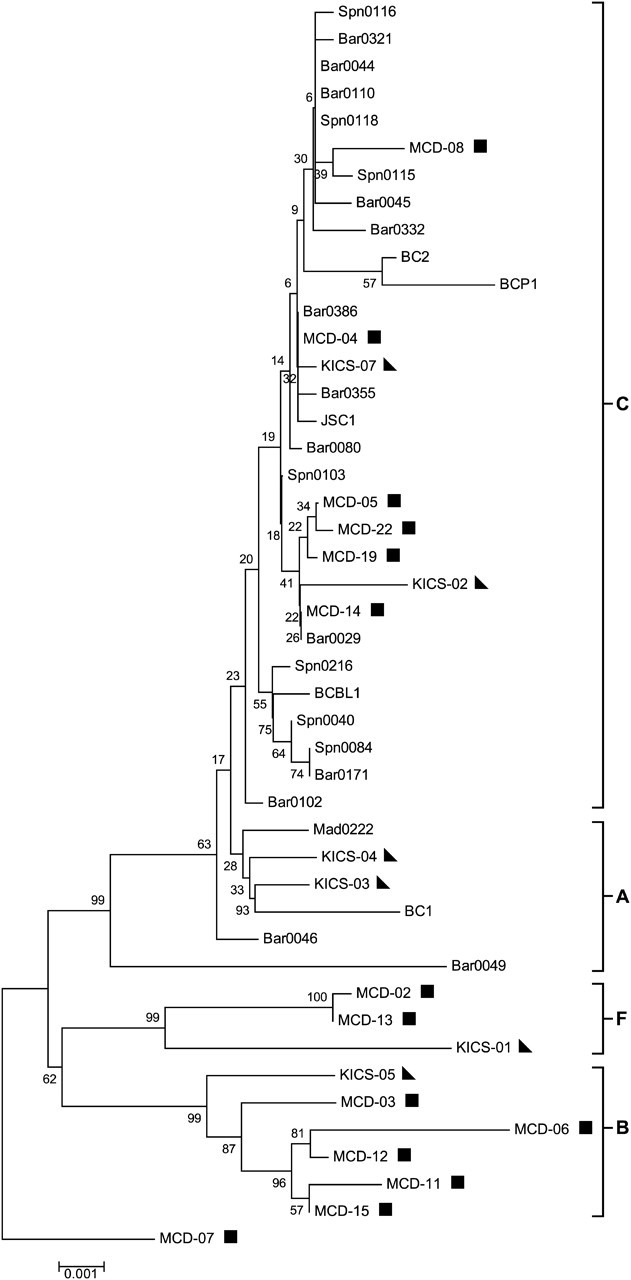
MicroRNA cluster region phylogenetic tree. MicroRNA cluster region phylogenies were determined using bootstrap neighbor-joining analysis and rooted with outlier patient MCD-07 using MEGA (version 4.0.2). Subtype nomenclature is consistent with previous reports [1, 35]. Kaposi sarcoma–associated herpesvirus (KSHV)–multicentric Castleman disease (MCD) patients are indicated by a square, and KSHV-associated inflammatory cytokine syndrome (KICS) patients are indicated by a triangle. AIDS–Kaposi sarcoma cases and controls are unmarked. Conserved sequences are shown as subtypes A and C and variant sequences are shown as subtypes F and B.
Figure 3.
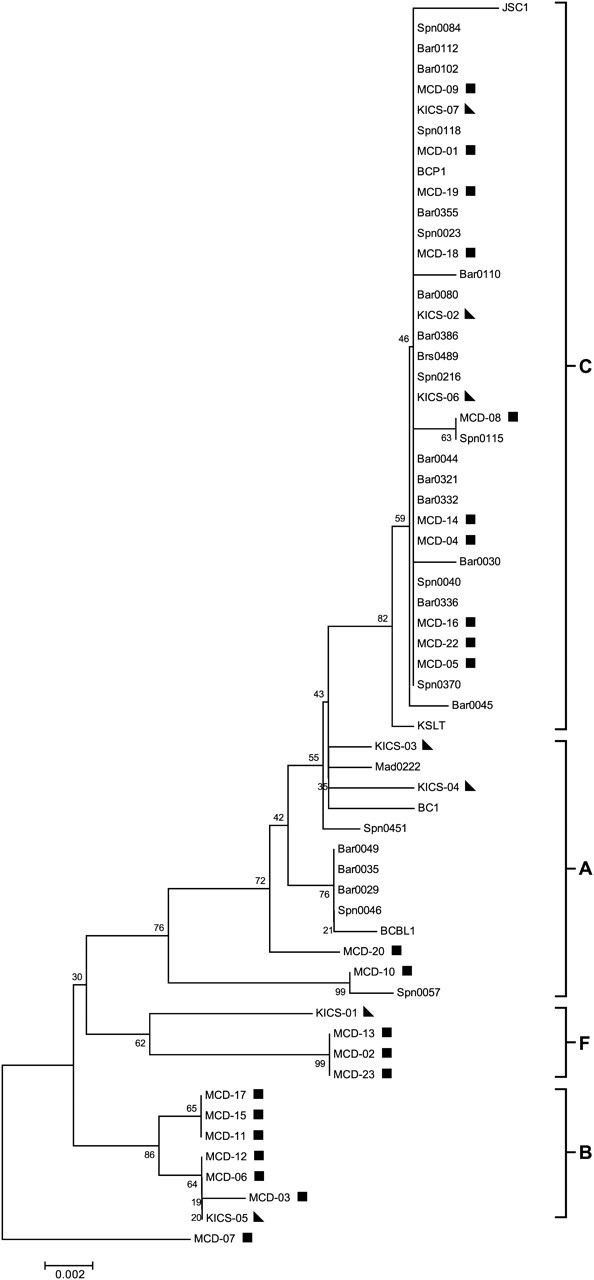
T0.7/K12 Phylogenetic tree. Subtype nomenclature for the present trees are consistent with that established for T0.7/K12. Neighbor-joining bootstrap analysis was performed using 100 bootstrap replicates and rooted with outlier patient MCD-07 using MEGA (version 4.0.2). Kaposi sarcoma–associated herpesvirus (KSHV)–multicentric Castleman disease (MCD) patients are indicated by a square, and KSHV-associated inflammatory cytokine syndrome (KICS) patients are indicated by a triangle. AIDS–Kaposi sarcoma cases and controls are unmarked. Conserved sequences are shown as subtypes A and C and variant sequences are shown as subtypes F and B.
Statistical Analysis of SNPs in the T0.7 and MicroRNA Cluster Region
The prevalence of identified SNPs at each position in the microRNA coding region within each of the 4 disease groups was calculated. To rule out PCR artifacts, only SNPs present in ≥2 samples were included in the analysis. Sequencing of the 2.8-kbp microRNA cluster and 646-bp T0.7 region identified 77 SNPs. The percentage of patients within each disease group with SNPs at each locus was examined (Supplementary Figure 2). Overall, a higher percentage of KSHV-MCD and KICS patients had SNPs in identified loci than did the AIDS-KS patients and KSHV controls. In 15 of the 18 (83.3%) identified SNP loci in the T0.7 region, KSHV-MCD and KICS patients showed a higher percentage of SNPs than did AIDS-KS cases and controls. The same pattern was seen in the microRNA cluster region: a higher percentage of KSHV-MCD and KICS patients had SNPs in 10 of 10 loci (100%) in the fragment coding for microRNAs 1–5, 16 of 16 loci (100%) in the fragment coding for microRNAs 6, 11, and 7, and 29 of 31 loci (93.5%) in the fragment coding for microRNAs 7–9.
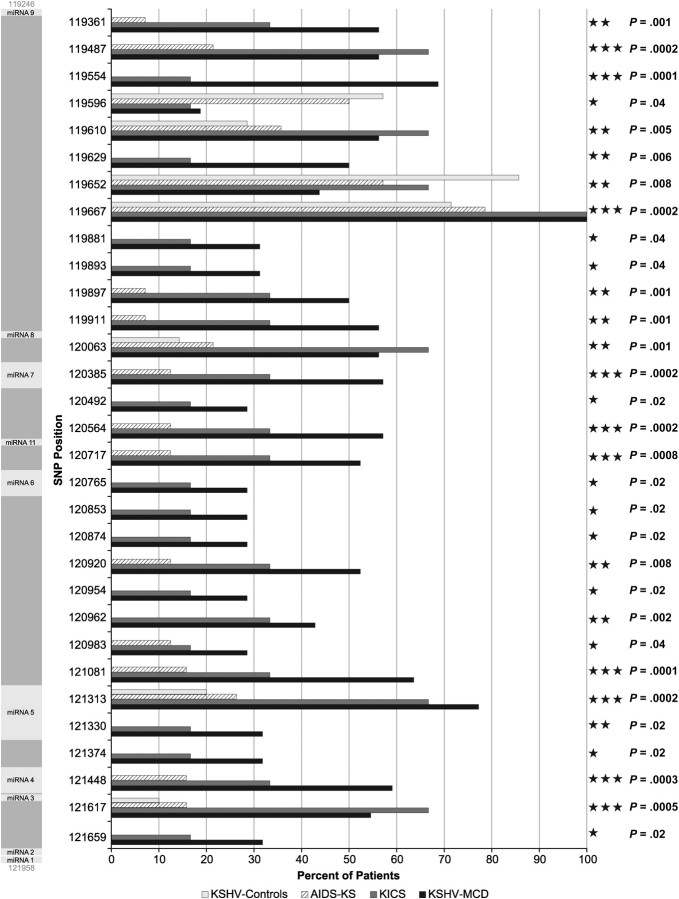
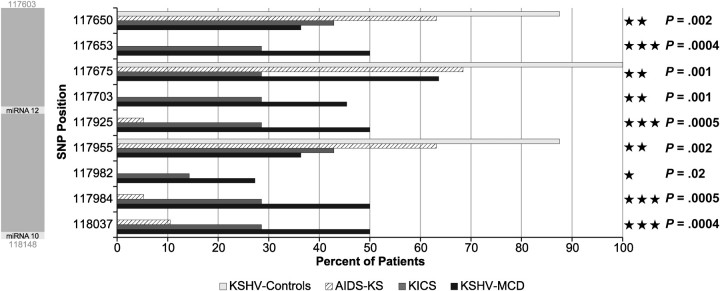
Pearson χ2 test and classification tree analysis were performed on the collected sequence data. The individual SNPs that differentiate KSHV-MCD/KICS from AIDS-KS cases and KSHV controls with at least marginal significance are displayed in Figures 4 and 5. In total, 40 of 77 SNPs were at least of marginal significance (P < .05), with 14 approaching significance (P < .005), and 14 being significant (P < .0005).
Figure 4.
Summary of significant single-nucleotide polymorphisms in microRNA cluster region. Kaposi sarcoma–associated herpesvirus (KSHV)–multicentric Castleman disease (MCD) and KSHV-associated inflammatory cytokine syndrome (KICS) patients were compared to AIDS–Kaposi sarcoma cases and controls using Pearson χ2 test. Because multiple comparisons were performed, P < .05 was deemed to be a threshold for marginal significance, P < .005 to be approaching significance, and P < .0005 to be significant. P values are listed to the right. The microRNA cluster region is shown to the left as a reference, and locations of KSHV pre-microRNAs are indicated by light gray shading.
Figure 5.
Summary of significant single-nucleotide polymorphisms (SNPs) in T0.7/K12 region. SNPs identified in the T0.7/K12 region were analyzed in the same manner as those identified in the microRNA cluster region. The P values are listed to the right and the T0.7/K12 region is shown to the left with corresponding microRNAs in light gray shading.
Classification tree analysis was next used to determine combinations of SNPs that predict KSHV-MCD and KICS with the precedence that a combination of SNPs may affect microRNA biogenesis. This exploratory model displayed 4 combinations of 6 SNPs as putative indicators of KSHV-MCD and KICS and is listed in Table 2. Classification trees are shown in Supplementary Figure 3. Logit model analysis determined the probabilities of being KSHV-MCD or KICS vs AIDS-KS or KSHV control from the combinations of SNPs identified from classification tree analysis (Table 2).
Table 2.
Logit Model Analysis of Putative Combinations of Single-Nucleotide Polymorphisms Predicting Kaposi Sarcoma–Associated Herpesvirus (KSHV)–Multicentric Castleman Disease/KSHV-Associated Inflammatory Cytokine Syndrome
| Loci Combination |
Conditional Probability of KSHV-MCD/KICS |
||||||
|---|---|---|---|---|---|---|---|
| 117650 | 117675 | 117984 | 119585 | 119630 | 120983 | P Value | (95% CI) |
| WT | WT | SNP | .00% | (.0–100.0) | |||
| SNP | WT | SNP | .00% | (.0–100.0) | |||
| WT | WT | WT | 2.50% | (.1–32.8) | |||
| WT | WT | SNP | 2.70% | (.0–100.0) | |||
| WT | WT | WT | 7.40% | (1.4–31.4) | |||
| WT | WT | WT | 9.70% | (2.3–32.4) | |||
| WT | WT | WT | 11.80% | (4.4–28.1) | |||
| SNP | WT | WT | 15.80% | (4.0–45.7) | |||
| SNP | WT | WT | 18.40% | (5.1–48.4) | |||
| WT | WT | SNP | 20.80% | (1.5–81.9) | |||
| WT | SNP | WT | 26.80% | (6.4–66.2) | |||
| SNP | SNP | WT | 34.10% | (15.8–58.9) | |||
| SNP | WT | WT | 47.40% | (6.8–91.8) | |||
| SNP | SNP | SNP | 50.00% | (1.6–98.4) | |||
| SNP | SNP | SNP | 59.90% | (13.2–93.6) | |||
| SNP | SNP | WT | 68.80% | (8.4–98.2) | |||
| SNP | WT | WT | 69.20% | (26.7–93.3) | |||
| SNP | SNP | WT | 74.40% | (27.0–95.8) | |||
| SNP | SNP | SNP | 89.20% | (31.8–99.3) | |||
| SNP | WT | SNP | 89.70% | (31.5–99.4) | |||
| WT | SNP | WT | 92.70% | (47.9–99.4) | |||
| WT | WT | SNP | 93.20% | (44.9–99.6) | |||
| WT | SNP | SNP | 100.00% | (2.9–100.0) | |||
| SNP | SNP | WT | 100.00% | (.0–100.0) | |||
| SNP | WT | SNP | 100.00% | (4.4–100.0) | |||
| WT | SNP | WT | 100.00% | (.7–100.0) | |||
| WT | SNP | SNP | 100.00% | (10.0–100.0) | |||
| SNP | SNP | SNP | 100.00% | (10.0–100.0) | |||
| WT | SNP | SNP | 100.00% | (3.6–100.0) | |||
| WT | WT | SNP | 100.00% | (.4–100.0) | |||
Abbreviations: CI, confidence interval; KICS, KSHV-associated inflammatory cytokine syndrome; KSHV, Kaposi sarcoma–associated herpesvirus; MCD, multicentric Castleman disease; SNP, single-nucleotide polymorphism; WT, wild type.
DISCUSSION
We previously reported phylogenetic analysis of the KSHV microRNA encoding region from KS, PEL, and KSHV-MCD patients and showed a distinct phylogenetic branch including 2 MCD patients distinct from a conserved branch [34]. M-fold prediction has also shown that SNPs in the pre-microRNA sequence can have dramatic effects on secondary structure [34]. We also reported that SNPs in the microRNA-coding region of KSHV are associated with disease risk in AIDS-KS patients [35]. SNPs in human microRNAs as well as their respective targets have been shown to play a role in disease pathogenesis [45, 46]. In the current study we extended our previous observations to include a greater number of KSHV-MCD patients, as well as patients with the newly described KICS.
To evaluate the association with individual SNPs or combinations of SNPs in the KSHV microRNA-coding region with KSHV-MCD or KICS, both phylogenetic and statistical analyses were performed. These analyses showed significant differences in the KSHV microRNA-coding region of many of the KSHV-MCD and KICS sequences, which differentiated them from AIDS-KS cases and KSHV controls. Consistent with phylogenetic analysis, KSHV-MCD and KICS patients showed an overall higher number of polymorphisms in sequences of the microRNA-coding region than AIDS-KS cases and controls, with SNPs being more common in patients with KSHV-MCD or KICS in all of the 14 statistically significant SNP loci. To our knowledge, this is the first study linking KSHV viral genetic variation to KSHV-MCD or KICS risk.
The overall phylogenetic pattern seen in samples from KSHV-MCD and KICS patients is rather striking. In all trees examined, 2 major branches were observed, a conserved group and a variant group. All sequences from AIDS-KS cases and KSHV controls branched in the conserved group and every variant branch observed consisted exclusively of KSHV-MCD and KICS patients. Although there were sequences from KSHV-MCD and KICS patients present in the conserved group, a majority of them branched in the variant group away from the AIDS-KS cases and KSHV controls in all trees examined (Table 2). In addition, sequences from patients who were Hispanic, black, or of African origin were more likely to cluster in the variant group; however, all ethnicities were present in both the conserved and variant groups.
Analysis of the sequences from the KSHV-MCD and KICS patients showed a higher frequency of polymorphisms in their microRNA coding region compared to that from AIDS-KS cases and KSHV controls. This was evident not only by the phylogenetic pattern described above but also by the presence of SNPs in the microRNA coding region as well as the higher percentage of KSHV-MCD and KICS patients with these SNPs. Notably, for 72 of the 77 SNP loci, an SNP was present at a higher percentage in sequences from KSHV-MCD and KICS patients. This also held true for all of the statistically significant SNPs determined by Pearson χ2 analysis. Furthermore, although the region encoding microRNAs 6, 11, and 7 was confirmed to be the most conserved of the microRNA cluster region, 11 of the 16 SNPs detected in this region were exclusively observed in the sequences from KSHV-MCD and KICS patients. Fourteen other SNPs, including 4 present in the T0.7 region, were shown to be statistically significantly associated with KSHV-MCD and KICS risk.
Few targets of KSHV-encoded microRNAs have been determined. Interestingly, several microRNAs for which targets have been determined have observed polymorphisms in or near their pre-microRNA sequences. For example, miR-K12-9, miR-K12-7, and miR-K12-5 are involved in downregulating RTA (which plays an important role in the latent-lytic switch), and miR-K12-7 is involved in the expression of IL-6 and IL-10 [26, 28, 47, 48]. The higher prevalence of polymorphisms in this area of the KSHV genome in KSHV-MCD and KICS patients may be playing a role in disease by altering expression of the microRNAs and causing changes in downstream gene regulation. This may help explain the increased KSHV loads and elevated inflammatory cytokine levels observed in KSHV-MCD and KICS patients.
A majority of the SNPs detected in examined sequences in this study were present in the pri-microRNA sequence. Several studies have shown that SNPs present in the pri-microRNA sequence lead to altered levels of mature microRNA expression. Expression of the human microRNAs let-7e and miR-16 has been shown to be altered due to SNPs present in their respective pri-microRNA sequences [45, 49]. Furthermore, it has been demonstrated that pri-microRNAs can fold into compact tertiary structures, which may sequester individual pre-microRNA sequences and affect their processing [50]. Presumably, SNPs may alter the tertiary structure of pri-microRNA structures in KSHV and affect KSHV microRNA processing and biogenesis. Further research into the processing of KSHV pri-microRNA and downstream mature microRNA expression is warranted.
There are several limitations to this study. For some patients with low KSHV viral load, complete microRNA sequences could not be obtained. The absence of these sequences may have led to conservative bias in the statistical analysis. Further sample size limitations are due to the rare nature of KSHV-MCD and the recent characterization of KICS. Yet, despite a small sample size, the presence of a much higher frequency of polymorphism in KSHV-MCD and KICS patients’ microRNA sequences compared to those of AIDS-KS cases and KSHV controls suggests that the potential importance of these SNPs cannot be disregarded.
A distinguishing characteristic of KSHV-MCD and KICS is a higher KSHV viral load in the peripheral blood as well as the expression of genes involved in lytic replication. Recent reports have shown that when KSHV microRNA processing is disrupted, lytic gene expression is induced [29]. Additionally, the observation that an SNP immediately preceding KSHV miR-K12-5 increases expression levels of this microRNA also highlights how alteration in microRNA processing can affect microRNA expression. The presence of higher viral loads in the peripheral blood of MCD and KICS patients, as well as the presence of a higher degree of polymorphisms in the microRNA-coding region, suggests that variations in the sequence of KSHV-encoded microRNA region may play a role in the pathogenesis of KSHV-MCD and KICS.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Tammy Schroyer for help producing the figures, Sasha McClain and Marshall Thompson for technical assistance in the lab, and Rachel Bagni for helpful bioinformatic and phylogenetic discussions. We also especially thank the patients who participated in this study.
Author contributions. A. R., V. M., and D. W. designed the study. A. R. and V. M. collected the data. T. U., K. W., K. A., M. N. P., R. F. L., and R. Y. cared for patients. R. L. and N. L. did statistical analysis. A. R. and D. W. wrote the manuscript.
Financial support. This work was supported by the National Cancer Institute, National Institutes of Health (contract number HHSN261200800001E).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–9. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–91. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Soulier J, Grollet L, Oksenhendler E, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–80. [PubMed] [Google Scholar]
- 4.Oksenhendler E, Duarte M, Soulier J, et al. Multicentric Castleman's disease in HIV infection: a clinical and pathological study of 20 patients. AIDS. 1996;10:61–7. [PubMed] [Google Scholar]
- 5.Uldrick TS, Polizzotto MN, Aleman K, et al. High-dose zidovudine plus valganciclovir for Kaposi sarcoma herpesvirus-associated multicentric Castleman disease: a pilot study of virus-activated cytotoxic therapy. Blood. 2011;117:6977–86. doi: 10.1182/blood-2010-11-317610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt SJ, Bodine DM, Dunbar CE, Nienhuis AW. Dysregulated interleukin 6 expression produces a syndrome resembling Castleman's disease in mice. J Clin Invest. 1990;86:592–9. doi: 10.1172/JCI114749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoki Y, Jaffe ES, Chang Y, et al. Angiogenesis and hematopoiesis induced by Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93:4034–43. [PubMed] [Google Scholar]
- 8.Oksenhendler E, Carcelain G, Aoki Y, et al. High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric Castleman disease in HIV-infected patients. Blood. 2000;96:2069–73. [PubMed] [Google Scholar]
- 9.Aoki Y, Yarchoan R, Wyvill K, Okamoto S, Little RF, Tosato G. Detection of viral interleukin-6 in Kaposi sarcoma-associated herpesvirus-linked disorders. Blood. 2001;97:2173–6. doi: 10.1182/blood.v97.7.2173. [DOI] [PubMed] [Google Scholar]
- 10.Choi J, Means RE, Damania B, Jung JU. Molecular piracy of Kaposi's sarcoma associated herpesvirus. Cytokine Growth Factor Rev. 2001;12:245–57. doi: 10.1016/s1359-6101(00)00029-0. [DOI] [PubMed] [Google Scholar]
- 11.Uldrick TS, Wang V, O'Mahony D, et al. An interleukin-6-related systemic inflammatory syndrome in patients co-infected with Kaposi sarcoma-associated herpesvirus and HIV but without multicentric Castleman disease. Clin Infect Dis. 2010;51:350–8. doi: 10.1086/654798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook PM, Whitby D, Calabro ML, et al. Variability and evolution of Kaposi's sarcoma-associated herpesvirus in Europe and Africa. International Collaborative Group. AIDS. 1999;13:1165–76. doi: 10.1097/00002030-199907090-00004. [DOI] [PubMed] [Google Scholar]
- 13.Lacoste V, Judde JG, Briere J, et al. Molecular epidemiology of human herpesvirus 8 in Africa: both B and A5 K1 genotypes, as well as the M and P genotypes of K14.1/K15 loci, are frequent and widespread. Virology. 2000;278:60–74. doi: 10.1006/viro.2000.0629. [DOI] [PubMed] [Google Scholar]
- 14.Meng YX, Spira TJ, Bhat GJ, et al. Individuals from North America, Australasia, and Africa are infected with four different genotypes of human herpesvirus 8. Virology. 1999;261:106–19. doi: 10.1006/viro.1999.9853. [DOI] [PubMed] [Google Scholar]
- 15.Zong JC, Ciufo DM, Alcendor DJ, et al. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–70. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayward GS. KSHV strains: the origins and global spread of the virus. Semin Cancer Biol. 1999;9:187–99. doi: 10.1006/scbi.1998.0116. [DOI] [PubMed] [Google Scholar]
- 17.Staskus KA, Zhong W, Gebhard K, et al. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–9. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:9301–5. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–50. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YT, Kincaid RP, Arasappan D, Dowd SE, Hunicke-Smith SP, Sullivan CS. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA. 2010;16:1540–58. doi: 10.1261/rna.1967910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41:130–4. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–85. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–5. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei X, Bai Z, Ye F, et al. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat Cell Biol. 2010;12:193–9. doi: 10.1038/ncb2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J Virol. 2010;84:2697–706. doi: 10.1128/JVI.01997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu CC, Li Z, Chu CY, et al. MicroRNAs encoded by Kaposi's sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. 2010;11:784–90. doi: 10.1038/embor.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abend JR, Uldrick T, Ziegelbauer JM. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J Virol. 2010;84:12139–51. doi: 10.1128/JVI.00884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman AE, Zheng T, Yi C, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–7. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B, Rao L, Peng Y, et al. Common genetic polymorphisms in pre-microRNAs were associated with increased risk of dilated cardiomyopathy. Clin Chim Acta. 2010;411:1287–90. doi: 10.1016/j.cca.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 33.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 34.Marshall V, Parks T, Bagni R, et al. conservation of virally encoded microRNAs in Kaposi sarcoma-associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J Infect Dis. 2007;195:645–59. doi: 10.1086/511434. [DOI] [PubMed] [Google Scholar]
- 35.Marshall V, Martro E, Labo N, et al. Kaposi sarcoma (KS)-associated herpesvirus microRNA sequence analysis and KS risk in a European AIDS-KS case control study. J Infect Dis. 2010;202:1126–35. doi: 10.1086/656045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. Kaposi's sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J Virol. 2011;85:2620–30. doi: 10.1128/JVI.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan CC, Miley W, Waters D. A quantification of human cells using an ERV-3 real time PCR assay. J Virol Methods. 2001;91:109–17. doi: 10.1016/s0166-0934(00)00244-5. [DOI] [PubMed] [Google Scholar]
- 38.de Sanjose S, Marshall V, Sola J, et al. Prevalence of Kaposi's sarcoma-associated herpesvirus infection in sex workers and women from the general population in Spain. Int J Cancer. 2002;98:155–8. doi: 10.1002/ijc.10190. [DOI] [PubMed] [Google Scholar]
- 39.Mehta CR, Patel NR. A network algorithm for performing Fisher's exact test in rxc contingency tables. J Am Stat Assoc. 1983;78:427–34. [Google Scholar]
- 40.Breiman L, Friedman J, Olshen R, Stone C. Classification and regression trees. New York, NY: Chapman and Hall; 1984. [Google Scholar]
- 41.Little RJA, Rubin DB. Statistical analysis with missing data. Hoboken, NJ: John Wiley and Sons, Inc; 1987. [Google Scholar]
- 42.Schafer JL. Analysis of incomplete multivariate data. London: Chapman and Hall; 1997. [Google Scholar]
- 43.Schimert J, Scafer JL, Hesterberg T, Fraley C, Clarkson DB. Analyzing data with missing values in S-Plus. Seattle, WA: Insightful Corporation; [Google Scholar]
- 44.Spiegelhalter D, Thomas A, Best N, Lunn D. BUGS: Bayesian Inference Using Gibbs Sampling. Cambridge, UK: MRC Biostatistics Unit; 2002. Version 1.4. [Google Scholar]
- 45.Wu M, Jolicoeur N, Li Z, et al. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis. 2008;29:1710–6. doi: 10.1093/carcin/bgn073. [DOI] [PubMed] [Google Scholar]
- 46.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin ZKP, Plaisance K, Parsons CH. Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukoc Biol. 2009;87:25–34. doi: 10.1189/jlb.0409251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin X, Liang D, He Z, Deng Q, Robertson ES, Lan K. miR-K12–7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS One. 2011;6:e16224. doi: 10.1371/journal.pone.0016224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 50.Chaulk S, Thede GL, Kent OA, et al. Role of pri-miRNA tertiary structure in miR-17∼92 miRNA biogenesis. RNA Biology. 2011;8:1105–14. doi: 10.4161/rna.8.6.17410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.