Abstract
Fibrosis is the end result of pathologic wound healing and is characterized by inflammation, excessive proliferation of fibroblasts, and abnormal deposition of extracellular matrix (ECM) proteins. Despite the advanced treatments for the fibrotic diseases, as well as the researches on the fibrosis, pathologic fibrotic diseases remain to be hard cured and the molecular mechanism of fibrosis is still unclear. In our previous studies we found ITGB4BP was involved in the myofibroblast differentiation. However there were no studies about the roles of ITGB4BP in fibrosis. On this background this review explores the basic features and the biological function of ITGB4BP which might imply the underlying cellular and molecular mechanism in the regulation of fibrotic diseases.
Keywords: ITGB4BP, fibrosis
Introduction
Fibrotic diseases are caused by the excessive deposition of extracellular matrix including collagen, which leads to the overgrowth, hardening, and scarring of various tissues. Long-term irritation of chronic inflammation, such as continuous infection, autoimmune disease, allergic response, chemical injury, radiation, and tissue damage, induces fibroblasts to differentiate into myofibroblasts. The fibrosis of a lot of connective tissues leads to ultimately organ failure and death [1-3]. Hypertrophic scar is one of fibrotic diseases, arising from fibroproliferation disorder which occurs after the damage of deep dermis by thermal or traumatic injuries, and patients often have to suffer serious injuries to the appearance and body function [4]. In clinic, antagonizing the effects of high expressed proteins is the main medical treatment for tissue fibrosis and scarring [5], however, the effect of this therapy is not good [1]. On this background, we are looking forward to finding the low expressed proteins related to the high expressed proteins in hypertrophic scar so that we could upregulate the target proteins to control and/or inhibit the formation of hypertrophic scar. P311 has been suggested to be a potent cytokine in inducing myofibroblast differentiation, a key pathologic change [6]. We previously identified the upregulation of p311 may promote the formation of hypertrophic scar [7,8], but it is rarely reported about the mechanism of p311 function. In order to balance fibrosis formation, a potent negative cytokine might be needed to interact with the positive cytokine to regulate the fibrosis. Our previous studies found a potential interaction protein of P311 by both yeast two-hybrid system and Fluorescence Resonance Energy Transfer (FRET) screening, named integrin β4 binding protein (ITGB4BP), may function oppositely in the formation of hypertrophic scar. This protein mainly plays a very important role in ribosome formation, RNA-induced silencing complex (RISC), cytoskeleton, and apoptosis, as well as the process of tissue fibrosis in hypertrophic scar. With this review, we gave an overview of ITGB4BP function.
Coding gene of ITGB4BP
Francesca Sanvito et al. found that ITGB4BP gene locates on the 20q11.2 region of the long arm of 21st chromosome by fluorescence in situ hybridization, its mRNA length is 1108bp, and its open reading frame consists of 735 nucleotides [9]. ITGB4BP gene is comprised of 7 exons and 6 introns, and there are no TATA promoter binding domain and CpG islands [10]. In eukaryote cells, the sequence of ITGB4BP gene is conservative. ITGB4BP gene is found not only in Homo sapiens, mice, rats, Xenopus laevis, and other vertebrates, but also in Drosophila, sepia, molluscum, and other invertebrates, and the domains of ITGB4BP gene are highly conservative between vertebrates and invertebrates [11].
Coding protein of ITGB4BP
ITGB4BP protein is called as Eukaryotic translation initiation factor 6 (eIF6), p27 beta 4 binding protein (p27BBP), CAB and Eukaryotic translation initiation factor 3A (eIF3A) etc. It was first discovered by Biffo Sanvito in 1997 [12]. When Biffo and other researchers conducted research on the relationship between β4 integrin and the formation of hemidesmosomes by yeast two-hybrid system, they found a protein was interacting with β4, so they named it Integrin beta 4 binding protein, ITGB4BP. ITGB4BP protein consists of 245 amino acids with a molecular weight of 27 kDa [11]. In mammals and yeasts, the homology of ITGB4BP protein is 72%, of which the sequence of ITGB4BP protein is 85% similar to each other [13].Through Protein Kinase C (PKC) and/or Casein Kinase C (CK1), the serine residues at positions of 174, 175 and 235 on ITGB4BP are phosphorylated and may play a critical role in the regulation of translation along a developmental pathway [13,14]. Current studies on ITGB4BP are focused on protein synthesis, the formation of cytoskeleton, the differentiation and transformation of cancer cells
Expression and distribution of ITGB4BP
ITGB4BP protein is widely distributed and can be expressed in a variety of tissues, or different cells within a kind of tissue, or different cell cycles for the same cells. ITGB4BP protein expression can be found in epitheliums, fibroblasts, cancer cells, activated T cells, activated mast cells, and muscle fibers. ITGB4BP protein is also expressed in all the cells that express α6β4 integrin [15]. ITGB4BP protein is mainly expressed in the cytoplasm (near intermediate filaments) and nucleus (mainly at the periphery of nucleus, apparently attached to the nuclear membrane) [16]. ITGB4BP is highly expressed in rapidly cycling cells and decreased in villous cells committed to apoptotic cell death [17]. Apparently ITGB4BP protein is related to cytoskeleton and cell proliferation.
Regulation of protein synthesis
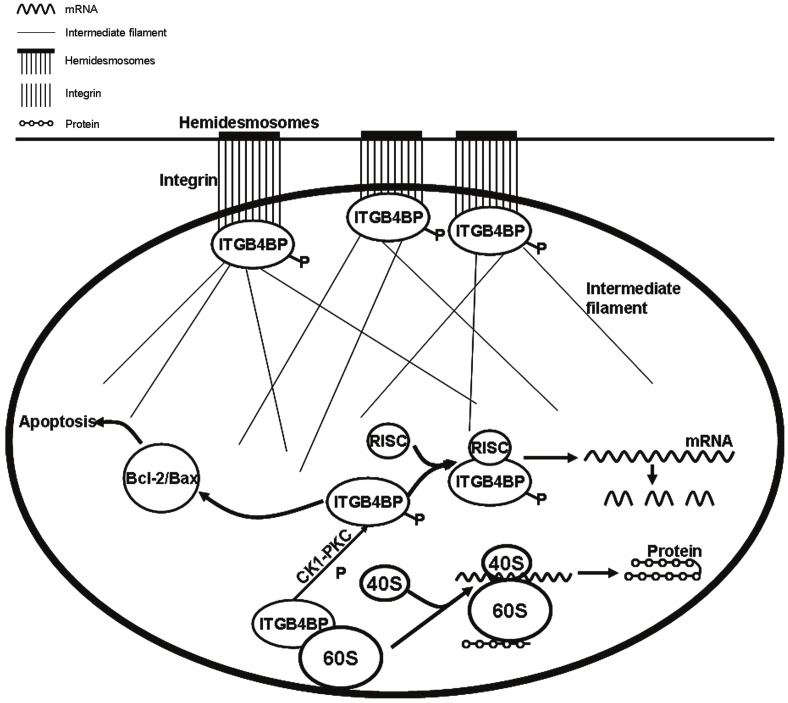
During the growth and differentiation of eukaryote cells, most genes in the genome are not transcribed and only a few genes can be expressed. After the process of transcription and post-transcription, genes turn into mature mRNA, and are translated into polypeptide chains on ribosomes in the cytoplasm and then into the cell needed proteins. The regulation of mRNA translation is based on many levels, among which, the regulation of initial stage in eukaryotes is very important. In the nucleus, the binding of ITGB4BP with 60s subunit inhibits the formation of 80s subunit (Figure 1). In the initial stage of protein translation, as the eukaryotic initiation factor (eIF), ITGB4BP is phosphorylated under the action of CK1-PKC, and depolymerized down from 60s subunit, which enhances the formation of 80s subunit through the binding of 40s subunit with 60s subunit, and at last the protein translation is initiated [18-21].
Figure 1.
The regulation of ITGB4BP in cells. With the phosphorylation of ITGB4BP, its dissociation from 60s subunit allows the formation of 80s subunit and further induces the protein synthesis as a translation initiation factor. Secondly, ITGB4BP downregulates the special protein expression via RISC. Thirdly, ITGB4BP inhibits cell apoptosis through Bcl-2/Bax signal pathway. Fourthly, ITGB4BP regulates cell adhesion and cytoskeleton formation via connecting integrin and intermediate filaments.
On the other hand, the synthesis of ITGB4BP protein is relevant to the regulation on microRNA. By binding ITGB4BP and RNA-induced silencing complex (RISC), the function of the corresponding microRNA is promoted, the mRNA formation is interfered from the transcriptional level,and the expression of the corresponding target protein is inhibited (Figure 1). On the contrary, if the ITGB4BP is removed, the inhibition of microRNA towards the target protein expression is relieved, and as a result, the expression of the target protein is enhanced [22,23].
Biological formation of ribosomes
The formation and translation of ribosomes are two interrelated processes involved in cell growth. Although the formation and translation of ribosomes are regulated by different factors, ITGB4BP can regulate both at the same time. It plays a role in translation as well as in the formation of pre-ribosomes and 60s subunit [24]. Some studies proved that if ITGB4BP gene was removed from yeasts and mice, the loss of ribosome 60s subunit in the nucleus would inhibit ribosome formation, leading to their death [15,25,26]. Therefore, ITGB4BP gene is particularly important in the biological formation of ribosomes.
Role in apoptosis
Apoptosis is a basic biological phenomenon, which plays an important role in evolution, homeostasis, and the development of many systems. Apoptosis is also called programmed cell death (PCD), which is an active cell suicide process regulated by genes. The process of cell apoptosis is complicated and mainly regulated by the Bcl-2 family and Caspase family (Figure 1). ITGB4BP is reported to be able to react on the upstream of Bcl-2/Bax signal pathway and inhibit cell apoptosis [27].
Regulation of cell adhesion and cytoskeleton formation
Integrin family is the receptor of adhesion molecules, which mediate the interactions between cell and extracellular matrix, and the interactions between cells. Cell adhesion includes conjugation, desmosome connection and hemidesmosome connection etc. The main extracellular matrix of hemidesmosome connection is laminin. β4 integrin is the acceptor of laminin, so β4 integrin is closely related to the formation of desmosome and hemidesmosome. ITGB4BP is β4 integrin binding protein, and it binds with intermediate filaments and the intracellular domain of α6β4 integrin at the same time. It acts as the bridge to mediate hemidesmosome formation and cell adhesion [12]. In addition, ITGB4BP also exists in nuclear matrix and involves in the formation of cytoskeleton through the connection to nuclear matrix, intermediate filaments, and other cytoskeletal structure (Figure 1). ITGB4BP has already been proved to be highly expressed in intermediate filaments and nuclear matrix [15]. ITGB4BP can also regulate the expression of β-catenin in Wnt signal pathway, which affects cytoskeleton formation [28].
Effect on the formation and metastasis of tumors
Abnormal cell proliferation, apoptosis inhibition and easy metastasis are the main characteristics of tumors. ITGB4BP participates in cell proliferation as a translation regulator. The subcellular proteomics analysis of nuclear matrix components in hepatoma cells before and after differentiation induction showed that ITGB4BP is highly expressed in differentiated cells, so it is likely to be involved in the differentiation of cancer cells [29]. ITGB4BP can be detected in normal mucosal cells, but it is only highly expressed in cancer cells, for example, ITGB4BP is upregulated in colorectal cancer cells [16]. Interestingly, the same kind of trend of ITGB4BP has been observed in the metastasis of lymph node [30]. It is concluded that the highly expression of ITGB4BP maybe play a part in the formation and metastasis of tumors.
In our previous studies, we have found that ITGB4BP was involved in the fibrotic diseases. ITGB4BP expression was detected to downregulate in hypertrophic scar compared with the normal skin. The upregulation of ITGB4BP could decrease the expression of TGF-β1 and inhibit the TGF-β1 signal pathway. According to ITGB4BP biological roles, ITGB4BP might be a negative regulator participating in the formation of fibrosis via RISC inhibiting the expression of TGF-β1. It implies that ITGB4BP could be a novel target in the control of fibrotic disorders.
Although ITGB4BP may play a very important role in protein synthesis, ribosome formation, cell apoptosis, cytoskeleton formation, the formation and metastasis of tumors, few literatures are related to ITGB4BP. To date, no studies about the function of ITGB4BP in fibrotic diseases have been reported. Therefore, further intensive studies of ITGB4BP function in fibrotic diseases may provide deep insights into the mechanism of fibrosis formation and possibly be useful to find new approaches to treat fibrotic diseases.
References
- 1.Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest. 2007;117:524–529. doi: 10.1172/JCI31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Veer WM, Bloemen MC, Ulrich MM, Molema G, van Zuijlen PP, Middelkoop E, Niessen FB. Potential cellular and molecular causes of hypertrophic scar formation. Burns. 2009;35:15–29. doi: 10.1016/j.burns.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Li JT, Liao ZX, Ping J, Xu D, Wang H. Molecular mechanism of hepatic stellate cell activation and antifibrotic therapeutic strategies. J Gastroenterol. 2008;43:419–428. doi: 10.1007/s00535-008-2180-y. [DOI] [PubMed] [Google Scholar]
- 4.Brown JJ, Bayat A. Genetic susceptibility to raised dermal scarring. Br J Dermatol. 2009;161:8–18. doi: 10.1111/j.1365-2133.2009.09258.x. [DOI] [PubMed] [Google Scholar]
- 5.Howell JE, McAnulty RJ. TGF-beta: its role in asthma and therapeutic potential. Curr Drug Targets. 2006;7:547–565. doi: 10.2174/138945006776818692. [DOI] [PubMed] [Google Scholar]
- 6.Pan D, Zhe X, Jakkaraju S, Taylor GA, Schuger L. P311 induces a TGF-beta1-independent, nonfibrogenic myofibroblast phenotype. J Clin Invest. 2002;110:1349–1358. doi: 10.1172/JCI15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Ma B, Yi S, Wang Z, He W, Luo G, Chen X, Wang X, Chen A, Barisoni D. Gene expression of early hypertrophic scar tissue screened by means of cDNA microarrays. J Trauma. 2004;57:1276–1286. doi: 10.1097/01.ta.0000108997.49513.dc. [DOI] [PubMed] [Google Scholar]
- 8.Tan J, Peng X, Luo G, Ma B, Cao C, He W, Yuan S, Li S, Wilkins JA, Wu J. Investigating the role of P311 in the hypertrophic scar. Plos One. 2010;5:e9995. doi: 10.1371/journal.pone.0009995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanvito F, Arrigo G, Zuffardi O, Agnelli M, Marchisio PC, Biffo S. Localization of p27 beta 4 binding protein gene (ITGB4BP) to human chromosome region 20q11.2. Genomics. 1998;52:111–112. [PubMed] [Google Scholar]
- 10.Donadini A, Giodini A, Sanvito F, Marchisio PC, Biffo S. The human ITGB4BP gene is constitutively expressed in vitro, but highly modulated in vivo. Gene. 2001;266:35–43. doi: 10.1016/s0378-1119(01)00370-5. [DOI] [PubMed] [Google Scholar]
- 11.Si K, Chaudhuri J, Chevesich J, Maitra U. Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6. Proc Natl Acad Sci USA. 1997;94:14285–14290. doi: 10.1073/pnas.94.26.14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biffo S, Sanvito F, Costa S, Preve L, Pignatelli R, Spinardi L, Marchisio PC. Isolation of a novel beta4 integrin-binding protein (p27(BBP)) highly expressed in epithelial cells. J Biol Chem. 1997;272:30314–30321. doi: 10.1074/jbc.272.48.30314. [DOI] [PubMed] [Google Scholar]
- 13.Basu U, Si K, Deng H, Maitra U. Phosphorylation of mammalian eukaryotic translation initiation factor 6 and its Saccharomyces cerevisiae homologue Tif6p: evidence that phosphorylation of Tif6p regulates its nucleocytoplasmic distribution and is required for yeast cell growth. Mol Cell Biol. 2003;23:6187–6199. doi: 10.1128/MCB.23.17.6187-6199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carotenuto R, De Marco N, Biffo S, Wilding M, Vaccaro MC, Marchisio PC, Capriglione T, Russo GL, Campanella C. Phosphorylation of p27 (BBP)/eIF6 and its association with the cytoskeleton are developmentally regulated in Xenopus oogenesis. Cell Mol Life Sci. 2005;62:1641–1652. doi: 10.1007/s00018-005-5153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanvito F, Piatti S, Villa A, Bossi M, Lucchini G, Marchisio PC, Biffo S. The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J Cell Biol. 1999;144:823–837. doi: 10.1083/jcb.144.5.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balbo A, Bozzaro S. Cloning of Dictyostelium eIF6 (p27BBP) and mapping its nucle(ol)ar localization subdomains. Eur J Cell Biol. 2006;85:1069–1078. doi: 10.1016/j.ejcb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Sanvito F, Vivoli F, Gambini S, Santambrogio G, Catena M, Viale E, Veglia F, Donadini A, Biffo S, Marchisio PC. Expression of a highly conserved protein, p27BBP, during the progression of human colorectal cancer. Cancer Res. 2000;60:510–516. [PubMed] [Google Scholar]
- 18.Basu U, Si K, Warner JR, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol. 2001;21:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 20.Benelli D, Marzi S, Mancone C, Alonzi T, la Teana A, Londei P. Function and ribosomal localization of aIF6, a translational regulator shared by archaea and eukarya. Nucleic Acids Res. 2009;37:256–267. doi: 10.1093/nar/gkn959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455:684–688. doi: 10.1038/nature07267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 23.Flavin RJ, Smyth PC, Finn SP, Laios A, O'Toole SA, Barrett C, Ring M, Denning KM, Li J, Aherne ST, Aziz NA, Alhadi A, Sheppard BL, Loda M, Martin C, Sheils OM, O'Leary JJ. Altered eIF6 and Dicer expression is associated with clinicopathological features in ovarian serous carcinoma patients. Mod Pathol. 2008;21:676–684. doi: 10.1038/modpathol.2008.33. [DOI] [PubMed] [Google Scholar]
- 24.Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 2009;10:459–465. doi: 10.1038/embor.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Si K, Maitra U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol Cell Biol. 1999;19:1416–1426. doi: 10.1128/mcb.19.2.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood LC, Ashby MN, Grunfeld C, Feingold KR. Cloning of murine translation initiation factor 6 and functional analysis of the homologous sequence YPR016c in Saccharomyces cerevisiae. J Biol Chem. 1999;274:11653–11659. doi: 10.1074/jbc.274.17.11653. [DOI] [PubMed] [Google Scholar]
- 27.De Marco N, Iannone L, Carotenuto R, Biffo S, Vitale A, Campanella C. p27(BBP)/eIF6 acts as an anti-apoptotic factor upstream of Bcl-2 during Xenopus laevis development. Cell Death Differ. 2009;17:360–372. doi: 10.1038/cdd.2009.128. [DOI] [PubMed] [Google Scholar]
- 28.Ji Y, Shah S, Soanes K, Islam MN, Hoxter B, Biffo S, Heslip T, Byers S. Eukaryotic initiation factor 6 selectively regulates Wnt signaling and beta-catenin protein synthesis. Oncogene. 2008;27:755–762. doi: 10.1038/sj.onc.1210667. [DOI] [PubMed] [Google Scholar]
- 29.Tang J, Niu JW, Xu DH, Li ZX, Li QF, Chen JA. Alteration of nuclear matrix-intermediate filament system and differential expression of nuclear matrix proteins during human hepatocarcinoma cell differentiation. World J Gastroenterol. 2007;13:2791–2797. doi: 10.3748/wjg.v13.i20.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosso P, Cortesina G, Sanvito F, Donadini A, Di Benedetto B, Biffo S, Marchisio PC. Overexpression of p27BBP in head and neck carcinomas and their lymph node metastases. Head Neck. 2004;26:408–417. doi: 10.1002/hed.10401. [DOI] [PubMed] [Google Scholar]