Abstract
Anaplastic plasmacytomas (APCTs) from NFS.V+ congenic mice and pristane-induced plasmacytic PCTs from BALB/c mice were previously shown to be histologically and molecularly distinct subsets of plasma cell neoplasms (PCNs). Here we extended these comparisons, contrasting primary APCTs and PCTs by gene expression profiling in relation to the expression profiles of normal naïve, germinal centre, and memory B cells and plasma cells. We also sequenced immunoglobulin genes from APCT and APCT-derived cell lines and defined surface phenotypes and chromosomal features of the cell lines by flow cytometry and by spectral karyotyping and fluorescence in situ hybridization. The results indicate that APCTs share many features with normal memory cells and the plasma cell-related neoplasms (PLs) of FASL-deficient mice, suggesting that APCTs and PLs are related and that both derive from memory B cells.
Keywords: plasmacytoma, anaplastic plasmacytoma, memory B cell, plasma cell, oligonucleotide array
Introduction
Plasma cell neoplasms (PCNs) are the second most common human haematological malignancies but are rare spontaneous diseases in mice. Human PCNs include plasma cell myeloma (PCM), commonly known as multiple myeloma (MM); plasmacytomas (PCTs); and immunoglobulin (Ig) deposition diseases [1]. Because MM is currently a uniformly fatal disease, there are wide-ranging efforts to develop new treatments and pre-clinical systems to model the disease.
The most common spontaneous mouse PCNs are designated PCTs, but models in which PCTs are readily induced at high frequencies, such as plasmacytic PCTs of pristane-treated BALB/cPt mice, are providing important information on pathogenetic mechanisms in the transformation of mature plasma cells [2]. Pristane-induced tumours are characterized by Ig/Myc-activating translocations and their requirement for interleukin (IL)-6, a cytokine expressed in the peritoneal granulomas in which they develop. MYC-activating translocations also occur in ~15% of primary MM tumours, in 50% of advanced primary cases, and in almost all cultured lines generated from terminal plasma cell leukaemias [3]. Autocrine or paracrine expression of IL-6 also appears to play a major role in the pathogenesis of MM [4]. Recent studies have demonstrated previously unappreciated parallels between MM and various mouse models of PCT, including bone marrow (BM) localization with associated lytic lesions [5,6] and involvement of NOTCH [7], MYC, and other signalling pathways in pathogenesis [8].
Nonetheless, MM and pristane-induced PCTs of mice differ in a number of important respects. Immunoglobulins produced by most cases of MM have undergone class switch recombination (CSR) and have Ig variable (V) regions with high levels of somatic hypermutation (SHM), indicating their origin from activation-induced cytidine deaminase (AID)-experienced germinal centre (GC) B cells. In addition, the majority of translocations in MM that involve the Ig heavy chain locus (IgH) do not activate MYC but rather D-type cyclins, MAF family members, or FGFR3 and MMSET. Although most pristane-induced PCTs have undergone CSR, their Ig V regions are usually minimally mutated. In addition, Myc-activating translocations are present in more than 95% of cases, with AID being implicated in the chromosomal breaks in both Ig and Myc [9–11]. Thus, while both human and mouse PCNs derive from cells with genetic signatures of AID activity, GC passage can be argued strongly for MM but less forcefully for pristane-induced PCT. Indeed, the demonstration that BTK-deficient mice, which lack B1a cells, are PCT-resistant suggests that B1a rather than GC B cells are the cells of origin for pristane-induced PCT [12].
Variations on these themes occur in PCNs of both species. Subsets of mouse PCNs that do not bear Ig/Myc translocations and express Myc at low levels include plasmacytoid lymphomas (PLs) of autoimmune mice mutant for Fas or Fasl [13,14], BM-associated spontaneous PCTs of C57BL/KaLwRij mice [15], pristine-induced PCTs of C57BL/6 mice [16], and the plasmablastic and anaplastic PCTs identified in NFS.V+ congenic mice [17], which we will refer to collectively as anaplastic PCTs (APCTs). PLs and APCTs are distinct from mature plasmacytic PCTs, which we will refer to simply as PCTs, both cytologically and for gene expression profiles [14,17]. Nonetheless, APCTs and PLs have cytological similarities to post-GC immunoblasts; both express cytoplasmic Ig and PLs are secretory, indicating that they are well progressed towards terminal plasma cell differentiation. In addition, the Ig genes of PL are heavily mutated; those of APCT have not been studied. This suggests that the origins of APCT and PL may be from cells arrested at a stage of differentiation less mature than those giving rise to PCT. Alternatively, they might reflect a process of de-differentiation from PCT to a less mature, more aggressive form of PCN, as sometimes seen in MM [18,19].
Whether PCTs derive from GC-experienced or B1a cells, there are several AID-experienced alternative pathways to plasma cell development from which APCTs and PLs might arise. They include extrafollicular B-cell responses initiated by marginal zone (MZ) or follicular B cells, B cells in isolated lymphoid follicles, and memory B cells [20,21]. Here we show that APCTs and cell lines derived from primary APCTs are more closely related to normal memory B and naïve B cells than to plasma cells or GC B cells and that they share many features with PLs.
Materials and methods
Mice, primary tumours, and cell lines
NFS.V+ mice [22], the source of primary APCT, were maintained under NIAID protocol LIP-4. The B6-1710 B cell line [23] originated from a B6 mouse with murine AIDS (MAIDS) diagnosed at necropsy with APCT. The B6-207 B cell line was cultured from tissues of a B6 mouse diagnosed with APCT. The origins of primary PCT have been described previously [7].
Microarray and quantitative real-time RT-PCR (qPCR) analyses
Microarray experiments were performed as described previously [7] with material generated from 27 primary APCTs and 25 primary PCTs using chips printed by the NIAID Microarray Research Facility comprising ~ 18 000 genes represented by 70 mer oligonucleotides. After raw data were normalized with the lowess smoothing function, 1018 genes distinguishing APCTs and PCTs at p < 0.05 were identified with significance analysis of microarray (SAM) (Supporting information, Supplementary Table 1). From published microarray data on purified subsets of normal naïve B cells, germinal centre (GC) B cells, memory B cells, and plasma cells [24], we identified 4700 non-redundant genes that matched genes assessed by our microarray analyses of PCNs. To quantify more precisely gene expression differences between PCT and APCT, we generated a customized quantitative real-time RT-PCR (qPCR) array that surveyed 92 genes selected from among those that best distinguished the PCN subsets and that were differentially expressed among the normal B-cell populations, both as determined by microarrays (Supporting information, Supplementary Table 2). qPCR analyses were performed as described previously [7].
Immunohistochemical and western blot analyses
Immunohistochemical (IHC) studies of sections from formalin-fixed, paraffin-embedded tissues were performed by the avidin–biotin peroxidase complex method using the panel of antibodies and procedures listed in the Supporting information, Supplementary Table 3.
Proteins extracted from primary tumours were separated by SDS-PAGE on 10% gels (30 μg/lane) and electroblotted to nitrocellulose membranes (Amersham, Arlington Heights, IL, USA). Membranes were processed and proteins detected by standard enhanced chemiluminescence methods. Antibodies are listed in the Supporting information, Supplementary Table 3.
Flow cytometry
Cells were blocked with anti-mouse FcγR antibody (2.4G2), stained with the indicated antibodies (Supporting information, Supplementary Table 3), and analysed on a FACSCalibur (BD Bioscience, San José, CA, USA). Data were analysed by FlowJo software (Tree Star Inc, San Carlos, CA, USA).
Sequence analysis of Ig V genes
Total RNA, DNA extraction, and cDNA synthesis were performed according to standard procedures with V gene amplification performed as previously described [25]. PCR products were extracted from 1.5% agarose gels using QIAquick kits (Qiagen, Chatsworth, CA, USA) and cloned using TOPO TA kits (Invitrogen, San Diego, CA, USA). Plasmid DNA obtained from at least three individual clones was sequenced using BigDye terminator. Sequence alignment was performed using IgBLAST (http://www.ncbi.nlm.nih.gov/igblast).
Fluorescence in situ hybridization (FISH) and spectral karyotyping (SKY)
Metaphase chromosomes from APCT cell lines were prepared and hybridized to Igh, Cμ, Igκ, and c-Myc BAC-derived probes as previously described [26]. SKY was performed using a SKYPaint probe kit from Applied Spectral Imaging (ASI; Vista, CA, USA) according to the manufacturer’s instructions.
Results
Comparisons of the gene expression profiles of PCTs and APCTs with the expression patterns of normal B-cell subsets
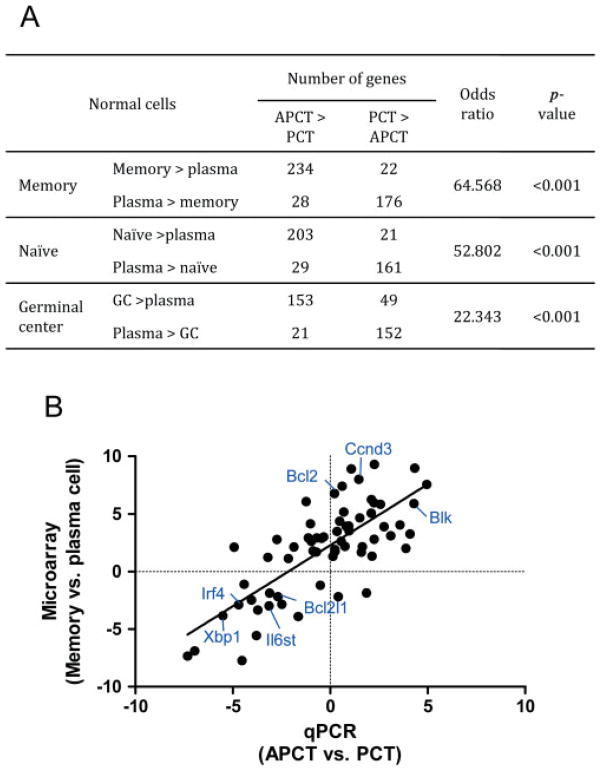
To extend our understanding of APCT as distinct from PCT and to identify their possible cellular origins, we used data from microarray analyses of the two PCN subsets to identify 1018 genes that best distinguished one from the other—445 for PCT and 573 for APCT (Supporting information, Supplementary Table 1). We then examined published microarray analyses of purified normal naïve B cells, GC B cells, memory B cells, and plasma cells [24], and identified 4700 non-redundant genes from the data set that could be matched with genes in our analyses of PCNs. Using Fisher’s exact test, we examined the relationships between the expression profiles of APCTs and normal naïve, memory, and GC cells (Figure 1A). Highly significant relationships were found between the expression profiles of APCTs and each of the normal B-cell subsets with a rank order, determined by odds ratios, of memory > naïve ≫ GC. The similarity of the APCT profile to both memory and naïve B-cell profiles is probably due to the fact that the transcriptional profiles of the two normal B-cell subsets overlap by 94% [24]. Similar comparisons of the transcriptional profiles of human tonsillar naïve, GC, and memory B cells also showed that the profile of memory B cells is closer to that of naïve than to that of GC B cells [27].
Figure 1.
Comparisons of the expression profiles for genes that distinguish PCTs and APCTs with genes that distinguish subsets of normal B-cell-lineage cells. (A) Genes that differ significantly for PCTs and APCTs determined from microarray analyses (Supporting information, Supplementary Table 1) and that overlap with non-redundant genes from the microarray data set of Battacharya et al [24] were compared with genes from the latter set that distinguish normal memory and plasma cells (top row), normal naïve and plasma cells (middle row), and normal germinal centre (GC) and plasma cells (bottom row). Odds ratio indicates the enrichment of genes differentially expressed in memory, naïve, and GC B cells compared with plasma cells. (B) Relationships between genes that distinguish PCTs from APCTs quantified by qPCR (X axis) and genes that distinguish normal memory and plasma cells quantified by microarrays (Y axis). Genes that characterize APCTs and memory B cells were assigned positive values, while genes that characterize PCTs and plasma cells were assigned negative values. The scale for each axis is fold change in log2.
To further assess the proposed relationship between APCT and normal memory B cells, we turned to more quantitative real-time RT-PCR (qPCR) analyses of 92 selected genes that best distinguished APCT from PCT or were shown by microarray analyses [24] to be useful in distinguishing subsets of normal B cells (Supporting information, Supplementary Table 2). Analyses of the comparative expression of these genes between PCTs and APCTs by qPCR and their pattern of expression among normal B-cell subsets is shown in Table 1; 30 genes differed significantly (p < 0.05) between APCTs and PCTs. Among 16 of these genes normally expressed by both naïve and memory cells or by memory cells alone, 11 (69%) were expressed preferentially by APCTs. All four plasma cell-specific genes were expressed preferentially by PCTs. We then examined the quantitative relationships between the gene expression patterns of APCTs and PCTs as determined by qPCR in comparison with the expression patterns of normal memory and plasma cells as determined by microarrays (Figure 1B). There were strong correlations between the expression patterns of memory B cells and APCTs, and between the expression patterns for plasma cells and PCT (1/slope = 0.95, r2 = 0.56), despite known problems in generating strong correlations between the differing formats of microarray and qPCR. This provides firm support for the suggestion that APCTs are more closely related to memory B cells than to plasma cells.
Table 1.
Quantitative real-time RT-PCR analyses of gene expression by PCTs and APCTs in relation to expression in normal B-cell subsets*
| Gene | Expression in normal B-cell subsets†
|
Fold change‡ (APCT/PCT) | |||
|---|---|---|---|---|---|
| Naïve | Plasma cell | GC cell | Memory cell | ||
| Spib | X | X | X | 20.06 | |
| Pax5 | X | X | X | 11.91 | |
| Sell | X | 5.93 | |||
| Irf8 | X | X | X | 4.72 | |
| Aicda | X | 4.29 | |||
| Klf2 | X | X | 2.85 | ||
| Ccnd3 | X | 2.75 | |||
| Casp3 | X | X | 2.42 | ||
| Relb | X | X | 1.95 | ||
| Mta3 | X | X | X | 1.92 | |
| Cd80 | X | 1.75 | |||
| Nfkb2 | X | X | 1.68 | ||
| Ets1 | X | X | 1.61 | ||
| Ung | X | X | 1.31 | ||
| Nfkb1 | X | X | 1.17 | ||
| Bcl2 | X | 1.16 | |||
| Runx3 | X | −1.27 | |||
| Rb1 | X | −1.36 | |||
| Ssbp3 | X | −1.85 | |||
| Foxp1 | X | X | −2.03 | ||
| Pml | X | −2.20 | |||
| Bcl6 | X | −2.37 | |||
| Pscdbp | X | X | X | −2.37 | |
| Il6ra | X | X | −4.44 | ||
| Xrcc3 | X | −8.65 | |||
| Atf6 | X | −8.88 | |||
| Bmi1 | X | −9.25 | |||
| Sdc1 | X | −23.07 | |||
| Irf4 | X | −26.13 | |||
| Xbp1 | X | −45.63 | |||
Gene transcript levels were determined by qPCR for the 96 genes listed in the Supporting information, Supplementary Table 2 in studies of ten APCTs and 10 PCTs. Data are for genes that differed significantly in expression (p < 0.05) between the two PCN subsets.
Expression patterns from ref 24.
Fold change.
Expression of proteins associated with maturational status, signalling, and survival in PCTs and APCTs
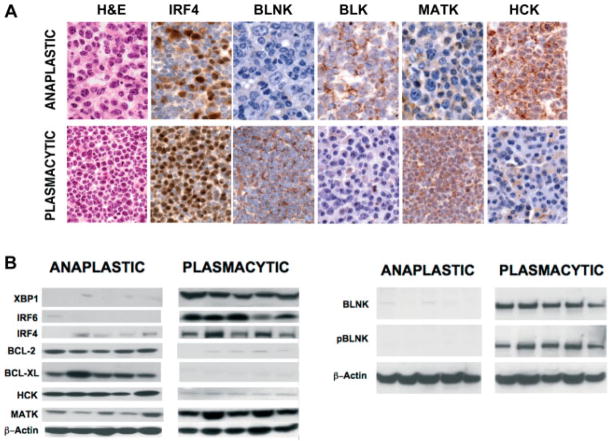
Based on the results from microarray and qPCR analyses, we selected a series of genes for validation by western blots of proteins extracted from primary tumours and by IHC. Predictably, PCTs uniformly expressed much higher levels of IRF4 and XBP1 than APCTs (Figures 2A and 2B, respectively). IRF6, known to be highly expressed at the transcript level in PCTs [7], was also expressed more strongly at the protein level by PCTs than by APCTs (Figure 2B).
Figure 2.
Immunohistochemical and western blot analyses of protein expression in PCTs and APCTs. (A) Comparative analyses of APCTs and PCTs on sections stained with H+E and with antibodies to proteins that differed significantly for expression between the tumour subsets. Immunohistochemical analyses show the significant difference between PCT and APCT. By H+E staining, PCT comprises almost all mature cells with a central nucleolus in a clock-face nucleus, basophilic cytoplasm, and a discernible Golgi. APCT is made up of a mixture of immunoblasts and anaplastic cells, as well as some plasmablasts. By IHC staining, higher levels of IRF4, BLNK, and MATK proteins are seen in PCT, and BLK and HCK in APCT. (B) Western blotting of protein extracts from the two tumour subsets using antibodies to the indicated proteins.
Several observations point to possible PCN subset-specific involvement of different Src kinase family members in disease. Two family members, BLK (Figure 2A) and HCK (Figures 2A and 2B), were expressed more highly by APCTs than by PCTs. BLK is rapidly activated after BCR ligation and signals downstream to promote proliferation and differentiation and to resist apoptosis [28]. From array analyses, Blk is among a small subset of genes expressed at high levels in both APCTs and memory B cells (Figure 1B). HCK interacts directly with the gp130 signal-transducing portion of the IL-6 receptor, encoded by Il6st, and mediates both proliferation and survival signals [29,30]. Although Il6st transcripts tended to be expressed at higher levels in PCTs than in APCTs (Figure 1B), the fact that both genes are active in APCT is consistent with BCR expression by normal memory B cells.
BLNK, a membrane-associated B-cell adaptor protein, promotes the coupling of BCR-associated Src kinases to downstream activation of PLCγ2 and RAC-JNK [31], as well as RAS [32]. By western blotting and IHC analyses, BLNK was expressed preferentially in PCTs (Figures 2A and 2B) and in an activated phosphorylated form (Figure 2B). Because PCTs lack BCR, the mechanisms responsible for BLNK activation remain to be determined. In contrast to the expression of Src kinases and signalling pathways that promote proliferation and survival in both classes of PCNs, MATK, a negative regulator of Src kinases [33], was expressed at higher levels in PCTs than in APCTs (Figures 2A and 2B). Understanding how these kinases and their downstream targets are regulated in mouse models of PCN may be of importance in view of the increasing interest in Src family kinases as targets for treatment of MM [34].
BCL2 and BCL-XL, anti-apoptotic members of the BCL2 family, were preferentially expressed in the subset of APCTs examined here (Figure 2B), although the microarray analyses indicated that expression by most APCTs is low, at least at the transcript level (Figure 1). BCL2 is reported to function in resting pre- and post-GC B cells, while BCL-XL is active in proliferating cells. Interestingly, and in keeping with the postulated origin of APCT from memory cells, quiescent human memory B cells in peripheral blood express both BCL2 and BCL-XL [35]. These findings indicate that the development of APCTs and PCTs may be differentially influenced by dissimilar Src signalling pathways and mechanisms employed to resist apoptosis, reinforcing the distinctness of the two forms of PCN studied here.
Relationships of APCTs from NFS.V+ mice to APCT-derived cell lines and PLs
All of the studies of APCTs described above and in a prior publication [17] were based on analyses of material saved at necropsy from primary cases in NFS.V+ mice. Previously, however, we established cultured lines from two mice diagnosed at necropsy with APCT: B6-1710, from a mouse with advanced MAIDS [23]; and B6-207, from a 1.5-year-old mouse.
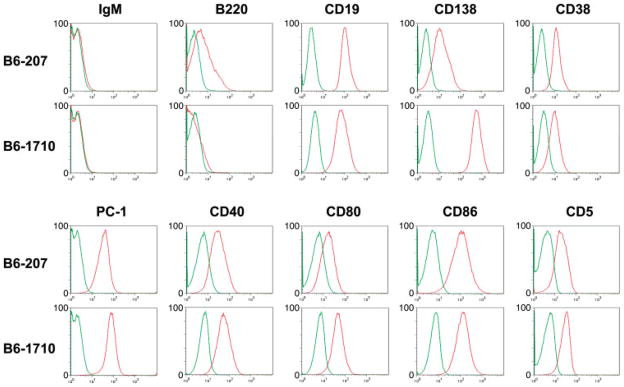
Analyses by flow cytometry (Figure 3) showed that both lines expressed B-cell markers (CD19, CD40, CD80, CD86) as well as plasma cell markers (CD138, PC-1), suggesting that they were arrested at a state of differentiation between B cells and plasma cells. Both also expressed CD5 at low levels. These expression patterns were quite similar to those reported for PLs from BALB/c or C3H mice mutant for Fasl (gld), tumours that are mostly class switched and express hypermutated Ig V genes [14]. By qPCR arrays, the cell lines from FAS-deficient mice also exhibited striking similarities to the APCT cell lines and to the consensus profile for NFS.V+ APCTs (data not shown). We conclude that the APCT and PL cell lines are very similar to one another and resemble primary APCTs much more closely than PCTs.
Figure 3.
Flow cytometric analyses of cell lines derived from primary APCTs. Single cell suspensions of the B6-207 and B6-1710 cell lines were stained with the indicated antibodies. Green, unstained cells; red, stained cells.
Sequences of Ig V regions of primary APCTs and APCT-derived cell lines
Memory B cells comprise several phenotypically and functionally distinct subpopulations that can be identified on the basis of their differential expression of Ig isotypes, tissue localization, and transcriptional profiles in both humans [36–38] and mice [24,39–41]. Although memory cells from both species have been thought of primarily as products of GC, and thus subject to AID-mediated CSR and SHM, it is clear that not all memory cells are switched or mutated [39,42,43].
To determine the mutational status of APCTs, we sequenced the Ig V regions of primary APCTs previously shown by Southern analyses to have prominent clonal rearrangements of IgH [17] and the APCT-derived cell lines. The results are summarized in Table 2, with full sequences in the Supporting information, Supplementary Table 5. Several tumours appeared to express single dominant clones: tumour 36 975 was significantly mutated for both IgH and IgK; 38 363 was minimally mutated for IgH and IgK but also expressed an unmutated IgL; and 38 397 was minimally mutated for IgH, IgK, and IgL. Two other tumours had clonal sequences for IgH—37 891 (mutated) and 36 896 (minimally mutated)—but both had two different IgK sequences. Clonal tumours expressing more than one light chain have been described [5]. Tumour 38 066 appears to be quite unusual in being germline for IgH but highly mutated for IgK. The IgH sequences from tumour 38 400 are indicative of oligoclonality, but the recovery of two identical IgL sequences suggests that one IgL-expressing clone may be more highly represented.
Table 2.
Immunoglobulin gene variable region sequences of primary APCTs and cell lines derived from primary APCTs*
| Primary APCT pathology number | Immunoglobulin heavy chain
|
Immunoglobulin light chain
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| V | D | J | C | % of GL | V | J | C | % of GL | |
| 36975 | J606.4.82 | DSP2.7 | 4 | ND | 92.7 | kh4 | 2 | Kappa | 95.4 and 95.8–both stop |
| 7183.7b | DSP2.9 | 4 | ND | 92.4–stop | ba9 | 2 | Kappa | 95.6 | |
| VL1 | 1 | Lambda | 98.3 | ||||||
| 37891 | 36–60.a1.85 | DQ52 | 3 | IgM | 97.8 | kb4 | 4 | Kappa | 98.8 |
| kb4 | 4 | Kappa | 98.1–different | ||||||
| VL2 | 2 | Lambda | 99.6 | ||||||
| 38006 | DQ52.8.22 | DFL16.1j | 4 | IgM | 100 | 4.57 | 5 | Kappa | 94.4 |
| VL2 | 2 | Lambda | 99.3 | ||||||
| 38363 | J588.20 | DSP2.24 | 4 | IgM | 99.6 | bb1 | 5 | Kappa | 100–stop |
| J558.20 | DSP2.24 | 4 | IgM | 100 | 4–50 | 5 | Kappa | 96.9–stop | |
| VL2 | 2 | Lambda | 99.7 | ||||||
| 38397 | DQ52.8.22 | DST4.3 | 3 | IgM | 99.6 | bv9 | 5 | Kappa | 99.6 |
| VL2 | 2 | Lambda | 99.3 | ||||||
| 38400 | SM.a2psi.88 | DSP2.9 | 3 | IgM | 97.7 | br9 | 1 | Kappa | 100–stop |
| DQ52.8.22 | DFL16.1j | 4 | IgM | 100 | kh4 | 5 | Kappa | 98.9 | |
| J558.83.189 | DSP2.6 | 2 | IgG | 95.9 | VL2 | 2 | Lambda | 99.7 | |
| Q52.a12.33 | DSP2.12 | 4 | IgG | 97.8 | VL2 | 2 | Lambda | 99.7 | |
| 38696 | DQ52.8.22 | DFL16.3 | 3 | IgM | 99.6 | bb1 | 2 | Kappa | 100 |
| kh4 | 5 | Kappa | 98.9 | ||||||
| VL2 | 2 | Lambda | 99.7 | ||||||
| APCT cell line | |||||||||
| B6-207 | J558.16.106 | DFL16.1j | 4 | IgG | 85 | ba9 | 2 | Kappa | 87.3 |
| B6-1710 | J558.12.102 | DFL16.1 | 4 | IgG | 97.4 | 23–39 | 5 | Kappa | 96.5–stop |
V(D)J and C region sequences for IgH and IgL. Pathology numbers for primary cases of APCT and cell line designations for cultured lines derived from primary APCTs are listed. Specific VH, DH, VK, and VL family members identified by sequencing are indicated by name together with J utilization and C region. For individual primary APCTs, cloned sequences that were not repeats of the top sequence listed are also given.
% of GL: per cent identity with germline sequences. Sequences differing by more than 2% from GL are considered to be significantly mutated. Stop: identification of a stop codon blocking expression of the heavy or light chain. ND: not determined as sequencing was done from genomic DNA.
Of the APCT-derived cell lines, B6-207 was heavily mutated for both IgH and IgK, and B6-1710 was significantly but not heavily mutated for both chains (Table 2 and Supporting information, Supplementary Table 4), findings similar to those for the Ig genes from PLs [14]. Together, the Ig sequence data from primary APCTs and the APCT-derived cell lines bear similarities to some PLs.
Chromosomal analyses of APCT-derived cell lines
Virtually all primary PCTs, PCT cell lines, and PCT precursors in BALB/c mice contain reciprocal Myc-activating translocations, either the common T(12;15) involving Igh or the variant T(6;15) involving Igk [2,44,45]. Prior SKY analyses showed that the vast majority of primary PCTs were tetraploid and contained additional alterations involving all chromosomes except the Y, reflecting generalized genomic instability [46].
Prior studies of primary APCTs by FISH using Igh and Myc probes on formalin-fixed, paraffin-embedded tissue sections revealed neither Igh/Myc co-localization nor gross abnormalities affecting these loci, indicating that canonical IgH/Myc translocations did not contribute to the pathogenesis of these tumours [17]. In the present study, we took advantage of the APCT-derived cell lines and analysed metaphase spreads by SKY or by FISH using probes for Igh, Igk, and Myc.
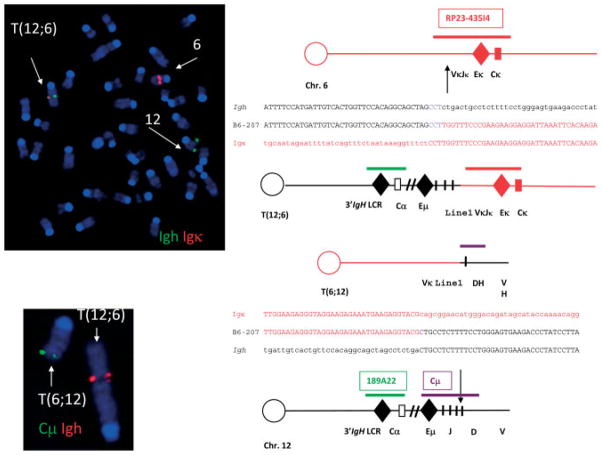
By SKY (Figure 4), the B6-1710 cell line had a near diploid karyotype of 42, X, T(4;12), + T(10;6), +Del(19), T(X;2). The Myc locus on chromosome 15 appeared normal and there were no gains of chromosome 15; however, as determined by SKY, the telomeric portion of chromosome 12 (band D1) was translocated to chromosome 4 (band D2.3), resulting in a non-reciprocal T(4;12) (Figure 4B).
Figure 4.
Chromosomal aberrations in cell lines derived from primary anaplastic PCTs detected by spectral karyotyping (SKY) studies of the B6-1710 and B6-207 cell lines. The Del(6E) is presumably the T(6;12) that could not be resolved by SKY.
SKY analyses of the B6-207 line (Figure 4) revealed a near diploid karyotype of 41, XY, T(3;1), Del(6E), T(10;5), +11, T(12;6) and no alterations in the Myc-containing region of chromosome 15. Notably, as in B6-1710, chromosome 12 was also translocated, but now to chromosome 6 (Figure 5, left), bringing the IgH locus in immediate proximity to Igk (Figure 5). These data confirm our previous findings that chromosomal translocations involving Myc are not involved in the pathogenesis of APCTs.
Figure 5.
Chromosomal studies of the B6-207 cell line. Top left: fluorescence in situ hybridization (FISH) using probes for Igh (green) and Igk (red) showing the translocation involving chromosome 12 and chromosome 6 (left side) that was present in all cells. Bottom left: FISH showing IgH (red) and Cμ (green) in abnormal locations. Diagrammatic structures of normal chromosome 12 and normal chromosome 6 with positions of probes for Igh, Cμ, and Igk and break-point positions give rise to the T(12;6) (right side).
The identification of a near reciprocal T(12;6), T(6;12) in B6-207 raised the possibility of identifying a gene that might be involved in the pathogenesis of these PCNs by virtue of its placement in proximity to Ig regulatory sequences. Analyses of the translocation by FISH using probes to IgH, kappa, and Cμ showed that the break on chromosome 12 occurred just downstream of Eμ and that the break on chromosome 6 occurred just upstream of Ek (Figure 5). Cloning of the breakpoint junction revealed the break on 12 to be in the J region and the break in 6 to be in the VκJκ region in a line 1 element. The translocation thus might be the result of aberrant V(D)J recombination.
Discussion
Classification has been described as ‘the language of medicine’, in that diseases must be clearly described, defined, and named to provide a basis for clinical practice and investigation, with a major component being class discovery [1]. Here and in a previous publication [17], we have utilized a variety of approaches to describe a subset of mouse PCNs, initially defined as APCTs based on cytological distinctions of primary tumours in NFS.V+ mice from the pristane-induced PCTs of BALB/c mice [46]. Previously, the definition of APCT was extended to include the absence of structural alterations in Myc detectable by Southern analyses [17], with Myc-activating translocations being a nearly invariant feature of PCT [47]. It is important to recognize, however, that APCTs were first described for PCNs in Eμ-v-abl TG mice [48] that regularly exhibit Igh/Myc translocations [49]. More recently, APCTs were identified in iMyc knockin [50], IgH 3′ LCR-Myc TG [51], Eμ-Ccnd1-T286A TG [52], and iMyc/BCL-XL double-TG mice [53]. These observations indicate that PCNs histologically indistinguishable from the APCTs of NFS.V+ mice can be induced by MYC-dependent and -independent pathways.
The present studies indicate that primary APCTs, APCT-derived cell lines, and PLs from FASL-deficient mice have many commonalities that readily distinguish them from pristane-induced PCTs. First, they all lack Myc-activating chromosomal translocations; second, their gene expression patterns are suggestive of a stage of differentiation between the GC and fully mature plasma cells; and finally, APCT-derived cell lines exhibit few of the chromosomal complexities characteristic of pristane-induced PCTs.
Clearly, APCTs, PLs, and APCT-derived cell lines are mostly AID-experienced, as evidenced by CSR and SHM. Importantly, Aicda transcripts are present in normal class-switched memory B cells as well as APCTs, and AID is active in T-cell-independent extrafollicular or mucosal B-cell responses [20,54] and following activation of B1a cells (K Hayakawa, personal communication). The APCT cases that have not undergone CSR but exhibit SHM resemble some IgM-secreting MMs that are somatically mutated and exhibit switch variants [55], and IgM-expressing Waldenstrom’s macroglobulinaemia tumour cells that also exhibit SHM [56]. An origin of both tumour types from memory cells has been postulated.
Based largely on comparative gene expression profiles of these PCNs with those of normal B-cell subsets, we postulate that a memory-like B cell is the normal cell of origin for APCT. The exact origin of the precursor is not fully clear, because memory B-cell subpopulations in mice are not as well defined as those in humans. Most characterizations of mouse memory B cells are based on studies of cells from immunized mice that retain antigen-binding specificity and stain with cocktails of monoclonal antibodies directed against panels of cell surface antigens [39–41,57]. How these carefully circumscribed cells relate to the full spectrum of normal mature plasma cells that can develop as immediate descendants of GC B cells, from GC-derived memory B cells, from extrafollicular B-cell responses, or from B1 cells is uncertain.
Supplementary Material
Microarray analyses of significant genes that distinguish PCT from APCT
Genes included in the qPCR array for comparing PCN subsets
Antibodies used for flow cytometry, western blot, and immunohistochemistry
Immunoglobulin gene variable region sequences from primary APCTs and cell lines derived from primary APCTs
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank Alfonso Macias for colony management and identification of tumour-bearing mice; NIAID intramural editor Brenda Rae Marshall for assistance; and Nicole McNeil, NCI, Genetics Branch for SKY interpretation.
Footnotes
No conflicts of interest were declared.
SUPPORTING INFORMATION ON THE INTERNET
The following supporting information may be found in the online version of this article.
References
- 1.Swerdlow SH, Campo E, Harris NL, Jaffe E, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. WHO Press; Geneva: 2008. [Google Scholar]
- 2.Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 3.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 4.Hong DS, Angelo LS, Kurzrock R. Interleukin-6 and its receptor in cancer: implications for translational therapeutics. Cancer. 2007;110:1911–1928. doi: 10.1002/cncr.22999. [DOI] [PubMed] [Google Scholar]
- 5.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boylan KL, Gosse MA, Staggs SE, Janz S, Grindle S, Kansas GS, et al. A transgenic mouse model of plasma cell malignancy shows phenotypic, cytogenetic, and gene expression heterogeneity similar to human multiple myeloma. Cancer Res. 2007;67:4069–4078. doi: 10.1158/0008-5472.CAN-06-3699. [DOI] [PubMed] [Google Scholar]
- 7.Shin DM, Shaffer DJ, Wang H, Roopenian DC, Morse HC., III NOTCH is part of the transcriptional network regulating cell growth and survival in mouse plasmacytomas. Cancer Res. 2008;68:9202–9211. doi: 10.1158/0008-5472.CAN-07-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park ES, Shaughnessy JD, Jr, Gupta S, Wang H, Lee JS, Woo HG, et al. Gene expression profiling reveals different pathways related to Abl and other genes that cooperate with c-Myc in a model of plasma cell neoplasia. BMC Genomics. 2007;8:302. doi: 10.1186/1471-2164-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Takizawa M, Tolarova H, Li Z, Dubois W, Lim S, Callen E, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter M, Wax JS, Hansen CT, Kenny JJ. BALB/c. CBA/N mice carrying the defective Btk(xid) gene are resistant to pristane-induced plasmacytomagenesis. Int Immunol. 1999;11:1059–1064. doi: 10.1093/intimm/11.7.1059. [DOI] [PubMed] [Google Scholar]
- 13.Davidson WF, Giese T, Fredrickson TN. Spontaneous development of plasmacytoid tumors in mice with defective Fas–Fas ligand interactions. J Exp Med. 1998;187:1825–1838. doi: 10.1084/jem.187.11.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JQ, Okumura C, McCarty T, Shin MS, Mukhopadhyay P, Hori M, et al. Evidence for selective transformation of autoreactive immature plasma cells in mice deficient in Fasl. J Exp Med. 2004;200:1467–1478. doi: 10.1084/jem.20041575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radl J, Croese JW, Zurcher C, Van den Enden-Vieveen MH, de Leeuw AM. Animal model of human disease. Multiple myeloma. Am J Pathol. 1988;132:593–597. [PMC free article] [PubMed] [Google Scholar]
- 16.Pumphrey JG, Hausner P, Rudikoff S. Characterization of C57BL/6 plasmacytomas lacking a c-myc translocation. Int J Cancer. 1997;72:892–897. doi: 10.1002/(sici)1097-0215(19970904)72:5<892::aid-ijc29>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 17.Qi CF, Zhou JX, Lee CH, Naghashfar Z, Xiang S, Kovalchuk AL, et al. Anaplastic, plasmablastic, and plasmacytic plasmacytomas of mice: relationships to human plasma cell neoplasms and late-stage differentiation of normal B cells. Cancer Res. 2007;67:2439–2447. doi: 10.1158/0008-5472.CAN-06-1561. [DOI] [PubMed] [Google Scholar]
- 18.Falini B, De Solas I, Levine AM. Emergence of B-immunoblastic sarcoma in patients with multiple myeloma: a clinicopathologic study of 10 cases. Blood. 1982;59:923–933. [PubMed] [Google Scholar]
- 19.Greipp PR, Raymond NM, Kyle RA, O’Fallon WM. Multiple myeloma: significance of plasmablastic subtype in morphological classification. Blood. 1985;65:305–310. [PubMed] [Google Scholar]
- 20.Cunningham AF, Gaspal F, Serre K, Mohr E, Henderson IR, Scott-Tucker A, et al. Salmonella induces a switched antibody response without germinal centers that impedes the extracellular spread of infection. J Immunol. 2007;178:6200–6207. doi: 10.4049/jimmunol.178.10.6200. [DOI] [PubMed] [Google Scholar]
- 21.MacLennan ICM, Toellner KM, Cunningham AF, Serre K, Sze DM, Zúñiga E, et al. Extrafollicular antibody responses. Immunol Rev. 2003;194:8–18. doi: 10.1034/j.1600-065x.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- 22.Hartley JW, Chattopadhyay SK, Lander MR, Taddesse-Heath L, Naghashfar Z, Morse HC, III, et al. Accelerated appearance of multiple B cell lymphoma types in NFS/N mice congenic for ecotropic murine leukemia viruses. Lab Invest. 2000;80:159–169. doi: 10.1038/labinvest.3780020. [DOI] [PubMed] [Google Scholar]
- 23.Klinken SP, Fredrickson TN, Hartley JW, Yetter RA, Morse HC., III Evolution of B cell lineage lymphomas in mice with a retrovirus-induced immunodeficiency syndrome, MAIDS. J Immunol. 1988;140:1123–1131. [PubMed] [Google Scholar]
- 24.Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, et al. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaw L, Siwarski D, Coleman A, Kim J, Jones GM, Dighiero G, et al. Restricted immunoglobulin variable region (Ig V) gene expression accompanies secondary rearrangements of light chain Ig V genes in mouse plasmacytomas. J Exp Med. 1999;190:1405–1415. doi: 10.1084/jem.190.10.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovalchuk AL, du Bois W, Mushinski E, McNeil NE, Hirt C, Qi CF, et al. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J Exp Med. 2007;204:2989–3001. doi: 10.1084/jem.20070882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsueh RC, Scheuermann RH. Tyrosine kinase activation in the decision between growth, differentiation, and death responses initiated from the B cell antigen receptor. Adv Immunol. 2000;75:283–316. doi: 10.1016/s0065-2776(00)75007-3. [DOI] [PubMed] [Google Scholar]
- 29.Hausherr A, Tavares R, Schäffer M, Obermeier A, Miksch C, Mitina O, et al. Inhibition of IL-6-dependent growth of myeloma cells by an acidic peptide repressing the gp130-mediated activation of Src family kinases. Oncogene. 2007;26:4987–4998. doi: 10.1038/sj.onc.1210306. [DOI] [PubMed] [Google Scholar]
- 30.Podar K, Mostoslavsky G, Sattler M, Tai YT, Hayashi T, Catley LP, et al. Critical role for hematopoietic cell kinase (Hck)-mediated phosphorylation of Gab1 and Gab2 docking proteins in interleukin 6-induced proliferation and survival of multiple myeloma cells. J Biol Chem. 2004;279:21658–21665. doi: 10.1074/jbc.M305783200. [DOI] [PubMed] [Google Scholar]
- 31.Ishiai M, Kurosaki M, Pappu R, Okawa K, Ronko I, Fu C, et al. BLNK required for coupling syk to PLCg2 and rac1-JNK in B cells. Immunity. 1999;10:117–125. doi: 10.1016/s1074-7613(00)80012-6. [DOI] [PubMed] [Google Scholar]
- 32.Imamura Y, Oda A, Katahira T, Bundo K, Pike KA, Ratcliffe MJ, et al. BLNK binds active H-Ras to promote B cell receptor-mediated capping and ERK activation. J Biol Chem. 2009;284:9804–9813. doi: 10.1074/jbc.M809051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B-C, Avraham S, Imamoto A, Avraham HK. Identification of the nonreceptor tyrosine kinase MATK/CHK as an essential regulator of immune cells using Matk/CHK-deficient mice. Blood. 2006;108:904–907. doi: 10.1182/blood-2005-12-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gertz MA. New targets and treatments in multiple myeloma: src family kinases as central regulators of disease progression. Leuk Lymphoma. 2008;49:2240–2245. doi: 10.1080/10428190802475311. [DOI] [PubMed] [Google Scholar]
- 35.Bovia F, Nabili-Tehrani AC, Werner-Favre C, Barnet M, Kindler V, Zubler RH. Quiescent memory B cells in human peripheral blood co-express bcl-2 and bcl-x(L) anti-apoptotic proteins at high levels. Eur J Immunol. 1998;28:4418–4423. doi: 10.1002/(SICI)1521-4141(199812)28:12<4418::AID-IMMU4418>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 36.Ehrhardt GRA, Hijikata A, Kitamura H, Ohara O, Wang JY, Cooper MD. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med. 2008;205:1807–1817. doi: 10.1084/jem.20072682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tangye SG, Good KL. Human IgM+ CD27+ B cells: memory B cells or ‘memory’ B cells? J Immunol. 2007;179:13–19. doi: 10.4049/jimmunol.179.1.13. [DOI] [PubMed] [Google Scholar]
- 38.Weller S, Mamani-Matsuda M, Picard C, Cordier C, Lecoeuche D, Gauthier F, et al. Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+ IgD+ CD27+ B cell repertoire in infants. J Exp Med. 2008;205:1331–1342. doi: 10.1084/jem.20071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson SM, Tomayko MM, Ahuja A, Haberman AM, Shlomchik MJ. New markers for murine memory B cells that define mutated and unmutated subsets. J Exp Med. 2007;204:2103–2114. doi: 10.1084/jem.20062571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chappell CP, Jacob J. Identification of memory B cells using a novel transgenic mouse model. J Immunol. 2006;176:4706–4715. doi: 10.4049/jimmunol.176.8.4706. [DOI] [PubMed] [Google Scholar]
- 41.Blink EJ, Light A, Kallies A, Nutt SL, Hodgkin PD, Tarlinton DM. Early appearance of germinal center-derived memory B cells and plasma cells in blood after primary immunization. J Exp Med. 2005;201:545–554. doi: 10.1084/jem.20042060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein U, Küppers R, Rajewsky K. Evidence for a large compartment of IgM-expressing memory B cells in humans. Blood. 1997;89:1288–1298. [PubMed] [Google Scholar]
- 43.Toyama H, Okada S, Hatano M, Takahashi Y, Takeda N, Ichii H, et al. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity. 2002;17:329–339. doi: 10.1016/s1074-7613(02)00387-4. [DOI] [PubMed] [Google Scholar]
- 44.Janz S, Müller J, Shaughnessy J, Potter M. Detection of recombinations between c-myc and immunoglobulin switch alpha in murine plasma cell tumors and preneoplastic lesions by polymerase chain reaction. Proc Natl Acad Sci U S A. 1993;90:7361–7365. doi: 10.1073/pnas.90.15.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovalchuk AL, Müller JR, Janz S. Deletional remodeling of c-myc-deregulating translocations. Oncogene. 1997;15:2369–2377. doi: 10.1038/sj.onc.1201409. [DOI] [PubMed] [Google Scholar]
- 46.Coleman AE, Ried T, Janz S. Chromosomes 1 and 5 harbor plasmacytoma progressor genes in mice. Genes Chromosomes Cancer. 2000;29:70–74. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1009>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 47.Fredrickson TN, Harris AW. Atlas of Mouse Hematopathology. Harwood Academic Publishers; Amsterdam: 2000. [Google Scholar]
- 48.Janz S. Myc translocations in B cell and plasma cell neoplasms. DNA Repair (Amst) 2006;5:1213–1224. doi: 10.1016/j.dnarep.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 49.Harris AW, Bath ML, Rosenbaum H, McNeall J, Adams JM, Cory S. Lymphoid tumorigenesis by v-abl and BCR-v-abl in transgenic mice. Curr Topics Microbiol Immunol. 1990;166:165–173. doi: 10.1007/978-3-642-75889-8_21. [DOI] [PubMed] [Google Scholar]
- 50.Park SS, Kim JS, Tessarollo L, Owens JD, Peng L, Han SS, et al. Insertion of c-Myc into IgH induces B-cell and plasma-cell neoplasms in mice. Cancer Res. 2005;65:1306–1315. doi: 10.1158/0008-5472.CAN-04-0268. [DOI] [PubMed] [Google Scholar]
- 51.Truffinet V, Pinaud E, Cogné N, Petit B, Guglielmi L, Cogné M, et al. The 3′ IgH locus control region is sufficient to deregulate a c-myc transgene and promote mature B cell malignancies with a predominant Burkitt-like phenotype. J Immunol. 2007;179:6033–6042. doi: 10.4049/jimmunol.179.9.6033. [DOI] [PubMed] [Google Scholar]
- 52.Gladden AB, Woolery R, Aggarwal P, Wasik MA, Diehl JA. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2006;25:998–1007. doi: 10.1038/sj.onc.1209147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung WC, Kim JS, Linden M, Peng L, Van Ness B, Polakiewicz RD, et al. Novel targeted deregulation of c-Myc cooperates with Bcl-X(L) to cause plasma cell neoplasms in mice. J Clin Invest. 2004;113:1763–1773. doi: 10.1172/JCI20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu W, Santini PA, Matthews AJ, Chiu A, Plebani A, He B, et al. Viral double-stranded RNA triggers Ig class switching by activating upper respiratory mucosa B cells through an innate TLR3 pathway involving BAFF. J Immunol. 2008;181:276–287. doi: 10.4049/jimmunol.181.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sahota SS, Garand R, Mahroof R, Smith A, Juge-Morineau N, Stevenson FK, et al. V(H) gene analysis of IgM-secreting myeloma indicates an origin from a memory cell undergoing isotype switch events. Blood. 1999;94:1070–1076. [PubMed] [Google Scholar]
- 56.Babbage G, Townsend M, Zojer N, Mockridge IC, Garand R, Barlogie B, et al. IgM-expressing Waldenstrom’s macroglobulinemia tumor cells reveal a potential for isotype switch events in vivo. Leukemia. 2007;21:827–830. doi: 10.1038/sj.leu.2404538. [DOI] [PubMed] [Google Scholar]
- 57.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray analyses of significant genes that distinguish PCT from APCT
Genes included in the qPCR array for comparing PCN subsets
Antibodies used for flow cytometry, western blot, and immunohistochemistry
Immunoglobulin gene variable region sequences from primary APCTs and cell lines derived from primary APCTs