Abstract
The rate of environmental niche evolution describes the capability of species to explore the available environmental space and is known to vary among species owing to lineage-specific factors. Trophic specialization is a main force driving species evolution and is responsible for classical examples of adaptive radiations in fishes. We investigate the effect of trophic specialization on the rate of environmental niche evolution in the damselfish, Pomacentridae, which is an important family of tropical reef fishes. First, phylogenetic niche conservatism is not detected in the family using a standard test of phylogenetic signal, and we demonstrate that the environmental niches of damselfishes that differ in trophic specialization are not equivalent while they still overlap at their mean values. Second, we estimate the relative rates of niche evolution on the phylogenetic tree and show the heterogeneity among rates of environmental niche evolution of the three trophic groups. We suggest that behavioural characteristics related to trophic specialization can constrain the evolution of the environmental niche and lead to conserved niches in specialist lineages. Our results show the extent of influence of several traits on the evolution of the environmental niche and shed new light on the evolution of damselfishes, which is a key lineage in current efforts to conserve biodiversity in coral reefs.
Keywords: trophic specialization, environmental niche, phylogenetic niche conservatism, evolutionary rate heterogeneity, evolutionary model
1. Introduction
The environment often plays a crucial role in species evolution and has been the focus of numerous studies since the description of Darwin's theory of natural selection [1,2]. What is still unclear is how particular phenotypes could influence, in return, environmental niche evolution [3]. It has been shown that species are restricted through both biotic and abiotic factors at their environmental margins [4], but the variability in their ability to adapt to previously unsuitable conditions is less well understood [5]. While rapid environmental niche shifts within radiating clades have been demonstrated [6,7], the idea that species retain their ancestral niche during diversification has also recently received support [8]. The variation in propensity of species to retain their niche can be linked with important differences in life-history traits, such as growth form in flowering plants, that influence the rate of climatic niche evolution [9]. In this context, the emergence of a novel trait that allows the exploitation of untapped resources may favour the colonization of novel environments and be followed by species radiation. By contrast, a trait conferring highly specialized use of a restricted range of resources in an environment may enhance species performance in this particular environment at the expense of adaptability to novel conditions. For instance, Cooper et al. [10] showed that in mammals, the thermal habitat niche of diet specialists has been more conserved during their evolution than the niche of diet generalists. This suggests feedback between evolutionary and ecological processes that potentially plays a role in shaping adaptation of species to their environment [3]. Investigating how the niche evolves in interaction with the emergence of novel characters can illustrate how key phenotypic traits constrain or amplify the rate of environmental niche evolution [4,9].
The trophic specialization of species can drive phenotypic evolution [11] and the adaptive radiation of the East African rift lakes cichlids exemplifies this spectacularly [12]. While cichlid species show great trophic diversity, there is exceptional morphological convergence among species that specialize on the same diet [13]. When a species radiation is driven by ecological opportunity [14,15], species environmental niches may be constrained because of adaptation for specific habitats [16]. Therefore, shifts in trophic specialization in additional groups of fishes could greatly influence the evolutionary history of lineages and, in particular, their rate of niche evolution.
The damselfishes (Pomacentridae) inhabit tropical and temperate near-shore waters around the globe, with the greatest species richness located along coral reefs [17]. Of the approximately 350 species, most are small (less than 30 cm), colourful and often numerically dominant in reef fish communities [18,19]. Damselfishes are usually separated into three main diet classes [20]: (i) herbivorous benthic feeders (e.g. the genera Dischistodus and Stegastes), (ii) an intermediate omnivorous group (e.g. Pomacentrus spp.), and (iii) pelagic feeders mainly eating planktonic prey (e.g. Chromis spp.). Other diet specializations exist, for example, species focusing on corals or sponges, but these comprise only a handful of species [20]. The three main diet types evolved independently many times along the phylogeny of the damselfish, and lineages sharing the same diet are characterized by convergent morphological evolution [20,21].
In damselfishes, trophic groups generally share patterns of social behaviour and degree of territoriality. Herbivores and omnivores are usually territorial and solitary, while planktivores form schools in the water column and actively guard territories only during reproduction. The territorial behaviour of herbivorous damselfishes is associated with the developement of cultivation mutualisms with algae [22,23]. Indeed, they maintain turfs composed of delicate filamentous algae (e.g. Polysiphonia sp.), which would be outcompeted by other algae or eaten by other reef herbivores, if it were not for the protection and careful tending provided by the damselfish. The algae found in such territories are the most easily eaten and digested by damselfishes, which actively select the algae species they cultivate [24]. The effect of territorial damselfishes on reef communities has led several authors to consider these as keystone species to support biodiversity in coral reef habitats [25–28]. The strong link between the herbivorous damselfishes and the algae makes this trophic group highly specialized, while planktivorous species are less specialized because they can feed on a wide variety of planktonic organisms. This difference in specialization level among trophic groups has probably influenced the rate of environmental niche evolution of damselfishes. If algal crop species have narrow distributions [23], specialized species could then be restricted by the environmental requirements of the algae they are tending. By contrast, planktivores that lack this constraint will probably have higher rates of environmental niche evolution. Some omnivorous damselfishes also tend algal crops, but they further use the algae to trap non-living organic matter, which represents a large fraction of their diet [26,29]. This suggests that omnivores are not ecologically intermediate between herbivores and planktivores, but are actually more similar to herbivorous damselfishes. The worldwide distribution of the damselfish family makes it an excellent group to study environmental niche evolution in relation to species trophic specialization. In this paper, we first assess whether the environmental niche of the damselfishes is phylogenetically conserved. We then compare the niche position and overlap between groups of species displaying distinct trophic specialization. Finally, we test whether the rate of environmental niche evolution varies significantly in the family and whether this variation is linked with the level of species trophic specialization.
2. Methods
(a). Phylogenetic analysis
We assembled a phylogenetic tree of the damselfish family by gathering DNA sequences from GenBank for five different DNA regions (three mitochondrial genes: 12S, 16S and cytB; and two nuclear genes: RAG1 and RAG2) for 175 fish species (169 damselfishes + 6 outgroup taxa; see the electronic supplementary material). The sequences were aligned with Muscle [30] to yield a matrix of a total length of 7383 nucleotides. We identified the GTR+G model as the best model by likelihood ratio tests in MrAIC [31], and used this substitution model for each gene region independently in MrBayes v. 3.1.2 [32]. We ran four chains in parallel for 107 generations and sampled trees every 1000 generations for a total of 10 000 trees. We checked model parameters for stabilization in Tracer [33] and removed the first 3000 trees as burn-in. From the posterior distribution, we randomly extracted 100 phylograms having their branch length relative to the expected number of substitution per site for subsequent analyses (see below).
We estimated divergence time with a relaxed clock model in BEAST [33], drawing rates from a lognormal distribution. Two fossils were available for calibration. We placed a lognormal prior (mean = 2, s.d. = 1, offset = 50 Myr) on the crown node of the damselfishes following the earliest pomacentrid record [34]. The second fossil was placed at the crown node of the cichlid outgroup [35] with a lognormal prior (mean = 2, s.d. = 1, offset = 5.33 Myr). The models of substitution were defined similarly as for the MrBayes analysis. We ran a single Markov chain Monte Carlo (MCMC) process for 5 × 107 generations, sampling trees every 1000 generations. Finally, the first 15 000 trees were removed as burn-in after checking the sampled model parameters in Tracer [33]. We then randomly extracted from the posterior distribution 100 chronograms and used their branch length relative to time in further analyses.
(b). Data retrieval and environmental niche similarity between trophic groups
We extracted dietary information on every damselfish included in this study from the literature [19]. We restricted our study to the three main trophic groups present in the damselfishes [20,21,36]: herbivores, omnivores and planktivores. The other trophic specializations that exist in the damselfishes are represented by less than 2 per cent of species [19] and are, thus, not informative for our comparative study.
We retrieved occurrence data for damselfishes from the Ocean Biogeographic Information System web site (www.iobis.org). The dataset had 62 380 occurrence records for the 169 damselfish species included in our study (average per species = 383; see the electronic supplementary material). We used the World Ocean Atlas 2009 [37] to extract at a global resolution of 1° annual mean values for salinity, temperature, apparent oxygen utilization, oxygen saturation, nitrate, oxygen, phosphate and silicate concentration. These variables are known to describe adequately the environmental niche of marine organisms [38]. We used outlying mean index (OMI) ordination [39], as implemented in the ade4 package [40] in R [41], to characterize species' environmental niches. The OMI analysis is an ordination technique that gives the species' average position within environmental space by maximizing the mean-squared distance between the centroid of the environmental space used by the species and the centroid of the available environment [39]. This measure has been used in many studies [42] to describe the average relative position of species in a multivariate environmental space. We used the axes of the OMI ordination that explained a meaningful proportion of the total variation to model the evolutionary dynamics of environmental niches of the damselfishes on phylogenetic trees.
We compared the environmental space used by the species belonging to each trophic specialization using a recently developed framework [43]. In contrast to previous methods that measure overlap in a geographical space [44], this approach measured the overlap between species in the relevant environmental space and, thus, related closely to the study of rates of environmental niche evolution. The robustness of the method has been demonstrated by its ability to accurately recover the known environmental niche overlap of simulated species [43]. We first ran a principal component analysis (PCA) on the total environmental niche available in the study area (here the world oceans). Then, environmental space occupied by each group of species was projected in the environmental space available. Next, a kernel density function was applied to determine for each group, the ‘smoothed’ density of occurrences of each pixel in the environmental space [43]. Finally, the density of occurrences was divided by the density of the environment in each focal pixel to obtain a measure of the density of the species relative to the availability of environmental space [43]. This measure was then used to test for environmental niche equivalency between trophic groups using Schoener's D metric [44], ranging from 0 (no niche overlap) to 1 (complete overlap). We used a phylogenetic multivariate analysis of variance (MANOVA) to test for significant difference in species mean environmental niche values among trophic groups. This test, as implemented in the R package geiger [45], took into account phylogenetic non-independence among species and used a single-rate Brownian motion (BM) model of character evolution on the first two axes of the OMI ordination.
(c). Phylogenetic signal and phylogenetic niche conservatism
Phylogenetic signal is defined as the level of dependency among species trait values owing to their phylogenetic relatedness [46]. We used the λ [47] and K [48] measures in the R package phytools [49] and samples of both phylograms and chronograms to assess phylogenetic signal in the environmental niche as described by OMI scores. Values of K or λ close to one are expected for data that follow a BM model of character evolution, while values close to 0 are diagnostic of weak or no phylogenetic structure. Because the comparative methods we want to apply are based on BM, analysing a set of phylogenies which has mean K or λ close to one conforms to the main assumption of the methods [50].
The amount of phylogenetic signal shown by an ecological character can also indicates presence of phylogenetic niche conservatism in the clade under investigation [51]. Under this definition, phylogenetic niche conservatism is present in a clade if the phylogenetic signal is significantly greater than one, the expected value of a BM fit. We performed simulations to test K > 1 by comparing the observed value of K with 1000 K values measured on characters simulated under perfect BM (i.e. K = 1). Then, we used a one-tailed test to estimate how many simulations could lead to a K value as small as the one obtained on the environmental niche described by the OMI axes. Such a test cannot be done with λ, which is usually bounded between 0 and 1 [45,49]. This definition of phylogenetic niche conservatism is disputed [8] and the use of Brownian rate of evolution is now usually preferred for assessing phylogenetic niche conservatism [10,52].
(d). Rates of environmental niche evolution among trophic levels
We measured the rate of environmental niche evolution in the damselfishes using the non-censored version of a Brownie analysis [53]. We compared using Akaike information criterion with a correction for a finite sample size (AICc) the likelihood of a single-rate BM model to that of a multiple rate model in which rates differ between the three trophic groups. We estimated the rates of evolution for values on the first two OMI axes. To assign internal branches of the phylogeny to a particular trophic specialization, we first estimated the transition matrix between trophic states using BayesTraits [54]. We used a non-symmetric model of evolution allowing different transition rates between trophic states. We ran a single Markov chain for 106 generations sampling each 1000 generations and used default uniform prior for the estimated parameters. After removal of the first 100 sampled generations as burn-in, we averaged the posterior distribution of the transition rates. The resulting transition matrix was used to map stochastically the trophic groups on the phylogenies [55]. We did this step using the R package phytools [49] that implements stochastic mapping of discrete characters. To account for uncertainty in reconstruction of character history, we replicated the stochastic mapping 100 times on each phylogenetic tree, we also performed the reconstruction using only two states by merging the herbivores and the omnivores together as both herbivores and omnivores tend algal crops (see the electronic supplementary material).
Current approaches (see above) that compare the rate of evolution of a continuous character along the branches of a phylogenetic tree need a priori information on where changes in rates happen along the phylogeny. To accommodate this uncertainty in the evolutionary process and illustrate potential heterogeneity in rates among trophic groups, we used a recently developed method that uses Bayesian inference to sample phenotypic evolutionary rates, implemented in the R package AUTEUR [56]. This approach models rate heterogeneity in a Bayesian framework by using reversible-jump MCMC [57,58]. During the optimization of parameters, the Markov chain jumps from models differing in number and position of rate shifts in the phylogeny [56,59]. The outcome is a posterior distribution of evolutionary rates for each branch in the phylogeny. A posterior distribution of model complexity is also produced and can be used to assess whether the multiple-rate model has a better fit than the single-rate model, the Bayesian equivalent of the likelihood ratio test used in Brownie.
To infer the rate of environmental niche evolution in the damselfishes, we used the species position on the first two axes of the OMI ordination and ran two parallel Markov chains sampling parameters every 1000 generations for a total of 4 × 106 generations. We assessed proper convergence of the MCMC chains using diagnostic metrics implemented in the R package coda [60]. After convergence of the chains and removal of the first 1000 samples as burn-in, we obtain marginalized distributions of relative rates for each branch of the tree. Following the authors' recommendations [56], we then computed the rates as a weighted average of posterior rate estimates. The weighting was determined by branch length, allowing longer branches to contribute for a greater weight to the scaled measure of evolutionary rate [56]. The relative rates of environmental niche evolution of every branch section corresponding to a particular trophic group could be extracted because trophic groups were previously mapped to branches using stochastic mapping. To account for phylogenetic uncertainty, we repeated the analysis on the sample of 100 phylograms obtained from MrBayes.
3. Results
(a). Environmental niche
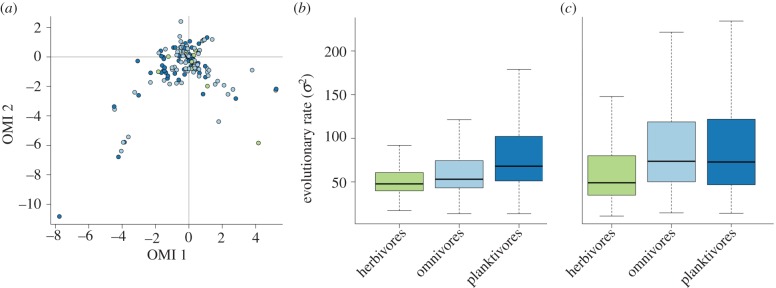
The first and second axis of the OMI ordination (figure 1a) explained, respectively, 35 and 31 per cent of the total variation. The other axes explained only small fractions of the variation and were not considered further in the analyses. We measured the environmental overlap between the environmental niches of species belonging to the three trophic groups (figure 2). Herbivorous damselfishes had less overlap with the planktivores (Schoener's D = 0.15) than with the omnivores (Schoener's D = 0.19), while omnivores and planktivores had the largest overlap (Schoener's D = 0.48). All tests of niche equivalency between trophic groups indicated that the niches occupied by the three groups of species were not equivalent (p < 0.01 for the three pairwise comparisons). However, when testing for a difference in mean niche position between species of different trophic levels, the phylogenetic MANOVA gave a non-significant result (p = 0.9).
Figure 1.
(a) Species environmental niche position in the OMI coordinates. Herbivores are in green, omnivores in light blue and planktivores in dark blue. The green outlier in the bottom right is Parma microlepis, the only herbivore that colonized temperate waters. Relative rates of environmental niche evolution of the three trophic groups inferred in AUTEUR for (b) the first and (c) second OMI axis. The boxplots represent the weighted average of posterior rate estimates per trophic group of each of the 100 phylograms.
Figure 2.
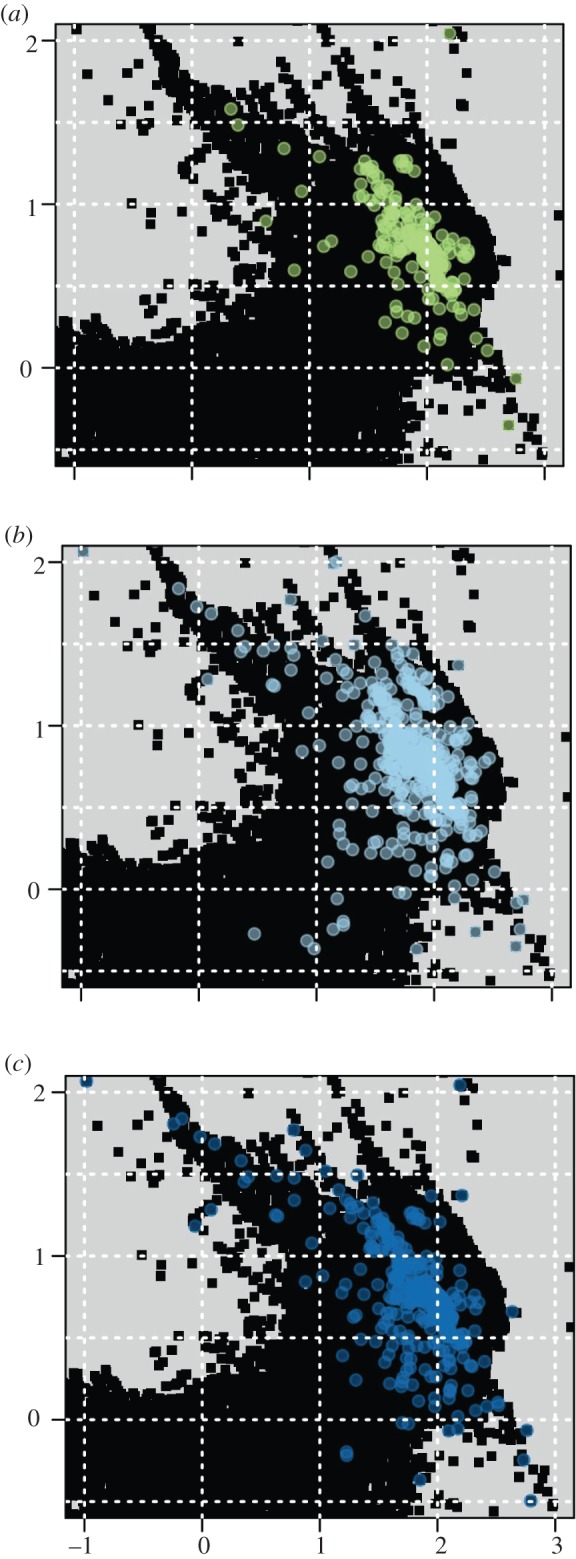
The environmental niche of the three trophic groups shown in the gridded environmental space of the PCA (the first principal components are on the x-axis and the second on the y-axis). The black squares illustrate the available environment. The environmental space occupied by each trophic group is shown in the following panels: (a) herbivores, (b) omnivores, and (c) planktivores.
(b). Phylogenetic signal
Posterior probabilities of nodes on both phylograms and chronograms were usually high and the topologies consistent with previous studies [17]. We measured K and λ for both the first and second axes of the OMI ordination on 100 phylograms and chronograms. The mean λ and K of the phylograms were closer to one than the λ and K of the chronograms (table 1). This suggests that, in our data, phylograms having branch lengths in expected number of substitutions per site are more suited for comparative methods than chronograms as they are closer to the assumed BM [50]. We thus performed further analyses on the sample of 100 phylograms, but provide the results from chronograms in the electronic supplementary material for completeness. The value of K was always lower and significantly different from K > 1 (table 1), suggesting that the environmental niche of damselfishes is not conserved under this definition [51].
Table 1.
Mean and standard deviation of the phylogenetic signal shown by the first two axes of the OMI and calculated across the 100 phylogenies drawn from each posterior distribution. (The p-values for K show if the phylogenetic signal is significantly different from K > 1 (* = p < 0.01).)
| OMI 1 |
OMI 2 |
|||
|---|---|---|---|---|
| K | λ | K | λ | |
| chronograms | 0.11 ± 0.03* | 0.07 ± 0.07 | 0.17 ± 0.04* | 0.47 ± 0.08 |
| phylograms | 0.14 ± 0.02* | 0.62 ± 0.14 | 0.25 ± 0.04* | 0.67 ± 0.07 |
(c). Environmental niche evolution
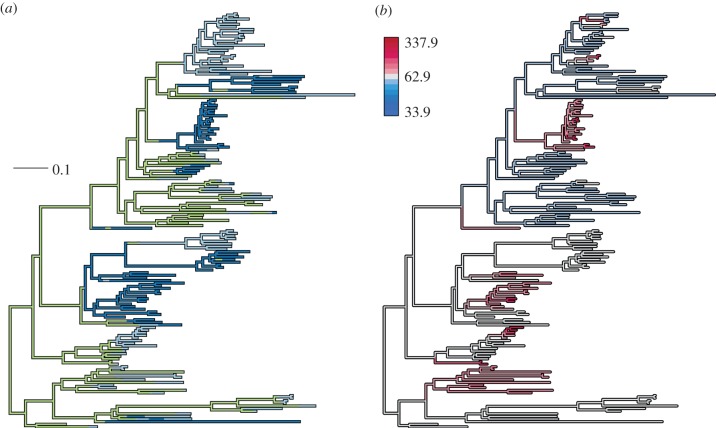
The multiple-rate model had better support than the single-rate null model for both the first OMI axis (ΔAICc = 16.43) and the second (ΔAICc = 4.83). When measured on OMI axis-1, herbivores had the slowest rate of environmental niche evolution (mean σ2 = 33.8). The omnivores were intermediate (mean σ2 = 78.9), while the planktivores were the fastest (mean σ2 = 96.1). The herbivores still had the slowest rate on the OMI axis-2 (mean σ2 = 50.44), but the pattern was reversed between omnivores (mean σ2 = 80.3) and planktivores (mean σ2 = 72). The AUTEUR analysis yielded similar results (figure 1b,c). After removal of the burn-in, the effective samples size of the rate parameter of the MCMC chains showed good convergence (mean effective sample size = 475 ± 192). The posterior distribution of model complexities always gave high support to multiple-rate models (cumulative posterior probabilities: OMI axis-1 = 0.99, OMI axis-2 = 1; see the electronic supplementary material), showing that the rate of environmental niche evolution is heterogeneous among damselfish species. On OMI axis-1, the model having the highest posterior probability (0.24) was a model with seven rates distributed among the branches of the phylogeny. Herbivores had the slowest rate and the planktivores the fastest, omnivores were situated in between on the OMI axis 1 (figure 1b) and had comparable rates with the planktivores on OMI axis-2 (figure 1c). Merging the herbivores and omnivores had no effect on the results as the herbivore/omnivore group still had a slower rate than the planktivores.
4. Discussion
Our results show high support for differential rates of environmental niche evolution among damselfish trophic groups (figures 1 and 3), as well as absence of phylogenetic niche conservatism in the family as indicated by phylogenetic signal. The approach we used to test for phylogenetic niche conservatism has been criticized as different evolutionary processes could lead to the same phylogenetic signal [8,46]. We nevertheless applied this test as it can be discussed in the light of the rate of environmental niche evolution that has also been proposed as a description of phylogenetic niche conservatism [8,10]. The three trophic groups had a relatively small total overlap in the environmental niche space (figure 2). As the availability of climate in the environmental space was taken into account during the overlap calculation, the overlap values obtained are small because points that occurred in rare climates were given a higher weight [43]. The phylogenetic MANOVA showed that the mean environmental niche was not significantly different between trophic groups. Indeed, most damselfishes occur in tropical reef ecosystems, while few species have colonized temperate waters. At the species level, the pattern of similar mean environmental niches among groups but small environmental overlap among species in different groups can be explained if trophic groups have different rates of expansion in environmental space. Our finding that the rates of environmental niche evolution of herbivores were slower than the rate of planktivores supports this (figure 1b,c). Our assessment of rates of environmental niche evolution indicates that although mean niche is similar between groups, planktivores diverged to occupy environmental space faster than did herbivores.
Figure 3.
Damselfish phylogeny with (a) one realization of the stochastic mapping that paints trophic groups on the branches (in expected substitution per site). Herbivores are in green (algivores), omnivores in light blue and planktivores in dark blue. (b) Inferred relative rates of environmental niche evolution that range from slow in blue to fast in red.
The highly specialized link between herbivore damselfishes and the algae species they tend can explain constraints on evolutionary rate. The environmental niche of herbivores is defined by their intrinsic physiological tolerances, but also by the ecological requirement of their algal crops [61]. This would suggest that the slower exploration of environmental space is linked with this additional constraint. By contrast, the planktivores that are more generalists in terms of the ecological requirements of their food could explore a more diverse environmental space. Unlike algae, plankton is a resource that is not linked with particular environments in reef ecosystems as it is composed of a multitude of species, and thus probably imposes weaker constraints on planktivore niche evolution. Omnivores are more similar to herbivores than planktivores in this regard because they exhibit similar territorial behaviour. Still our results show that their evolutionary rate is faster than that of herbivores (figure 1b,c). As the algae present in their territories are more used to trap sediments than as actual food, it is probable that the omnivorous damselfishes are less specialized on tending specific species of algae [26]. With a greater choice of potential crop species, the omnivores are less evolutionary constrained than herbivores. Such a pattern of specialists having a slower environmental niche evolutionary rate than generalists has also been found in mammals [10], suggesting that our conclusions could be true in a broader range of taxa. The relative species richness and abundance of marine herbivorous fishes were elsewhere negatively correlated with latitude, implying that temperature-related processes are probably important in their distribution pattern [62]. While such global pattern can inform us about general metabolic constraints on herbivory, it unlikely provides a complete explanation of our findings as the vast majority of the damselfishes we studied are found only in tropical waters.
Depending on the choice of definition of phylogenetic niche conservatism, we either could not detect it by testing for phylogenetic signal or found that specialist damselfishes had more conserved niches than the generalists, as shown by rates of environmental evolution. Indeed, if a clade shows a relatively slow rate of niche evolution, the divergence in niche values will be less between species and will consequently lead to more-conserved niches [8,10]. By integrating these two views, we suggest that the damselfishes, in general, do not show niche conservatism but whenever a lineage becomes specialized; niche evolution is then constrained and becomes more conserved over time than in generalist lineages. Trophic specialization could, therefore, impede the environmental niche evolution of a clade, and taking this effect into account is paramount when measuring phylogenetic niche conservatism. It also shows the effect of scale [8] as phylogenetic conservatism can only be detected in a localized region of the damselfish tree.
The evolution of environmental niche in a clade is usually assessed by measuring the level of phylogenetic niche conservatism. By looking at the relative effects of a particular phenotype, in this case, the level of trophic specialization of species, we obtain a clearer view of the processes shaping the evolution of environmental tolerance (figure 3). This approach can more closely relate to the extremely complex processes at work in nature. Our results suggest that biotic factors, such as the environmental requirements of food resources, can constrain the evolution of the environmental niche of the consumers’ dependent on the resource. A possible caveat of our study is that we defined trophic specialization using only three classes. Probably trophic strategies resolve more finely into a larger number of classes. This could explain why we find omnivores are more similar to herbivores on the OMI axis 1 and planktivores on the OMI axis 2. Nevertheless, our classification still allows an outline of the general pattern, even though other factors could also play a role in shaping the environmental niche evolution of damselfishes. Many ecological traits, such as territorial behaviour, are linked to trophic specialization and their potential effects are difficult to disentangle. However, this should not impact our results as specialization towards a food resource in a particular micro-habitat may constrain the rate of environmental niche evolution in a similar manner to diet specialization [8].
Biotic interactions are common and can restrict the realized niche of species [63]. In the case of the damselfishes, this could lead to the larger distribution of planktivores in environmental space compared with the other trophic groups. This suggests an important factor to take into account when trying to predict future distributions of species under climate-change scenarios [64]. In particular, failing to consider the distribution of the algae could yield spurious predicted distributions because biotic interactions might well here constrain the niche of herbivores. In conclusion, by integrating trophic specialization within a phylogenetic context, we were able to show that biotic interactions (here between herbivores and algae) are consistent with constraint of present species niches, a pattern that has developed over evolutionary time.
Acknowledgements
We thank Anne Dubuis, Anna Kostikova, Rafael Wüest, Blaise Petitpierre, Olivier Broennimann, Nils Arrigo and three anonymous reviewers for helpful discussions and comments on previous versions of this manuscript. This work was funded by grant no. CRS113-125240 from the Swiss National Science Foundation to N.S., P.B.P. and N.E.Z. This work received support from the Vital-IT facilities from the Swiss Institute of Bioinformatics.
References
- 1.Darwin C. 1859. On the origin of species. London, UK: John Murray [Google Scholar]
- 2.Barrett R. D. H., Schluter D. 2008. Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 10.1016/j.tree.2007.09.008 (doi:10.1016/j.tree.2007.09.008) [DOI] [PubMed] [Google Scholar]
- 3.Schoener T. W. 2011. The newest synthesis: understanding the interplay of evolutionary and ecological dynamics. Science 331, 426–429 10.1126/science.1193954 (doi:10.1126/science.1193954) [DOI] [PubMed] [Google Scholar]
- 4.Wiens J. J. 2011. The niche, biogeography and species interactions. Phil. Trans. R. Soc. B 366, 2336–2350 10.1098/rstb.2011.0059 (doi:10.1098/rstb.2011.0059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaston K. J. 2009. Geographical range limits of species. Proc. R. Soc. B 276, 1391–1393 10.1098/rspb.2009.0100 (doi:10.1098/rspb.2009.0100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans M. E. K., Smith S. A., Flynn R. S., Donoghue M. J. 2009. Climate, niche evolution, and diversification of the ‘bird-cage’ evening primroses (Oenothera, Sections Anogra and Kleinia). Am. Nat. 173, 225–240 10.1086/595757 (doi:10.1086/595757) [DOI] [PubMed] [Google Scholar]
- 7.Kozak K. H., Wiens J. J. 2010. Accelerated rates of climatic-niche evolution underlie rapid species diversification. Ecol. Lett. 13, 1378–1389 10.1111/j.1461-0248.2010.01530.x (doi:10.1111/j.1461-0248.2010.01530.x) [DOI] [PubMed] [Google Scholar]
- 8.Wiens J. J., et al. 2010. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324 10.1111/j.1461-0248.2010.01515.x (doi:10.1111/j.1461-0248.2010.01515.x) [DOI] [PubMed] [Google Scholar]
- 9.Smith S. A., Beaulieu J. M. 2009. Life history influences rates of climatic niche evolution in flowering plants. Proc. R. Soc. B 276, 4345–4352 10.1098/rspb.2009.1176 (doi:10.1098/rspb.2009.1176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper N., Freckleton R. P., Jetz W. 2011. Phylogenetic conservatism of environmental niches in mammals. Proc. R. Soc. B 278, 2384–2391 10.1098/rspb.2010.2207 (doi:10.1098/rspb.2010.2207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin C. H., Wainwright P. C. 2011. Trophic novelty is linked to exceptional rates of morphological diversification in two adaptive radiations of Cyprinodon pupfish. Evolution 65, 2197–2212 10.1111/j.1558-5646.2011.01294.x (doi:10.1111/j.1558-5646.2011.01294.x) [DOI] [PubMed] [Google Scholar]
- 12.Seehausen O. 2006. African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. B 273, 1987–1998 10.1098/rspb.2006.3539 (doi:10.1098/rspb.2006.3539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salzburger W. 2009. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol. Ecol. 18, 169–185 10.1111/j.1365-294X.2008.03981.x (doi:10.1111/j.1365-294X.2008.03981.x) [DOI] [PubMed] [Google Scholar]
- 14.Mahler D. L., Revell L. J., Glor R. E., Losos J. B. 2010. Ecological opportunity and the rate of morphological evolution in the diversification of greater Antillean Anoles. Evolution 64, 2731–2745 10.1111/j.1558-5646.2010.01026.x (doi:10.1111/j.1558-5646.2010.01026.x) [DOI] [PubMed] [Google Scholar]
- 15.Losos J. B. 2010. Adaptive radiation, ecological opportunity, and evolutionary determinism. American Society of Naturalists E. O. Wilson Award Address. Am. Nat. 175, 623–639 10.1086/652433 (doi:10.1086/652433) [DOI] [PubMed] [Google Scholar]
- 16.Veech J. A., Crist T. O. 2007. Habitat and climate heterogeneity maintain beta-diversity of birds among landscapes within ecoregions. Glob. Ecol. Biol. 16, 650–656 10.1111/j.1466-8238.2007.00315.x (doi:10.1111/j.1466-8238.2007.00315.x) [DOI] [Google Scholar]
- 17.Cooper J. W., Smith L. L., Westneat M. W. 2009. Exploring the radiation of a diverse reef fish family: phylogenetics of the damselfishes (Pomacentridae), with new classifications based on molecular analyses of all genera. Mol. Phylogenet. Evol. 52, 1–16 10.1016/j.ympev.2008.12.010 (doi:10.1016/j.ympev.2008.12.010) [DOI] [PubMed] [Google Scholar]
- 18.Allen G. R. 1975. Damselfishes of the South seas. Neptune City, NJ: T.F.H. Publications [Google Scholar]
- 19.Allen G. R. 1991. Damselfish of the world. Melle, Germany: Mergus Publishers [Google Scholar]
- 20.Cooper W. J., Westneat M. W. 2009. Form and function of damselfish skulls: rapid and repeated evolution into a limited number of trophic niches. BMC Evol. Biol. 9, 24 10.1186/1471-2148-9-24 (doi:10.1186/1471-2148-9-24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilar-Medrano R., Frédérich B., De Luna E., Balart E. F. 2011. Patterns of morphological evolution of the cephalic region in damselfishes (Perciformes: Pomacentridae) of the Eastern Pacific. Biol. J. Linn. Soc. 102, 593–613 10.1111/j.1095-8312.2010.01586.x (doi:10.1111/j.1095-8312.2010.01586.x) [DOI] [Google Scholar]
- 22.Hata H., Kato M. 2006. A novel obligate cultivation mutualism between damselfish and Polysiphonia algae. Biol. Lett. 2, 593–596 10.1098/rsbl.2006.0528 (doi:10.1098/rsbl.2006.0528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hata H., Watanabe K., Kato M. 2010. Geographic variation in the damselfish-red alga cultivation mutualism in the Indo-West Pacific. BMC Evol. Biol. 10, 185 10.1186/1471-2148-10-185 (doi:10.1186/1471-2148-10-185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hata H., Kato M. 2002. Weeding by the herbivorous damselfish Stegastes nigricans in nearly monocultural algae farms. Mar. Ecol. Prog. Ser. 237, 227–231 10.3354/meps237227 (doi:10.3354/meps237227) [DOI] [Google Scholar]
- 25.Ceccarelli D. M., Jones G. P., McCook L. J. 2005. Effects of territorial damselfish on an algal-dominated coastal coral reef. Coral Reefs 24, 606–620 10.1007/s00338-005-0035-z (doi:10.1007/s00338-005-0035-z) [DOI] [Google Scholar]
- 26.Ceccarelli D. M. 2007. Modification of benthic communities by territorial damselfish: a multi-species comparison. Coral Reefs 26, 853–866 10.1007/s00338-007-0275-1 (doi:10.1007/s00338-007-0275-1) [DOI] [Google Scholar]
- 27.Hoey A. S., Bellwood D. R. 2009. Damselfish territories as a refuge for macroalgae on coral reefs. Coral Reefs 29, 107–118 10.1007/s00338-009-0567-8 (doi:10.1007/s00338-009-0567-8) [DOI] [Google Scholar]
- 28.Jones G., Santana L., McCook L., McCormick M. 2006. Resource use and impact of three herbivorous damselfishes on coral reef communities. Mar. Ecol. Prog. Ser. 328, 215–224 10.3354/meps328215 (doi:10.3354/meps328215) [DOI] [Google Scholar]
- 29.Wilson S., Bellwood D. 1997. Cryptic dietary components of territorial damselfishes (Pomacentridae, Labroidei). Mar. Ecol. Prog. Ser. 153, 299–310 10.3354/meps153299 (doi:10.3354/meps153299) [DOI] [Google Scholar]
- 30.Edgar R. C. 2004. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 (doi:10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nylander J. A. A. 2004. MrAIC.pl. version 1.3. Program distributed by the author. Sweden: Evolutionary Biology Centre, Uppsala University; See http://www.abc.se/~nylander/ [Google Scholar]
- 32.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 33.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellwood D. R., Sorbini L. 1996. A review of the fossil record of the Pomacentridae (Teleostei: Labroidei) with a description of a new genus and species from the Eocene of Monte Bolca, Italy. Zool. J. Linn. Soc. 117, 159–174 10.1111/j.1096-3642.1996.tb02154.x (doi:10.1111/j.1096-3642.1996.tb02154.x) [DOI] [Google Scholar]
- 35.Genner M. J., Seehausen O., Lunt D. H., Joyce D. A., Shaw P. W., Carvalho G. R., Turner G. F. 2007. Age of cichlids: new dates for ancient lake fish radiations. Mol. Biol. Evol. 24, 1269–1282 10.1093/molbev/msm050 (doi:10.1093/molbev/msm050) [DOI] [PubMed] [Google Scholar]
- 36.Frédérich B., Fabri G., Lepoint G., Vandewalle P., Parmentier E. 2009. Trophic niches of thirteen damselfishes (Pomacentridae) at the Grand Récif of Toliara, Madagascar. Ichthyol. Res. 56, 10–17 10.1007/s10228-008-0053-2 (doi:10.1007/s10228-008-0053-2) [DOI] [Google Scholar]
- 37.Vanden Berghe E. (ed.) 2007. The ocean biogeographic information system: web pages. Available on http://www.iobis.org. Consulted on 27 July 2011. [Google Scholar]
- 38.Ready J., Kaschner K., South A. B., Eastwood P. D., Rees T., Rius J., Agbayani E., Kullander S., Froese R. 2010. Predicting the distributions of marine organisms at the global scale. Ecol. Model. 221, 467–478 10.1016/j.ecolmodel.2009.10.025 (doi:10.1016/j.ecolmodel.2009.10.025) [DOI] [Google Scholar]
- 39.Dolédec S., Chessel D., Gimaret-Carpentier C. 2000. Niche separation in community analysis: a new method. Ecology 81, 2914–2927 10.1890/0012-9658(2000)081[2914:NSICAA]2.0.CO;2 (doi:10.1890/0012-9658(2000)081[2914:NSICAA]2.0.CO;2) [DOI] [Google Scholar]
- 40.Chessel D., Dufour A.-B., Thioulouse J. 2004. The ade4 package. I. One-table methods. R News 4, 5–10 [Google Scholar]
- 41.R Development Core Team 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 42.Thuiller W., Lavorel S., Midgley G., Lavergne S., Rebelo T. 2004. Relating plant traits and species distributions along bioclimatic gradients for 88 leucadendron taxa. Ecology 85, 1688–1699 10.1890/03-0148 (doi:10.1890/03-0148) [DOI] [Google Scholar]
- 43.Broennimann O., et al. 2012. Measuring ecological niche overlap from occurrence and spatial environmental data. Glob. Ecol. Biol. 21, 481–497 10.1111/j.1466-8238.2011.00698.x (doi:10.1111/j.1466-8238.2011.00698.x) [DOI] [Google Scholar]
- 44.Warren D. L., Glor R. E., Turelli M. 2008. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62, 2868–2883 10.1111/j.1558-5646.2008.00482.x (doi:10.1111/j.1558-5646.2008.00482.x) [DOI] [PubMed] [Google Scholar]
- 45.Harmon L. J., Weir J. T., Brock C. D., Glor R. E., Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131 10.1093/bioinformatics/btm538 (doi:10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 46.Revell L. J., Harmon L. J., Collar D. C. 2008. Phylogenetic signal, evolutionary process, and rate. Syst. Biol. 57, 591–601 10.1080/10635150802302427 (doi:10.1080/10635150802302427) [DOI] [PubMed] [Google Scholar]
- 47.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–84 10.1038/44766 (doi:10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 48.Blomberg S. P., Garland T., Ives A. R. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 10.1111/j.0014-3820.2003.tb00285.x (doi:10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 49.Revell L. J. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 10.1111/j.2041-210X.2011.00169.x (doi:10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 50.Litsios G., Salamin N. 2012. Effects of phylogenetic signal on ancestral state reconstruction. Syst. Biol. 61, 533–538 10.1093/sysbio/syr124 (doi:10.1093/sysbio/syr124) [DOI] [PubMed] [Google Scholar]
- 51.Losos J. B. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003 10.1111/j.1461-0248.2008.01229.x (doi:10.1111/j.1461-0248.2008.01229.x) [DOI] [PubMed] [Google Scholar]
- 52.Ackerly D. 2009. Conservatism and diversification of plant functional traits: evolutionary rates versus phylogenetic signal. Proc. Natl Acad. Sci. USA 106(Suppl.), 19 699–19 706 10.1073/pnas.0901635106 (doi:10.1073/pnas.0901635106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Meara B. C., Ané C., Sanderson M. J., Wainwright P. C. 2006. Testing for different rates of continuous trait evolution using likelihood. Evolution 60, 922–933 10.1111/j.0014-3820.2006.tb01171.x (doi:10.1111/j.0014-3820.2006.tb01171.x) [DOI] [PubMed] [Google Scholar]
- 54.Pagel M., Meade A., Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673. [DOI] [PubMed] [Google Scholar]
- 55.Bollback J. P. 2006. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinf. 7, 88 10.1186/1471-2105-7-88 (doi:10.1186/1471-2105-7-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eastman J. M., Alfaro M. E., Joyce P., Hipp A. L., Harmon L. J. 2011. A novel comparative method for identifying shifts in the rate of character evolution on trees. Evolution 65, 3578–3589 10.1111/j.1558-5646.2011.01401.x (doi:10.1111/j.1558-5646.2011.01401.x) [DOI] [PubMed] [Google Scholar]
- 57.Metropolis N., Rosenbluth A. W., Rosenbluth M. N., Teller A. H., Teller E. 1953. Equation of state calculations by fast computing machines. J. Med. Phys. 21, 1087–1092 10.1002/qua.560560820 (doi:10.1002/qua.560560820) [DOI] [Google Scholar]
- 58.Hastings W. K. 1970. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109 10.1093/biomet/57.1.97 (doi:10.1093/biomet/57.1.97) [DOI] [Google Scholar]
- 59.Drummond A. J., Suchard M. A. 2010. Bayesian random local clocks, or one rate to rule them all. BMC Biol. 8, 114 10.1186/1741-7007-8-114 (doi:10.1186/1741-7007-8-114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plummer M., Best N., Cowles K., Vines K. 2010. CODA: convergence diagnosis and output analysis for MCMC. R News 6, 7–11 10.1159/000323281 (doi:10.1159/000323281) [DOI] [Google Scholar]
- 61.Ruttenberg B. I., Haupt A. J., Chiriboga A. I., Warner R. R. 2005. Patterns, causes and consequences of regional variation in the ecology and life history of a reef fish. Oecologia 145, 394–403 10.1007/s00442-005-0150-0 (doi:10.1007/s00442-005-0150-0) [DOI] [PubMed] [Google Scholar]
- 62.Floeter S. R., Behrens M. D., Ferreira C. E. L., Paddack M. J., Horn M. H. 2005. Geographical gradients of marine herbivorous fishes: patterns and processes. Mar. Biol. 147, 1435–1447 10.1007/s00227-005-0027-0 (doi:10.1007/s00227-005-0027-0) [DOI] [Google Scholar]
- 63.Lau J. A, McCall A. C., Davies K. F., McKay J. K., Wright J. W. 2008. Herbivores and edaphic factors constrain the realized niche of a native plant. Ecology 89, 754–762 10.1890/07-0591.1 (doi:10.1890/07-0591.1) [DOI] [PubMed] [Google Scholar]
- 64.Araújo M. B., Luoto M. 2007. The importance of biotic interactions for modelling species distributions under climate change. Glob. Ecol. Biol. 16, 743–753 10.1111/j.1466-8238.2007.00359.x (doi:10.1111/j.1466-8238.2007.00359.x) [DOI] [Google Scholar]