Abstract
Cryptosporidium hominis and Cryptosporidium parvum, which infect humans equally, are genetically/antigenically almost identical. It remains unclear, however, whether infection with C. hominis protects against C. parvum. Gnotobiotic piglets were used to investigate cross-protection. After ≥3 days of recovery from C. hominis infection, the piglets were completely protected against subsequent challenge with C. hominis but only partially against challenge with C. parvum, as compared with age-matched control animals challenged with either species. In conclusion, C. hominis–specific immunity was sufficient to completely protect against challenge with the same species but insufficient to provide the same level of protection against C. parvum.
Cryptosporidium species are enteric protozoa that infect most vertebrate animals. The 2 species associated predominantly with human disease are C. parvum and C. hominis. Cryptosporidium parvum is found in all mammalian species including humans, whereas C. hominis exclusively infects humans [1]. The 2 parasites are genetically (95%–97% sequence identity) and antigenically mostly indistinguishable. Some antigenic differences within the homologous antigens have been noted [2], but no unique antigen specific for any of the 2 parasites has been identified. The 2 parasites were separated into 2 species based on host predilection and the apparent absence in nature of recombinants among the 2 species [3, 4]. Despite the shared attributes between the 2 species to which humans are equally susceptible to, nothing is known about the immunological relatedness among them. The subject of this investigation was to assess whether infection with C. hominis protects against future infection with C. parvum. We conducted these studies in gnotobiotic piglets, which are equally susceptible to both C. hominis and C. parvum [1].
MATERIALS AND METHODS
Cryptosporidium Isolates
Cryptosporidium hominis isolate TU502 [4, 5] and C. parvum isolate MD [6] isolated originally from a Ugandan child and a red deer calf in Scotland, respectively, were used. Isolate MD belongs to a subspecific group of C. parvum isolates that infect humans and animals. This isolate has been characterized using multiple genetic markers [7] and has been shown to be infective to humans [6]. Both isolates were propagated in piglets and oocysts for challenge experiments purified from feces and genotyped as described elsewhere [5, 8]. The oocyst existation rate was estimated to determine the viability of the oocysts.
Gnotobiotic piglet Experiments
Piglets were derived by cesarean section and maintained in microbiological isolators as described elsewhere [9]. Piglets were divided into 4 groups. One day after birth 2 groups were orally infected with 1 × 106 oocysts per animal of C. hominis TU502 (primary infection), and the remaining 2 groups remained uninfected. The intensity of fecal oocyst shedding was estimated by counting oocysts in 30 evenly spaced microscopic fields (×100 magnification) of acid-fast-stained fecal smears [9]. A fecal sample was declared negative when no oocysts could be detected microscopically in 3 replicate smears (entire smears) from the same fecal specimen. An animal was declared uninfected or recovered from the infection if fecal oocysts could not be detected by microscopic examination and polymerase chain reaction (PCR) targeting the Cryptosporidium oocyst wall protein (COWP) [8] for ≥3 consecutive days. After recovery, 1 of the 2 primary-infection groups was orally challenged (challenge infection) with 1 × 107 oocysts per animal of C. hominis TU502 (C. hominis–C. hominis group), and the second group with 1 × 107 oocysts per animal of C. parvum MD (C. hominis–C. parvum group). The 2 previously uninfected groups were also infected orally with 1 × 107 oocysts per animal; 1 group with C. hominis TU502 (null–C. hominis group) and another with C. parvum MD (null–C. parvum group). The intensity of fecal oocyst shedding was again recorded daily. The species of the Cryptosporidium used to infect and then recovered from feces was determined by COWP PCR restriction fragment length polymorphism (RFLP) as described elsewhere [8]. After 12 days of the challenge infection, piglets in all 4 groups were euthanized. Before being euthanized, blood was drawn from all piglets to harvest serum for immunoblot analysis. Gut sections (n = 15) were taken from each animal from the small (sections 1–12) and large (sections 13–15) intestines. DNA was extracted from intestinal mucosa scraped from the gut sections and used to determine the Cryptosporidium species by COWP PCR-RFLP assay. Intestinal mucus was also used as a source of mucosal immunoglobulin A (IgA) for immunoblot analysis and prepared by adding 1 mL phosphate-buffered saline to 250 μL mucus, vortexing for 2 minutes, centrifuging at 13 000g for 2 minutes and collecting the supernatant (mucus supernatant). Twenty-one piglets from 3 litters were used for these experiments. Five piglets were used for each group with the exception of the C. hominis–C. hominis group (6 piglets).
Immunoblotting
Proteins of excysted oocysts (1 × 106 per lane) were separated on NuPAGE 4% to 12% Bis–Tris-polyacrylamide gradient gels (Invitrogen) and transferred to nitrocellulose membranes for immunoblotting. The membranes were blocked with 5% nonfat dried milk powder in TBS-T (Tris-buffered saline, 0.1% Tween-20) at room temperature for 1 hour, washed, and incubated with either serum (1:50) or mucus-supernatant (1:20) samples again at room temperature for 1 hour. Controls included preinfection serum samples and mucus supernatant from uninfected animals. Following washing, strips were incubated with horseradish peroxidase–conjugated goat anti-porcine immunoglobulin G (IgG) or IgA (1/1000; AbD Serotec) for 1 hour and then washed and developed with TMB Membrane Peroxidase-Substrate System (KPL).
Statistical Analysis
The Wilcoxon rank-sum test and Kolmogorov–Smirnov tests were utilized to test whether average shedding of null–C. hominis and null–C. parvum groups were significantly more than C. hominis–C. hominis and C. hominis–C. parvum groups, respectively. Resulting P values < .05 were considered significant.
RESULTS
Cross-Protection Studies in the Gnotobiotic Piglet Model
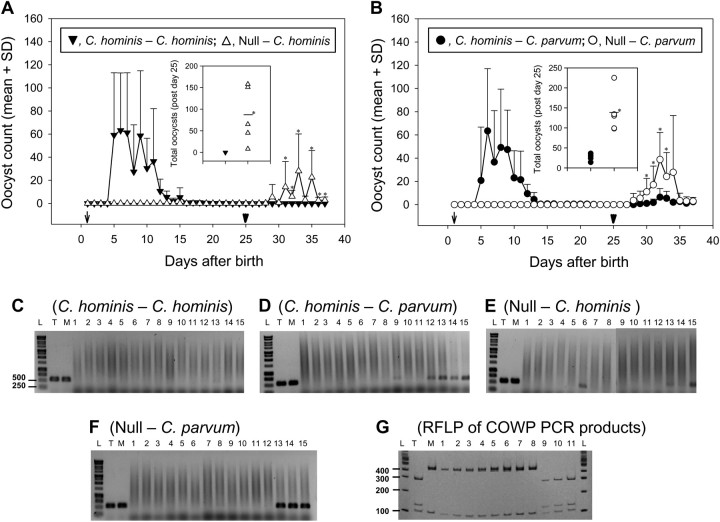
Four–five days after primary infection with C. hominis, all piglets started shedding oocysts in feces and continued shedding for 10–18 days (Figure 1). COWP PCR-RFLP showed that the oocysts shed in feces were of C. hominis genotype (results not shown). Uninfected piglets did not shed any oocysts (Figure 1). Following the challenge infection, the C. hominis–C. hominis group did not shed any oocysts for the duration of the experiment as determined by microscopic (Figure 1A) and COWP PCR analysis (results not shown) of feces. However, all C. hominis–C. parvum group piglets shed oocysts 3–4 days after challenge infection (Figure 1B); COWP PCR-RFLP confirmed that the oocysts were C. parvum (results not shown). All age-matched uninfected piglets that received the challenge infection with either C. hominis TU502 (null–C. hominis group, Figure 1A) or C. parvum MD (null–C. parvum group, Figure 1B) also excreted oocysts 3–4 days after the oral infection. The oocysts had the expected COWP genotype (results not shown).
Figure 1.
Cross-protection studies in the gnotobiotic piglet model. A, B, Piglets shed oocysts in feces following primary infection (arrows) with Cryptosporidium hominis (C. hominis–C. hominis, A; and C. hominis–Cryptosporidium parvum, B). Piglets that were not subjected to primary infection remained negative until infected (null–C. hominis, A; and null–C. parvum, B). Following challenge infection (arrowheads), piglets of all groups (A and B) except the C. hominis–C. hominis group (A) shed oocysts. The y-axis shows mean + standard deviation (SD) of oocyst count (in 30 microscopic fields at ×100 magnification). The statistically significant differences of oocyst count between the groups on a particular day are shown with asterisks. Insets of figure A and B show scatterplots (means represented by a bar) of total oocyst counts of each piglet during 12 days of postchallenge (days 26–37 after birth) from null–C. hominis/C. hominis–C. hominis groups and null–C. parvum/C. parvum–C. parvum groups, respectively. Statistically significant differences of total oocyst count between groups are indicated with asterisks. C–G, Cryptosporidium oocyst wall protein (COWP) polymerase chain recation (PCR) restriction fragment length polymorphism (RFLP) analysis of intestinal sections from a piglet of each of the 4 infection groups at necropsy 12 days after challenge infection. C–F, PCR analysis of the 15 intestinal sections; lanes 1–12 correspond to sections of the small intestine and lanes 13–15 to the large intestine. G, RFLP results of the 553-bp amplicon digested with RsaI. Lanes T and M are positive controls with C. hominis TU502 and C. parvum MD DNA, respectively. Lane L contains 1-kb ladder (C–F) or 100-bp ladder (G). The experimental group for panels C–F is shown above each figure. G, Lanes 1–5 contain RFLP products of the COWP PCR products of lanes 9 and 12–15 of the C. hominis–C. parvum group, respectively; lanes 6–8 contain COWP PCR-RFLP products of lanes 13–15 of the null–C. parvum group, respectively; and lanes 9–11 contain RFLP products of the COWP PCR products of lanes 6, 13, and 15 of the null–C. hominis group, respectively. Digestion with RsaI yields fragments of 285, 125, 106, and 34 bp for type C. hominis and 410, 106, and 34 bp for C. parvum. The 34-bp fragment is not shown as it runs through the gel.
Following challenge infection, total oocyst counts of the null–C. parvum group (range, 98–225) were significantly higher than that of the C. hominis–C. parvum group (range, 14–36) (Figure 1B). Counts for the former group were also significantly higher than those for the latter group on days 5–8 after challenge infection (Figure 1B). Because none of the piglets in the C. hominis–C. hominis group had oocyst shedding after challenge infection, but all piglets in the null–C. hominis group did, total oocysts shedding by the null–C. hominis group (range, 8–158) was significantly higher than that for the C. hominis–C. hominis group (Figure 1A). The former group shed more than the latter group on days 6–8 and 10–12 after challenge infection (Figure 1B). Statistically significant differences were determined by using the Wilcoxon rank and Kolmogorov-Smirnov tests.
COWP PCR-RFLP Analysis of Intestinal Sections
Cryptosporidium DNA was not present in any of the intestinal sections of the piglets of the C. hominis–C. hominis group (Figure 1C). Cryptosporidium DNA was detected in the other 3 groups almost entirely in the large intestine (Figure 1D–F). DNA isolated from intestinal sections of the C. hominis–C. parvum and null–C. parvum groups was of the C. parvum genotype (Figure 1G). In contrast, DNA isolated from intestinal sections of the null–C. hominis group was genotyped as C. hominis (Figure 1G).
Serum IgG and Mucosal IgA Responses
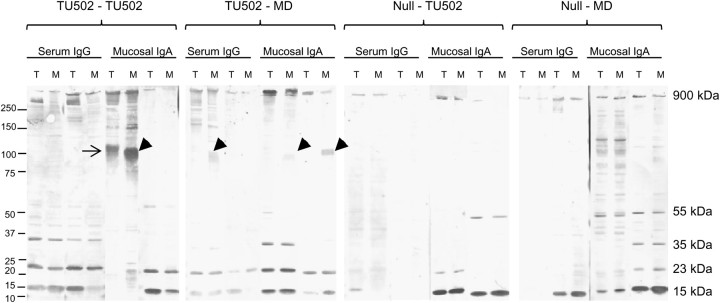
Almost all piglets generated serum IgG and mucosal IgA responses against a 900-kDa antigen, and a majority of animals generated responses against 15- and 23-kDa antigens (Figure 2). Antibodies of some animals reacted with a 35-kDa antigen and a few reacted with a 50–55-kDa antigen (Figure 2). If antibodies in any serum or mucus sample identified an antigen of a certain molecular weight of 1 parasite species, they almost always identified the same molecular weight antigen of the other species. The exception was mucosal IgA of 2 pigs in the C. hominis–C. hominis group, which recognized a 105-kDa antigen of C. hominis but a 100-kDa antigen and not 105-kDa antigen of C. parvum (C. hominis–C. hominis group, Figure 2). The 100-kDa antigen of C. parvum (Figure 2) was also identified by the serum and mucosal antibodies of C. hominis–C. parvum group animals. Preinfected serum samples and mucus from uninfected animals did not identify any antigen (not shown).
Figure 2.
Immunoblot reactivity of serum immunoglobulin G (IgG) and mucosal immunoglobulin A (IgA) (2 animals from each group) collected at necropsy 12 days after challenge infection with antigens of Cryptosporidium hominis TU502 (lane T) and Cryptosporidium parvum MD (lane M). A 105-kDa antigen of C. hominis and a 100-kDa antigen of C. parvum are indicated by the arrow and arrowhead, respectively. Molecular weight markers are shown on the left, and molecular weights of some of the Cryptosporidium antigens are on the right.
DISCUSSION
To our knowledge, this is the first study to show that the immunity generated against C. hominis provides complete protection against challenge with the same C. hominis isolate but is insufficient to provide complete protection against C. parvum. The results suggest that the antigenic similarities of the 2 parasite species induce a partially cross-protective immunity following C. hominis infection. The results also suggest that variability in C. hominis and C. parvum antigens plays an important role in inducing immunity that completely protects against the former but not the latter parasite. The GB piglet is the only laboratory animal model available in which to conduct such studies; like humans, GB piglets are equally susceptible to C. hominis and C. parvum. These observations reflect a credible trend. However, the high cost and the amount of labor associated with this animal model limits the number of animals that can be used. Nevertheless, the experiments were performed on piglets of 3 litters from 3 different sows and each litter had all 4 experimental groups; similar results were obtained for each experimental group from piglets of all litters.
Studies in human volunteers have shown that 1 year after recovery from cryptosporidiosis, the incidence, time of onset, and duration of clinical illness in volunteers rechallanged with the same C. parvum isolate were similar to that observed after the primary challenge [10]. However, fewer subjects excreted oocysts after challenge than after the first infection [10]. In the present study, the longevity of protective immunity induced by the primary infection could not be studied beyond 3–9 days as rapid growth of the piglets precluded us from keeping them for more than 5–6 weeks in microbiological isolators.
An earlier study has reported that Cryptosporidium infection in piglets infected 3–5 days after birth can be seen throughout the small and large intestine during initial period of the infection (5–6 days after infection) [9]. However, later during the infection, the large intestine is most heavily infected, with some infection present throughout the small intestine [9]. In the present study, 25-day-old piglets received challenge infection and were killed 12 days later; the infection was primarily localized in the large intestine with minimal infection in the small intestine. Changes in the gut physiology with age may lead to much greater localization of infection in the large intestine than the small intestine, especially during the later part of the infection.
The aim of the present study was to investigate cross-protection, rather than to identify actual protective mechanisms against Cryptosporidium species. Many studies have shown that T-cell–mediated responses, in particular Th1, are critical in the control of cryptosporidiosis [11, 12]. The precise role of antibodies remains unclear, but they seem to contribute to protection [11]. In the present study, local mucosal and serum antibody responses were analyzed to determine if antibodies from each experimental group of animals recognized any differences in antigenic profile between species. No such differences were identified except the recognition of 105-kDa and 100-kDa antigens of C. hominis and C. parvum by mucosal IgA from 2 animals of the C. hominis–C. hominis group, respectively. We believe these are homologous antigens and the differences in the molecular weights are contributed by the differences in glycosylation and/or the presence of polymorphic amino acid repeats.
Of the 3 immunodominant antigens of 15-, 23-, and 900-kDa molecular weight, the 15-kDa antigen was dominant. Previous studies have also shown prominent serum antibody reactivity with 15–17- and 23–27-kDa proteins of the parasite [11, 13, 14] and evaluated them as vaccine candidates [15].
In conclusion, the results suggest that C. hominis–specific immunity is sufficient to completely protect against challenge with the same species but insufficient to provide the same level of protection against C. parvum.
Notes
Acknowledgments.
We thank Rachel Sora for assistance with animal experiments.
Financial support.
This work was supported by the National Center for Research Resources of the National Institutes of Health (NIH) (grant RR17383) to A. S.; S. T. and G. W. acknowledge financial support from the National Institute of Allergy and Infectious Diseases of the NIH (grants AI050471 and AI052781, respectively).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Tzipori S, Widmer G. The biology of Cryptosporidium. Contrib Microbiol. 2000;6:1–32. doi: 10.1159/000060370. [DOI] [PubMed] [Google Scholar]
- 2.Sturbaum GD, Schaefer DA, Jost BH, Sterling CR, Riggs MW. Antigenic differences within the Cryptosporidium hominis and Cryptosporidium parvum surface proteins P23 and GP900 defined by monoclonal antibody reactivity. Mol Biochem Parasitol. 2008;159:138–41. doi: 10.1016/j.molbiopara.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Morgan-Ryan UM, Fall A, Ward LA, et al. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J Eukaryot Microbiol. 2002;49:433–40. doi: 10.1111/j.1550-7408.2002.tb00224.x. [DOI] [PubMed] [Google Scholar]
- 4.Xu P, Widmer G, Wang Y, et al. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–12. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 5.Akiyoshi DE, Feng X, Buckholt MA, Widmer G, Tzipori S. Genetic analysis of a Cryptosporidium parvum human genotype 1 isolate passaged through different host species. Infect Immun. 2002;70:5670–5. doi: 10.1128/IAI.70.10.5670-5675.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okhuysen PC, Rich SM, Chappell CL, et al. Infectivity of a Cryptosporidium parvum isolate of cervine origin for healthy adults and interferon-gamma knockout mice. J Infect Dis. 2002;185:1320–5. doi: 10.1086/340132. [DOI] [PubMed] [Google Scholar]
- 7.Tanriverdi S, Widmer G. Differential evolution of repetitive sequences in Cryptosporidium parvum and Cryptosporidium hominis. Infect Genet Evol. 2006;6:113–22. doi: 10.1016/j.meegid.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Akiyoshi DE, Mor S, Tzipori S. Rapid displacement of Cryptosporidium parvum type 1 by type 2 in mixed infections in piglets. Infect Immun. 2003;71:5765–71. doi: 10.1128/IAI.71.10.5765-5771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzipori S, Rand W, Griffiths J, Widmer G, Crabb J. Evaluation of an animal model system for cryptosporidiosis: therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulin. Clin Diagn Lab Immunol. 1994;1:450–63. doi: 10.1128/cdli.1.4.450-463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okhuysen PC, Chappell CL, Sterling CR, Jakubowski W, DuPont HL. Susceptibility and serologic response of healthy adults to reinfection with Cryptosporidium parvum. Infect Immun. 1998;66:441–3. doi: 10.1128/iai.66.2.441-443.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riggs MW. Recent advances in cryptosporidiosis: the immune response. Microbes Infect. 2002;4:1067–80. doi: 10.1016/s1286-4579(02)01631-3. [DOI] [PubMed] [Google Scholar]
- 12.Ehigiator HN, McNair N, Mead JR. Cryptosporidium parvum: the contribution of Th1-inducing pathways to the resolution of infection in mice. Exp Parasitol. 2007;115:107–13. doi: 10.1016/j.exppara.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Moss DM, Chappell CL, Okhuysen PC, et al. The antibody response to 27-, 17-, and 15-kDa Cryptosporidium antigens following experimental infection in humans. J Infect Dis. 1998;178:827–33. doi: 10.1086/515377. [DOI] [PubMed] [Google Scholar]
- 14.Reperant JM, Naciri M, Iochmann S, Tilley M, Bout DT. Major antigens of Cryptosporidium parvum recognised by serum antibodies from different infected animal species and man. Vet Parasitol. 1994;55:1–13. doi: 10.1016/0304-4017(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 15.Liu K, Zai D, Zhang D, et al. Divalent Cp15-23 vaccine enhances immune responses and protection against Cryptosporidium parvum infection. Parasite Immunol. 2010;32:335–44. doi: 10.1111/j.1365-3024.2009.01191.x. [DOI] [PubMed] [Google Scholar]