Abstract
Background. Human immunodeficiency virus (HIV) superinfection has been documented in high-risk individuals; however, the rate of superinfection among HIV-infected individuals within a general population remains unknown.
Methods. A novel next-generation ultra-deep sequencing technique was utilized to determine the rate of HIV superinfection in a heterosexual population by examining two regions of the viral genome in longitudinal samples from recent HIV seroconverters (n = 149) in Rakai District, Uganda.
Results. The rate of superinfection was 1.44 per 100 person years (PYs) (95% confidence interval [CI], .4–2.5) and consisted of both inter- and intrasubtype superinfections. This was compared to primary HIV incidence in 20 220 initially HIV-negative individuals in the general population in Rakai (1.15 per 100 PYs; 95% CI, 1.1–1.2; P = .26). Propensity score matching (PS) was used to control for differences in sociodemographic and behavioral characteristics between the HIV-positive individuals at risk for superinfection and the HIV-negative population at baseline and follow-up. After PS matching, the estimated rate of primary incidence was 3.28 per 100 PYs (95% CI, 2.0–5.3; P = .07) controlling for baseline differences and 2.51 per 100 PYs (95% CI, 1.5–4.3; P = .24) controlling for follow-up differences.
Conclusions. This suggests that the rate of HIV superinfection in a general population is substantial, which could have a significant impact on future public health and HIV vaccine strategies.
Human immunodeficiency virus (HIV) superinfection occurs when an HIV-infected individual acquires a new viral strain that is phylogenetically distinct from all detectable viral strains at a previous time point [1]. Inter- and intrasubtype HIV superinfections have been reported in high-risk individuals exposed through sexual or intravenous drug use [1–12]. The rate of HIV superinfection has often been found to be relatively frequent, particularly if multiple genomic sites are examined [6, 9, 13–15]. Other researchers have found no evidence of superinfection in both small- and large-scale studies; however, these studies utilized clonal analyses that were likely not sensitive enough to detect the levels of virus observed in some superinfection cases [16–18].
These discrepancies partly reflect differences in the techniques used to identify and verify superinfection [19]. Initial studies of the frequency of superinfection utilized heteroduplex mobility or multiregion hybridization assays followed by selective clonal analysis [6, 7, 16]. Other studies preselected high-risk individuals and employed in-depth cloning techniques to identify possible superinfections [6, 9, 18]. The sensitivity of these assays as possible screening techniques is determined by the number of clones amplified and the number of regions examined [9, 15, 20]. Therefore, to obtain the sensitivity needed to identify a variant circulating within an individual at approximately 1% of the total circulating viral population, over 100 clones would need to be examined per sample, an approach that becomes prohibitively labor intensive for large-scale population studies of superinfection [15, 19, 20]. We therefore recently designed and tested a novel, highly sensitive, high-throughput, next-generation, ultra-deep sequencing technique and sequence analysis protocol to identify HIV superinfection in 2 regions of the viral genome: the p24 region of the viral capsid and the gp41 region of the viral envelope [1]. Using this procedure, we examined HIV seroconverters from the Rakai Community Cohort Study (RCCS) in Rakai District, Uganda, to determine the incidence of HIV superinfection in this population.
METHODS
Superinfection Population
The RCCS is a rural, community-based, open cohort of persons aged 15–49 years in Rakai District in southwestern Uganda, which has been described in detail previously [21]. Since 1994, interviews and venous blood samples have been obtained annually from approximately 14 000 consenting adults living in 50 villages. All subjects provided written informed consent for sample storage and testing. The study was approved by the Science and Ethics Committee of the Uganda Virus Research Institute, the Uganda National Council for Research and Technology, Western Institutional Review Board, and the Committee on Human Research at Johns Hopkins Bloomberg School of Public Health.
Known seroconverters who had a positive HIV serological result within 2 years of a prior negative test between 1998 and 2004 and who had also provided at least 1 subsequent serological sample prior to 2009 were randomly selected for examination from the RCCS population (n = 203) [21, 22]. To identify HIV superinfection, viral RNA was isolated from serum obtained at the seroconverter's first HIV-positive time point (baseline) and the latest time point available prior to initiation of antiretroviral therapy (ART), loss to follow-up, or death [22]. Subjects were excluded from analysis if neither genomic region in the baseline sample could be amplified (n = 14). Subjects were also excluded if the follow-up genomic region corresponding to the amplified baseline sample failed to amplify (n = 40). The remaining subjects’ (n = 149) viral RNA extracts were initially amplified by reverse-transcription polymerase chain reaction (RT-PCR) in duplicate; the resulting products were pooled and subsequently amplified in a nested PCR strategy using barcoded primers specific for use on the 454 pyrosequencer platform [1].
Laboratory Analysis
Briefly, amplicons of p24 (approximately 390 base pair [bp]) and gp41 (approximately 324 bp) were amplified and sequenced as previously described [1]. The Amplicon Library Preparation Method was performed as recommended by the manufacturer (Roche), and all PCR products were purified with the following minor alterations. In an effort to eliminate the capture of primers, the bead-to-target ratio was reduced by incubating 30 μL of AMPure Beads XP (Agencourt; Beckman Coulter Genomics) with 25 μL of PCR product diluted in 25 μL of water. Purified PCR products were quantified using PicoGreen (Invitrogen), and each template was diluted to a 1 × 109 molecules/μL stock. The amplicon pools were made by combining 5 μL of each diluted barcoded template to make a final 1 × 109 molecules/μL stock containing 14 barcoded amplicons.
Preparation of templated beads for next generations sequencing (NGS) followed the emPCR Method Manual-Lib-L-MV (Roche). The 1 × 109 molecules/μL library pools were diluted to 1 × 105 molecules/μL for a target addition of 0.175 copies per bead to the DNA capture beads. Enriched DNA capture beads were sequenced on the 454 (Roche) per the manufacturer's instructions using a 4-region gasket [1].
Sequencing results were analyzed using the GS Amplicon Variant Analyzer version 2.5 (Roche). All sequence reads were compared, and similar sequences were combined into a single consensus sequence. Generated consensus sequences that were within 10 bases from both ends of the amplicon and comprised of a cluster of >10 individual, near-identical sequences were determined using the Roche Amplicon software and were classified as being consensus sequences of HIV variants. These consensus sequences were used for subsequent phylogenetic analysis [1]. The lower limit of detection of intersubtype minor variants for this assay was previously shown to be 1% [1].
HIV superinfection was defined in an individual whose follow-up serum sample demonstrated >2 distinct consensus sequences forming a monophyletic cluster that was phylogenetically unlinked from the individual's entire consensus sequences in the baseline sample and was of adequate genetic distance from the baseline sequences to rule out natural evolutionary drift [1]. In order to be considered a superinfection, the genetic distance of the new monophyletic cluster from the closest related viral sequences found at the earlier time point had to be ≥0.55% per year for the p24 region or ≥0.98% per year for the gp41 region [1]. All newly identified consensus sequences were phylogenetically compared with the most prominent strains of the other barcoded samples within the NGS runs to search for microcontamination, misclassification, or sequencing errors. If instances of these errors were found, these consensus sequences were eliminated. Possible superinfection events were reamplified and verified in a second 454 sequencing run [1]. The NGS consensus sequences for gp41 and p24 are available on the Los Alamos National Laboratory HIV-DB Next Generation Sequence Archive (http://www.hiv.lanl.gov/content/sequence/HIV/NextGenArchive/) and are also available upon request from the corresponding author. Total sequence reads and consensus sequences from the total population were each compared for both genomic regions at baseline and follow-up with the Kruskal–Wallis test.
Serum HIV type 1 RNA concentrations (viral loads) were determined by the Amplicor v1.5 (Roche Diagnostics).
Incidence Rate Population
The HIV incidence rate was estimated for the entire RCCS population among participants with a negative sample between 1998–2008 and at least 1 follow-up sample prior to 2009 (n = 20 220), providing 100 550 person years (PYs) at risk [23]. Human immunodeficiency virus infection was assumed to have occurred at the midway point between the last negative and first positive HIV test.
Statistical Analysis
The rate of superinfection among the amplified HIV-positive samples was initially compared with the rate of primary HIV infection among the general population using a univariate Poisson log-linear model. Because the sociodemographic characteristics and risk behavior profiles of the individuals tested for superinfection were different from those of the general HIV-negative population at risk of primary HIV infection, propensity score matching was used to compare the rates of superinfection and primary infection in the general HIV-negative population matched for the sociodemographics and behaviors of the HIV-positive population at risk of superinfection [24, 25]. The baseline and follow-up sociodemographic and behavior variables were compared between the two populations using χ2 tests, and any variable that differed significantly between the groups at P = .05 was entered in the logistic regression model to estimate the propensity score. The objective of constructing a propensity score is to use a 1-dimension balance score (ie, the propensity score) to summarize all the measured variables that differ between the two populations [26]. Additionally, because the follow-up time differed between the two populations, duration of follow-up was also entered in the propensity score model in order to balance the exposure times between the populations. One-nearest neighbor matching on the propensity score was applied to select the subjects in the general population who had similar profiles to the subjects in the HIV-positive population at risk of superinfection. Further analyses were conducted to check the balance in each sociodemographic and risk behavior variable between the populations. A Poisson log-linear model was then used to estimate the expected incidence in the general population given the characteristics and behaviors of the HIV-positive population at risk of superinfection. The propensity score matching was conducted using the MatchIt R package [24, 25].
RESULTS
Of the 149 individuals examined for superinfection, valid pyrosequencing results were obtained for the p24 and gp41 genomic regions at both time points for 109 individuals (73.2%). Thirty-one individuals had valid results at both time points for the gp41 region only (20.8%), and 9 had p24 results only (6.0%). There were no significant differences in the total reads between the baseline p24 (median, 10 998; interquartile range [IQR], 8262–14 549), baseline gp41 (median, 10 581; IQR, 8309–12 713), and follow-up p24 regions (median, 11 386; IQR, 9488–13 638). However, the follow-up gp41 total reads (median, 8923; IQR, 7186–11 617) were significantly lower than all 3 (P < .05). There were no significant differences between the consensus sequence totals for the baseline p24 (median, 74; IQR, 43–108), baseline gp41 (median, 70; IQR, 54–87), follow-up p24 (median, 79; IQR, 56–104), and follow-up gp41 regions (median, 72; IQR, 48–91) (P = .26).
At initial seroconversion, 91 individuals were infected with a single subtype D viral population (61.1%), 24 individuals were infected with a single subtype A viral population (16.1%), 1 individual was infected with subtype C viral population (0.7%), and 33 individuals were infected with either recombinant viruses or multiple viral populations (22.1%). This is consistent with the HIV subtype distribution previously observed in Rakai [27]. The median time interval between the baseline and follow-up time points was 2.84 years (IQR, 1.64–5.08; range, 0.99–7.47).
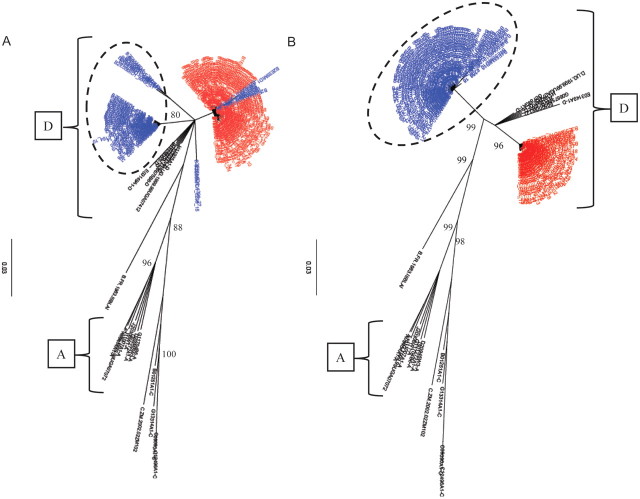
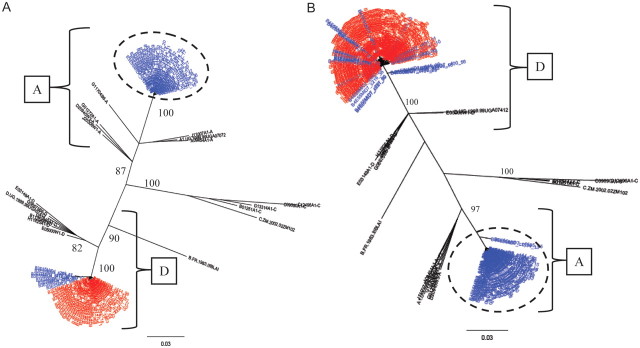
Seven cases of HIV superinfection were identified over 485.7 PYs of follow-up, for an HIV superinfection incidence of 1.44 per 100 PYs (95% confidence interval [CI], .37–2.51) (Figures 1 and 2; Supplementary Figure 1). All superinfection events were detected in the gp41 region. In 3 cases, there were no sequences available for the p24 region, and in the remaining cases there were no new sequences detected in the p24 region. In addition, all 7 superinfected individuals were initially infected with HIV subtype D at baseline (Table 1). Four of the superinfection events were intrasubtype events with initial subtype D–infected individuals superinfected with a novel subtype D strain(s) (Figure 1; Supplementary Figure 1A and 1B). The other 3 cases were intersubtype superinfection events with new subtype A viral populations being found in the follow-up sample (Figure 2; Supplementary Figure 1C). There was no consistent change in the viral loads before and after superinfection (Table 1).
Figure 1.
A and B, Phylogenetic trees of consensus gp41 viral sequences (≥10 reads) derived from 454 pyrosequences of baseline serum sample (red) and follow-up sample (blue) are shown for 2 representative intrasubtype human immunodeficiency virus type 1 (HIV-1) superinfection events. All consensus sequences were combined in a joint tree for identification of superinfecting strains as indicated (dashed circle). The HIV subtypes A and D are indicated by brackets. The number of repeated sequences represented by each consensus sequence is shown at the end of the consensus identifier. Distance is indicated for each individual tree with scale at bottom of the tree, and samples are grouped with a selection of subtype reference sequences and random sequences from individuals in Rakai (black) [1].
Figure 2.
A and B, Phylogenetic trees of consensus gp41 viral sequences (≥10 reads) derived from 454 pyrosequencs of baseline serum sample (red) and follow-up sample (blue are shown for 2 representative inter-subtype HIV-1 superinfection events. All consensus sequences were combined in a joint tree for identification of superinfecting strains as indicated (dashed circle). The HIV subtypes A and D are indicated by brackets. The number of repeated sequences represented by each consensus sequence is shown at the end of the consensus identifier. Distance is indicated for each individual tree with scale at bottom of the tree, and samples are grouped with a selection of subtype reference sequences and random sequences from individuals in Rakai (black) [1].
Table 1.
Next-Generation Sequencing Total Reads, Consensus Sequence Data, and Viral Load for Superinfection Cases
| ID | Baseline Subtype | Baseline Total Reads | Baseline Consensus Sequences | Superinfecting (SI) Subtype | Follow-up Total Reads | Follow-up Consensus Sequences | Consensus Sequences Identified as SI Strain, % | Baseline Viral Load | Follow-up Viral Load |
|---|---|---|---|---|---|---|---|---|---|
| 1 | D | 10 150 | 90 | D | 6007 | 51 | 82.4 | 5.72a | 5.34 |
| 2 | D | 13 628 | 124 | D | 2666 | 25 | 12.0 | 4.89a | 5.41 |
| 3 | D | 11 128 | 78 | D | 2950 | 9 | 100 | 5.36a | 3.33 |
| 4 | D | 11 784 | 44 | D | 12 212 | 85 | 100 | 3.51 | 6.22 |
| 5 | D | 13 798 | 93 | A | 11 592 | 93 | 69.9 | 4.18 | 4.27 |
| 6 | D | 25 049 | 119 | A | 14 559 | 92 | 69.6 | 3.36a | 3.83 |
| 7 | D | 8263 | 42 | A | 5516 | 43 | 81.4 | 3.26 | 5.19 |
a Due to sample availability, 4 baseline viral load levels were determined from samples taken <1 month after the baseline sample.
The HIV incidence rate was estimated for the entire RCCS population among participants with a negative sample between 1998–2008 and at least one follow-up sample in the same time period (n = 20 220 with 100 550 PYs of follow-up) (Table 2). In this population, 1152 HIV seroconversion events were identified for an unadjusted HIV incidence of 1.15 per 100 PYs (95% CI, 1.08–1.21). The rate of HIV superinfection did not differ from this unadjusted primary incidence rate (P = .26).
Table 2.
Sociodemographic and Behavioral Characteristics of At-Risk Populations Examined for Superinfection and Primary Incidence at Baseline and Follow-up
| Baseline |
Follow-Up |
|||||
|---|---|---|---|---|---|---|
| Superinfection | HIV Infection | Superinfection | HIV Infection | |||
| n = 149, No. (%) | n = 20 220, No. (%) | P Value | n = 149, No. (%) | n = 20 220, No. (%) | P Value | |
| Age, years | ||||||
| ≤20 | 14 (9) | 7611 (38) | <.001* | 5 (3) | 3375 (17) | <.001* |
| 21–25 | 46 (31) | 4494 (22) | 22 (15) | 4274 (21) | ||
| 26–30 | 32 (22) | 3140 (16) | 38 (26) | 3973 (20) | ||
| 31–35 | 23 (15) | 1843 (9) | 34 (23) | 2961 (15) | ||
| ≥36 | 34 (23) | 3132 (15) | 50 (34) | 5637 (28) | ||
| Gender | ||||||
| Female | 96 (64) | 11 032 (55) | .02* | 96 (64) | 11 032 (55) | .02* |
| Male | 53 (36) | 9188 (45) | 53 (36) | 9188 (45) | ||
| Marital statusa | ||||||
| Never married | 24 (16) | 7033 (35) | <.001* | 15 (10) | 4853 (24) | <.001* |
| Currently married | 97 (65) | 11 507 (57) | 82 (55) | 12 928 (64) | ||
| Divorced, separated, widowed | 28 (19) | 1678 (8) | 52 (35) | 2433 (12) | ||
| Number of sex partners past yeara | ||||||
| None | 9 (6) | 4153 (21) | <.0001* | 26 (17) | 2855 (14) | .30 |
| 1 | 106 (71) | 12 313 (61) | 99 (66) | 13 266 (66) | ||
| >2 | 34 (23) | 3744 (19) | 24 (16) | 4084 (20) | ||
| Condom use with all partnersa | ||||||
| Never | 88 (63) | 10 391 (65) | .04* | 68 (55) | 10 598 (61) | .40 |
| Inconsistent | 46 (33) | 4175 (26) | 39 (32) | 4917 (28) | ||
| Always | 6 (4) | 1491 (9) | 16 (13) | 1835 (11) | ||
| Alcohol use with sexa | ||||||
| Never | 65 (46) | 10 242 (64) | <.0001* | 66 (54) | 7662 (44) | .04* |
| Sometimes | 75 (54) | 5815 (36) | 57 (46) | 9688 (56) | ||
| Person-year follow-up | ||||||
| <2.5 | 56 (38) | 6942 (34) | <.0001* | 56 (38) | 6942 (34) | <.0001* |
| 2.5–5 | 67 (45) | 5224 (26) | 67 (45) | 5224 (26) | ||
| 5–7.5 | 24 (16) | 2959 (15) | 24 (16) | 2959 (15) | ||
| ≥7.5 | 2 (1) | 5095 (25) | 2 (1) | 5095 (25) | ||
* indicates significant differences.
a Individuals who were not sexually active or refused to answer questionnaire were excluded.
At baseline and follow-up, the sociodemographic and behavioral characteristics of the HIV-infected population at risk for superinfection were markedly and significantly different from the HIV-negative population at risk of primary incident infection (Table 2). At baseline, members of the HIV-infected population at risk of superinfection were older and more likely to be female, to have experienced marital dissolution, to be sexually active, to use condoms inconsistently, and to consume alcohol before sex, all of which are known risk factors for HIV acquisition in this population. At follow-up, members of the the population tested for superinfection were older and more likely to be female, to have marital dissolution, and to consume alcohol with sex, but they were not more sexually active than the general population. Propensity score matching was used to estimate HIV incidence in the general population adjusted for these differences in sociodemographic and behavioral characteristics. When the general population was matched to the HIV-infected population's characteristics at baseline, the estimated HIV incidence rate was 3.28 per 100 PYs (95% CI, 2.0–5.3), which was higher than the observed rate of superinfection, and the difference was of borderline statistical significance (P = .07). When the populations were propensity score matched by sociodemographic and behavioral characteristics at follow-up, the estimated primary incidence rate was 2.51 per 100 PYs (95% CI, 1.5–4.3), which was not significantly different from the observed rate of superinfection (P = .24).
DISCUSSION
This is the first large-scale study of the rate of HIV superinfection in a general heterosexual population using a validated, highly sensitive, next-generation, ultra-deep sequencing technique. The finding of inter- and intrasubtype superinfection events supports earlier work in a group of high-risk female sex-workers in Kenya [6, 9, 15]. Other studies have found few or no cases of superinfection, even in European homosexual male populations reporting high-risk exposures, but this may be due to the less-sensitive clonal analyses used [17, 18].
In addition, this is the first study to compare the rate of HIV superinfection to primary HIV incidence in a general heterosexual population in sub–Saharan Africa. The rate of HIV superinfection (1.44 per 100 PYs) did not differ significantly from the unadjusted primary HIV incidence rate (1.15 per 100 PYs). However, after matching for the baseline characteristics and behaviors, the adjusted primary incidence (3.28 per 100 PYs) was higher than the observed incidence of superinfection, and this was of borderline statistical significance. When the follow-up behavioral characteristics were used to derive the propensity score matching, the adjusted primary HIV incidence (2.51 per 100 PYs) was nonsignificantly higher than the rate of HIV superinfection. This difference in estimated rates based on baseline or follow-up propensity score matching may reflect change in behaviors among HIV-infected individuals after learning their status and decreased risky behavior due to HIV disease progression [28]. In addition, our analysis is somewhat constrained by the relatively small number of superinfections detected, which limited the power of the study to detect a small significant difference in unadjusted or propensity score–matched incidence rates. Nonetheless, these data suggest that the rate of superinfection in a generalized heterosexual epidemic is substantial and could have a significant impact on the ongoing epidemic.
Almost all HIV transmission in the RCCS population occurs through vaginal heterosexual intercourse, and therefore the findings may not be generalizable to populations with other modes of transmission. Prior studies in men who have sex with men and in commercial sex-workers in Kenya suggest that rates of HIV superinfection may be increased in populations with higher primary HIV incidence [9, 14]. It has also been shown that cellular components of the natural immune response to HIV do not seem to be protective against superinfection [2, 29]. However, there is some evidence that neutralizing antibody responses may be associated with protection [30].
There is previous evidence that HIV superinfection can have detrimental clinical effects, even in individuals who were previously controlling their HIV infection [2, 11, 14, 31]. These data suggest that post-test counseling of HIV-infected individuals needs to emphasize the risk of HIV superinfection and the possible clinical implications of continued unsafe behaviors. These results also have significant implications for estimations of the age of the HIV epidemic and for phylogenetic modeling of viral evolution because many of these models assume that superinfection is not occurring [32]. In addition, the finding that superinfection is common and occurs within and between HIV subtypes suggests that the immune response elicited by primary infection confers limited protection and raises concerns that vaccine strategies designed to replicate the natural anti-HIV immune response may have limited effectiveness. In summary, the finding that superinfection is a relatively frequent event has substantial implications for HIV prevention, clinical management, and future vaccine development.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors would like to thank all the participants of the Rakai cohort and the staff of the Rakai health science program. We would like to especially thank Susanna Lamers for her assistance in sequence submission.
Financial support. This study was supported in part by funding from the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH); Office of AIDS Research, NIH; Division of AIDS, NIAID (R01 A134826 and R01 A134265); National Institute of Child Health & Human Development (R01 HD 050180 and 5P30HD06826); HPTN Network Lab grant (U01-AI-068613); the Gates Foundation (22006.03), Henry M. Jackson Foundation; and the Fogarty Foundation (grant 5D43TW00010). A. A. R. T. was supported by the NIH (1K23AI093152-01A1) and Doris Duke Charitable Foundation Clinician Scientist Development Award (2011036).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Redd AD, Collinson-Streng A, Martens C, et al. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda using next generation deep sequencing. J Clin Microbiol. 2011;49: 2859–67 doi: 10.1128/JCM.00804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld M, Allen TM, Yu XG, et al. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature. 2002;420:434–9. doi: 10.1038/nature01200. [DOI] [PubMed] [Google Scholar]
- 3.Jost S, Bernard MC, Kaiser L, et al. A patient with HIV-1 superinfection. N Engl J Med. 2002;347:731–6. doi: 10.1056/NEJMoa020263. [DOI] [PubMed] [Google Scholar]
- 4.Ramos A, Hu DJ, Nguyen L, et al. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J Virol. 2002;76:7444–52. doi: 10.1128/JVI.76.15.7444-7452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braibant M, Xie J, Samri A, Agut H, Autran B, Barin F. Disease progression due to dual infection in an HLA-B57-positive asymptomatic long-term nonprogressor infected with a nef-defective HIV-1 strain. Virology. 2010;405:81–92. doi: 10.1016/j.virol.2010.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Chohan B, Lavreys L, Rainwater SM, Overbaugh J. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J Virol. 2005;79:10701–8. doi: 10.1128/JVI.79.16.10701-10708.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCutchan FE, Hoelscher M, Tovanabutra S, et al. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J Virol. 2005;79:11693–704. doi: 10.1128/JVI.79.18.11693-11704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbinger KH, Gerhardt M, Piyasirisilp S, et al. Frequency of HIV type 1 dual infection and HIV diversity: analysis of low- and high-risk populations in Mbeya Region, Tanzania. AIDS Res Hum Retroviruses. 2006;22:599–606. doi: 10.1089/aid.2006.22.599. [DOI] [PubMed] [Google Scholar]
- 9.Piantadosi A, Chohan B, Chohan V, McClelland RS, Overbaugh J. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 2007;3:e177. doi: 10.1371/journal.ppat.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streeck H, Li B, Poon AF, et al. Immune-driven recombination and loss of control after HIV superinfection. J Exp Med. 2008;205:1789–96. doi: 10.1084/jem.20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunthard HF, Huber M, Kuster H, et al. HIV-1 superinfection in an HIV-2-infected woman with subsequent control of HIV-1 plasma viremia. Clin Infect Dis. 2009;48:e117–20. doi: 10.1086/598987. [DOI] [PubMed] [Google Scholar]
- 12.van der Kuyl AC, Kozaczynska K, Arien KK, et al. Analysis of infectious virus clones from two HIV-1 superinfection cases suggests that the primary strains have lower fitness. Retrovirology. 2010;7:60. doi: 10.1186/1742-4690-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor JE, Korber BT. HIV-1 intra-subtype superinfection rates: estimates using a structured coalescent with recombination. Infect Genet Evol. 2005;5:85–95. doi: 10.1016/j.meegid.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Smith DM, Wong JK, Hightower GK, et al. Incidence of HIV superinfection following primary infection. JAMA. 2004;292:1177–8. doi: 10.1001/jama.292.10.1177. [DOI] [PubMed] [Google Scholar]
- 15.Piantadosi A, Ngayo MO, Chohan B, Overbaugh J. Examination of a second region of the HIV type 1 genome reveals additional cases of superinfection. AIDS Res Hum Retroviruses. 2008;24:1221. doi: 10.1089/aid.2008.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rachinger A, Stolte IG, van de Ven TD, et al. Absence of HIV-1 superinfection 1 year after infection between 1985 and 1997 coincides with a reduction in sexual risk behavior in the seroincident Amsterdam cohort of homosexual men. Clin Infect Dis. 2010;50:1309–15. doi: 10.1086/651687. [DOI] [PubMed] [Google Scholar]
- 17.Gonzales MJ, Delwart E, Rhee SY, et al. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J Infect Dis. 2003;188:397–405. doi: 10.1086/376534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rachinger A, Manyenga P, Burger JA, et al. Low incidence of HIV-1 superinfection even after episodes of unsafe sexual behavior of homosexual men in the Amsterdam Cohort Studies on HIV Infection and AIDS. J Infect Dis. 2011;203:1621–8. doi: 10.1093/infdis/jir164. [DOI] [PubMed] [Google Scholar]
- 19.Rachinger A, van de Ven TD, Burger JA, Schuitemaker H, van't Wout AB. Evaluation of pre-screening methods for the identification of HIV-1 superinfection. J Virol Methods. 2010;165:311–7. doi: 10.1016/j.jviromet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Pacold M, Smith D, Little S, et al. Comparison of methods to detect HIV dual infection. AIDS Res Hum Retroviruses. 2010;26:1291–6. doi: 10.1089/aid.2010.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wawer MJ, Gray RH, Sewankambo NK, et al. A randomized, community trial of intensive sexually transmitted disease control for AIDS prevention, Rakai, Uganda. AIDS. 1998;12:1211–25. doi: 10.1097/00002030-199810000-00014. [DOI] [PubMed] [Google Scholar]
- 22.Laeyendecker O, Redd AD, Lutalo T, et al. Frequency of long-term nonprogressors in HIV-1 seroconverters From Rakai Uganda. J Acquir Immune Defic Syndr. 2009;52:316–9. doi: 10.1097/QAI.0b013e3181bc08f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wawer MJ, Sewankambo NK, Berkley S, et al. Incidence of HIV-1 infection in a rural region of Uganda. BMJ. 1994;308:171–3. doi: 10.1136/bmj.308.6922.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho D, Imai K, King G, Stuart E. Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Political Analysis. 2007;15:199–236. [Google Scholar]
- 25.Ho D, Imai K, King G, Stuart E. Matchit: nonparametric preporcessing for parametric causal inference. J Stat Softw. 2011;42:1–28. [Google Scholar]
- 26.Rosenbaum P, Rubin D. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- 27.Conroy SA, Laeyendecker O, Redd AD, et al. Changes in the distribution of HIV type 1 subtypes D and A in Rakai District, Uganda between 1994 and 2002. AIDS Res Hum Retroviruses. 2010;26:1087–91. doi: 10.1089/aid.2010.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClelland RS, Hassan WM, Lavreys L, et al. HIV-1 acquisition and disease progression are associated with decreased high-risk sexual behaviour among Kenyan female sex workers. AIDS. 2006;20:1969–73. doi: 10.1097/01.aids.0000247119.12327.e6. [DOI] [PubMed] [Google Scholar]
- 29.Blish CA, Dogan OC, Jaoko W, et al. Cellular immune responses and susceptibility to HIV-1 superinfection: a case-control study. AIDS. 2012;26(5):643–6. doi: 10.1097/QAD.0b013e3283509a0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith DM, Strain MC, Frost SD, et al. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology. 2006;355:1–5. doi: 10.1016/j.virol.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Smith DM, Wong JK, Hightower GK, et al. HIV drug resistance acquired through superinfection. AIDS. 2005;19:1251–6. doi: 10.1097/01.aids.0000180095.12276.ac. [DOI] [PubMed] [Google Scholar]
- 32.Volz EM. Complex population dynamics and the coalescent under neutrality. Genetics. 2012;190:187–201. doi: 10.1534/genetics.111.134627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.