Abstract
The ability of the Escherichia coli protein, BirA, to function as both a metabolic enzyme and a transcription repressor relies on use of a single surface for two distinct protein:protein interactions. BirA forms a heterodimer with the biotin acceptor protein of acetyl-CoA carboxylase and catalyzes post-translational biotinylation. Alternatively, it forms a homodimer that binds sequence-specifically to DNA to repress transcription initiation at the biotin biosynthetic operon. Several surface loops on BirA, two of which exhibit sequence conservation in all biotin protein ligases and the remainder of which are highly variable, are located at the two interfaces. The function of these loops in both homodimerization and biotin transfer was investigated by characterizing alanine-substituted variants at 18 positions of one constant and three variable loops. Sedimentation equilibrium measurements reveal that 11 of the substitutions, which are distributed throughout conserved and variable loops, significantly alter homodimerization energetics. By contrast, steady-state and single turnover kinetic measurements indicate that biotin transfer to BCCP is impacted by 7 substitutions, the majority of which are in the constant loop. Furthermore, constant loop residues that function in biotin transfer also support homodimerization. The results reveal clues about the evolution of a single protein surface for use in two distinct functions.
Keywords: Protein:protein interactions, bispecificity, surface loops, evolution
Introduction
Multispecificity in protein:protein interactions provides a mechanism for integration of seemingly unrelated cellular processes. For example, through its ability to bind to many distinct protein partners ubiquitin functions in protein turnover, DNA repair, cellular trafficking and chromatin remodeling.1 Although general mechanisms, including conformational flexibility and intrinsic disorder2, for achieving multispecific protein binding have been identified, the evolution of multispecificity in protein interactions remains obscure. One pressing question is how multispecificity is achieved without severely impacting function.
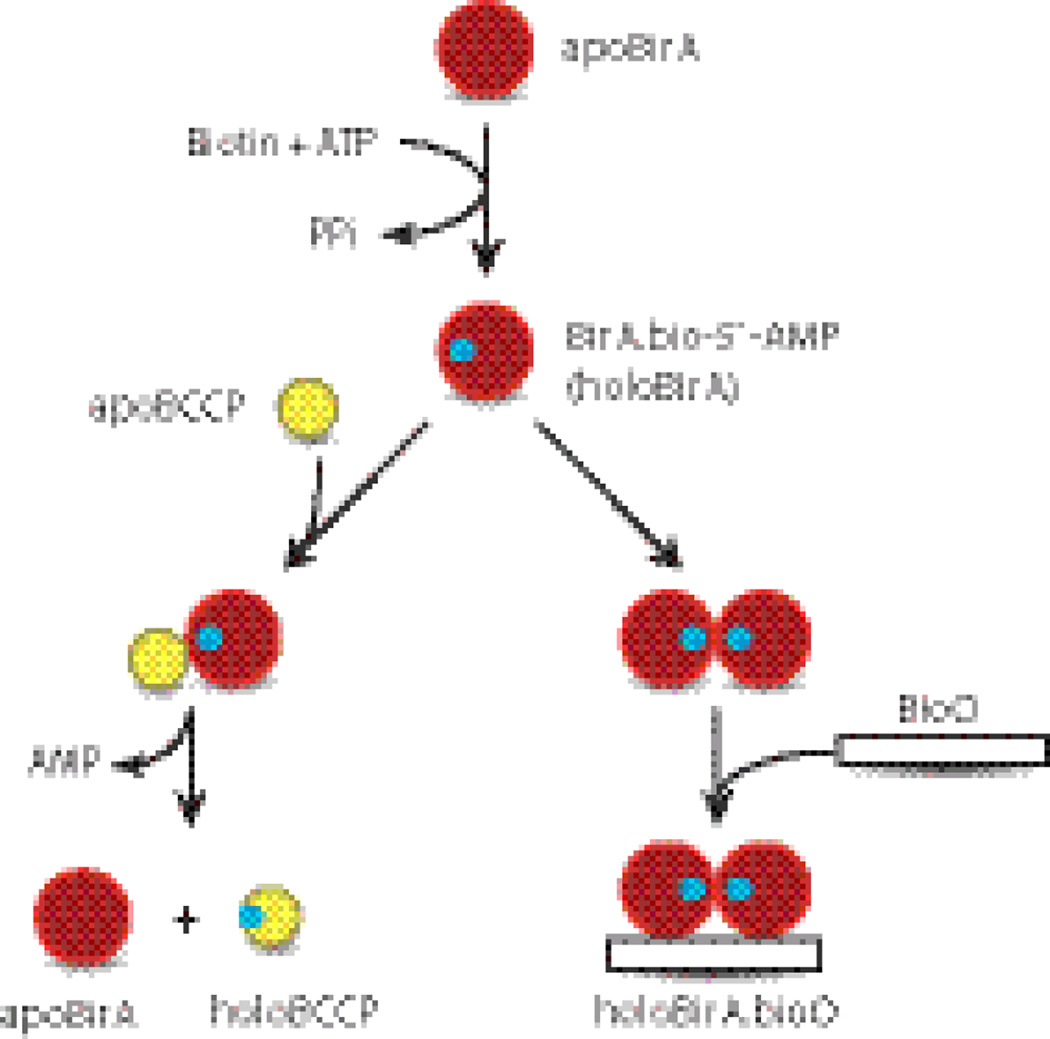
The Escherichia coli biotin protein ligase, BirA, provides an ideal system for investigation of a multispecific protein binding surface.3; 4 BirA binds biotin and ATP to catalyze synthesis of bio-5`-AMP, and the resulting enzyme-intermediate complex (holoBirA) partitions to one of two functions in response to cellular biotin demand (Figure 1). During rapid growth, when demand is high, holoBirA transfers biotin to the biotin carboxyl carrier protein (BCCP) subunit of acetyl-CoA carboxylase, the enzyme that catalyzes the first committed step of fatty acid synthesis.5 ApoBCCP provides the direct signal for biotin demand6 and only upon its depletion can holoBirA homodimerize and bind sequence-specifically to the biotin operator DNA, bioO, to repress transcription initiation at the biotin biosynthetic operon.7 The enzyme intermediate, bio-5’-AMP, binding is required for tight homodimerization.8; 9 Although post-translational biotinylation is essential for viability, only a subset of biotin protein ligases are also repressors.10
Fig. 1.
The Escherichia coli Biotin Regulatory System: BirA catalyzes bio-5`-AMP synthesis from biotin and ATP, and the resulting holoBirA either channels biotin to metabolism by linking it to BCCP or homodimerizes and binds sequence-specifically to the biotin operator to repress transcription of the biotin biosynthetic operon.
A single surface on BirA is used for both hetero- and homodimerization(Figure 2A, B) with both dimers forming by extension of a β-sheet in the BirA central domain.11; 12 Additionally, several BirA surface loops are located in the two interfaces. Previous studies reveal that substitution or deletion of residues in a subset of these loops results in both decreased repression in vivo and impaired DNA binding and homodimerization in vitro.3; 4; 13; 14 A number of these sequence changes also affect the interaction with BCCP.15 Due to their limited scope, these results were insufficient to draw conclusions about the relationship between the structure of the bispecific BirA surface and its two functions.
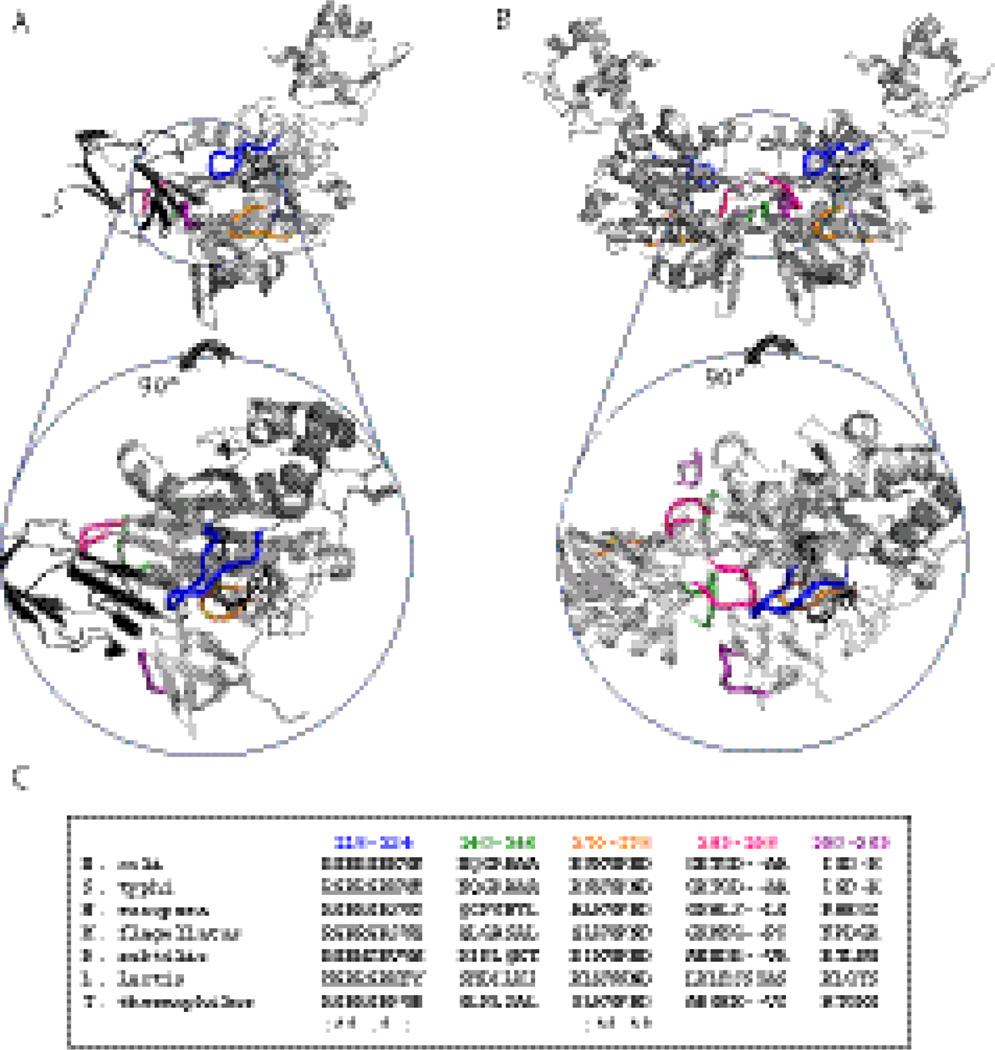
Fig. 2.
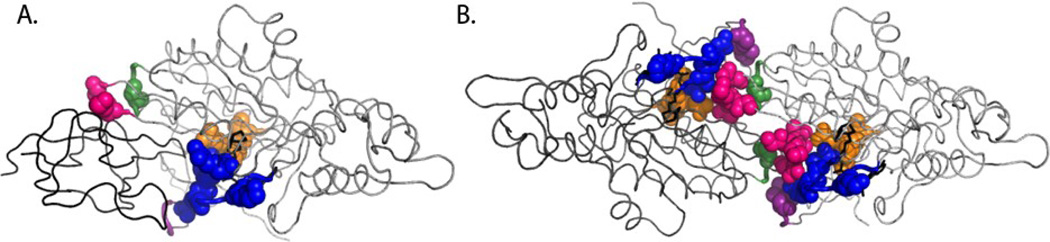
Models of holoBirA-BCCP87 heterodimer (A) and holoBirA homodimer (B). Rotation of the molecules by 90° displays cross-sections of the heterodimer interface (bottom left) and homodimer interface (bottom right), highlighting the surface loops involved in the two interactions. All models were created using Pymol39 with input file 2EWN for the homodimer and a file constructed by Zachary Wood for the heterodimer. This model, which uses the experimentally determined structure of the Pyroccocushorikoshi biotin protein ligase:BCCP complex as a template (2EJG)19, was constructed using the holoBirA monomer coordinates from 2EWN12 and the coordinates for apoBCCP87 (1BDO)40 C. Multiple sequence alignment of several bifunctional microbial biotin protein ligases performed using Clustal W.16 The color codes for loop sequences correspond to the coloring of loops in the models in A and B.
Alignment of sequences of bifunctional bacterial ligases provides clues to the evolution of bispecific protein:protein interactions in BirA (Figure 2C, Supplementary Figure16). The average sequence identity among these ligases ranges from 90 to 21%. However, alignment of several interface loop sequences reveals that they fall into two classes designated “conserved and variable.” The sequences of conserved loops 116–124 and 170–176 are similar in biotin ligases from bacteria to humans, including mono-functional ligases, which either do not form homodimers or do so using surfaces distinct from that used by Ec BirA.17; 18; 19 Consequently, this conservation likely reflects the critical roles of these loops in heterodimerization and/or biotin transfer. This does not, however, preclude roles for some of the constant loop residues in homodimerization.3; 4; 13; 14; 15 By contrast, the variable loop sequences, comprising residues 140–146, 193–199, and 280–283, likely evolved to support homodimerization while not compromising the essential heterodimerization function. Furthermore, the sequence variability in these loops for the bifunctional ligases indicates degenerate solutions to evolution of homodimerization function.
The hypothesized roles of four of the BirA surface loops, including three variable and one constant loop, in bispecific dimerization were investigated by measuring the functional consequences of single alanine substitutions for the two reactions. Sedimentation equilibrium measurements of alanine variants reveal that substitutions in any of the four loops can impact homodimerization and yield dimerization free energies spanning an 8.0 kcal/mole range. By contrast, steady-state and single turnover kinetic measurements of biotin transfer indicate large perturbations primarily for alanine substitutions in the conserved loops. Additionally, the constant loop residues that function in biotin transfer to BCCP also support the homodimerization reaction. These results reveal a strategy for acquisition of additional protein interaction potential in a protein surface that is evolutionarily constrained to function in an essential metabolic reaction.
Results
Homodimerization is sensitive to alanine substitution in variable and constant loops
The effects of alanine substitutions in the variable loops 140–146, 193–199 and 280–283 and constant loop 170–176 on homodimerization were measured using equilibrium analytical ultracentrifugation. The conserved loop comprising residues 116–124 was not included in the present work because previously published data indicate roles for this loop in both homodimerization and the interaction with BCCP.13; 14; 15 The total number of variants in the four remaining loops was limited to 18 either because only non-alanine residues were replaced or the variant protein could not be purified. Since stable dimerization requires binding of the allosteric effector, bio-5’-AMP, all measurements were performed on complexes of proteins bound to the adenylate. Saturation was ensured by preparing solutions at a 1.5:1 molar excess of bio-5’-AMP:protein in the micro molar range of concentration, at which adenylate binding is stoichiometric. Samples prepared at three protein concentrations were centrifuged at three rotor speeds (Figure 3) and global analysis of the resulting data using a monomer-dimer model provided the equilibrium dimerization constant. For each measurement the agreement between the data and the model was assessed from the distributions of the residuals of the fits as well as the magnitude of the square root of the variance of the fit (Figure 3).
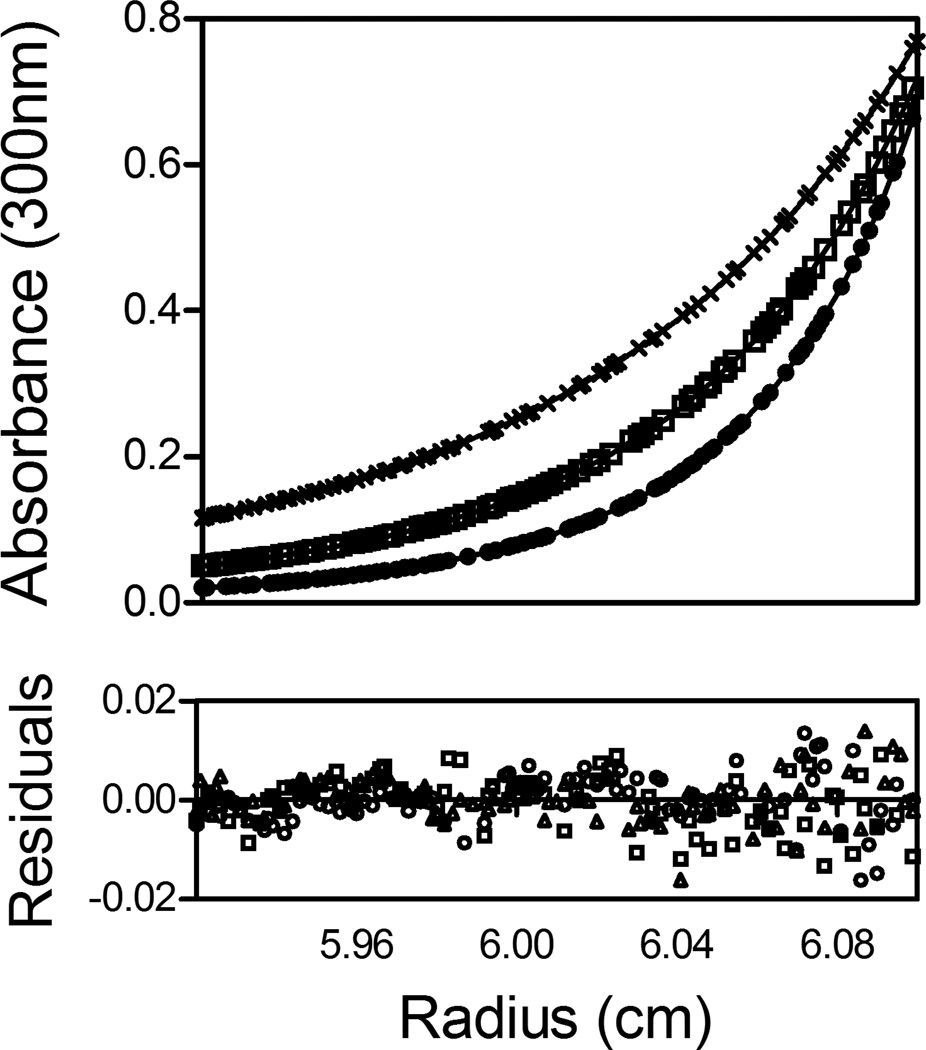
Fig. 3.
Absorbance versus radius profiles of E140A-bio-5`-AMP obtained at 40 µM protein concentration and centrifuged at 18 (×), 22 (□) and 26 (○) k rpm. The lines represent the best-fit curves obtained from global analysis of three data sets to a monomer-dimer model. The residuals of the fits are shown in the lower panel.
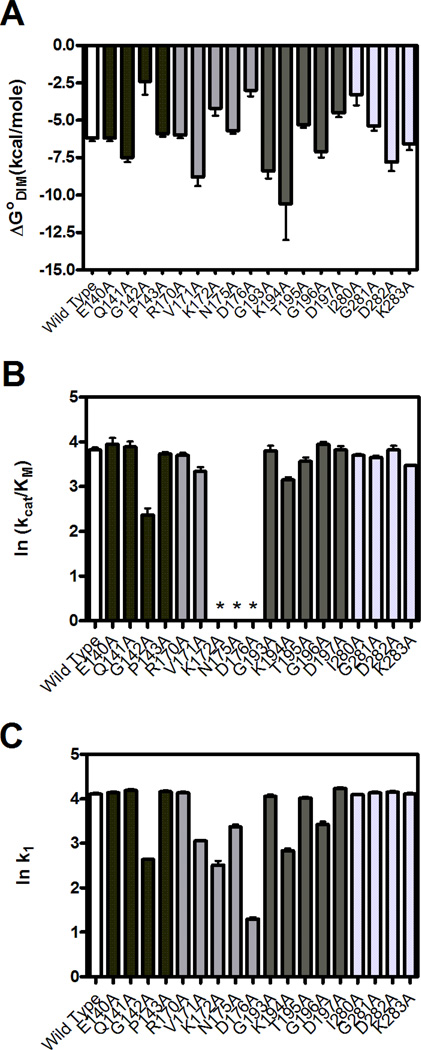
Dimerization measurements for all alanine-substituted variants in the four loops indicate a broad range of equilibrium constants (Table 1). With the exception of the G142A, D176A, I280A, K172A and K194A variants, the sedimentation equilibrium data for the proteins are well-described by a monomer-dimer model. The equilibrium constants for the reaction range from 17×10−3 to 1×10−8 M (Table 1) corresponding to Gibbs free energies of −2.5 to −10.6 kcal/mol (Figure 4A). For the G142A, K172A, D176A, and I280A variants, which exhibit weak dimerization (Table 1, Figure 4A), the concentration versus radius profiles yielded only the monomer at the highest loading concentration of 150 µM. Hence, the reported dimerization constants are lower limits. The K194A variant shows the tightest dimerization with a KD of approximately 10 nM, which corresponds to an energetic enhancement to dimerization of approximately −3.5 kcal/mol. The large errors in the equilibrium constant and free energy of dimerization for this variant (Table 1) reflect the necessity, due to the detection limits of the instrument’s absorption optics, of using protein concentrations at which the monomer population was low. Therefore, the reported equilibrium constant and free energy are upper limits.
Table 1.
Equilibrium Parameters for Dimerization of BirA Variants
| BirA variant | Kdim (M)a,b | ΔGodim (kcal/mol)c |
|---|---|---|
| Wild Type | 7 ( ± 3) × 10−6 | −6.8 ( ± 0.3) |
| E140A | 22 ( ± 5)× 10−6 | −6.2 ( ± 0.2) |
| Q141A | 3 ( ± 2) × 10−6 | −7.5 ( ± 0.3) |
| G142A | 17 (± 52) × 10−3 | −2.4 ( ± 0.9) |
| P143A | 4 ( ± 1) × 10−5 | −5.9 ( ± 0.2) |
| R170A | 4 ( ± 1) × 10−5 | −6.0 ( ± 0.2) |
| V171A | 3 ( ± 2) × 10−7 | −8.8 ( ± 0.6) |
| K172A | 7 ( ± 4) × 10−4 | −4.2 ( ± 0.5) |
| N175A | 6 ( ± 2) × 10−5 | −5.7 ( ± 0.2) |
| D176A | 6 ( ± 6) × 10−3 | −3.0 ( ± 0.4) |
| G193A | 5 ( ± 4) × 10−7 | −8.4 ( ± 0.5) |
| K194A | 1 ( ± 7)× 10−8 | −10.6 ( ± 2.4) |
| T195A | 10 ( ± 4) × 10−5 | −5.3 ( ± 0.2) |
| G196A | 5 ( ± 4))c × 10−6 | −7.1 ( ± 0.4) |
| D197A | 5 ( ± 3) × 10−4 | −4.5 ( ± 0.3) |
| I280A | 4 ( ± 3) × 10−3 | −3.3 ( ± 0.7) |
| G281A | 9 ( ± 4) × 10−5 | −5.4 ( ± 0.3) |
| D282A | 2 ( ± 2) × 10−6 | −7.8 ( ± 0.6) |
| K283A | 12 ( ± 7) × 10−6 | −6.6 ( ± 0.4) |
The equilibrium constants were measured in Standard Buffer (10 mM TrisHCl, pH 7.50±0.02 at 20°C, 200 mM KCl, 2.5 mM MgCl2) as described in “Materials and Methods.” In all cases the protein was saturated with bio-5’-AMP. The Gibbs free energies were calculated using the equation ΔGo=RTlnKdim.
The errors represent the standard error of two independent experiments
The Kdim values reported for G142A, K172A, D176A, and I280A are lower limits and that for K194A is an upper limit.
The reported uncertainties for each variant were calculated using standard error propagation methods
Fig. 4.
Homodimerization energetics and kinetic parameters for biotin transfer to BCCP. A. Gibbs free energies of homodimerization. B. The kcat/KM values for biotin transfer to BCCP87 obtained from steady-state measurements. *Not Determined. C. The bimolecular rates of association, k1, of variants with BCCP87 obtained from single turnover kinetic measurements. The kinetic parameters obtained for B and C are shown on a natural logarithmic scale to place them on an energy scale. Error bars for each graph represent the 68% confidence intervals. The bar color coding in each panel is wt BirA (white) and variable loop 140–146 (black), constant loop 170–176 (medium-gray), variable loop 193–199 (dark gray) and variable loop 280–283 (light gray).
Two-step biotin transfer to BCCP is sensitive to alanine substitutions in primarily the constant loops
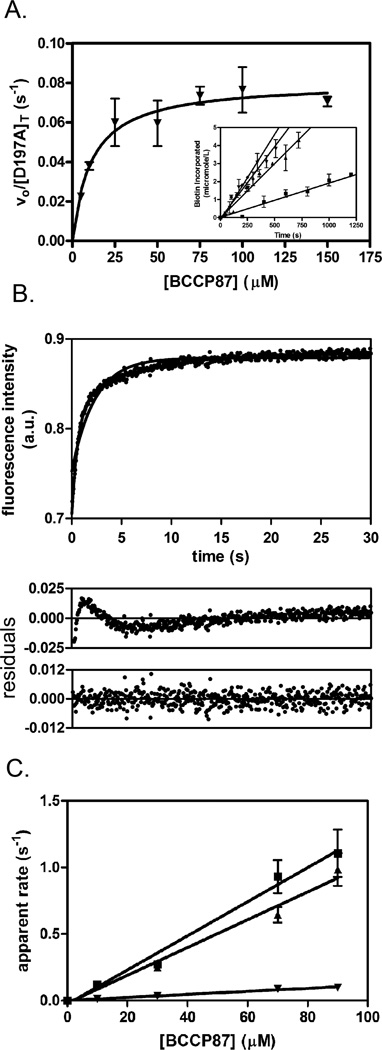
BirA-catalyzed biotin transfer is a two-step reaction in which the adenylated intermediate, bio-5’-AMP, is first synthesized from biotin and ATP and the biotin is then transferred from the adenylate to BCCP. Since any alanine substitution in the loops can, in principle, affect either or both of these steps, two assays, one of the overall reaction and the second of biotin transfer from the intermediate alone, were employed to evaluate the variants. The impact of each substitution was first measured using steady state kinetic measurements of BirA-catalyzed 3H-biotin incorporation into a C-terminal fragment of the E. coli acetyl Co-A carboxylase biotin carboxyl carrier protein, BCCP87, which is functionally equivalent to the full-length BCCP in BirA-catalyzed biotin transfer.20 In the assay3H-biotin incorporation into the acceptor protein is monitored as a function of time in reactions in which ATP and total biotin (labeled plus unlabeled) concentrations were constant and BCCP87 concentration was varied. Biotin and ATP concentration were at saturating and KM, respectively.21 For each variant, the dependence of the initial rate, obtained from the linear portion of the product versus time curves (Figure 5A, inset), on BCCP87 concentration was analyzed using the Michaelis-Menten formalism to obtain the values of KM and kcat (Figure 5A). The parameters obtained for the variant D197A are 12±2 µM and 0.08±0.01 s−1 for KM and kcat, respectively, identical to those obtained for wt BirA, of 9±2 µM and 0.060±0.006 s−1, respectively.
Fig. 5.
Kinetics of biotin transfer to BCCP. A. Steady state kinetic analysis of the two-step BirA-catalyzed biotin transfer reaction. Inset: initial rates of biotin incorporated at 5 µM (■), 25 µM (▲), 75 µM (▼) and 100 µM (♦) BCCP concentrations. For clarity only four of seven lines are shown. Each data point is an average of two independent measurements with error bars representing the standard error. Initial rates versus substrate concentration were subjected to nonlinear least squares analysis using the Michaelis-Menten model. Each initial rate represents the average of values measured in two independent experiments, with error bars representing the standard error. B. Stopped-flow measurements of biotin transfer to BCCP in the second half reaction. The fluorescence intensity versus time trace for D197A at 90 µM BCCP is shown with residuals for a single exponential model (middle panel) or a double exponential model (lower panel). C. The dependence of the apparent rate of biotin transfer on apoBCCP concentration. The plots obtained for wt BirA (■), T195A (▲) and V171A (▼) are shown. Each data point is the average of rates obtained from at least four stopped flow traces and the lines represent the best fits of the rate versus concentration profiles to a linear equation.
Steady-state measurements of biotin transfer catalyzed by the entire set of variants indicate relative insensitivity of the variable loop sequences to amino acid replacement (Table 2, Figure 4B). By contrast alanine substitutions in the 170–176 loop yielded three proteins, K172A, N175A and D176A, that have no activity in catalyzing biotin transfer to BCCP in the steady-state assay. Additional measurements indicate that these proteins can, when provided with synthetic intermediate, bio-5’-AMP, catalyze biotin transfer to BCCP87 (see below). Among the alanine-substituted variants in the variable loops only two, K194A and G142A, yielded KM values that differ significantly from that measured for the parent wild type protein. The changes in KM for the two proteins are roughly 5 and 20-fold, respectively. The kcat values for biotin transfer catalyzed by all variable loop variants are identical to that measured for BirA wt.
Table 2.
The steady-state and single turnover kinetic parameters for BirA-catalyzed biotin transfer to BCCP87
| BirA variant | KM(BCCP) (µM)a |
kcat (s−1)a | kcat/KM(M−1s−1)b | k1 (M−1s−1)c |
|---|---|---|---|---|
| Wild Type | 9 ± 1 | 0.060 ± 0.003 | 6700± 800 | 12,800 ± 800 |
| E140A | 8 ± 3 | 0.07 ± 0.01 | 8800± 3500 | 13,900 ± 800 |
| Q141A | 9 ± 1 | 0.07 ± 0.02 | 7800 ± 2400 | 16,000 ± 1000 |
| G142A | 214 ± 9 | 0.05 ± 0.02 | 230 ± 90 | 440 ± 10 |
| P143A | 13 ± 1 | 0.070 ± 0.005 | 5400 ± 600 | 14,500 ± 1200 |
| R170A | 16 ± 2 | 0.080 ± 0.002 | 5000± 600 | 13,700 ± 800 |
| V171A | 23 ± 6 | 0.050 ± 0.003 | 2200 ± 600 | 1100 ± 100 |
| K172A | N.D.d | N.D. | N.D. | 320 ± 50 |
| N175A | N.D. | N.D. | N.D. | 4100 ± 100 |
| D176A | N.D. | N.D. | N.D. | 20 ± 2 |
| G193A | 11 ± 3 | 0.070 ± 0.005 | 6400 ± 1800 | 12,000 ± 1000 |
| K194A | 64 ± 9 | 0.090 ± 0.005 | 1400 ± 200 | 700 ± 100 |
| T195A | 19 ± 3 | 0.07 ± 0.01 | 3700 ± 800 | 10,400 ± 800 |
| G196A | 8 ± 1 | 0.070 ± 0.005 | 8700 ± 1300 | 5000 ± 100 |
| D197A | 12 ± 2 | 0.08 ± 0.01 | 6700 ± 1400 | 17,000 ± 1000 |
| I280A | 12 ± 1 | 0.0600 ± 0.0003 | 5000± 400 | 12,500 ± 600 |
| G281A | 20 ± 2 | 0.09000 ± 0.00005 | 4500± 500 | 14,000 ± 1000 |
| D282A | 9 ± 2 | 0.060 ± 0.003 | 6700 ± 1500 | 14,400 ± 800 |
| K283A | 17 ± 1 | 0.050 ± 0.001 | 2900 ± 200 | 10,100 ± 500 |
The kcat, KM and k1 values were measured as described in Materials and Methods in Standard Buffer (10 mM Tris HCl, pH 7.50±0.02 at 20°C, 200 mM KCl, 2.5 mM MgCl2)
The errors represent the standard error of two independent experiments
The reported uncertainties were calculated using standard error propagation
The errors represent the uncertainties in the linear regression of the kobs versus [apoBCC87] profiles.
N.D.-Not Determined because these variants were incapable of catalyzing bio-5’-AMP synthesis.
Effects of alanine substitutions on the heterodimer interaction were also assessed using single turnover measurements of the second half reaction. This assay allows separation of effects on the interaction of the enzyme intermediate complex with BCCP in the second half reaction from those on the overall reaction, which is particularly important for this system because the rate determining step in the overall reaction corresponds to the first half-reaction, bio-5’-AMP synthesis.20; 21; 22 In these measurements pre-formed BirA-adenylate is rapidly mixed with apoBCCP87 and the resulting time-dependent increase in the intrinsic BirA fluorescence signal is monitored (Figure 5B).20 This increase, which accompanies the conversion of adenylate-bound BirA to apoBirA that occurs upon biotin transfer, has been shown to parallel product, bioBCCP, accumulation.22 Analysis of each transient was performed using either a single exponential or double exponential model (Figure 5B). For variants that exhibit double exponential behavior the apparent rate of the first, faster phase increases with acceptor protein concentration and that of the second phase is substrate-concentration independent.20 Apparent rate versus BCCP concentration data show a linear dependence with no leveling off (Figure 5C). The simplest interpretation of this behavior is that it reflects collision of the enzyme˙ intermediate complex with apoBCCP, which, in the kinetic scheme shown below, is governed by the bimolecular rate constant k1:
| (Equation 1) |
The slope obtained from linear regression of the rate versus acceptor protein concentration profiles yields the bimolecular association constant governing ternary complex formation, k1 (Figure 5C), from BirA˙bio-5’-AMP and BCCP.22 The concentration-independent phase, which occurs with a rate of approximately 0.2 s−1, is assigned to the product dissociation step governed by k2.20; 22
Rate versus concentration profiles for all 18 variants indicate that the majority of the proteins behave identically to wild type BirA (Table 2, Figure 4C). Of the seven variants that deviate, four are proteins with substitutions in the constant loop. Only three variants with substitutions in variable loops including G142A, K194A and G196A differ from wtBirA in biotin transfer from the adenylate. All variants exhibit the same linear dependence of apparent rate on substrate concentration that is observed for wild type BirA. However, the magnitudes of k1 values obtained for these proteins are 2.5-600 fold slower than that of the wild type protein (Table 2), indicating that they are compromised in the rate at which they associate with apoBCCP. Moreover, with the exception of G196A, for variants that could be assayed using both steady-state and single turnover methods, decreases in k1 correlate with increases in KM, consistent with the Michaelis constant reflecting, in part, binding of the enzyme˙intermediate complex to the acceptor protein. Two of the variants, N175A and G196A, exhibit a hyperbolic dependence of rate on acceptor protein concentration and analysis of the rate versus substrate concentration for these proteins was, consequently, performed using a Michaelis-Menten model to obtain kcat/KM for the reactions. The maximal biotin transfer rate exhibited by these two variants of 0.2 s−1 is equivalent to the rate of product dissociation from the wild type enzyme.20 Thus, the hyperbolic dependence reflects the similarity in the magnitudes of the apparent rates for the two kinetic phases at the substrate concentrations employed for these two variants.
Discussion
The roles of BirA loop sequences in its two functions were assessed using combined sedimentation equilibrium and kinetic analysis of alanine substituted proteins. Consistent with the hypothesis that the variable loop sequences support the homodimerization required for transcription repression, alanine substitutions in the 140–146, 193–199, and 280–283 loops have large energetic consequences for homodimerization but only minor impact on heterodimerization. Contrary to the hypothesized role of constant loops in biotin transfer only, the 170–176 loop sequence functions in both interactions.
Previous studies of the thermodynamics of BirA homo- and heterodimerization indicate that although the Gibbs free energies measured for the two interactions at 20°C are similar, the underlying enthalpic and entropic contributions are very different,15 consistent with distinct structural determinants for the two interactions. The measured effects of alanine substitutions in the four surface loops on homodimerization and biotin transfer allow mapping of these structural differences (Figure 6). Highlighting of loop residues at which alanine substitution results in large perturbations to the homo or hetero interaction indicates distinct gross features of the two interfaces. Data obtained previously on the 116–124 loop, which functions in both homodimerization and biotin transfer, are included in this analysis.13–15 The homodimer interface consists of two dyad symmetric sub-interfaces, each of which is characterized by continuous interaction between the variable loop residues of one monomer with the constant loop residues of the second. By contrast, the heterodimer interface is bipartite with the constant loops, which constitute the enzyme active site, forming one locus of interaction and the 193–199 loop forming the second.
Fig. 6.
The BirA heterodimer (A) and homodimer (B) interfaces with residues that exhibit sensitivity to alanine substitution (with k1 rates at least 20 times less than wt BirA, or the |ΔΔGdim|≥1kcal/mole) highlighted. The color-coding is identical to that used for the loops in Figure 2. The data for the 116–124 loop (in blue) were obtained from previously published results.13; 14; 15
Detailed structural interpretation of the energetic consequences of alanine substitutions for homodimerization is complicated by the known inherent flexibility of variable loops 140–146 and 193–199 and constant loop 116–124, which are disordered in the apoBirA monomer structure.23 Thus alanine substitutions can potentially result in altered conformations of any of these loops. Furthermore, for several variants the functional effects may reflect a disruption of intermolecular and/or intramolecular interactions. For example, replacement of V171 with alanine renders dimerization more favorable by −2 kcal/mole. In the wild type BirA dimer structure the V171 side chain is packed in the interior of each protein monomer far from the interface. Disruption of another apparently purely intramolecular interaction between I280 and W310 leads to a 3.5 kcal/mole penalty for dimerization when the isoleucine is replaced by alanine. The D197 side chain interacts electro statically with R119 in an intermolecular interaction and substitution of the D with A results in a 2.3 kcal/mole energetic penalty to dimerization. The stabilizing effect of the K194 replacement reflects the relief of charge repulsion between the positively charged lysine and multiple arginine side chains in 116–124 loop of the opposing monomer. Relative to homodimerization, alanine replacement of a small number of residues impacts biotin transfer. The inability of constant loop variants K172A, N175A and D176A to synthesize bio-5`-AMP and their slow rates of biotin transfer to BCCP likely reflects roles for these side chains in positioning of biotin and ATP in the active site, as well as associating with BCCP to transfer the biotin from the adenylate. Interpretation of the slow rate of biotin transfer observed for the K194A variant is complicated by the fact that it exists as a dimer, which competes with apoBCCP binding, at the 500nM concentration required for detection in the stopped-flow fluorescence measurements. Nevertheless, in the holoBirA˙BCCP model the distance between the epsilon amino group of K194 on BirA and the carboxylate of E128 of BCCP is consistent with formation of a salt bridge. This interaction may, in part, form the basis of the selectivity that is observed in the biotin transfer reaction.22
The results reported in this work when combined with those of previous studies suggest a model for how the surface loops on BirA support bispecific protein:protein interactions. First, structural information in constant loops is multi-functional. Alanine substitutions in the 170–176 loop indicate perturbations to bio-5’-AMP synthesis, biotin transfer to the acceptor protein, and homodimerization. Previous studies of the second constant loop comprising residues 116–124 indicate similar complexity in its function with variants exhibiting defects in biotin and bio-5’-AMP binding, hetero- and homodimerization.13; 14; 15 The sequence conservation in these loops reflects their critical roles in the multi-step post-translational biotin addition reaction that is required for viability. Second, in the homodimer interface the variable loop sequences complement the constant loops. This is illustrated in Figure 6, which shows that in the homodimer interface the majority of the variable loop side chains that are functionally sensitive to alanine substitution contact constant loop residues. Thus, it is the complementarity of variable loop side chains to the constrained residues of the constant loops that enables use of a single surface on BirA for two distinct protein:protein interactions.
The range of homodimerization energetics measured for the loop variants illustrates the ease with which the interaction can be significantly altered through minor structural changes. These large energetic changes reflect, in part, the two-fold symmetry of the homodimer interface, with each alanine substitution perturbing at least two interactions. The large impact on homodimerization coupled with the absence of impact of several amino acid changes in the variable loops on heterodimerization/biotin transfer prompts consideration of why wild type BirA did not evolve to homodimerize more tightly. From the standpoint of transcription repression alone, tighter dimerization would lead to energetically more favorable assembly of the BirA˙bioO complex and resulting greater repression. It is possible that if homodimerization were too tight, heterodimerization could not compete, with resulting deleterious consequences for viability. Comparison of the results of steady state and single turnover kinetic measurements for the K194A variant supports this possibility. The lack of an effect on kcat in the steady state measurements reflects the experimental design in which reactions are initiated by enzyme addition. Upon bio-5’-AMP synthesis the significantly higher apoBCCP concentration ensures that the enzyme intermediate complex collides with acceptor protein rather than itself. By contrast, in the single turnover measurements pre incubation of the K194A variant in its complex with the adenylate results in significantly slower biotin transfer to BCCP than is observed for the wild type enzyme. This slower transfer may be limited by homodimer dissociation. An alternative explanation for the relatively weak homodimerization is that very tight dimerization would render the repression sensitive to biotin concentrations that are insufficient to support viability. Assembly of the repression complex occurs via coupled dimerization of the adenylate-bound monomer and binding of the pre-formed dimer to bioO.7 Simulations of the dependence of transcription repression on biotin concentration indicate that as the equilibrium dimerization constant for holoBirA becomes tighter, repression occurs at lower biotin concentrations.24 These predictions are currently being tested.
In general, protein surface loops can serve important roles in mediating interactions with other proteins. Structure-based sequence alignments of common domains have led to the proposal that surface loops evolve to either allow (enable) or prevent (disable) an interaction.25 A surface loop on the E. coli guanylate kinase has been experimentally demonstrated to promote its oligomerization.26 In some cases loop length, and not sequence, is the most important property for self-association.27 Alternatively, as observed in BirA, the particular loop sequence is important for tuning dimerization energetics.28 However, the variability in 140–146, 193–199 and 280–283 loop sequences among bifunctional ligases in general suggests that many sequences can support homodimerization. This apparent tolerance of homodimerization to sequence variation, which may be facilitated by the intrinsic structural flexibility of the interface loops, is intriguing in light of the complementarity between variable and constant loop side chains in the Ec BirA homodimer interface (Figure 6).
This work demonstrates that variable surface loops evolved to support BirA function in the homodimerization required for transcription repression by complementing the constant loops, which are constrained because of their essential role in post-translational biotin addition. The results further illustrate that relatively subtle changes in surface loop sequence can elicit large energetic changes in protein:protein interactions. The observations have implications for both evolution of protein:protein interactions and the design of novel interactions.
Materials and Methods
Chemicals and Biochemicals
All chemicals used in buffer preparation were at least reagent grade. The isopropyl-β-D-thiogalactoside (IPTG), phenylmethylsulfonyl fluoride (PMSF), polyethyleneimine (PEI), unlabeled biotin and ATP were purchased from Sigma. The 1,4-dithio-DL-threitol (DTT) was purchased from Research Organics. Biotin [8, 9- 3H] was purchased from Perkin Elmer and stored under argon at −70°C. ATP was prepared by dissolving the disodium salt in water and adjusting the pH to 7.5 and its concentration was determined using UV absorbance with an extinction coefficient of 15400 M cm−1 at 259 nm. The unlabeled biotin stock solution was prepared by dissolving biotin in 10 mM Tris HCl, 200 mM KCl, 2.5 mM MgCl2 and adjusting the pH to 7.5 at 20°C. The bio-5`-AMP was synthesized and purified as previously described.29; 30 The 87 amino acid C-terminal fragment (BCCP87) of biotin carboxyl carrier protein from acetyl-CoA carboxylase was purified as previously described.20
Mutagenesis and expression and purification of BirA variants
Site directed mutagenesis was performed using the Quik Change II XL Site-Directed Mutagenesis kit (Stratagene) using a pBtac2 (Boehringer Mannheim) derivative that contains the BirA coding sequence under transcriptional control of the tac promoter. A C-terminal (His)6 tag, which has no effect on function, enabled purification of the variant proteins away from the chromosomally encoded wt BirA. After DpnI digestion of the mutagenized plasmid, the DNA was transformed into XL10-Gold cells (Stratagene) and several single colonies were screened for expression. The sequences were confirmed for the entire gene. Proteins were purified as previously described31, with the exception that a final Q-Sepharose column step was added. The absorbance at 280 nm was used to calculate the concentration of each protein using the molar extinction coefficient of 47510 M−1 cm−1 calculated from the amino acid composition.32 Each protein was approximately 95% pure, as assessed by SDS-PAGE analysis, and all proteins were >90% active as determined by stoichiometric titrations with bio-5`-AMP monitored by fluorescence spectroscopy.29
Sedimentation equilibrium measurements
Homo-dimerization of each protein bound to bio-5’-AMP was determined by sedimentation equilibrium using a Beckman Coulter Optima XL-1 analytical ultra centrifuge with a four-hole An-60 rotor. Standard 12 mm six-hole or 3 mm two-hole cells with charcoal-filled Epon centerpieces and sapphire windows were used in all experiments. Prior to centrifugation, proteins (~ 400 µl) were exhaustively dialyzed against Assay Buffer (10 mM Tris-HCl, pH 7.5, 20°C, 200 mM KCl and 2.5 mM MgCl2). The bio-5`-AMP, which was diluted into the dialysis buffer immediately before sample preparation, was added at a 1.5:1 molar ratio to protein. Samples prepared at three protein concentrations were centrifuged at three rotor speeds33 (ranging from 18K to 26K rpm) at 20°C. After reaching equilibrium at each speed the absorbance at 295 or 300 nm was measured with a step size of 0.001 cm and five replicates per step.
Steady State Kinetic Measurements
Kinetics of biotin transfer as a function of BCCP87 concentration was measured by monitoring the incorporation of [3H]-biotin into BCCP87. Reactions containing 5 µM cold biotin, 60 nM [3H]-biotin (specific activity 32.6 Ci/mmol), 2.5 mM ATP, 0.1 mg/ml acetylated bovine serum albumin carrier protein (Invitrogen) and a range of apoBCCP87 concentrations were performed at 20°C in Assay Buffer. After initiation by addition of BirA (wt or variant) that had been pre incubated at 20°C to a final concentration of 100 nM (G193A, 50 nM) 20 µl volumes of the reaction were quenched at various time points by spotting onto What man filter paper that had been pretreated with a solution containing 1 mM biotin and 10% trichloroacetic acid (Alfa Aesar) in water, and washed as previously described.34; 35 Each dried filter paper was transferred to a scintillation vial containing Ready Protein+ cocktail (Beckman Coulter) and radioactivity was measured by scintillation counting for 10 minutes.
Initial Rate Measurements of Biotin Transfer from the Intermediate, bio-5’-AMP
The rates of BirA-catalyzed biotin transfer from bio-5`-AMP to the BCCP87 acceptor substrate were measured using a Kintek SF-2001 stopped-flow instrument equipped with fluorescence detection. A solution of the enzyme, wt or alanine variant, at a concentration of 1µM was first pre-incubated for at least 15 minutes with either 800 nM biotin and 500 µM ATP or 800 nM bio-5`-AMP at 20°C. The resulting enzyme˙intermediate complex was rapidly mixed at a 1:1, vol:vol ratio with BCCP87 to achieve final concentrations of 400 nM and 10–90µM of BirA˙bio-5’-AMP and apoBCCP87, respectively. Bio-5`-AMP was used for variants K172A, N175A and D176A, which are incapable of bio-5`-AMP synthesis. The excitation wavelength was 295 nm and emission was measured above 340 nm using a cutoff filter (Corion Corp.). At least four traces were obtained at each BCCP87 concentration.
Data Analysis
Sedimentation equilibrium
The individual absorbance versus radius profiles were analyzed separately and globally using a single species model to obtain the reduced molecular weight, σ, using the program Win Non Lin,36 from which the molecular weight was calculated using the following relationship,
| (2) |
where M is the molecular weight, ν̅ is the partial specific volume of the protein, ρ is the density of the buffer, ω is the angular velocity of the rotor, R is the gas constant and T is the absolute temperature. The nine data sets were then globally analyzed using a monomer-dimer model37 to obtain an association constant, Ka, using the equation,
| (3) |
where ct is the total concentration at position r, δ is the baseline offset, cm(ro) is the concentration at the reference radial position ro, and σm is the reduced molecular weight of the monomer. The association constants, which are obtained in absorbance units from the analysis, are reported as molar equilibrium dissociation constants or KDIM. The quality of each analysis was assessed by the square root of variance of the fit and the distribution of residuals.
Steady-state kinetic measurements
The initial velocity, obtained as the slope of the cpm versus time plots, versus BCCP87 concentration data were subjected to non-linear least squares analysis using the Michaelis-Menten equation in GraphPad Prism 438 to obtain Vmax and KM. The Vmax and kcat values were converted from counts/min to micro molar biotin using the relation between counts/min and micromoles obtained from a control reaction performed at conditions in which the 3H biotin is quantitatively incorporated into BCCP87.
Single turnover kinetic measurements
The fluorescence intensity versus time traces, obtained for each variant at a range of BCCP87 concentrations, were analyzed using either a single or double exponential model to obtain apparent rate(s). Further analysis of the rate versus concentration profiles was performed as described in Results.
Supplementary Material
Supplementary Figure. Multiple sequence alignment of five surface loops from bifunctional biotin protein ligases.16
We investigated the roles a of single protein surface in two distinct functions.
Alanine replacements in loops on the protein surface were constructed.
The protein variants were subjected to functional analysis.
Co-evolution of surface loop sequences underlies bi-functionality.
Acknowledgments
This work was supported by NIH grants R01GM46511 and S10RR15899 to DB. The authors thank Dr. Zachary Wood for constructing the holoBirA-BCCP87 model.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Erijman A, Aizner Y, Shifman JM. Multispecific recognition: mechanism, evolution, and design. Biochemistry. 2011;50:602–611. doi: 10.1021/bi101563v. [DOI] [PubMed] [Google Scholar]
- 3.Barker DF, Campbell AM. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzymesynthetase in repression of the biotin operon in Escherichia coli. J. Mol. Biol. 1981;146:469–492. doi: 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- 4.Barker DF, Campbell AM. The birA gene of Escherichia coli encodes a biotin holoenzymesynthetase. J. Mol. Biol. 1981;146:451–467. doi: 10.1016/0022-2836(81)90042-5. [DOI] [PubMed] [Google Scholar]
- 5.Cronan JE. Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J. Biol. Chem. 1988;263:10332–10336. [PubMed] [Google Scholar]
- 6.Li SJ, Cronan JE. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J.Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Streaker ED, Beckett D. Coupling of protein assembly and DNA binding: biotin repressor dimerization precedes biotin operator binding. J. Mol. Biol. 2003;325:937–948. doi: 10.1016/s0022-2836(02)01308-6. [DOI] [PubMed] [Google Scholar]
- 8.Eisenstein E, Beckett D. Dimerization of the Escherichia coli biotin repressor: corepressor function in protein assembly. Biochemistry. 1999;38:13077–13084. doi: 10.1021/bi991241q. [DOI] [PubMed] [Google Scholar]
- 9.Prakash O, Eisenberg MA. Biotinyl 5'-adenylate: corepressor role in the regulation of the biotin genes of Escherichia coli K-12. Proc. Natl Acad. Sci. USA. 1979;76:5592–5595. doi: 10.1073/pnas.76.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodionov DA, Mironov AA, Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLano WL. The PyMOL Molecular Graphics 696 System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 12.Wood ZA, Weaver LH, Brown PH, Beckett D, Matthews BW. Co-repressor induced order and biotin repressor dimerization: a case for divergent followed by convergent evolution. J. Mol. Biol. 2006;357:509–523. doi: 10.1016/j.jmb.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 13.Kwon K, Streaker ED, Ruparelia S, Beckett D. Multiple disordered loops function in corepressor-induced dimerization of the biotin repressor. J. Mol. Biol. 2000;304:821–833. doi: 10.1006/jmbi.2000.4249. [DOI] [PubMed] [Google Scholar]
- 14.Kwon K, Beckett D. Function of a conserved sequence motif in biotin holoenzymesynthetases. Protein Sci. 2000;9:1530–1539. doi: 10.1110/ps.9.8.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Naganathan S, Beckett D. Thermodynamic and structural investigation of bispecificity in protein–protein interactions. J. Mol. Biol. 2009;389:336–348. doi: 10.1016/j.jmb.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 17.Tron CM, McNae IW, Nutley M, Clarke DJ, Cooper A, Walkinshaw MD, Baxter RL, Campopiano DJ. Structural and functional studies of the biotin protein ligase from Aquifexaeolicus reveal a critical role for a conserved residue in target specificity. J. Mol. Biol. 2009;387:129–146. doi: 10.1016/j.jmb.2008.12.086. [DOI] [PubMed] [Google Scholar]
- 18.Gupta V, Gupta RK, Khare G, Salunke DM, Surolia A, Tyagi AK. Structural ordering of disordered ligand-binding loops of biotin protein ligase into active conformations as a consequence of dehydration. PLoS ONE. 2010;5:e9222. doi: 10.1371/journal.pone.0009222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagautdinov B, Matsuura Y, Bagautdinova S, Kunishima N. Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate. J. Biol. Chem. 2008;283:14739–14750. doi: 10.1074/jbc.M709116200. [DOI] [PubMed] [Google Scholar]
- 20.Nenortas E, Beckett D. Purification and characterization of intact and truncated forms of the Escherichia coli biotin carboxyl carrier subunit of acetyl-coA carboxylase. J. Biol. Chem. 1996;271:7559–7567. doi: 10.1074/jbc.271.13.7559. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Beckett D. Kinetics of biotinyl-5'-adenylate synthesis catalyzed by the Escherichia coli repressor of biotin biosynthesis and the stability of the enzyme-product complex. Biochemistry. 1994;33:7354–7360. doi: 10.1021/bi00189a041. [DOI] [PubMed] [Google Scholar]
- 22.Ingaramo M, Beckett D. Biotinylation, a post-translational modification controlled by the rate of protein-protein association. J. Biol. Chem. 2011;286:13071–13078. doi: 10.1074/jbc.M110.183624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia coli biotin holoenzymesynthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc.Natl Acad. Sci. USA. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tungtur S, Skinner H, Zhan H, Swint-Kruse L, Beckett D. In vivo tests of thermodynamic models of transcription repressor function. Biophys. Chem. 2011;159:142–151. doi: 10.1016/j.bpc.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akiva E, Itzhaki Z, Margalit H. Built-in loops allow versatility in domain–domain interactions: lessons from self-interacting domains. Proc. Natl Acad. Sci. USA. 2008;105:13292–13297. doi: 10.1073/pnas.0801207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hible G, Renault L, Schaeffer F, Christova P, Zoe Radulescu A, Evrin C, Gilles A-M, Cherfils J. Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase. J. Mol. Biol. 2005;352:1044–1059. doi: 10.1016/j.jmb.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 27.Sato K, Li C, Salard I, Thompson AJ, Banfield MJ, Dennison C. Metal-binding loop length and not sequence dictates structure. Proc. Natl Acad. Sci. USA. 2009;106:5616–5621. doi: 10.1073/pnas.0811324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakurai K, Goto Y. Manipulating monomer-dimer equilibrium of bovine β-lactoglobulin by amino acid substitution. J. Biol. Chem. 2002;277:25735–25740. doi: 10.1074/jbc.M203659200. [DOI] [PubMed] [Google Scholar]
- 29.Abbott J, Beckett D. Cooperative binding of the Escherichia coli repressor of biotin biosynthesis to the biotin operator sequence. Biochemistry. 1993;32:9649–9656. doi: 10.1021/bi00088a017. [DOI] [PubMed] [Google Scholar]
- 30.Lane MD, Rominger KL, Young DL, Lynen F. The enzymatic synthesis of holotranscarboxylase from apotranscarboxylase and (+)-biotin. J. Biol. Chem. 1964;239:2865–2871. [PubMed] [Google Scholar]
- 31.Naganathan S, Beckett D. Nucleation of an allosteric response via ligand-induced loop folding. J. Mol. Biol. 2007;373:96–111. doi: 10.1016/j.jmb.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal.Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 33.Roark DE. Sedimentation equilibrium techniques: multiple speed analyses and an overspeed procedure. Biophys.Chem. 1976;5:185–196. doi: 10.1016/0301-4622(76)80034-8. [DOI] [PubMed] [Google Scholar]
- 34.Chapman-Smith A, Morris TW, Wallace JC, Cronan JE. Molecular recognition in a post-translational modification of exceptional specificity. J. Biol. Chem. 1999;274:1449–1457. doi: 10.1074/jbc.274.3.1449. [DOI] [PubMed] [Google Scholar]
- 35.Ingaramo M, Beckett D. Distinct amino termini of two human HCS isoforms influence biotin acceptor substrate recognition. J. Biol. Chem. 2009;284:30862–30870. doi: 10.1074/jbc.M109.046201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laue TM. Sedimentation equilibrium as thermodynamic tool. Methods Enzymol. 1995;259:427–452. doi: 10.1016/0076-6879(95)59055-2. [DOI] [PubMed] [Google Scholar]
- 38.Motulsky H, Christopoulos A. Fitting Models to Biological Data Using Linear and Nonlinear Regression. A Practical Guide to Curve Fitting. New York: Oxford University Press; 2004. [Google Scholar]
- 39.DeLano WL. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- 40.Athappilly FK, Hendrickson WA. Structure of the biotinyl domain of acetyl-coenzyme Acarboxylase determined by MAD phasing. Structure. 1995;3:1407–1419. doi: 10.1016/s0969-2126(01)00277-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure. Multiple sequence alignment of five surface loops from bifunctional biotin protein ligases.16