Abstract
Alterations of the genital mucosal barrier may influence the number of viruses transmitted from a human immunodeficiency virus–infected source host to the newly infected individual. We used heteroduplex tracking assay and single-genome sequencing to investigate the effect of a tenofovir-based microbicide gel on the transmission bottleneck in women who seroconverted during the CAPRISA 004 microbicide trial. Seventy-seven percent (17 of 22; 95% confidence interval [CI], 56%–90%) of women in the tenofovir gel arm were infected with a single virus compared with 92% (13 of 14; 95% CI, 67%–>99%) in the placebo arm (P = .37). Tenofovir gel had no discernable impact on the transmission bottleneck.
It is well established that there is a severe genetic bottleneck associated with human immunodeficiency virus (HIV) transmission, resulting in low genetic diversity in early infection. Recently, larger studies that sequenced single HIV genomes from mainly heterosexual cohorts were able to more accurately quantitate this bottleneck [1–4]. It is estimated that approximately 80% (141 of 175) of sexually transmitted HIV infections are the result of only a single variant crossing the mucosal barrier to establish clinical infection. However, HIV and simian immunodeficiency virus studies have shown that the virus transmission bottleneck is affected by both the dose and the route of infection [4–7]. In macaques, a lower-dose inoculum resulted in a reduced number of transmitted variants [6]. Although in both macaques and humans injection drug use (IDU) resulted in an increased frequency of multiple-variant infections [5, 7], a separate study from Russia found that IDU did not relax the genetic bottleneck [8]. This conflict in findings about IDU could be due to differences in cohort characteristics, including risk behavious. Men who have sex with men are also at higher risk of infection than heterosexuals [5] and were found to be twice as likely to acquire multiple-variant HIV infections, possibly due to anatomical and histological differences between the 2 modes of transmission [4]. Similarly, inflammatory genital infections have also been shown to be associated with both increased risk of transmission and multivariant infection [2]. Together these studies suggest that breaching or bypassing the mucosal barrier can mitigate the viral transmission bottleneck and that characterizing the diversity following transmission can provide a surrogate marker of increased transmission risk.
Microbicides for women aim to reinforce the vaginal epithelial barrier against infection, although some microbicides, such as nonoxyl-9, compromise the mucosal barrier and increase the risk of transmission [9]. The recent pivotal CAPRISA 004 microbicide trial showed that women in urban and rural KwaZulu-Natal, South Africa, who used 1% tenofovir-based gel had a significantly lower incidence of HIV acquisition (5.6 per 100 women-years) than women on the placebo arm (9.1 per 100 women-years Africa) [10], with an overall 39% protective effect. These results demonstrate the utility of having tenofovir at the site of transmission to inhibit the early rounds of viral replication [10]. We studied intrapatient genetic diversity of HIV infection in the CAPRISA 004 trial participants with breakthrough infections to determine whether the use of the 1% tenofovir gel affected the genetic bottleneck.
METHODS
Samples were provided from participants who seroconverted while participating in the CAPRISA004 trial [10]. Date of infection was defined as the midpoint between the last HIV-negative test and the first HIV-positive test or as 14 days prior in the event of polymerase chain reaction (PCR)–positive, antibody-negative result. In CAPRISA 004, 98 study participants became infected with HIV (tenofovir gel, n = 38; placebo gel, n = 60). Two blinded panels of 43 and 7 samples were provided, of 7 seven were excluded because participants had been infected for >12 weeks, making it difficult to differentiate between single and multiple variant infections due to viral diversification [11]. A further 7 were not amplifiable, 5 of which had low viral load copy number.
To differentiate between single- and multiple-variant HIV transmission, we first screened for heterogeneity in 2 regions, V1/V2 and V4/V5, using heteroduplex tracking assay (HTA) because these variable regions of env are most likely to harbor the highest sequence diversity. Each amplicon was annealed to 2 separate probes to further enhance the chance of detecting sequence differences. Samples were classified as single-variant infection if there was a dominant band for all 4 comparisons, with the presence of a second band in 1 region accepted if it was faint and suggestive of recent diversification from a single founder. All samples classified as single-variant infections contained ≥30 amplifiable copies, which is sufficient to reproducibly quantify a 10% minority population [12]. Samples were classified as potential multiple-variant infections if >1 dominant band was visualized in any region. Multiple bands on a gel may be due to either multiple-variant transmission or viral evolution from a single founder virus infection. To differentiate these 2 scenarios, we used single genome amplification (SGA) and sequencing [11].
Human immunodeficiency virus type 1 RNA was extracted using the MagNA Pure RNA isolation kit (Roche Diagnostics). Samples with viral loads ≤5000 copies/mL were concentrated by centrifugation. For the HTA, the V1/V2 (HXB2: 6555–6981) and V4/V5 (HXB2: 7341–7715) regions were PCR amplified as described [13]. Amplicons were assayed using 2 sets of subtype C–radiolabelled probes to each of the V1/V2 and V4/V5 env domains [13]. For HIV-1 env SGA analysis, participant complementary DNAs were diluted to a single copy and amplified as described [1, 3]. Sequences were excluded if they contained >1 double peak in the sequence chromatogram or deletions >100 nucleotides. A single double peak in the sequence chromatogram was permitted only if it was not located in the same position as other double peaks from the same participant.
Pairwise DNA distances and neighbor-joining phylogenetic trees were computed using MEGA 4.1. [14]. Sequence differences were visualised using Highlighter nucleotide transition and transversion plots (http://www.hiv.lanl.gov). Time of divergence from the most recent common ancestor (tMRCA) was estimated using Bayesian Evolutionary Analysis Sampling Trees version 1.4.7 [15], as described previously [1, 3].
All P values are 2-sided and calculated using Fisher's exact test, and all confidence intervals (CIs) are 95% and calculated using GraphPad Prism version 5.00.
This study was approved by both the University of Cape Town Research Ethics Committee (025/2004) and the University of KwaZulu-Natal Biomedical Research Ethics Committee.
RESULTS
To determine the genetic complexity of HIV following transmission in women exposed to tenofovir vs placebo, we screened 29 of the 36 samples by HTA. This method is sensitive to detecting small sequence differences between variants, which are visualized as altered migration patterns of heteroduplexes in a gel (Figure 1). Eighteen participants were classified as being infected with a single-variant infection based on the presence of a single dominant band in both env variable domains assayed; in 6 of these participants, there was evidence of diversification from a single founder detected as faint additional bands in 1 region (Table 1; Figure 1B and 1C). Eleven individuals were identified with high-diversity infection based on multiple bands.
Figure 1.
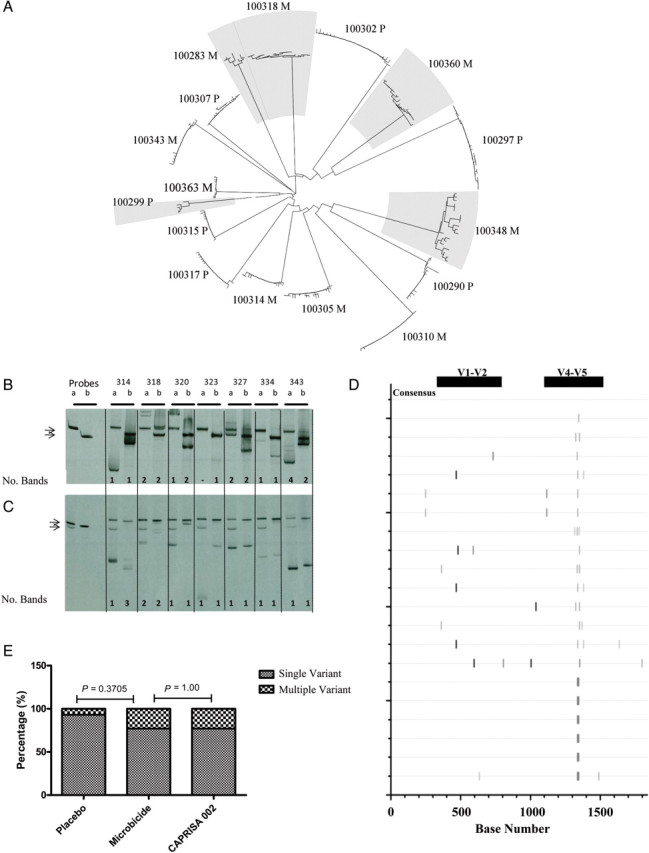
Envelope sequence analysis. (A) Neighbour-joining tree of envelope sequences for all participants analysed by SGA. Shaded branches indicate multiple variant infections. (B-C) Typical V1/V2 and V4/V5-HTA results respectively for study participants labelled according to the last 3 digits of the participant identification number. The number of viral variants are estimated by counting the number of heteroduplexes for each probe set (shown below each gel). (D) Highlighter plot illustrating nucleotide mismatches within sequences compared to a consensus sequence for participant 314 who displayed discordant results between HTA and SGA analyses. Horizontal lines represent HIV envelope sequences, while the vertical ticks indicate the nucleotide mismatches or sequence gaps. Black bars indicate the position of the V1/V2 (∼330 – 760 bp) (HXB2: 6555-6981) and V4/V5 (∼1120 – 1500 bp) (HXB2: 7341-7715) probe domains analysed in the HTA. (E) Comparison of the incidence of multiple variant infections between individuals using the tenofovir-based microbicide trial and individuals not receiving treatment in the CAPRISA 002 acute infection cohort [1]
Table 1.
Patient Data and Complexity of Infecting Viruses in the 1% Tenofovir-based Microbicide Gel Group Compared With the Placebo Gel Group
| PID/Study Arm | Time Post Infection (days) | Viral Load (copies/mL) | No. of Bands (HTA) | Max. DNA Distance (%) | Mean No. of Days Since MRCA (95% CI)b | Multiplicity of Infection | STIc at First HIV-Positive Diagnosis |
|---|---|---|---|---|---|---|---|
| Microbicide | |||||||
| 304 M | 42 | 94 900 | 1 | Single | |||
| 372 M | 22 | 107 000 | 1 | Single | |||
| 323 M | 24 | 22 000 | 1 | Single | Yes | ||
| 367 M | 24 | 3 360 000 | 1 | Single | Yes | ||
| 310 M | 30 | 305 000 | 1a | 0.10 | 9 (1–19) | Single | Yes |
| 362 M | 30 | 44 200 | 1 | Single | |||
| 355 M | 32 | 19 700 | 1 | Single | Yes | ||
| 370 M | 34 | 28 800 | 1 | Single | Yes | ||
| 320 M | 35 | 37 500 | 1a | Single | |||
| 375 M | 38 | 192 000 | 1a | Single | |||
| 363 M | 38 | 97 500 | >1 | 0.30 | 93 (41–153) | Single | |
| 334 M | 44 | 21 200 | 1 | Single | Yes | ||
| 352 M | 51 | 107 000 | 1 | Single | |||
| 314 M | 57 | 166 000 | >1 | 0.40 | 111 (53–176) | Single | |
| 343 M | 63 | 130 000 | >1 | 0.40 | 75 (42–112) | Single | |
| 358 M | 67 | 717 | 1 | Single | |||
| 305 M | 78 | 45 600 | >1 | 0.50 | 120 (74–173) | Single | |
| 325 M | 22 | 127 000 | >1 | NA | NA | Multiple | Yes |
| 360 M | 25 | 80 600 | >1 | 1.60 | 883 (291–1794) | Multiple | Yes |
| 318 M | 31 | 22 300 | >1 | 1.00 | 573 (242–1083) | Multiple | |
| 283 M | 45 | 6650 | >1 | 1.30 | 870 (398–1443) | Multiple | |
| 348 M | 58 | 305 000 | >1 | 2.60 | 1615 (901–2559) | Multiple | Yes |
| Placebo | |||||||
| 302 P | 21 | 1 980 000 | NA | 0.30 | 49 (27–73) | Single | |
| 303 P | 21 | 506 000 | NA | 0.50 | 12 (1–32) | Single | |
| 290 P | 21 | 84 500 | NA | 0.70 | 23 (4–55) | Single | Yes |
| 315 P | 21 | 108 000 | NA | 0.50 | 147 (87–215) | Single | |
| 327 P | 21 | 91 600 | 1a | Single | |||
| 351 P | 22 | 104 000 | 1 | Single | |||
| 312 P | 23 | 1850 | 1a | Single | Yes | ||
| 307 P | 28 | 155 000 | NA | 0.30 | 55 (25–90) | Single | Yes |
| 289 P | 28 | 142 000 | NA | 0.50 | 38 (4–84) | Single | Yes |
| 297 P | 38 | 8000 | >1 | 0.20 | 58 (16–115) | Single | |
| 292 P | 39 | 458 000 | 1 | 0.20 | 9 (1–25) | Single | |
| 317 P | 49 | 351 000 | NA | 0.40 | 81 (45–122) | Single | |
| 308 P | 62 | 37 100 | 1a | Single | |||
| 299 P | 16 | 5080 | >1 | 1.20 | Multiple | Yes |
Abbreviations: CI, confidence interval; HIV, human immunodeficiency virus; HTA, heteroduplex tracking assay; MRCA, most recent common ancestor; NA, not available; PID, participant identification number; STI, sexually transmitted infection.
a Single dominant band in both variable env regions (V1/V2, V3/V4), with a faint band in 1 region indicative of recent diversification.
b MRCA calculated using Bayesian evolutionary analysis by sampling trees.
c STI screen (Karim et al, 2010): syphilis, Trichomonas vaginalis, Neisseria gonorrhoea, Chlamydia trachomatis, Mycoplasma genitalium, bacterial vaginosis, and herpes simplex virus type 2.
Twenty samples were sequenced using the single genome amplification approach: in addition to the 11 samples identified as putative multiple-variant infections by HTA, a further 7 were sequenced because there was no HTA analysis available, and 2 samples were sequenced as controls (participant-292, HTA detected no diversity; and participant-310, HTA detected diversity in 1 variable region). In total, 363 sequences were generated (median of 20 per participant) spanning the first approximately 1950 nucleotides of gp160, including the entire gp120. All 20 participants were infected with HIV-1 subtype C (Figure 1A).
Classification of single-variant transmission was based on low intraparticipant sequence diversity with no or limited structure in the phylogenetic tree and genetic differences within the expected range for the duration of infection [3]. Based on these criteria, 11 individuals were classified as having single-variant infections, of which 5 had been identified as having putative multiple-variant transmissions using HTA. Heteroduplex tracking assay is very sensitive to sequence changes, and the accuracy of defining the number of transmitted viruses is greatest when sampling is close to the date of transmission. The samples with multiple bands were acquired at >6 weeks postinfection and had evidence of immune pressure, including sequence gaps in the V1/V2 env domains, which accounted for the diversification (Table 1; Figure 1D). The mean of the maximum sequence diversity in the single-variant infections was 0.2% and ranged 0%–0.3%. The classification as single-variant transmission was supported by the estimated time to tMRCA, which was within the range of the duration of infection (Table 1). Combining the HTA and sequencing results, 30 participants were identified as being infected with a single variant.
Five participants were classified as infected with multiple variants based on high pairwise distance of the sequences, structure in the phylogenetic tree, and tMRCA predating the estimated date of infection (Table 1). In 1 of the 5 participants (participant-299), we were not able to generate sufficient amplicons to estimate tMRCA; however, the phylogenetic analysis and maximum genetic distance of 1.3% were sufficient to confidently classify this individual as having a multiple-variant infection (Table 1). A sixth participant (participant-325) was classified as infected with multiple variants because, although we were unable to sequence the virus, there were clear multiple dominant bands on HTA gels in both genomic regions, a banding pattern that exceeded that expected for the short duration of infection (approximately 3 weeks). Of these 6 participants, 5 were from the tenofovir gel arm, and 1 was from the placebo arm.
In summary, to determine if the use of the 1% tenofovir gel affected the mucosal barrier, thereby altering the genetic bottleneck associated with transmission, we defined multiplicity of infection in 36 participants: 22 were from the tenofovir gel arm and 14 were from the placebo arm. We found that the frequency of single-variant infection was not significantly different between the tenofovir gel and placebo arms (P = .37), with a single virus responsible for 17 (77%; 95% CI, 56%–90%) infections in the tenofovir arm and 13 (93%; 95% CI, 67%–>99%) infections in the placebo arm. In a previous study of 69 men and women from Malawi and South Africa, we found 78% of infections involved single-variant transmission. Of these, 26 were women from the CAPRISA 002 cohort, which recruited from the same population and geographical region. The percentage of single-virus transmission in the tenofovir arm was identical to that of gel-unexposed participants reported for the CAPRISA 002 cohort (20 of 26; 77%) [1] (Figure 1E).
DISCUSSION
No alteration in the transmission bottleneck in the breakthrough HIV infections in the tenofovir gel arm of the CAPRISA 004 microbicide trial was detected. The single-virus transmission rates were not significantly different between the tenofovir gel and placebo gel: 77% of the women from the tenofovir gel arm (17 of 22) and 93% from the placebo gel arm (13 of 14) (P = .37) had HIV infections that were a result of a single variant. Further, the single-virus transmission rate in the tenofovir gel arm was similar to that from a previous study [1] in a similar group of women not exposed to the microbicide or placebo gel.
These results provide further insights into the dynamics of transmission in the CAPRISA 004 tenofovir trial [10]. Past studies in African heterosexual cohorts found significantly increased (50%) incidence of multiple-variant infections in individuals with compromised vaginal mucosal barriers [2]. In the CAPRISA004 trial, genital samples were tested for sexually transmitted infections (STIs) at first diagnosis of HIV-1 infection (Table 1) [10]. In the subset of participants included in the current study, no difference (P = 1.00) in the incidence of STIs was observed between the 2 study arms, and the presence of STIs, albeit in a limited sample size, did not appear to impact the incidence of multiple-variant transmission (P = .21).
The results presented here from the CAPRISA 004 tenofovir gel trial show no significant increase in the incidence of multiple-variant infections in the tenofovir gel– or placebo-using arm of the trial and an identical frequency in the incidence of multiple-variant infections to individuals recruited from similar communities who did not use the gel (Figure 1E). Although we included all but 1 of the amplifiable samples from the tenofovir gel users, it should be noted that this limited sample size would only have detected a large effect of the microbicide on the transmission bottleneck. In conclusion, the protective effect of tenofovir gel [12], together with the maintenance of the multiplicity of infection between different groups (1% for the tenofovir gel group, the placebo gel group, and the non-gel users from a similar community), suggests that the microbicide did not compromise the vaginal mucosa barrier.
Notes
Acknowledgments. We thank the women who participated in the CAPRISA 004 trial and the CAPRISA004 study personnel. We are grateful for support from the CAPRISA laboratory.
Financial support. This work was supported by CAPRISA; the United States Agency for International Development (USAID); Family Health International (FHI) (cooperative agreement GPO-A-00-05-00022-00, contract 132119); and LIFElab, a biotechnology center of the South African Department of Science & Technology. These studies were also supported by the TRAPS (Tenofovir Gel Research for AIDS Prevention Science) Program, which is funded by CONRAD (cooperative grant GP00-08-00005-00, subproject agreement # PPA-09-046O). We thank the US National Institutes for Health's Comprehensive International Program of Research on AIDS (CIPRA grant AI51794) for the research infrastructure. Ziyaad Valley-Omar was funded by the Columbia University-Southern African Fogarty AIDS International Training and Research Programme (AITRP grant D43TW00231). Ronald Swanstrom, Jeffrey Anderson and Leslie Arney were funded by the US Natoinal Institute of Health (NIH grant R37 AI44667).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Abrahams MR, Anderson JA, Giorgi EE, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol. 2009;83:3556–67. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haaland RE, Hawkins PA, Salazar-Gonzalez J, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Bar KJ, Wang S, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar KJ, Li H, Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84:6241–7. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Keele BF, Li H, et al. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J Virol. 2010;84:10406–12. doi: 10.1128/JVI.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenier JL, Miller CJ, Lu D, et al. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J Virol. 2001;75:3753–65. doi: 10.1128/JVI.75.8.3753-3765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masharsky AE, Dukhovlinova EN, Verevochkin SV, et al. A substantial transmission bottleneck among newly and recently HIV-1-infected injection drug users in St Petersburg, Russia. J Infect Dis. 2010;201:1697–702. doi: 10.1086/652702. [DOI] [PubMed] [Google Scholar]
- 9.Van DL, Ramjee G, Alary M, et al. Effectiveness of COL-1492, a nonoxynol-9 vaginal gel, on HIV-1 transmission in female sex workers: a randomised controlled trial. Lancet. 2002;360:971–7. doi: 10.1016/s0140-6736(02)11079-8. [DOI] [PubMed] [Google Scholar]
- 10.Karim QA, Karim SS, Frohlich JA, et al. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329:1168–74. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salazar-Gonzalez JF, Bailes E, Pham KT, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–70. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Resch W, Parkin N, Stuelke EL, Watkins T, Swanstrom R. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc Natl Acad Sci USA. 2001;98:176–81. doi: 10.1073/pnas.011511298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riddle TM, Shire NJ, Sherman MS, Franco KF, Sheppard HW, Nelson JA. Sequential turnover of human immunodeficiency virus type 1 env throughout the course of infection. J Virol. 2006;80:10591–9. doi: 10.1128/JVI.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 15.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]


