Abstract
Prospects for a comprehensive inventory of global biodiversity would be greatly improved by automating methods of species delimitation. The general mixed Yule–coalescent (GMYC) was recently proposed as a potential means of increasing the rate of biodiversity exploration. We tested this method with simulated data and applied it to a group of poorly known bats (Hipposideros) from the Philippines. We then used echolocation call characteristics to evaluate the plausibility of species boundaries suggested by GMYC. In our simulations, GMYC performed relatively well (errors in estimated species diversity less than 25%) when the product of the haploid effective population size (Ne) and speciation rate (SR; per lineage per million years) was less than or equal to 105, while interspecific variation in Ne was twofold or less. However, at higher but also biologically relevant values of Ne × SR and when Ne varied tenfold among species, performance was very poor. GMYC analyses of mitochondrial DNA sequences from Philippine Hipposideros suggest actual diversity may be approximately twice the current estimate, and available echolocation call data are mostly consistent with GMYC delimitations. In conclusion, we consider the GMYC model useful under some conditions, but additional information on Ne, SR and/or corroboration from independent character data are needed to allow meaningful interpretation of results.
Keywords: biodiversity discovery, cryptic species, echolocation, Hipposideros, species delimitation, taxonomy
1. Introduction
Achieving a taxonomy that reflects evolutionary history is critical for effective conservation [1], projecting responses of biodiversity to climate change [2], and illuminating the underlying evolutionary and ecological forces that generate biotic communities [3,4]. Thus, rapid and accurate methods of species delimitation are urgently needed to expedite biodiversity discovery and documentation [5,6]. Recently, new species delimitation methods have been developed that are relatively automated and take advantage of DNA sequence datasets [7–10]. These methods offer the promise of making species delimitation both more efficient and less subjective. One example is the general mixed Yule–coalescent (GMYC) [7] that uses maximum-likelihood statistics and a time-calibrated gene tree to delimit species. GMYC analyses typically involve the collection of DNA sequences from a single locus (often mitochondrial) for a sample of closely related organisms. An ultrametric phylogeny is estimated from the sequences, and the GMYC aims to identify a time in the tree when the branching rate shifts (in forward time) from a Yule (species) to a coalescent (population) process. The GMYC originally estimated the time of a single threshold [7], but was later extended to allow independent transition times on different branches of the phylogeny [11]. GMYC has been used to delimit species in several poorly known groups of organisms [11–14], but few studies have tested its efficacy using either simulated data or by analysing clades with well-established species boundaries. Two studies have explored GMYC performance in the context of an island system, focusing on the effects of migration and sampling density [15,16]. However, a more general test of GMYC on gene trees simulated under a range of biologically relevant conditions has not been undertaken.
Here, we assess GMYC performance using simulated data, and apply the same method to an empirical system from the Philippines: round-leaf bats of the genus Hipposideros. The Philippine archipelago is an astounding centre of biodiversity with nearly 1200 species of terrestrial vertebrates [17]. Many new species have been described recently [18–21], and an unknown portion of the country's biodiversity may be threatened with extinction before it is discovered [22]. Several recent phylogeographic studies of Philippine vertebrates documented underappreciated diversity within currently recognized species [23–26], but others found surprisingly little genetic diversity within morphologically variable animals [27]. Thus, analytical techniques such as the GMYC, which offer the promise of rapid and objective biodiversity documentation, are potentially very useful.
Eight species of Hipposideros have been reported from the Philippines [28], comprising five that are widespread in southeast Asia (Hipposideros ater, H. bicolor, H. cervinus, H. diadema, H. lekaguli), two that are endemic to and widespread within the Philippines (H. obscurus and H. pygmaeus), and one that is endemic to the Greater Mindanao Pleistocene aggregate island complex [29–32] in the southern Philippines (H. coronatus). Taxonomically, these bats were last revised nearly half a century ago [33] when museum collections of insectivorous bats were more limited and morphological species definitions often failed to recognize ‘cryptic’ taxa. Murray et al. [34] recently noted the presence of taxonomic problems in the family Hipposideridae, including multiple polyphyletic species. Many species of Hipposideros are difficult to identify with morphology alone, necessitating the use of other information such as genetic characters and echolocation call traits [34–36].
In this study, we first use simulated data to explore the effects of per lineage speciation rate (SR), extinction, effective population size (Ne) and interspecific variation in Ne on GMYC accuracy and precision. We then conduct phylogenetic and GMYC analyses on a single-locus dataset for Philippine round-leaf bats (Hipposideros) to gain a sense of whether the current taxonomy of this group reflects its evolutionary history. Finally, we examine echolocation call frequencies in these bats to evaluate their congruence with the unrecognized species inferred by the GMYC analyses.
2. Methods
(a). General mixed Yule–coalescent and simulated data
Using Dendropy v. 3.9.0 [37], we simulated species trees with a final diversity of 20 species using a Yule [38], or pure birth, model. A single gene tree (with 10 alleles sampled per species) was then simulated under standard coalescent conditions within each species tree. Simulated gene trees were ultrametric, with branch lengths in generations. In these simulations, we employed a range of species birth rates, using values of 0.1, 1 and 10 births per lineage per million years (Myr), spanning the range of observed SR for a variety of organisms [39–43]. We also considered a range of biologically relevant Ne values for the coalescent simulations (haploid population sizes of 104, 105 and 106) that correspond with empirical observations of species [44,45]. We further considered the effect of interspecific variation in Ne, which has been documented in numerous clades [44,45]. To do so, we increased Ne by either twofold or tenfold on 19 randomly selected edges within the species tree (a rooted tree with 20 terminals contains 39 edges). We also considered the effect of extinction. Extinction rates were set at half of the SR, and applied to all 27 combinations described earlier, resulting in a total of 54 parameter combinations. Under each scenario, 200 gene trees were simulated, each derived from an independent species tree.
Species were delimited directly on each simulated gene tree. Our simulations thus present ‘best-case scenarios’ for GMYC in that they lack uncertainty in the topology and branch lengths—these uncertainties are encountered when gene trees are estimated from sequence data. Our GMYC analyses used the R package SPLITS [7,11], employing both the single- and multiple-threshold models. For each simulation parameter combination, we calculated the mean and standard deviation of the number of species estimated by the GMYC across the 200 gene trees. To characterize the shape of simulated gene trees, we also calculated the proportion of species that are monophyletic and a measure of gene flow, Slatkin & Maddison's ‘s’ [46], using Dendropy v. 3.9.0 [37].
(b). General mixed Yule–coalescent and species delimitation in Philippine round-leaf bats
We sequenced two protein-coding mitochondrial DNA fragments, including the entire NADH dehydrogenase subunit 2 gene (1044 bp) and 508 nucleotides of cytochrome b. Taxon sampling included 413 individuals, representing 13 currently recognized species of Hipposideros and two specimens of Rhinolophus virgo for use as an outgroup [34]. Among the 413 Hipposideros samples, 399 individuals are from the Philippines (2–173 of each species) and 14 are from neighbouring regions (Indochina, Java, Borneo, Sulawesi and Papua New Guinea). All sequences were deposited in the GenBank under accession numbers JQ915217–JQ916034.
Detailed methods of DNA sequencing and phylogenetic analyses are provided in the electronic supplementary material. Briefly, we used Bayesian inference of phylogeny with relaxed clock methods on an arbitrary timescale, using beast [47]. Analyses were completed on the entire character matrix and on a reduced matrix that included only unique haplotypes. Phylogenies were summarized as maximum clade credibility trees (MCCT) using median node ages, with branch lengths in substitutions per site. Because the GMYC relies on relative branch lengths to estimate species boundaries, the units of branch lengths do not affect the results.
We delimited species with the GMYC model on the MCCT derived from the complete character matrix. Separate analyses used the single- and multiple-threshold methods. We compared alternative models (single versus multiple thresholds) to a null model with only a coalescent branching rate (single species) and to each other using likelihood ratio tests. To assess the influence of branch-length uncertainty, we repeated the single shift-point analysis on the last 1000 trees in the posterior distribution (250 from each run).
To roughly place our empirical GMYC delimitations in the context of our simulation tests, we estimated Ne for putative species for which we had sequences of at least 10 individuals. We considered both taxonomically defined species that were monophyletic in our phylogenetic analyses and GMYC-delimited species. We calculated Ne using π for synonymous sites from seven putative species and the approximate minimum (0.01 per site per Myr) and maximum (0.14 per site per Myr) synonymous substitution rates observed in cytochrome b for bats by Nabholz et al. [48].
We recorded echolocation calls of adult Hipposideros from three sites on Luzon Island (Mt Makiling, Laguna; Mt Banahaw, Quezon; and Mt Isarog, Camarines Sur), and one site on Bohol Island that is part of the Greater Mindanao Pleistocene complex (see the electronic supplementary material for details).
3. Results
(a). Simulation results
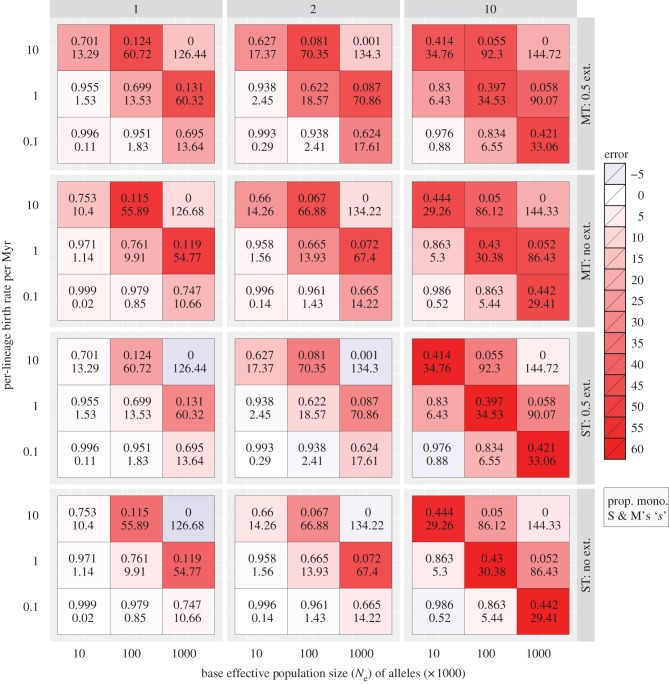
The GMYC method performed relatively well when Ne was small and the SR slow (figure 1). Under these conditions, species tend to be monophyletic in their gene trees. However, with increasing Ne and/or SR, both accuracy and precision declined (see figure 1 and electronic supplementary material, figure S1). Variation in Ne further decreased accuracy. In our simulations with stable and twofold variation in Ne, GMYC accuracy was poorest when Ne × SR = 106. In simulations with tenfold variation in Ne, accuracy was the lowest when Ne × SR = 105. Once Ne and SR were increased beyond these points, GMYC recovered some accuracy in terms of estimating the correct number of species. However, because under these conditions most species are not monophyletic (figure 1), the true assignment of individuals to species was not possible. Thus, accurate estimates, in terms of species number and assignment of individuals to species, were recovered only when SR and Ne were both low.
Figure 1.
A heat map showing errors in the estimated number of species derived from GMYC species delimitation on simulated gene trees. Columns labelled 1, 2 and 10 indicate the magnitude of interspecific variation in Ne (i.e. zero, twofold and tenfold variation); rows are arranged by the model used (single-threshold, ST; or multiple-threshold, MT), and the presence (0.5 ext.) or absence of extinction (no ext.). Numbers inside cells indicate the average proportion of monophyletic species (upper) and Slatkin & Maddison's ‘s’ (lower) in simulated trees.
For the single-threshold model, the number of species was underestimated when Ne × SR ≤ 104, overestimated from 105 to 106, and then underestimated again at the upper limit of our simulations (Ne × SR = 107) with zero or twofold variation in Ne. With a tenfold variation in Ne, the estimated number of species was low at the lowest values of Ne and SR, but overestimated at all other parameter combinations. These patterns were consistent between simulations with and without extinction (figure 1). By contrast, the multiple-threshold model always overestimated the number of species, though the relative magnitude of errors followed a similar pattern, with the greatest errors at intermediate values of Ne × SR (figure 1). The variance in estimates of species diversity was generally high when the error in the estimates was high, and it was nearly always greater for the single-threshold model than for the multiple-threshold model (see electronic supplementary material, figure S1).
(b). Species delimitation in Philippine round-leaf bats
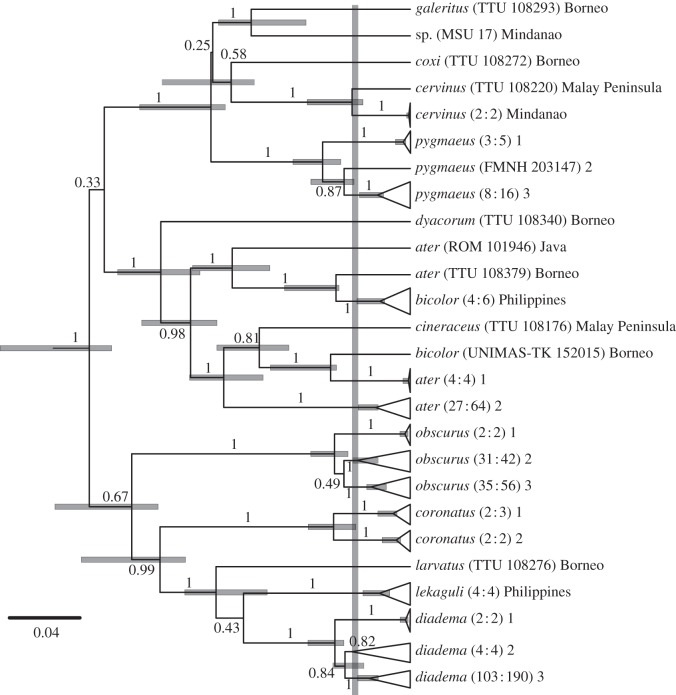
The phylogenetic analysis of all sequences resulted in very broad 95 per cent highest posterior densities (HPDs) of node ages; HPDs were much narrower in the analysis of unique haplotypes (figure 2). Phylogenetic results revealed extensive genetic variation across the Philippines in H. ater, H. coronatus, H. obscurus and H. pygmaeus (figure 2), but did not detect substantial genetic variation within our limited samples of Philippine H. bicolor, H. cervinus or H. lekaguli. Genetic variation in H. diadema, the largest and best-sampled species, occurs at a broader geographical scale, with somewhat distinct mitochondrial lineages occurring in the Philippines, Borneo, Indochina, Sulawesi and Papua New Guinea. Hipposideros ater and H. bicolor, as currently recognized, have polyphyletic mitochondrial DNA (figure 2).
Figure 2.
Maximum clade credibility tree of Hipposideros, primarily sampled from the Philippines. Numbers at nodes are posterior probabilities, and horizontal bars show the 95% highest posterior density of node ages on an arbitrary time scale from the analysis of unique haplotypes. The vertical grey bar shows the transition point from a Yule to a coalescent branching process in the analysis of all sequences, as estimated by the single-threshold model. Terminals are labelled with species identifications, followed by either the museum catalogue number, or the number of unique haplotypes and number of individuals enclosed in parentheses, and finally by either the geographical origin of the samples or a number, which refers to a location in figure 3.
GMYC analyses suggest the presence of several unrecognized species. Single- and multiple-threshold models with multiple branching rates (i.e. multiple species) were favoured over the null models with only coalescent branching rates (i.e. a single species; p-value ≪ 0.001). The single-threshold analysis of the MCCT (generated from 13 currently recognized ingroup species) suggested the presence of 26 species (95% CI: 9–43 species), whereas the multiple-threshold model suggested the presence of 54 species (CI: 16–188). However, a likelihood ratio test did not favour the multiple-threshold model (χ2 = 5.238; d.f. = 15; p-value = 0.99). Thus, we analysed the sample of 1000 trees from the posterior distribution using only the single-threshold model, which resulted in a mean of 31.86 species (sample s.d. = 10.72).
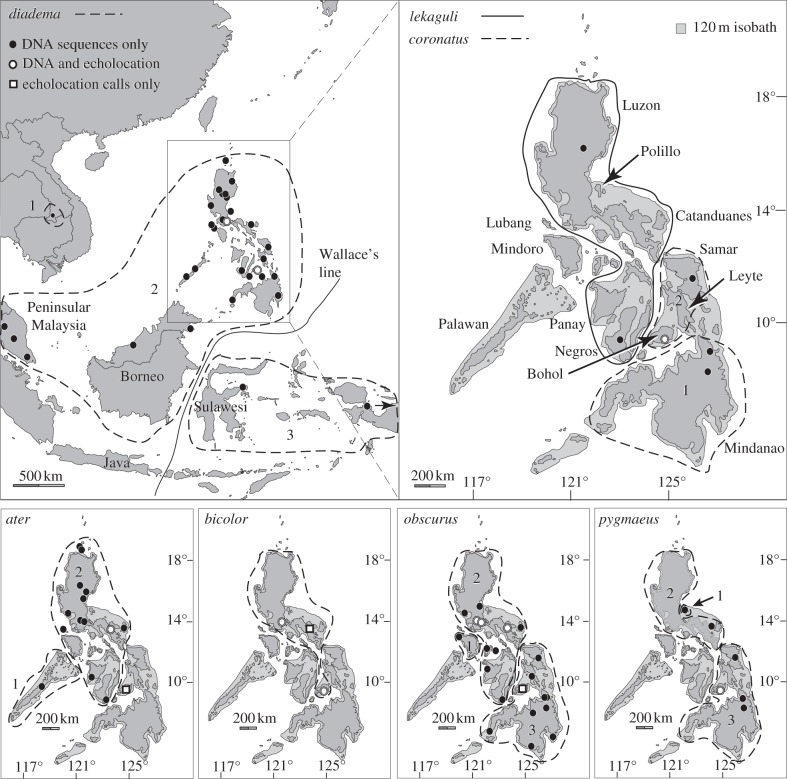
Among the widespread species, GMYC analyses of the MCCT suggest there are four species within H. ater, two within H. bicolor and three within H. diadema, though the geographical ranges of the latter are much larger than those of the other taxa. When restricted geographically to Philippine populations, these analyses suggest there are two species currently referred to H. ater, one to H. bicolor, two to H. coronatus, one to H. diadema, one to H. lekaguli, two to H. obscurus, three to H. pygmaeus and one unknown species most closely related to H. galeritus (figures 2 and 3).
Figure 3.
Maps showing the inferred geographical ranges of species, as delimited with the general mixed Yule–coalescent. Panels showing only the Philippines show modern islands and the 120 m isobath that approximates the extent of dry land during Pleistocene glacial maxima. Symbols indicate sample localities and data types available. Solid and dashed lines delimit the presumed ranges of species, by enclosing all DNA sequence sample localities within a species and any additional islands that were connected during periods of low sea level.
Estimates of Ne for the seven putative species of Hipposideros (taxonomically defined and GMYC defined) based on pairwise nucleotide diversity and segregating sites ranged approximately from 3 × 105 to 1.5 × 107, suggesting the data from these bats fall in the mid- to high range of Ne used in the simulations.
We evaluated the results of the GMYC analyses using echolocation calls from up to four populations of six currently recognized species, and recovered results that were mostly consistent. The echolocation calls of H. ater and H. diadema from Luzon and Bohol islands were similar (see electronic supplementary material, table S1); each of these taxonomic species was clustered as a single species by GMYC across the oceanic Philippines (i.e. excluding Palawan). Calls of H. coronatus and H. pygmaeus were available only from Bohol Island; however, they were distinct from the others and results from the GMYC analysis were consistent in the sense that these lineages were inferred to be distinct species. Distinct phonic groups were observed among individuals identified as H. obscurus and H. bicolor (electronic supplementary material, table S1). Calls of H. obscurus were 24 kHz higher on Bohol than on Luzon, and GMYC analyses suggested that a distinct species occurs on each of these Pleistocene island complexes. Calls of H. bicolor from Quezon on Luzon Island were distinct from H. bicolor from Camarines Sur (also on Luzon) and H. bicolor on Bohol Island, whereas calls from the latter two populations were similar. However, the GMYC analyses grouped H. bicolor individuals from Quezon and Bohol as a single species. Unfortunately, sequences from Camarines Sur H. bicolor were not obtained, so the acoustic data from this population could not be compared with GMYC inferences. In conclusion, analysis of echolocation calls was mostly consistent with results from GMYC analyses, except that two populations of H. bicolor with distinct calls were grouped into a single species.
4. Discussion
(a). Testing the general mixed Yule–coalescent
The GMYC method of species delimitation attempts to exploit a transition in gene trees between a Yule process that governs branching times during speciation, and a coalescent process that governs branching times within populations. However, the Yule and coalescent models make several assumptions that are unrealistic for free-living species [15,49–52], potentially limiting their value in this context. For instance, the Yule [38] model assumes a constant rate of speciation and no extinction. These assumptions may explain the failure of Yule models to fit observed phylogenies owing to the prevalence of unbalanced tree shapes [49,50,53] and declines in branching rates near the present [42]. Similarly, the coalescent assumes panmixia and constant population size [51,52], neither of which is likely to prove true for real species. The GMYC uses scaling parameters to relax some assumptions, but the analyses still failed to estimate diversity accurately over a wide range of parameter space.
In our simulations, GMYC was relatively accurate at low Ne and SR, but performance declined dramatically as these parameter values increased (figure 1). At low values of Ne and SR, gene trees tended to have high proportions of monophyletic species, but this proportion declined as Ne and SR increased (figure 1). Because the GMYC draws a threshold transition across the phylogeny, it can capture the true species delimitation only when all species are monophyletic. Thus, although the analysis cannot correctly assign individuals to species when species are not monophyletic, it may nevertheless correctly estimate the total number of species.
GMYC analyses produced reasonable results when Ne × SR was less than 105, but grossly overestimated the number of species at Ne × SR = 105 or 106. The analyses returned to more reasonable estimates of species number when Ne × SR extended above 106, apparently because the increasing number of deep coalescences made more of the tree look coalescent to the method (see electronic supplementary material, figure S2). However, because high Ne and SR cause non-monophyly of species, accurate assignment of individuals to species was impossible. In general, our results indicate that the analysis has a tendency to overestimate species diversity across a wide range of parameter space, but as the number of deep coalescences increases, the estimated number of species decreases, eventually resulting in underestimates of species number (see figure 1 and electronic supplementary material, S2). The same factors that make GMYC delimitation inaccurate also increase the variance in the number of species estimated (see electronic supplementary material, figure S1). Thus, a single gene tree drawn from a particular parameter set or empirical system could result in either an accurate or a grossly inaccurate estimate of species diversity.
When a gene tree samples species and populations, and when species are monophyletic with all population-level lineages coalescing rapidly, a lineage-through-time (LTT) plot may show a pronounced shift in the rate of lineage accumulation. Whether this inflection point corresponds with the species-population boundary is debatable [15,16]. However, waiting times between ancient coalescent events are expected to be long, relative to waiting times of recent coalescent events [51,52]. Thus, if the branching rate deep in the coalescent process is similar to the Yule branching rate, inflection points in LTTs may tend to occur more recently than the actual transition to a population-level process. Therefore, if we take the inflection point in an LTT as approximating species boundaries, we may overestimate the number of species, as happened with many of our simulated gene trees. However, as the number of deep coalescences in gene trees increases (e.g. when population size is large and/or the rate of speciation is high), speciation and coalescent processes intermingle on the gene tree, and the inflection point may become less pronounced, potentially causing the GMYC to underestimate the diversity of species (electronic supplementary material, figure S2). This may explain why the GMYC tends to overestimate the number of species when ancestral polymorphism is low, but underestimate it when ancestral polymorphism is extremely high.
Extinction is also relevant to the use of gene trees in species delimitation because many empirical schemes involve incomplete sampling of clades. For example, by focusing our sampling on Philippine Hipposideros, which is a polyphyletic assemblage [33,34], rather than Hipposideros in its entirety, we added extinction-like effects to the inferred phylogeny. However, our simulations suggest that extinction is far less important than other factors, such as the magnitude and variance of Ne. Nevertheless, Lohse [15] showed that incomplete sampling of demes could result in overestimates of diversity through the artificial construction of sequence clusters.
Because many methods that rely on gene trees to quantify diversification have low performance when Ne and SR are high [54], it is not surprising that the GMYC is inaccurate and imprecise under these circumstances. Consideration of the assumptions associated with Yule and coalescent models along with the results from our simulations argues that interpreting GMYC delimitations will require additional information on Ne and SR, and/or corroboration from independent character data.
(b). Species delimitation in Philippine round-leaf bats
Analysis of mitochondrial DNA sequences and echolocation call recordings indicates inconsistencies between current taxonomy and evolutionary relationships, highlighting the need for improvements in our capacity to quantify biodiversity rapidly. Two of the taxonomically defined species (H. ater and H. bicolor) have polyphyletic mitochondrial DNA [34]. Several of the currently recognized species for which we sampled multiple individuals from multiple populations were inferred to contain multiple species. Most Philippine species contained endemic lineages within the country, and one taxonomic species (H. diadema) was divided into three species at a larger geographical scale. With the caveats suggested by our simulations and other studies [15] in mind, these putative species are worthy of further taxonomic investigation.
Our data from echolocation calls were limited, but mostly consistent with GMYC species boundaries. For instance, the GMYC model suggested H. obscurus from the Greater Luzon and Greater Mindanao Pleistocene complexes are distinct species, and we found that they have echolocation calls differing by 20 kHz. GMYC also suggested H. ater and H. diadema are each single species across the oceanic Philippines (i.e. excluding Palawan); in each case, their echolocation calls on Bohol and Luzon differed by less than 4 kHz. However, in H. bicolor, the situation is less clear, where two populations were lumped in a single species by GMYC but their echolocation calls differed by approximately 23 kHz. A second Luzon Island sampling locality, however, had a peak frequency similar to the Bohol Island population, differing by only 3 kHz. This apparent inconsistency may stem from small sample sizes in the genetic and echolocation call datasets, from H. bicolor having multiple distinct echolocation calls, or from failure of the methods.
We estimate a plausible range of Ne for species of Hipposideros to be approximately 3 × 105–1.4 × 107, suggesting species of Hipposideros have Ne values from the mid to high range of our simulations. However, estimates of Ne from mitochondrial DNA should be treated with caution [55]. Qualitatively, the shape of the inferred mitochondrial DNA phylogeny (electronic supplementary material, figure S3) resembles simulated trees with moderate values of Ne (electronic supplementary material, figure S2), perhaps suggesting that GMYC has overestimated diversity in Philippine Hipposideros.
(c). Implications of biological complexity for general mixed Yule–coalescent species delimitation
The biogeography of Philippine plants and animals has been influenced by many factors, including ecological gradients associated with elevation, accretion of groups of islands into single islands, and the history of connectivity and isolation among islands currently separated by shallow seas [17,31,56]. This geographical system provides a challenging test case for methods of species delimitation. Island area, for instance, which varies over at least three orders of magnitude (i.e. those islands large enough to harbour endemic species), is presumably positively correlated with species' Ne. Hence, tenfold variation in Ne among closely related species—a level that resulted in highly inaccurate GMYC delimitations on simulated gene trees—may be common. Island age also varies greatly from less than 1 Myr to more than 20 Myr [57]. This potentially affects GMYC analyses by producing species of very different ages. Assuming equal Ne, old species are more likely to have monophyletic gene trees than young species—the GMYC can correctly assign individuals to species only when species are monophyletic in their gene trees. Furthermore, many islands were connected during periods of low sea level at different times and for different durations [29–31]. Thus, intermittent gene flow might produce yet another layer of complexity on the branching rates of gene trees. These complicating factors argue for corroboration of species limits derived from independent character sets.
5. Conclusions
The potential value of the GMYC lies with the hope of delimiting species with limited information, for example using mitochondrial DNA sequences. However, performance of the GMYC varies across biologically relevant values of Ne, variation in Ne and in the rate of speciation. When species have low Ne, limited variation in Ne and low SR, the method performs well. However, when Ne is high and/or highly variable, or when the rate of speciation is fast, GMYC has low accuracy and precision. Thus, interpreting the results from a GMYC analysis requires additional sources of information, such as estimates of Ne and SR, sequences of independent loci, and corroboration from morphology. As Marshall et al. [13] and others have stated, delimiting species is difficult and unlikely ever to be completely automated, or entirely free of arbitrary decisions. Ultimately, large datasets that integrate multiple character types will be most powerful [15]. Multilocus coalescent methods [8] are promising, but need to account for complicating factors such as genetic structure owing to isolation by distance and mild or intermittent barriers to gene flow.
In our application of the GMYC to Philippine Hipposideros, several putative new species are delimited. We consider these lineages important targets for future research, but our analyses are insufficient to demonstrate their status as species. Expanded geographical and genetic sampling, examinations of morphological variation, and collection of additional echolocation call data will all facilitate progress towards a taxonomy that adequately captures evolutionary history and informs conservation efforts.
Acknowledgements
The Philippine DENR and PAWB provided research permits. This research was funded by the NSF through grant nos DEB 0743491 and 0640737 to Rafe Brown and OISE 0965856 to J.A.E., and by the Field Museum's Brown Fund, the Negaunee Foundation and an Ontario Ministry of Research and Innovation fellowship to J.A.E. Phillip Alviola, Nonito Antoque, Danilo Balete, Rafe Brown, Jerry Cantil, Carl Oliveros, Joel Sarmiento, Cameron Siler and many others helped with fieldwork. The Universiti Kebangsaan Malaysia, Universiti Malaysia Sarawak, Museum of Texas Tech University, Museum of Southwestern Biology, Mindanao State University, Royal Ontario Museum, Cincinnati Museum Center and U.S. National Museum all provided tissue loans. Finally, Eric Rickart, Bryan Carstens and an anonymous reviewer gave helpful comments on earlier drafts.
References
- 1.McCloud D. S. 2010. Of least concern? Systematics of a cryptic species complex: Limnonectes kuhlii (Amphibia: Anura: Dicroglossidae). Mol. Phylogenet. Evol. 56, 991–1000 10.1016/j.ympev.2010.04.004 (doi:10.1016/j.ympev.2010.04.004) [DOI] [PubMed] [Google Scholar]
- 2.Peterson A. T., Ortega-Huerta M. A., Bartley J., Sánchez-Cordero V., Soberón J., Buddemeier R. H., Stockwell D. R. B. 2002. Future projections for Mexican faunas under global climate change scenarios. Nature 416, 626–629 10.1038/416626a (doi:10.1038/416626a) [DOI] [PubMed] [Google Scholar]
- 3.Esselstyn J. A., Maher S. P., Brown R. M. 2011. Species interactions during diversification and community assembly in an island radiation of shrews. PLoS ONE 6, e21885. 10.1371/journal.pone.0021885 (doi:10.1371/journal.pone.0021885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Setiadi M. I., McGuire J. A., Brown R. M., Zubairi M., Iskandar D. T., Andayani N., Supriatna J., Evans B. J. 2011. Adaptive radiation and ecological opportunity in Sulawesi and Philippine fanged frog (Limnonectes) communities. Am. Nat. 178, 221–240 10.1086/660830 (doi:10.1086/660830) [DOI] [PubMed] [Google Scholar]
- 5.Kim K. C., Byrne L. B. 2006. Biodiversity loss and the taxonomic bottleneck: emerging biodiversity science. Ecol. Res. 21, 794–810 10.1007/s11284-006-0035-7 (doi:10.1007/s11284-006-0035-7) [DOI] [Google Scholar]
- 6.Sites J. W., Marshall J. C. 2003. Delimiting species: a renaissance issue in systematic biology. Trends Ecol. Evol. 18, 462–470 10.1016/S0169-5347(03)00184-8 (doi:10.1016/S0169-5347(03)00184-8) [DOI] [Google Scholar]
- 7.Pons J., Barraclough T. G., Gomez-Zurita J., Cardoso A., Duran D. P., Hazell S., Kamoun S., Sumlin W. D., Vogler A. P. 2006. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609 10.1080/10635150600852011 (doi:10.1080/10635150600852011) [DOI] [PubMed] [Google Scholar]
- 8.Yang Z., Rannala B. 2010. Bayesian species delimitation using multilocus sequence data. Proc. Natl Acad. Sci. USA 107, 9264–9269 10.1073/pnas.0913022107 (doi:10.1073/pnas.0913022107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ence D. D., Carstens B. C. 2011. SpedeSTEM: a rapid and accurate method for species delimitation. Mol. Ecol. Res. 11, 473–480 10.1111/j.1755-0998.2010.02947.x (doi:10.1111/j.1755-0998.2010.02947.x) [DOI] [PubMed] [Google Scholar]
- 10.Knowles L. L., Carstens B. C. 2007. Delimiting species without monophyletic gene trees. Syst. Biol. 56, 887–895 10.1080/10635150701701091 (doi:10.1080/10635150701701091) [DOI] [PubMed] [Google Scholar]
- 11.Monaghan M. T., et al. 2009. Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Syst. Biol. 58, 298–311 10.1093/sysbio/syp027 (doi:10.1093/sysbio/syp027) [DOI] [PubMed] [Google Scholar]
- 12.Barraclough T. G., Hughes M., Ashford-Hodges N., Fujisawa T. 2009. Inferring evolutionary significant units of bacterial diversity from broad environmental surveys of single-locus data. Biol. Lett. 5, 425–428 10.1098/rsbl.2009.0091 (doi:10.1098/rsbl.2009.0091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall D. C., Hill K. B. R., Cooley J. R., Simon C. 2011. Hybridization, mitochondrial DNA phylogeography, and prediction of the early stages of reproductive isolation: lessons from New Zealand cicadas (Genus Kikihia). Syst. Biol. 60, 482–502 10.1093/sysbio/syr017 (doi:10.1093/sysbio/syr017) [DOI] [PubMed] [Google Scholar]
- 14.Vuataz L., Sartori M., Wagner A., Monaghan M. T. 2011. Toward a DNA taxonomy of alpine Rhithrogena (Ephemeroptera: Heptageniidae) using mixed Yule–coalescent analysis of mitochondrial and nuclear DNA. PLoS ONE 6, e19728. 10.1371/journal.pone.0019728 (doi:10.1371/journal.pone.0019728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohse K. 2009. Can mtDNA barcodes be used to delimit species? A response to Pons et al. (2006). Syst. Biol. 58, 439–442 10.1093/sysbio/syp039 (doi:10.1093/sysbio/syp039) [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulou A., Monaghan M. T., Barraclough T. G., Vogler A. P. 2009. Sampling error does not invalidate the Yule–coalescent model for species delimitation. A response to Lohse (2009). Syst. Biol. 58, 442–444 10.1093/sysbio/syp038 (doi:10.1093/sysbio/syp038) [DOI] [Google Scholar]
- 17.Brown R. M., Diesmos A. C. 2009. Philippines, biology. In Encyclopedia of islands (eds Gillespie R. G., Clague D. A.), pp. 723–732 Berkeley, CA: University of California Press [Google Scholar]
- 18.Balete D. S., Rickart E. A., Rosell-Ambal R. G. B., Jansa S., Heaney L. R. 2007. Descriptions of two new species of Rhynchomys Thomas (Rodentia: Muridae: Murinae) from Luzon Island, Philippines. J. Mammal. 88, 287–301 10.1644/06-MAMM-A-090R.1 (doi:10.1644/06-MAMM-A-090R.1) [DOI] [Google Scholar]
- 19.Esselstyn J. A. 2007. A new species of stripe-faced fruit bat (Chiroptera: Pteropodidae: Styloctenium) from the Philippines. J. Mammal. 88, 951–958 10.1644/06-MAMM-A-294R.1 (doi:10.1644/06-MAMM-A-294R.1) [DOI] [Google Scholar]
- 20.Heaney L. R., Balete D. S., Rickart E. A., Alviola P. A., Duya M. R. M. D., Duya M. V., Veluz M. J., VandeVrede L., Steppan S. J. 2011. Seven new species and a new subgenus of forest mice (Rodentia: Muridae: Apomys) from Luzon Island. Fieldiana Life Earth Sci. 2, 1–60 10.3158/2158-5520-2.1.1 (doi:10.3158/2158-5520-2.1.1) [DOI] [Google Scholar]
- 21.Siler C. D., Brown R. M. 2010. Phylogeny-based species delimitation in Philippine slender skinks (Reptilia: Squamata: Scincidae: Brachymeles): taxonomic revision of pentadctyl species groups and description of three new species. Herp. Monogr. 24, 55–85 10.1655/HERPMONOGRAPHS-D-10-00005.1 (doi:10.1655/HERPMONOGRAPHS-D-10-00005.1) [DOI] [Google Scholar]
- 22.Catibog-Sinha C., Heaney L. R. 2006. Philippine biodiversity: principles and practice. Quezon City, The Philippines: Haribon Foundation [Google Scholar]
- 23.Brown R. M., Guttman S. I. 2001. Phylogenetic systematics of the Rana signata complex of Philippine and Bornean stream frogs: reconsideration of Huxley's modification of Wallace's Line at the Oriental–Australian faunal zone interface. Biol. J. Linn. Soc. 76, 393–461 10.1111/j.1095-8312.2002.tb01704.x (doi:10.1111/j.1095-8312.2002.tb01704.x) [DOI] [Google Scholar]
- 24.Esselstyn J. A., Timm R. M., Brown R. M. 2009. Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in Southeast Asian shrews. Evolution 63, 2595–2610 10.1111/j.1558-5646.2009.00743.x (doi:10.1111/j.1558-5646.2009.00743.x) [DOI] [PubMed] [Google Scholar]
- 25.Oliveros C. H., Moyle R. G. 2010. Origin and diversification of Philippine bulbuls. Mol. Phylogenet. Evol. 54, 822–832 10.1016/j.ympev.2009.12.001 (doi:10.1016/j.ympev.2009.12.001) [DOI] [PubMed] [Google Scholar]
- 26.Siler C. D., Oaks J. R., Esselstyn J. A., Diesmos A. C., Brown R. M. 2010. Phylogeny and biogeography of Philippine bent-toed geckos (Gekkonidae: Cyrtodactylus) contradict a prevailing model of Pleistocene diversification. Mol. Phylogenet. Evol. 55, 699–710 10.1016/j.ympev.2010.01.027 (doi:10.1016/j.ympev.2010.01.027) [DOI] [PubMed] [Google Scholar]
- 27.Brown R. M., et al. 2010. Phylogeography and historical demography of Polypedates leucomystax in the island of Indonesia and the Philippines: evidence for recent human-mediated range expansion? Mol. Phylogenet. Evol. 57, 598–619 10.1016/j.ympev.2010.06.015 (doi:10.1016/j.ympev.2010.06.015) [DOI] [PubMed] [Google Scholar]
- 28.Heaney L. R., Dolar M. L., Balete D. S., Esselstyn J. A., Rickart E. A., Sedlock J. L. 2010. Synopsis of Philippine mammals. See http://archive.fieldmuseum.org/philippine_mammals/ (accessed 15 August 2011)
- 29.Dickerson R. E. 1928. Distribution of life in the Philippines. Manila, The Philippines: Philippine Bureau of Science [Google Scholar]
- 30.Kloss B. 1929. The zoo-geographical boundaries between Asia and Australia and some Oriental sub-regions. Bull. Raffles Mus. 2, 1–10 [Google Scholar]
- 31.Heaney L. R. 1985. Zoogeographic evidence for middle and late Pleistocene land bridges to the Philippine islands. Mod. Quatern. Res. SE Asia 9, 127–143 [Google Scholar]
- 32.Voris H. K. 2000. Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeogr. 27, 1153–1167 10.1046/j.1365-2699.2000.00489.x (doi:10.1046/j.1365-2699.2000.00489.x) [DOI] [Google Scholar]
- 33.Hill J. E. 1963. A revision of the genus Hipposideros. Bull. Br. Mus. Nat. Hist. (Zool.) 2, 1–129 [Google Scholar]
- 34.Murray S. W., Campbell P., Kingston T., Zubaid A., Francis C. M., Kunz T. H. 2011. Molecular phylogeny of hipposiderid bats from Southeast Asia and evidence of cryptic diversity. Mol. Phylogenet. Evol. 62, 597–611 10.1016/j.ympev.2011.10.021 (doi:10.1016/j.ympev.2011.10.021) [DOI] [PubMed] [Google Scholar]
- 35.Bates P. J. J., Rossiter S. J., Suyanto A., Kingston T. 2007. A new species of Hipposideros (Chiroptera: Hipposideridae) from Sulawesi. Acta Chiropterol. 9, 13–26 10.3161/1733-5329(2007)9[13:ANSOHC]2.0.CO;2 (doi:10.3161/1733-5329(2007)9[13:ANSOHC]2.0.CO;2) [DOI] [Google Scholar]
- 36.Douangboubpha B., Bumrungsri S., Soisook P., Satasook C., Thomas N. M., Bates P. J. J. 2010. A taxonomic review of the Hipposideros bicolor species complex and H. pomona (Chiroptera: Hipposideridae) in Thailand. Acta Chiropterol. 12, 415–438 10.3161/150811010X537990 (doi:10.3161/150811010X537990) [DOI] [Google Scholar]
- 37.Sukumaran J., Holder M. T. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26, 1569–1571 10.1093/bioinformatics/btq228 (doi:10.1093/bioinformatics/btq228) [DOI] [PubMed] [Google Scholar]
- 38.Yule G. U. 1924. A mathematical theory of evolution, based on the conclusions of Dr J. C. Willis. Phil. Trans. R. Soc. Lond. B 213, 21–87 10.1098/rstb.1925.0002 (doi:10.1098/rstb.1925.0002) [DOI] [Google Scholar]
- 39.Baldwin B. G., Sanderson M. J. 1998. Age and rate of diversification of the Hawaiian silversword alliance (Compositae). Proc. Natl Acad. Sci. USA 95, 9402–9406 10.1073/pnas.95.16.9402 (doi:10.1073/pnas.95.16.9402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendelson T. C., Shaw K. L. 2005. Rapid speciation in an arthropod. Nature 433, 375–376 10.1038/433375a (doi:10.1038/433375a) [DOI] [PubMed] [Google Scholar]
- 41.Moyle R. G., Filardi C. E., Smith C., Diamond J. M. 2009. Explosive Pleistocene diversification and hemispheric expansion of a ‘great speciator’. Proc. Natl Acad. Sci. USA 106, 1863–1868 10.1073/pnas.0809861105 (doi:10.1073/pnas.0809861105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillimore A. B., Price T. D. 2008. Density-dependent cladogenesis in birds. PLoS Biol. 6, 483–489 10.1371/journal.pbio.0060071 (doi:10.1371/journal.pbio.0060071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowe K. C., Aplin K. P., Baverstock P. R., Moritz C. 2011. Recent and rapid speciation with limited morphological disparity in the genus Rattus. Syst. Biol. 60, 188–203 10.1093/sysbio/syq092 (doi:10.1093/sysbio/syq092) [DOI] [PubMed] [Google Scholar]
- 44.Burgess R., Yang Z. 2008. Estimation of hominoid ancestral population sizes under Bayesian coalescent models incorporating mutation rate variation and sequencing errors. Mol. Biol. Evol. 25, 1979–1994 10.1093/molbev/msn148 (doi:10.1093/molbev/msn148) [DOI] [PubMed] [Google Scholar]
- 45.Russell A. L., Cox M. P., Brown V. A., McCracken G. F. 2011. Population growth of Mexican free-tailed bats (Tadarida brasiliensis mexicana) predates human agricultural activity. BMC Evol. Biol. 11, 88. 10.1186/1471-2148-11-88 (doi:10.1186/1471-2148-11-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slatkin M., Maddison W. P. 1989. A cladistic measure of gene flow inferred from the phylogeny of alleles. Genetics 123, 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nabholz B., Glémin S., Galtier N. 2008. Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol. Biol. Evol. 25, 120–130 10.1093/molbev/msm248 (doi:10.1093/molbev/msm248) [DOI] [PubMed] [Google Scholar]
- 49.Guyer C., Slowinski J. B. 1993. Adaptive radiation and the topology of large phylogenies. Evolution 47, 253–263 10.2307/2410133 (doi:10.2307/2410133) [DOI] [PubMed] [Google Scholar]
- 50.Mooers A. Ø., Page R. D. M., Purvis A., Harvey P. H. 1995. Phylogenetic noise leads to unbalanced cladistic tree reconstructions. Syst. Biol. 44, 332–342 [Google Scholar]
- 51.Nordborg M. 2001. Coalescent theory. In Handbook of statistical genetics (eds Balding D. J., Bishop M. J., Cannings C.), pp. 179–212 Chichester, UK: John Wiley & Sons, Inc [Google Scholar]
- 52.Wakeley J. 2009. Coalescent theory: an introduction. Greenwood Village, CO: Roberts & Company Publishers [Google Scholar]
- 53.Mooers A. Ø., Heard S. B. 1997. Inferring evolutionary process from phylogenetic tree shape. Q. Rev. Biol. 72, 31–54 10.1086/419657 (doi:10.1086/419657) [DOI] [Google Scholar]
- 54.Burbrink F. T., Pyron R. A. 2011. The impact of gene-tree/species-tree discordance on diversification rate estimation. Evolution 65, 1851–1861 10.1111/j.1558-5646.2011.01260.x (doi:10.1111/j.1558-5646.2011.01260.x) [DOI] [PubMed] [Google Scholar]
- 55.Nabholz B., Glémin S., Galtier N. 2009. The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol. Biol. 9, 54. 10.1186/1471-2148-9-54 (doi:10.1186/1471-2148-9-54) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heaney L. R., Rickart E. A. 1990. Correlations of clades and clines: geographic, elevational, and phylogenetic distribution patterns among Philippine mammals. In Vertebrates in the tropics (eds Peters G., Hutterer R.), pp. 321–332 Bonn, Germany: Alexander Koenig Zoological Research Institute and Zoological Museum [Google Scholar]
- 57.Yumul G., Dimalanta C., Queaño K., Marquez E. 2009. Philippines, geology. In Encyclopedia of islands (eds Gillespie R. G., Clague D. A.), pp. 732–738 Berkeley, CA: University of California Press [Google Scholar]