Abstract
Background
Genetic variation influences susceptibility or resistance to tuberculosis. Interleukin 6 (IL-6) contributes to protection against tuberculosis in mice. However, its role in regulating susceptibility or resistance to tuberculosis in humans is unclear.
Methods
Genotyping of polymorphisms in IL-6 and IL-6R (CD126) genes was performed in 2 independent cohorts, an experimental population (495 cases and 358 controls) and a validation population (1383 cases and 1149 controls). The associations of the variants with tuberculosis were tested using 2 case-control association studies. In addition, the regulatory effects of single-nucleotide polymorphism rs1800796 (-572C > G) on IL-6 production in plasma and CD14+ monocyte cultures stimulated with a Mycobacterium tuberculosis (M. tuberculosis) product were assessed.
Results
The rs1800796 polymorphism is associated with increased resistance to tuberculosis (odds ratio [OR], 0.771; 95% confidential interval, .684–.870). The rs1800796GG genotype is strongly associated with reduced risk to tuberculosis (OR, 0.621; 95% CI, .460–.838). Interestingly, CD14+ monocytes isolated from individuals with rs1800796GG genotype produced significantly less IL-6 in response to M. tuberculosis 19-kDa lipoprotein than those with CC or CG genotype.
Conclusions
We identified a genetic polymorphism in the IL-6 promoter that regulates cytokine production and host resistance to pulmonary tuberculosis in Chinese populations.
Tuberculosis, an infectious disease caused by Mycobacterium tuberculosis (M. tuberculosis), is an ongoing major public health problem in many developing countries, including China [1]. Interestingly, only 10% of the individuals infected with M. tuberculosis will develop active disease, whereas 90% of infected individuals will remain latently infected without clinical symptom [2], suggesting that host genetics play an important role in regulating disease progression. In support of this view, a recent genome-wide association study performed on Ghana and Gambia populations identified that a locus located on chromosome 18q11.2 is associated with tuberculosis [3]. Unraveling the mechanisms underlying the genetic variations that influence the susceptibility or resistance to tuberculosis may lead to better understanding of the pathogenesis of mycobacterial infection and development of novel strategies for prevention and treatment of tuberculosis.
It has been well established that both innate and adaptive immune responses are required for host control of M. tuberculosis infection [4, 5]. Therefore, it is expected that the genetic variants of molecules involved in innate host-defense mechanisms, such as vitamin D receptor, Toll-like receptors (TLR1/TLR6), and cytokines and chemokines and their receptors, are associated with host susceptibility to tuberculosis [6–8]. Interleukin 6 (IL-6), through interaction with its receptor (IL-6R), is expressed by a variety of cell lineages and has diverse functions. IL-6 has been shown to contribute to the differentiation and activation of cells that are and cells that are not part of the immune response [9]. As a key proinflammatory cytokine, IL-6 is associated with the pathogenesis of many chronic inflammatory diseases, including tuberculosis [10–12]. Accordingly, genetic variants in IL-6/IL-6R have been linked to the susceptibility to and/or severity of a wide range of diseases, including chronic hepatitis C virus infection, meningococcal disease, respiratory tract infection, asthma, and rheumatoid arthritis [13–17]. In mycobacterial infection, IL-6–deficient mice have been shown to exhibit an altered immune response and increased susceptibility to M. tuberculosis infection when compared to wild-type mice [18, 19]. However, the role of IL-6 in regulating host resistance to M. tuberculosis infection in humans is unknown.
Previously, we reported that downregulation of IL-6R expression on CD4 T cells in patients with active pulmonary tuberculosis is associated with decreased helper T cell 17 response [20], suggesting a role for IL-6 in the regulation of T cell effector functions, as well as in the progression of tuberculosis in humans. To investigate whether genetic variations in IL-6 or its receptor determine the outcome of M. tuberculosis infection in humans, in this study we examined 11 single-nucleotide polymorphisms (SNPs) in IL-6 and IL-6R genes for their association with tuberculosis in 2 independent experimental cohorts. We found that, while no association was observed for IL-6R (CD126) SNPs, a rare genetic variation in the IL-6 gene, rs1800796, is significantly associated with tuberculosis disease in the Chinese Han population.
METHODS
Study Population and Samples
Patients with different clinical features of tuberculosis were recruited at clinics in Shenzhen Third People's Hospital. The diagnosis of tuberculosis was based on clinical symptoms, radiological evidence, and findings from bacterial examination, as described elsewhere [20]. Healthy controls with normal chest radiograph findings and without a clinical history of tuberculosis were recruited from Shenzhen City, China. All participants were tested for human immunodeficiency virus (HIV) by serological analysis, and individuals with HIV infection were excluded. Characteristics of the study population are shown in Table 1. Whole blood samples collected before February 2010 were used as our experimental cohort (495 cases with tuberculosis and 358 healthy controls), which has been used in previous IL-22 polymorphisms study [21], and samples collected after that date were used as our validation cohort (1383 cases with tuberculosis and 1149 healthy controls). Peripheral blood samples were collected by venipuncture from the populations mentioned above. The study was approved by the Institutional Review Board of Shenzhen Third People's Hospital, and informed consent was obtained from each participant.
Table 1.
Characteristics of Healthy Controls and Cases With Tuberculosis in the 2 Cohort Populations
| Cohort, Arma | No. of Subjects | Age, Years, Mean ± SD) | Sex, Male:Female |
|---|---|---|---|
| Experimental | |||
| Controls | 358 | 36.92 ± 16.52 | 173:185 |
| Cases | 495 | 38.64 ± 18.44 | 356:139 |
| Validation | |||
| Controls | 1149 | 38.71 ± 27.82 | 467:682 |
| Cases | 1383 | 40.58 ± 20.79 | 907:476 |
Data are no. of subjects, unless otherwise indicated.
DNA Extraction, SNP Selection, and Genotyping
Genomic DNA was extracted from blood samples, using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA purities and final concentrations were determined spectrophotometrically. SNPs were selected according to methods described previously, with focuses on their potential regulatory roles, such as transcription binding sites in the promoter region and microRNA target sites in the 3′ untranslated region (UTR) of the selected genes [21]. For the experimental cohort, SNP genotypes were determined by primer-extension mass spectrometry, using the Sequenom MassARRAY system (San Diego, CA). For the validation cohort, rs1800796 was genotyped by TaqMan assays in 50 μL reaction volume, using ABI 7500 real-time polymerase chain reaction systems (ABI, Carlsbad, CA).
Cells Preparation and In Vitro Stimulation
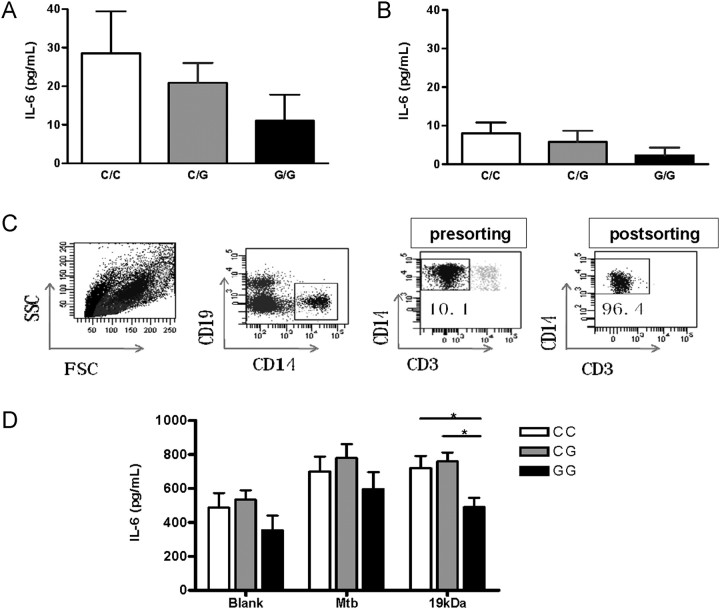
Peripheral blood mononuclear cells (PBMCs) were isolated as described previously [22]. PBMCs were then cryopreserved in liquid nitrogen. For functional assays, PBMCs were thawed and stained with antibodies against CD3, CD14, and CD19. CD14+ monocytes were sorted as a CD14+CD3−CD19− population, using a BD FACSAria II cell sorter (BD Biosciences, San Jose, CA). The purity of CD14+ monocytes was >95%, as determined by postsort analysis (Figure 1C).
Figure 1.
Influence of single nucleotide polymorphism rs1800796 on interleukin 6 (IL-6) production in plasma and CD14+ monocytes. Plasma levels of IL-6 in subjects with different IL-6 rs1800796 genotypes were determined by enzyme-linked immunosorbant assay (ELISA). A, Plasma IL-6 levels in patients with tuberculosis, by rs1800796 genotype. A total of 36 cases had genotype CC, 46 had genotype CG, and 16 had genotype GG. B, Plasma IL-6 levels in healthy controls. A total of 15 controls had genotype CC, 17 controls had genotype CG, and 15 controls had genotype GG. C, Purification of monocyte populations. Monocytes (CD3−CD19−CD14+) were isolated from peripheral blood mononuclear cells of healthy controls with different rs1800796 genotypes. FACS plots show the sorting strategy of a representative experiment. D, IL-6 production in purified CD14+ monocyte cultures. Monocytes (2 × 106/mL) were left unstimulated or were cultured with heat-killed Mycobacterium tuberculosis (M. tuberculosis) lysate (20 μg/mL) or M. tuberculosis 19-kDa lipoprotein (0.5 μg/mL) for 48 hours. The concentration of secreted IL-6 was determined by ELISA. Six specimens had genotype CC, 6 had genotype CG, and 5 had genotype GG. Data are mean ± standard error. *P < .05.
Purified CD14+ monocytes were cultured in a 96-well plate, with 2 × 105 cells/well, in Roswell Park Memorial Institute 1640 medium supplemented with 2 mM L-glutamine, antibiotics, and 10% heat-inactivated fetal bovine serum, in the absence or presence of heat-killed M. tuberculosis lysate (20 μg/mL) or M. tuberculosis 19-kDa lipoprotein (0.5 μg/mL; Lionex GmbH, Braunschweig, Germany). After 48 hours, culture supernatants were harvested, and cytokine levels were determined.
Measurements of IL-6, Interleukin 10 (IL-10), and Interleukin 12p70 (IL-12p70)
The concentrations of IL-6, IL-10, and IL-12p70 protein in supernatants were measured by an enzyme-linked immunosorbant assay, following the manufacturer's instruction (R&D, Minneapolis, MN).
Statistical Analysis
The allele and genotype frequencies were determined by direct counting. The Pearson χ2 test was used to compare allele and genotype distribution in patients with tuberculosis and healthy control subjects. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated with Miettinen's method. The Hardy-Weinberg equilibrium of each polymorphism was determined by the program HWE. One-way analysis of variance with Bonferroni correction was used to compare IL-6 levels between participants with different genotypes. All computations were done using GraphPad Prism software (version 4.0) (GraphPad Software, La Jolla, CA). A P value of <.05 was considered significant.
RESULTS
Analysis of IL-6/IL-6R SNPs in the Experimental Cohort
A total of 11 SNPs, including 6 in the IL-6 promoter region, 2 in IL-6R (α chain, CD126) promoter region, and 3 in the IL-6R 3′ UTR region were selected using the strategy described previously [21]. Genotypes of these 11 SNPs were determined using the MassARRAY platform in an experimental cohort of 495 cases and 358 controls (Table 2). Of the 11 SNPs, only 1 in the IL-6 promoter region (rs1800796) and 3 in the IL-6R 3′ UTR region (rs2229238, rs4379670, and rs4379670) showed frequencies of variants of >5% in the control populations. The SNP rs1800196, which is located 572 base pairs upstream from the IL-6 gene transcription start site, was in Hardy–Weinberg equilibrium. The minor allele frequency of 0.261 in the control population is consist with a previous report involving the Chinese Han population but significantly different from the annotations of other populations in the HapMap Han Chinese in Beijing, 0.23; Utah residents with ancestry from northern and western Europe, 0.96; Japanese in Tokyo, 0.14). The G allele is associated with a decreased risk of tuberculosis susceptibility (OR, 0.716 [95% CI, .57–.899]; P = .004). Interestingly, no genetic variants were found in other SNPs in the IL-6 promoter region, including the functional SNP rs1800795 (G-174C), which exists widely, with a minor allele frequency of approximately 40% in the Western white population [23, 24]. This indicates the significant difference of IL-6 gene polymorphisms among different populations.
Table 2.
Association Between IL6/IL6R Single Nucleotide Polymorphisms (SNPs) and Tuberculosis Susceptibility in the Experimental Cohort
| Allele Frequency, % |
Genotype |
Hardy–Weinberg Equilibrium |
Allelic Comparison |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID and Polymorphism, Arm | A | a | AA | Aa | aa | χ2 | Pa | χ2 | Pb | ORc | 95% CI |
| rs13306436, G/A | |||||||||||
| Controls | 692 (96.7) | 24 (3.4) | 334 | 24 | 0 | 0.643 | .725 | … | … | … | |
| Cases | 955 (96.5) | 35 (3.5) | 460 | 35 | 0 | 0.641 | .725 | 0.042 | .838 | 1.057 | .623–1.793 |
| rs2069824, T/C | |||||||||||
| Controls | 711 (99.3) | 5 (0.7) | 353 | 5 | 0 | 0.018 | .991 | … | … | … | |
| Cases | 983 (99.3) | 7 (0.7) | 488 | 7 | 0 | 0.025 | .988 | 0.000 | .983 | 1.013 | .320–3.203 |
| rs36215814, G/A | |||||||||||
| Controls | 716 (100) | 0 (0.0) | 358 | 0 | 0 | NA | … | … | … | ||
| Cases | 988 (99.8) | 2 (0.2) | 493 | 2 | 0 | 0.002 | .999 | 1.448 | .229 | 3.624 | .174–75.660 |
| rs1800797, G/A | |||||||||||
| Controls | 714 (99.7) | 2 (0.3) | 356 | 2 | 0 | 0.003 | .999 | … | … | … | |
| Cases | 986 (99.6) | 4 (0.4) | 491 | 4 | 0 | 0.008 | .996 | 0.184 | .668 | 1.448 | .264–7.932 |
| rs1800796, C/G | |||||||||||
| Controls | 529 (73.9) | 187 (26.1) | 202 | 125 | 31 | 1.548 | .461 | … | … | … | |
| Cases | 790 (78.7) | 200 (21.3) | 319 | 152 | 24 | 0.534 | .766 | 8.289 | .004 | 0.716 | .570–.899 |
| rs1800795, G/C | |||||||||||
| Controls | 715 (99.7) | 1 (0.3) | 357 | 1 | 0 | 0.007 | .997 | … | … | … | |
| Cases | 986 (99.6) | 4 (0.4) | 491 | 4 | 0 | 0.008 | .996 | 0.994 | .319 | 2.901 | .323–26.020 |
| rs56077270, T/C | |||||||||||
| Controls | 716 (100) | 0 (0.0) | 358 | 0 | 0 | NA | … | … | … | ||
| Cases | 988 (99.8) | 2 (0.2) | 493 | 2 | 0 | 0.002 | .999 | 1.448 | .229 | 3.624 | .174–75.660 |
| rs1552481, A/G | |||||||||||
| Controls | 704 (99.2) | 6 (0.8) | 351 | 2 | 2 | 3.893 | .143 | … | … | … | |
| Cases | 987 (99.7) | 3 (0.3) | 492 | 3 | 0 | 0.005 | .998 | 2.307 | .129 | 0.357 | .089–1.431 |
| rs2229238, A/G | |||||||||||
| Controls | 586 (81.8) | 130 (18.2) | 242 | 102 | 14 | 0.290 | .865 | … | … | … | |
| Cases | 819 (82.7) | 171 (17.3) | 339 | 141 | 15 | 0.003 | .999 | 0.223 | .637 | 0.941 | .732–1.210 |
| rs3887104, G/A | |||||||||||
| Controls | 679 (94.8) | 37 (5.2) | 321 | 37 | 0 | 1.008 | .604 | … | … | … | |
| Cases | 951 (96.1) | 39 (3.9) | 456 | 39 | 0 | 0.679 | .712 | 1.473 | .225 | 0.753 | .475–1.193 |
| rs4379670, A/T | |||||||||||
| Controls | 594 (83.0) | 122 (17.0) | 249 | 96 | 13 | 0.442 | .802 | … | … | … | |
| Cases | 827 (83.5) | 163 (16.5) | 348 | 131 | 16 | 0.336 | .845 | 0.099 | .754 | 0.960 | .742–1.241 |
Abbreviations: CI, confidence interval; NA, not available; OR, odds ratio.
a Hardy–Weinberg P value for tests of deviation from Hardy-Weinberg equilibrium.
b Compares the difference in allele frequency between cases and controls.
c OR for Tuberculosis, with the minor allele in controls as the reference allele.
Replication of Association Between Tuberculosis and rs1800796 in the Validation Cohort
The association of rs1800796 with tuberculosis was further confirmed in the validation cohort, which had 1383 cases and 1149 controls. A significantly lower frequency of the G allele was observed among patients with active pulmonary tuberculosis, compared with controls (OR, 0.800 [95% CI, .701–.913]; P = .0009) (Table 3). Analysis of the pooled data (1878 cases and 1507 controls) showed an even more significant association (OR, 0.771 [95% CI, .684–.870]; P = .0001). Since the G allele is associated with increased resistance to tuberculosis, we further analyzed the genotype difference between those with and those without the G allele. The frequency of the GG genotype among patients with tuberculosis was significant lower than that among controls in the experimental cohort (P = .025), controls in the validation cohort (P = .018), and the overall population of participants (P = .0017) (Table 4).
Table 3.
Replication of the Association Between rs1800796 and Tuberculosis in the Validation Cohort
| Allele Frequency, % |
Genotype |
Hardy–Weinberg Equilibrium |
Allelic Comparison |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort, Arm | Subjects, No. | A | a | AA | Aa | aa | χ2 | Pc | χ2 | Pa | ORb | 95% CI |
| Validation | ||||||||||||
| Controls | 1149 | 1733 (75.4) | 565 (24.6) | 655 | 423 | 71 | 0.030 | .985 | … | … | … | |
| Cases | 1383 | 2194 (79.3) | 572 (20.7) | 868 | 458 | 57 | 0.062 | .969 | 11.011 | .0009 | 0.800 | .701–.913 |
| Pooled | ||||||||||||
| Controls | 1507 | 1845 (74.8) | 619 (25.1) | 857 | 548 | 102 | 0.623 | .733 | … | … | … | |
| Cases | 1878 | 2984 (79.4) | 772 (20.6) | 1187 | 610 | 81 | 0.027 | .986 | 17.880 | .0001 | 0.771 | .684–.870 |
Abbreviations: CI, confidence interval; OR, odds ratio.
a Compares the difference in allele frequency between cases and controls.
b OR for Tuberculosis, with the minor allele in controls as the reference allele.
c Hardy–Weinberg P value for tests of deviation from Hardy–Weinberg equilibrium.
Table 4.
Rs1800796 Genotype Comparison Between Cases With Tuberculosis and Healthy Controls in Multiple Stages
| Genotype Distribution |
Genotype Comparison |
||||||
|---|---|---|---|---|---|---|---|
| Cohort, Arm | Subjects, No. | CC + CG | GG | χ2 | Pa | ORb | 95% CI |
| Experimental | |||||||
| Controls | 358 | 327 | 31 | … | … | … | |
| Cases | 495 | 471 | 24 | 5.001 | .025 | 0.538 | .310–.933 |
| Validation | |||||||
| Controls | 1149 | 1078 | 71 | … | … | … | |
| Cases | 1383 | 1326 | 57 | 5.537 | .018 | 0.653 | .456–.933 |
| Pooled | |||||||
| Controls | 1507 | 1405 | 102 | … | … | … | |
| Cases | 1878 | 1797 | 81 | 9.856 | .0017 | 0.621 | .460–.838 |
Abbreviations: CI, confidence interval; OR, odds ratio.
a Compares the difference in allele frequency between cases and controls.
b OR for Tuberculosis, with the minor allele in controls as the reference allele.
Association Between the rs1800796 SNP and Tuberculosis, Stratified by Sex and Age
Since male subjects constituted a higher proportion in the active tuberculosis group, compared with the healthy control group, in both cohorts (Table 1), we further analyzed the difference in the distribution of the rs1800796 allele between controls and tuberculosis patients, stratified by sex. As shown in Table 5, the frequency of allele G in the tuberculosis group was significantly lower than that in the control group for both male subjects (OR, 0.753 [95% CI, .643–.881]; P = .0004) and female subjects (OR, 0.778 [95% CI, .651–.930]; P = .0057). We further stratified our pooled data according to the age recorded at enrollment. A significant association was observed in both young patients (age, ≤25 years; OR, 0.746 [95% CI, .607–.908]; P = .0054) and older patients (age, >25 years; OR, 0.361 [95% CI, .306–.426]; P < .0001) (Table 6).
Table 5.
Rs1800796 Allele Comparison, Stratified by Sex, Between Cases With Tuberculosis and Healthy Controls
| Allele Frequency, % |
Genotype |
Allelic comparison |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sex, Arm | Subjects, No. | C | G | CC | CG | GG | χ2 | Pa | ORb | 95% CI |
| Male | ||||||||||
| Controls | 640 | 948 (74.1) | 332 (25.9) | 348 | 252 | 40 | … | … | … | |
| Cases | 1263 | 1999 (79.1) | 527 (20.9) | 772 | 455 | 36 | 12.521 | .0004 | 0.753 | .643–.881 |
| Female | ||||||||||
| Controls | 867 | 1314 (75.8) | 420 (24.2) | 509 | 296 | 62 | … | … | … | |
| Cases | 615 | 985 (80.1) | 245 (19.9) | 415 | 155 | 45 | 7.655 | .0057 | 0.778 | .651–.930 |
Abbreviations: CI, confidence interval; OR, odds ratio.
a Compares the difference in allele frequency between cases and controls.
b OR for Tuberculosis, with the minor allele in controls as the reference allele.
Table 6.
Rs1800796 Allele Comparison, Stratified by Age, Between Cases With Tuberculosis and Healthy Controls
| Allele Frequency, % |
Genotype |
Allelic comparison |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age, Arm | Subjects, No. | C | G | CC | CG | GG | χ2 | Pa | ORb | 95% CI |
| ≤25 years | ||||||||||
| Controls | 497 | 750 (75.5) | 244 (24.5) | 288 | 174 | 35 | … | … | … | |
| Cases | 563 | 906 (80.5) | 220 (19.5) | 360 | 186 | 17 | 7.748 | .0054 | 0.746 | .607–.918 |
| >25 years | ||||||||||
| Controls | 1010 | 1512 (74.8) | 508 (25.2) | 569 | 374 | 67 | … | … | … | |
| Cases | 1315 | 2078 (79.0) | 252 (21.0) | 827 | 424 | 64 | 154.210 | <.0001 | 0.361 | .306–.426 |
Abbreviations: CI, confidence interval; OR, odds ratio.
a Compares the difference in allele frequency between cases and controls.
b OR for Tuberculosis, with the minor allele in controls as the reference allele.
Effects of the rs1800796 Genotype on IL-6 Production
Previous studies have shown that SNPs in the IL-6 promoter region influence IL-6 expression [25, 26]. In particular, individuals with rs1800795GG and rs1800795CC genotypes were found to be high and low producers of IL-6, respectively [25–27]. Since only rs1800796 was found with variants in our study populations and since the rs1800796G allele was associated with increased resistance to tuberculosis, we further examined whether rs1800796 is involved in IL-6 production and thereby regulates host resistance. We first compared plasma IL-6 levels in healthy controls and tuberculosis patients with different rs1800796 genotypes. Consistent with previous reports [12, 20], an increased plasma IL-6 level was observed in patients with tuberculosis, compared with healthy controls (Figure 1). In both healthy controls and tuberculosis cases, the plasma IL-6 level in individuals with rs1800796GG genotypes was consistently lower than the level in individuals with CC and CG genotypes. However, the difference did not reach statistically significance, which is probably due to limited sample size (Figure 1).
Although many cells, including CD4+ T cells, can produce IL-6, it has been well established that monocytes are the major source of IL-6 production. Therefore, we compared IL-6 production by CD14+ monocytes in the absence or presence of M. tuberculosis lysate and M. tuberculosis 19-kDa lipoprotein, a known TLR2/TLR6 ligand derived from M. tuberculosis. To ensure the purity of monocytes, we FACS-sorted CD19−CD3−CD14+ monocytes (purity, >95%) from healthy individuals carrying different alleles (Figure 1C). We observed that IL-6 production in response to the 19-kDa lipoprotein is significantly lower for CD14+ monocytes isolated from individuals with GG genotype than for CD14+ monocytes from individuals with CC and CG genotypes (Figure 1D). Low levels of IL-10 and IL-12p70 were detected in the supernatants of CD14+ monocyte cultures. However, no difference in IL-10 and IL-12p70 production was observed regardless rs1800796 genotype (data not shown).
DISCUSSION
In this study, we analyzed DNA samples collected from 2 independent Chinese cohorts with >3000 subjects in total and identified a strong association between increased resistance to pulmonary tuberculosis and a genetic variation (rs1800796) in the promoter region of IL-6. In contrast, no IL-6R (CD126) SNPs examined were found to be associated with disease susceptibility in the same populations. Unexpectedly, we observed that, upon stimulation with M. tuberculosis products in vitro, CD14+ monocytes isolated from individuals with protective allele rs1800795GG secreted lower levels of IL-6 than those with CC or CG allele, arguing that a tightly regulated IL-6 response to M. tuberculosis may have contributed to the increased resistance to tuberculosis progression in this population.
SNPs other than rs1800796 in the IL-6 gene have been reported to be associated with active tuberculosis in previous studies performed in non-Chinese populations. When compared to Caucasians, Canadian aboriginal populations (Dene and Cree) showed a significantly higher frequency of the rs1800195G allele, suggesting an association between rs1800795G and host susceptibility to tuberculosis [26]. Interestingly, the same association was not observed in 2 other separate studies involving Indian and Colombian populations [28, 29]. The underlying causes for the discrepancy among the studies are unknown. While it could be a result of technical shortfalls, such as small sample size, a more likely explanation stems from the distinct ethnic makeup of each study.
Consistent with this hypothesis, SNP rs1800796 identified here has been reported by several independent genetic studies to be associated with the susceptibility to inflammatory conditions in the Chinese Han population, which include type 2 diabetes, idiopathic membranous nephropathy, coronary heart disease, and chronic periodontitis [30–33]. In contrast, variants at rs1800195 and rs1800797 in the IL-6 gene identified previously in other ethnic populations are relatively rare in the Chinese Han population [31–35], suggesting that rs1800796 is a common genetic variant in the Chinese Han population that underlies susceptibility to a broad spectrum of inflammatory diseases, including mycobacterial infection.
It has been reported that IL-6 plays an important role in protection against murine M. tuberculosis infection [19, 36], probably through orchestration of the CD4+ T cells response [37]. Compared with IL-6–competent mice, M. tuberculosis-infected IL-6–deficient animals show an impaired helper T cell 1 response and increased bacterial loads, indicating a requirement for IL-6 in host resistance to M. tuberculosis infection [18, 19]. On the other hand, IL-6 has also been suggested to downregulate macrophage microbicidal activity. Thus, IL-6 inhibits the production of tumor necrosis factor α and promotes in vitro growth of Mycobacterium avium [38, 39]. Moreover, IL-6 secreted by M. tuberculosis-infected macrophages sup presses the responses of uninfected macrophages to interferon γ (IFN-γ) [40]. Finally, increased levels of IL-6 in the lung, along with increased levels of IL-1β and IL-11, is significantly correlated with tuberculosis progression in a panel of genetically susceptible mice [11], although a causal relationship between the bacterial loads and IL-6 levels cannot be determined in this setting. Together, these mouse studies indicate that IL-6 may play multiple roles and contribute both positively and negatively to host control of M. tuberculosis infection.
In this study, we found that the frequencies of the G allele and the GG genotype are significantly lower among individuals with pulmonary tuberculosis than among control subjects. Interestingly, CD14+ monocytes sorted from individuals with the protective rs1800796GG genotype produced lower levels of IL-6 in response to the M. tuberculosis 19-kDa lipoprotein than those with the CC or CG genotype. The association between the M. tuberculosis 19-kDa lipoprotein and IL-6 production by CD14+ monocytes from individuals with the rs1800796 polymorphism is likely due to the polymorphism's direct effect, instead of to other causal variants in linkage disequilibrium with this polymorphism, since the C > G variation at rs1800796 has been reported to reduce transcriptional activity of the IL-6 promoter in both U-937 and K562 cell lines [35]. Therefore, our data argue that the rs1800796G allele plays a role in regulating IL-6 production in inflammation, at least in the Han Chinese population.
Although the exact function of IL-6 in human tuberculosis has yet to be defined, our current study suggests that hyper IL-6 production may be detrimental rather than beneficial to the host resistance to M. tuberculosis infection [11]. It has been reported that reduced production of IL-6 and IL-10 resulting from the hyporesponsive TLR1 or TLR6 allele is associated with enhanced IFN-γ and IL-2 production by PBMCs isolated from BCG-vaccinated infants, suggesting a possible regulatory role for IL-6 in the development of the helper T cell 1 response in infection [41]. Understanding cellular and molecular mechanisms underlying the regulatory function of IL-6 in human tuberculosis will be the focus of future investigations.
In summary, we have identified that the rs1800796GG genotype in the IL-6 promoter contributes to protection against tuberculosis in the Chinese Han population through downregulation of IL-6 production. Our findings may assist in identifying individuals at high risk of developing active disease. Finally, these observations emphasize the importance of host genetics in resistance to tuberculosis and have important implications for understanding the role of IL-6 in other inflammatory conditions in humans.
Notes
Acknowledgments. We thank Drs Hong Yuan, Jie Zhou, Cishi Shen, Xiuming Wen, Guanggui Ding, and Yingjian Song for their assistance in recruiting patients and volunteers.
Financial support. This work was supported by Eleven-Fifth Mega-Scientific Project on the “prevention and treatment of AIDS, viral hepatitis and other infectious diseases” (2008ZX10003-005), the Natural Science Foundation of China (grants 30872258 and 81172732), the Intramural Research Program, Department of Health and Human Services, National Institute of Allergy and Infectious Diseases, National Institutes of Health (to C. G. F.), and the Food and Health Bureau of Hong Kong (10091262 to J. W.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bauer T, Eils R, Konig R. RIP: the regulatory interaction predictor—a machine learning-based approach for predicting target genes of transcription factors. Bioinformatics. 2011;27:2239–47. doi: 10.1093/bioinformatics/btr366. [DOI] [PubMed] [Google Scholar]
- 2.Murray CJ, Styblo K, Rouillon A. Tuberculosis in developing countries: burden, intervention and cost. Bull Int Union Tuberc Lung Dis. 1990;65:6–24. [PubMed] [Google Scholar]
- 3.Thye T, Vannberg FO, Wong SH, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2011;42:739–41. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper AM. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natarajan K, Kundu M, Sharma P, Basu J. Innate immune responses to M. tuberculosis infection. Tuberculosis (Edinb) 2011;91:427–31. doi: 10.1016/j.tube.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Mamtani M, Mummidi S, Ramsuran V, et al. Influence of variations in CCL3L1 and CCR5 on tuberculosis in a northwestern Colombian population. J Infect Dis. 2011;203:1590–4. doi: 10.1093/infdis/jir145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Yang Y, Zhou F, et al. SLC11A1 (NRAMP1) polymorphisms and tuberculosis susceptibility: updated systematic review and meta-analysis. PLoS One. 2011;6:e15831. doi: 10.1371/journal.pone.0015831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol. 2011;2011 doi: 10.1155/2011/405310. 405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishimoto T, Hibi M, Murakami M, Narazaki M, Saito M, Taga T. The molecular biology of interleukin 6 and its receptor. Ciba Found Symp. 1992;167:5–16. doi: 10.1002/9780470514269.ch2. discussion 16–23. [DOI] [PubMed] [Google Scholar]
- 10.Barnes TC, Anderson ME, Moots RJ. The many faces of interleukin-6: the role of IL-6 in inflammation, vasculopathy, and fibrosis in systemic sclerosis. Int J Rheumatol. 2011;2011 doi: 10.1155/2011/721608. 721608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyadova IV, Tsiganov EN, Kapina MA, et al. In mice, tuberculosis progression is associated with intensive inflammatory response and the accumulation of Gr-1 cells in the lungs. PLoS One. 2010;5:e10469. doi: 10.1371/journal.pone.0010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law K, Weiden M, Harkin T, Tchou-Wong K, Chi C, Rom WN. Increased release of interleukin-1 beta, interleukin-6, and tumor necrosis factor-alpha by bronchoalveolar cells lavaged from involved sites in pulmonary tuberculosis. Am J Respir Crit Care Med. 1996;153:799–804. doi: 10.1164/ajrccm.153.2.8564135. [DOI] [PubMed] [Google Scholar]
- 13.Cussigh A, Falleti E, Fabris C, et al. Interleukin 6 promoter polymorphisms influence the outcome of chronic hepatitis C. Immunogenetics. 2011;63:33–41. doi: 10.1007/s00251-010-0491-7. [DOI] [PubMed] [Google Scholar]
- 14.Rantala A, Lajunen T, Juvonen R, et al. Association of IL-6 and IL-6R gene polymorphisms with susceptibility to respiratory tract infections in young Finnish men. Hum Immunol. 2011;72:63–8. doi: 10.1016/j.humimm.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Balding J, Healy CM, Livingstone WJ, et al. Genomic polymorphic profiles in an Irish population with meningococcaemia: is it possible to predict severity and outcome of disease? Genes Immun. 2003;4:533–40. doi: 10.1038/sj.gene.6364020. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira MA, Matheson MC, Duffy DL, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamas JR, Rodriguez-Rodriguez L, Varade J, et al. Influence of IL6R rs8192284 polymorphism status in disease activity in rheumatoid arthritis. J Rheumatol. 2010;37:1579–81. doi: 10.3899/jrheum.091455. [DOI] [PubMed] [Google Scholar]
- 18.Saunders BM, Frank AA, Orme IM, Cooper AM. Interleukin-6 induces early gamma interferon production in the infected lung but is not required for generation of specific immunity to Mycobacterium tuberculosis infection. Infect Immun. 2000;68:3322–6. doi: 10.1128/iai.68.6.3322-3326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladel CH, Blum C, Dreher A, Reifenberg K, Kopf M, Kaufmann SH. Lethal tuberculosis in interleukin-6-deficient mutant mice. Infect Immun. 1997;65:4843–9. doi: 10.1128/iai.65.11.4843-4849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Zhang M, Liao M, et al. Reduced Th17 response in patients with tuberculosis correlates with IL-6R expression on CD4+ T Cells. Am J Respir Crit Care Med. 2010;181:734–42. doi: 10.1164/rccm.200909-1463OC. [DOI] [PubMed] [Google Scholar]
- 21.Zhang G, Chen X, Chan L, et al. An SNP selection strategy identified IL-22 associating with susceptibility to tuberculosis in Chinese. Sci Rep. 2011:1–20. doi: 10.1038/srep00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Zhou B, Li M, et al. CD4(+)CD25(+)FoxP3(+) regulatory T cells suppress Mycobacterium tuberculosis immunity in patients with active disease. Clin Immunol. 2007;123:50–9. doi: 10.1016/j.clim.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Li MJ, Wang P, Liu X, et al. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1182. 40(Database issue):D1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr798. 40(Database issue):D1308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–76. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larcombe LA, Orr PH, Lodge AM, et al. Functional gene polymorphisms in Canadian aboriginal populations with high rates of tuberculosis. J Infect Dis. 2008;198:1175–9. doi: 10.1086/592049. [DOI] [PubMed] [Google Scholar]
- 27.Doyle WJ, Casselbrant ML, Li-Korotky HS, et al. The interleukin 6 -174 C/C genotype predicts greater rhinovirus illness. J Infect Dis. 2010;201:199–206. doi: 10.1086/649559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvaraj P, Alagarasu K, Harishankar M, Vidyarani M, Nisha Rajeswari D, Narayanan PR. Cytokine gene polymorphisms and cytokine levels in pulmonary tuberculosis. Cytokine. 2008;43:26–33. doi: 10.1016/j.cyto.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Henao MI, Montes C, Paris SC, Garcia LF. Cytokine gene polymorphisms in Colombian patients with different clinical presentations of tuberculosis. Tuberculosis (Edinb) 2006;86:11–9. doi: 10.1016/j.tube.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Chen SY, Chen CH, Huang YC, Chuang HM, Lo MM, Tsai FJ. Effect of IL-6 C-572G polymorphism on idiopathic membranous nephropathy risk in a Han Chinese population. Ren Fail. 2010;32:1172–6. doi: 10.3109/0886022X.2010.516857. [DOI] [PubMed] [Google Scholar]
- 31.Fan WH, Liu DL, Xiao LM, Xie CJ, Sun SY, Zhang JC. Coronary heart disease and chronic periodontitis: is polymorphism of interleukin-6 gene the common risk factor in a Chinese population? Oral Dis. 2011;17:270–6. doi: 10.1111/j.1601-0825.2010.01736.x. [DOI] [PubMed] [Google Scholar]
- 32.Gao SP, Pan M, Chen C, et al. The G to A polymorphism at -597 of the interleukin-6 gene is extremely rare in southern Han Chinese. Cytokine. 2011;55:1–3. doi: 10.1016/j.cyto.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Ma L, Peng F, et al. The endothelial dysfunction in patients with type 2 diabetes mellitus is associated with IL-6 gene promoter polymorphism in Chinese population. Endocrine. 2011;40:124–9. doi: 10.1007/s12020-011-9442-9. [DOI] [PubMed] [Google Scholar]
- 34.Pan M, Gao SP, Jiang MH, Guo J, Zheng JG, Zhu JH. Interleukin 6 promoter polymorphisms in normal Han Chinese population: frequencies and effects on inflammatory markers. J Investig Med. 2011;59:272–6. doi: 10.231/JIM.0b013e318206ffad. [DOI] [PubMed] [Google Scholar]
- 35.Gu W, Du DY, Huang J, et al. Identification of interleukin-6 promoter polymorphisms in the Chinese Han population and their functional significance. Crit Care Med. 2008;36:1437–43. doi: 10.1097/CCM.0b013e31816a0adb. [DOI] [PubMed] [Google Scholar]
- 36.Appelberg R. Protective role of interferon gamma, tumor necrosis factor alpha and interleukin-6 in Mycobacterium tuberculosis and M. avium infections. Immunobiology. 1994;191:520–5. doi: 10.1016/S0171-2985(11)80458-4. [DOI] [PubMed] [Google Scholar]
- 37.Dienz O, Rincon M. The effects of IL-6 on CD4 T cell responses. Clin Immunol. 2009;130:27–33. doi: 10.1016/j.clim.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindler R, Mancilla J, Endres S, Ghorbani R, Clark SC, Dinarello CA. Correlations and interactions in the production of interleukin-6 (IL-6), IL-1, and tumor necrosis factor (TNF) in human blood mononuclear cells: IL-6 suppresses IL-1 and TNF. Blood. 1990;75:40–7. [PubMed] [Google Scholar]
- 39.Shiratsuchi H, Johnson JL, Ellner JJ. Bidirectional effects of cytokines on the growth of Mycobacterium avium within human monocytes. J Immunol. 1991;146:3165–70. [PubMed] [Google Scholar]
- 40.Nagabhushanam V, Solache A, Ting LM, Escaron CJ, Zhang JY, Ernst JD. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J Immunol. 2003;171:4750–7. doi: 10.4049/jimmunol.171.9.4750. [DOI] [PubMed] [Google Scholar]
- 41.Randhawa AK, Shey MS, Keyser A, et al. Association of human TLR1 and TLR6 deficiency with altered immune responses to BCG vaccination in South African infants. PLoS Pathog. 2011;7:e1002174. doi: 10.1371/journal.ppat.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]