Abstract
The glutathione thiol/disulfide couple is the major redox buffer in the endoplasmic reticulum (ER); however, mechanisms by which it contributes to the tightly regulated redox environment of this intracellular organelle are poorly understood. The recent development of genetically encoded, ratiometric, single green fluorescent protein-based redox-sensitive (roGFP) sensors adjusted for more oxidative environments enables non-invasive measurement of the ER redox environment in living cells. In turn, Förster resonance energy transfer (FRET) sensors based on two fluorophore probes represent an alternative strategy for ratiometric signal acquisition. In previous work, we described the FRET-based redox sensor CY-RL7 with a relatively high midpoint redox potential of −143 mV, which is required for monitoring glutathione potentials in the comparatively high oxidative environment of the ER. Here, the efficacy of the CY-RL7 probe was ascertained in the cytosol and ER of live cells with fluorescence microscopy and flow cytometry. The sensor was found to be fully reduced at steady state in the cytosol and became fully oxidized in response to treatment with 1-chloro-2,4-dinitrobenzene, a depletor of reduced glutathione (GSH). In contrast, the probe was strongly oxidized (88%) upon expression in the ER of cultured cells. We also examined the responsiveness of the ER sensor to perturbations in cellular glutathione homeostasis. We observed that the reductive level of the FRET sensor was increased two-fold to about 28% in cells pretreated with N-acetylcysteine, a substrate for GSH synthesis. Finally, we evaluated the responsiveness of CY-RL7 and roGFP1-iL to various perturbations of cellular glutathione homeostasis to address the divergence in the specificity of these two probes. Together, the present data generated with genetically encoded green fluorescent protein (GFP)-based glutathione probes highlight the complexity of the ER redox environment and indicate that the ER glutathione pool may be more oxidized than is currently considered.
Keywords: live cell imaging, endoplasmic reticulum, redox-sensitive probe, green fluorescent protein, glutathione, Förster resonance energy transfer
Introduction
Glutathione (GSH) is the major thiol-disulfide redox buffer of the cell. Therefore, the redox potential of the reduced/oxidized glutathione (GSH/GSSG) couple can be used as an indicator of the redox environment of the cytosol or subcellular compartments.1 Recent evidence indicates that the glutathione couple is present in different concentrations and that the GSH/GSSG ratio varies considerably among subcellular organelles.2–5 Glutathione concentrations in the cytosol and lumen of the endoplasmic reticulum (ER) are similar (i.e. 1–10 mmol/L), but the ER GSH/GSSG ratio is nearly 20 times lower than that of the cytosol.6 In contrast to the reduced redox environment of the cytosol, the ER redox potential ranges from approximately −170 to −185 mV, assuming the total glutathione concentration in the secretory pathway is 8 mmol/L, or approximately −133 to −165 mV if the concentration is 1 mmol/L.7–9
Analyzing redox potentials within subcellular compartments is inherently more difficult than total cellular or tissue measurements due to the time and methods necessary to extract the subcellular fractions.5 To address these limitations, novel molecular tools have been developed to enable studies of the redox conditions in intracellular organelles. The most reliable information has been obtained with green fluorescent protein-based redox-sensitive (roGFP) probes targeted to subcellular compartments.10–12 The ratiometric, genetically encoded sensors allow non-invasive and extended assessments of reversible redox changes in the cytosol, mitochondria, ER and peroxisomes of mammalian and plant cells.10,11,13–20 However, the roGFPs are limited by their relatively reducing midpoint redox potentials, the probe roGFP2 being at least 97% oxidized in the ER of plant and animal cells.17,21 Measurements in epithelial cells with roGFP1 targeted to the ER revealed the intraorganellar redox potential to be approximately −217 mV at pH 7.4, which is decidedly more reduced than previous estimates with invasive protocols.7,8,18
The rough ER, the site of synthesis for membrane-bound and secreted proteins, harbors an intracellular redox environment conducive to disulfide bond formation.7 Protein folding in the ER is a tightly regulated process relying on many quality control measures that examine both the overall structural integrity and cell-specific factors that serve for specific protein export.22 As such, the ER redox environment is a complex milieu of multiple redox couples among which the GSH/GSSG couple is the major contributor to the thiol-oxidizing machinery required for oxidative protein folding.6,23–25 The driving force for disulfide formation is provided by the activity of ER-resident endoplasmic oxidoreducin 1 (Ero1) enzymes, which function to oxidize cysteine side chains at the expense of molecular oxygen, as well as the relatively oxidizing GSH/GSSG ratio.25–27 A complementary mechanism of ER redox regulation, mediated by GSSG acting as a specific oxidant of protein disulfide isomerase (PDI), has been proposed recently in opposition to the currently considered notion that the relatively low luminal GSH/GSSG ratio is the consequence rather than the cause of oxidative protein folding.6,24,28 In addition to a protein relay involving Ero1 and PDI, several other less characterized pathways exist that may affect the ER GSH/GSSG ratio.24,29 Apart from its essential role in protein folding and secretion, the ER’s oxidative state is proposed to be responsive to cellular stress signals and elicit an ‘unfolded protein response’, which generates cell death decisions.30 Understanding molecular mechanisms that shape the unique redox environment of the ER is essential for the development of successful therapeutic strategies for the treatment of ER-stress-related diseases.31–33
Thus, a new family of roGFP1-iX probes with a less reducing midpoint potential (as low as −229 mV) has been developed and successfully assessed in the ER of live yeast and mammalian cells.29,34,35 The roGFP1-iL probe with a half point redox potential of −229 mV was recently reported near its midpoint potential in the ER of yeast and mammalian cells at steady state, thereby enabling full sensitivity to both oxidative and reductive insults.29,35 The roGFP1-iL probe responded not only to acute insults with exogenous reductants and oxidants, but it was able to monitor changes in the ER redox environment in response to downregulation of Ero1α as well as to the release of nascent polypeptides from ribosomes triggered by puromycin.29 The ER glutathione redox potential was found to be in the range of −230 to −235 mV at pH 7.0 with the roGFP1-iL probe. Notably, this potential is close to the −240 mV value of the standard redox potential of the GSH/GSSG couple, at which glutathione is present in a mainly reduced state.2 As an alternative strategy in offering ratiometric signal acquisition, Förster resonance energy transfer (FRET)-based redox probes composed of two fluorophores were developed.19,36 FRET is a distance-dependent physical process, in which the excitation energy of a fluorescent donor molecule is transferred to an acceptor molecule through non-radiative dipole–dipole coupling. In FRET-based redox probes, redox-sensitive polypeptide domains are inserted between donor/acceptor pairs to gain increased responsiveness to glutathione redox potentials.12 Recently, we described a novel redox-sensitive probe with an oxidative midpoint redox potential of −143 mV (pH 7.0), which is ideal for probing the ER environment.37
In present work, we report the validation of the CY-RL7 probe expressed in two subcellular compartments, the cytosol and the ER of mammalian cells. The dynamic range of the CY-RL7 probe targeted to the ER of mammalian cells was evaluated in response to the oxidant diamide and the reductant dithiothreitol (DTT). We also targeted both the CY-RL-7 and the roGFP1-iL probes to the ER of Chinese hamster ovary (CHO) and HCT116 cells and studied their responses to perturbations in glutathione homeostasis with subsequent immediate monitoring of intracellular glutathione content. The two independently developed probes specific to glutathione redox potentials revealed different read-outs in glutathione redox status at steady states. Thus, these sensors may complement each other and should be beneficial in monitoring and decoupling of individual redox pathways involved in regulation of ER glutathione homeostasis.
Materials and methods
Materials
Reagents were purchased from Sigma (St Louis, MO, USA), unless otherwise specified. Enzymes for the modification of DNA as well as transformation reagent Lipofectamine 2000 were from Invitrogen (Carlsbad, CA, USA). The oligonu-cleotides used in sensor design were obtained from Integrated DNA Technologies (Coralville, IA, USA). CHO K1 fibroblasts were from ATCC (Manassas, VA, USA) and human colon epithelial (HCT116) cells were a generous gift from Dr Bert Vogelstein of Johns Hopkins Medicine (Baltimore, MD, USA). QIAprep spin miniprep and QIAquick PCR purification kits were from Qiagen (Valencia, CA, USA). The EKAR construct in pRK5 was purchased from Addgene (Cambridge, MA, USA),38 pEYFP-N1 was from BD Biosciences (Palo Alto, CA, USA; YFP, yellow fluorescent protein) and roGFP1-iL in pQE-30 was a kind gift from Dr James Remington (University of Oregon, OR, USA).
Cell culture, transfection and cell sorting
Tissue culture supplies were from Falcon (Franklin Lakes, NJ, USA) and Corning (Corning, NY, USA). CHO and HCT116 cells were maintained at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) and McCoy’s 5A media, respectively (Cell Media Facility, University of Illinois, Urbana, IL, USA). Both media were supplemented with 10% fetal bovine serum (Tissue Culture Biologicals, Tulare, CA, USA), streptomycin (50 mg/L), penicillin (50,000 U/L) and fungizone (0.25 mg/L). Streptomycin, penicillin and fungizone were purchased from Gibco (Grand Island, NY, USA), phosphate-buffered saline (PBS) from Lonza (Walkersville, MD, USA), and Hank’s buffered saline solution (HBSS) and trypsin from Mediatech (Manassas, VA, USA). Trypan blue and dimethyl sulfoxide, used respectively in cell counting and cryogen storage, were from MP Biomedicals (Solon, OH, USA) and other tissue culture reagents were obtained from Invitrogen. Transfection of CHO cells with Lipofectamine 2000, deriving stable CHO cell lines, and cell sorting were performed as described elsewhere.36 HCT116 cells were transfected with the Neon Transfection System from Invitrogen, according to the manufacturer’s protocol.
Genetic constructs
To create ER-targeted sensors, an ER leader sequence and KDEL retention signal were derived from human calreticulin.39 The DNA fragment encoding the ER leader signal was prepared by annealing and extending the oligonucleotide primer set LS-Forward 5′ CCTCTAGAGCTAGCGCTACC GGTCGCCACCATGCTGCTATCCGTGCCGCTGCTGCT 3′ and LS-Reverse 5′ TGCCATGGACTCGGCGACGGCCAGG CCGAGGAGGCCGAGCAGCAGCGGCACGGATAGCA 3′ with the cleavage sites NcoI/XbaI. This DNA fragment was confirmed by automated sequencing and ligated upstream of CY-RL7 in pET19b as described earlier (Supplementary Figure 1a).37 For all supplementary figures, please see http://ebm.rsmjournals.com/lookup/suppl/doi:10.1258/ebm.2012.011436/-/DC1. The ER retention signal was added downstream of the EYFP gene by polymerase chain reaction (PCR) with the primer set YFP-Forward 5′ GCGAATTCATGGTGAGCAAGGGCGAGGA 3′ and YFP-Reverse 5′ GGATCCTACAGCTCGTCCTTCTTGTACA GCTCGTCCATGCCGA 3′. The pEYFP-N1 served as a template. Next, the EYFP previously cloned in the cytosolic CY-RL7 construct was replaced by the newly obtained sequence EYFP + KDEL with the cleavage sites EcoRI/BamHI. Finally, the integrated CY-RL7 targeting ER (erCY-RL7) was cloned into NheI/BamHI sites of pECFP-C1 (Supplementary Figure 1c; CFP, cyan fluorescent protein). The ECFP-RL7-mVenus (mutated Venus) targeting ER was obtained by replacement of EYFP (Supplementary Figure 1d). For that purpose, the mVenus with attached retention KDEL signal was amplified by PCR with the YFP primer set from a 5.8-kb template obtained by EcoRI digest of the EKAR construct. The roGFP1-iL targeting the ER was prepared by two sequential PCR reactions with the primer set iL-Forward1 5′ ATCCGTGCCGCTGCTGCT CGGCCTCCTCGGCCTGGCCGTCGCCGAGATGAGTAA AGGAGAAGAACTTTTCACT 3′ with iL-Reverse 5′ GGAT CCTACAGCTCGTCCTTTTTGTATAGTTCATCCATGCCAT GTGT 3′ and iL-Forward2 5′ CCTCTAGAGCTAGCGCTA CCGGTCGCCACCATGCTGCTATCCGTGCCGCTGCTGCT 3′ with iL-Reverse. The final PCR product was cloned in pCR2.1 and confirmed by automated sequencing. Next, the ER-roGFP1-iL construct was subcloned in the NheI/BamHI sites of pECFP-C1 backbone plasmid (Supplementary Figure 1e).
Fluorescence microscopy
Time-resolved imaging of live cells expressing FRET constructs was performed using cells seeded in eight-well, ibiTreat microscopy chambers (Ibidi, Munich, Germany) and cultured in phenol red-free DMEM or McCoy’s 5A media for 24–48 h. The adherent cells in the wells were washed with PBS just before data acquisition. Cells were imaged on a Zeiss Axiovert 200M (Zeiss, Thornwood, NY, USA) inverted microscope with a Zeiss Plan-Apochromat × 63 oil immersion objective (1.4NA). The fluorescence microscope was also equipped with an environmental chamber with temperature and CO2 controlled to 37°C and 5%, respectively. Images were taken with a cooled CCD camera (Cascade 512b; Photometrics, Tucson, AZ, USA) controlled by AxioVision 4.8 software (Zeiss). FRET imaging was performed with FRET, CFP and YFP channels using ‘three-cube FRET’ fluorescence microscopy.40 All three channels were purchased as filter sets (Zeiss). Briefly, the FRET channel was defined by a 436/20 nm excitation filter, a 458 nm dichroic mirror and an emission 535/30 nm filter. Similarly, the CFP channel filter parameters were 436/20 nm excitation, 458 nm dichroic and 480/40 nm emission. The YFP cube contained a 500/20 nm excitation filter with a 515 nm dichroic mirror and an emission filter at 535/30 nm. The excitation light was provided by an EXFO X-Cite 120 metal halide lamp operated at 12% iris to reduce photobleaching. Exposure times were set at 100–400 ms, and images were taken every 15 or 30 s. Fluorescence images were background-corrected by manual selection of cell-free regions and processed as described earlier.41
Cells expressing roGFP1-iL were cultured as described above and optical sections were captured using a Zeiss LSM700 confocal microscope controlled by ZEN 2010 software. Dual excitation ratio imaging at room temperature was performed with 405 nm and 488 nm laser lines and a Plan-Apochromat ×63 oil immersion objective (1.4NA) and compiled using AxioVision 4.8 software. Each image was corrected for background by subtracting the intensity of an adjacent cell-free region.
Validation of redox probes for ER localization
CHO cells expressing erCY-RL7 or ERroGFP1-iL were cultured in ibiTreat microscopy chambers (Ibidi) in phenol red-free DMEM medium for 48 h. Before imaging, cells were washed twice with 1 × HBSS and incubated with 1 μmol/L ER-Tracker Red (Invitrogen) at 37°C for 30 min. Finally, cells were washed and imaged on a Zeiss LSM710 confocal microscope equipped with a Plan-Apochromat × 63 oil immersion objective (1.4NA) and controlled by ZEN 2010 software (Zeiss). To confirm spatial localization, images of the sensors were taken with a 488 nm laser line and emission with 500–590 nm and images of the ER-Tracker Red were taken with a 594 nm laser line and emission over 600 nm. Images were deconvoluted with AutoQuant X software (MediaCybernetics, Bethesda, MD, USA) and analyzed with Imaris software (Bitplane, South Windsor, CT, USA). A single optical slice out of 46 is present in the figures.
Intracellular analysis by flow cytometry
CHO cells were cultured in 75 cm2 tissue culture flasks to confluence. Control flasks went untreated while others were pretreated either with 5 mmol/L N-acetylcysteine (NAC) or 0.1 mmol/L l-buthionine-sulfoximine (BSO) for 24 h. Cells were then trypsinized, centrifuged and resuspended in HBSS to 2.0 ×106 cells/mL. HBSS was supplemented with NAC or BSO at concentrations used in pretreatments. Cells were treated with either 2 mmol/L diamide, 2 mmol/L DTT, 1 mmol/L 1-chloro-2,4-dinitrobenzene (CDNB) or 1 mmol/L H2O2 for 5–30 min at room temperature before data collection. FRET measurements were taken after each treatment using the LSR II (Becton Dickinson, San Jose, CA, USA) instrument as described earlier.36 The relative amount of reduced erCY-RL7 (RRL7) was calculated according to the following equation:
where FRET is the ratio of FRET at steady state (excited at 405 nm) to YFP fluorescence (excited at 488 nm). YFP fluorescence serves as a normalizing factor. FRETox and FRETred are normalized FRET signals of fully oxidized or reduced erCY-RL7 that were determined by incubating CHO cells with diamide or DTT, respectively.
Ratiometric measurements (405/488 nm) for roGFP1-iL were collected using the same LSR II with emission of 530/30 bandpass filters in the primary 405 and 488 nm laser pathways. All data were analyzed using FACS Express Version 3 (De Novo Software, Thornhill, Ontario, Canada; FACS, fluorescence-activated cell sorting). The relative amount of reduced ERroGFP1-iL (RiL) was calculated as described elsewhere.29,34 Briefly, the relative amount of reduced ERroGFP1-iL was calculated from the ratio of reduced to oxidized ERroGFP1-iL according to the following equation:
where F, Fox and Fred are the 405:488 nm excitation ratios at steady state, fully oxidized and fully reduced, respectively. Iox and Ired are the fluorescence intensities at 488 nm for the fully oxidized and fully reduced samples. The fully oxidized and fully reduced states of the roGFP1-iL were obtained after adding diamide or DTT at designated concentrations.
Determination of reduction potential
The reduction potential (E0′) for given redox sensor can then be calculated by using the Nernst equation as described elsewhere.34 Briefly, by substitution RT/nF evaluated at 298 K, the reduction potential of CY-RL7 was calculated according to the equation:
where E0′CY-RL7 is the midpoint potential for CY-RL7 equal to −143 mV.37
GSH measurement
To measure GSH content in cultured cells, membrane-permeant monochlorobimane (mBCl) was used as described elsewhere.42 The method is based on the formation of specific fluorescent GSH-mBCl adducts by intracellular glutathione-S-transferases.43 Briefly, cells in HBSS were incubated with 40 μmol/L mBCl at room temperature for 10 min followed by immediate cytometric analysis on the BD LSR II instrument. A violet 405 nm laser line at 25 mV and a 450/50 bandpass filter were used to collect geometric mean cellular fluorescence.
Statistical analysis
Results are expressed as mean ± SD. Duncan’s and Bonferroni tests for multiple pairwise comparisons were used to determine the significant differences between the treatment means. These analyses were carried out using XLStat program (v 2011.4.01; Addinsoft, New York, NY, USA). Differences were considered significant if P < 0.05.
Results
Sensitivity of the FRET-based redox sensor in live cells
Previous studies demonstrated the efficacy of α-helical polypeptide linkers containing redox switches composed of dispersed cysteine residues.36 Recently, a novel FRET-based redox-sensitive sensor ECFP-RL7-EYFP (CY-RL7) with improved dynamic range in vitro was developed (Supplementary Figure 1a).37 Here, we present the validation of the CY-RL7 probe by live cell imaging and demonstrate that the sensitivity of the intracellular FRET-based thiol biosensor is exclusively due to the nature of the redox linker (Figures 1a and c). For that purpose, the construct ECFP-P14-EYFP (CY-P14) was used as a control (Figures 1b and d). The P14 linker is comprised of 14 proline residues and the CY-P14 construct has been previously used in vitro assays (Supplementary Figure 1b).36 To generate oxidative or reductive insult, stably transfected CHO cells were treated sequentially with diamide and DTT at concentrations of 1 mmol/L (Figures 1c and d). The CY-RL7 biosensor at steady state is characterized by relatively low normalized net FRET (NFRET), reflective of a fully reduced state that cannot change after reductive insult with DTT (data not shown).
Figure 1.
Time-lapse FRET response of the CY-RL7 sensor (a, c) and CY-P14 construct (b, d) expressed in CHO cells to sequential treatment with exogenous oxidant and reductant. Each trace within panels (c) and (d) indicates a separate cell. Diamide (vertical solid line) and DTT (vertical dashed line) were added to cells at concentrations of 1 mmol/L. The data are representative of six independent experiments using a minimum of three ROIs. CHO, Chinese hamster ovary; DTT, dithiothreitol; FRET, Förster resonance energy transfer. (A color version of this figure is available in the online journal)
In order to visualize the full dynamic range, an oxidative insult was provided by diamide followed by DTT (Figure 1c). Cell oxidation with diamide occurred very fast, in the time frame of mere seconds, which is evidenced by the rapid FRET increase of the sensor (Figure 1c). The slope of the sensor’s response to DTT treatment is dose-dependent to this reductant (not shown). Responses to diamide and DTT were not observed for the negative control CY-P14 construct, confirming that all observed changes are due to the specific redox linker RL7 (Figure 1d). These data demonstrate that CY-RL7 expressed in the cytosol is fully reduced, as would be expected in a normally reductive intracellular environment.
Sensor targeting cytosol responds to endogenous modulation of GSH level
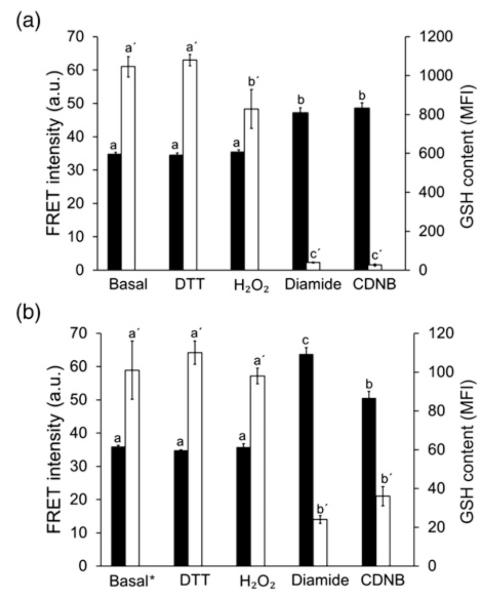
While diamide significantly depletes intracellular GSH levels in a short time, it may also directly oxidize the biosensor in live cells as it does in vitro.37 To test whether the sensor CY-RL7 responds to changes of cytosolic glutathione status, the cellular GSH pool was depleted by incubation with 1 mmol/L CDNB, an electrophilic xenobiotic. Both the changes in the FRET signal and the relative intracellular GSH pool were monitored. In the cell, CDNB is enzymatically detoxified via conjugation to GSH, resulting in rapid and complete depletion of the cellular GSH pool.17 Full oxidation of the biosensor with CDNB (Figure 2a) may highlight direct specificity of CY-RL7 sensor to the glutathione redox potential. Both diamide and CDNB treatments were similar in action for both biosensor response and depletion of intracellular GSH. On the other hand, no significant change in FRET signal was observed in cells exposed to oxidation with hydrogen peroxide (H2O2). This result is in agreement with a relatively modest decrease in the GSH level in response to H2O2 treatment (Figure 2a).
Figure 2.
Effects of various treatments on both FRET signal response (black bars, left y-axis) of the CY-RL7 probe targeting cytosol and GSH content (white bars, right y-axis) in CHO cells cultured (a) at standard conditions and (b) in the presence of 0.1 mmol/L BSO for 24 h. Intracellular GSH content, determined with mBCl, is expressed by the geometric mean fluorescence intensity (MFI). Note that GSH content at basal* state in the bar graph (b) corresponds to cells pretreated with BSO and differs from content at basal state in (a). The data represent means ± SD (n = 3). Statistical differences are indicated by different letters. Bars without a common letter are significantly different (P < 0.05). CHO, Chinese hamster ovary; FRET, Förster resonance energy transfer; GSH, glutathione; mBCl, monochlorobimane; BSO, l-buthionine-sulfoximine; DTT, dithiothreitol; CDNB, 1-chloro-2,4-dinitrobenzene
In addition to CDNB, intracellular GSH content was modulated with BSO, another commonly used glutathione depletor, which inhibits GSH synthesis with high specificity.44 In spite of the 10-fold drop in GSH levels of CHO cells pretreated with 0.1 mmol/L BSO for 24 h, the FRET signal was unchanged at basal state (Figure 2b). Additional treatment with H2O2 had no or little effect on both GSH level and the CY-RL7 response. The CY-RL7 sensor also effectively responded to diamide and CDNB in BSO-pretreated cells. Surprisingly, the cytosolic content of GSH affected the dynamic range of the sensor. The decreased GSH level caused by BSO significantly altered the FRET signal in response to diamide (Figure 2b). The increase in the FRET signal was almost doubled in comparison to cells not treated with BSO and consequently the dynamic range of the probe was nearly doubled in BSO-treated cells. At the same time, the magnitude of the FRET signal increase in response to CDNB treatment was independent of BSO pretreatment.
Analysis of the ER redox environment with the FRET probe
To observe time-resolved redox changes in the ER in response to altered environmental conditions, the calreticulin targeting signal and KDEL retention signal were added to the CY-RL7 gene. To verify that CY-RL7 was in fact targeted to the ER (erCY-RL7), the construct was expressed in CHO cells that were subsequently incubated with ER-Tracker Red and examined on a laser scanning confocal microscope (Figures 3a–d). Indeed, our FRET sensor exits the ribosome in the reduced state with post-translational delivery to the ER where the probe reacts with ER constituents to attain a steady-state level of oxidation. In contrast to the fully reduced state of the CY-RL7 sensor expressed in the cytosol, the erCY-RL7 sensor was found in a highly oxidized state. That is evidenced by the treatment of cells with 1 mmol/L diamide resulting in only a moderately increased FRET signal (Figure 3g). The alteration in FRET was not very pronounced because the oxidative ER environment presumably promotes photolysis of EYFP, which could be neutralized by the spike of reductant (Figure 3h). Next, to improve performance of CY-RL7 in the ER, we replaced EYFP with Venus, the brighter YFP variant which has a more efficient maturation as well as a reduced pH and halide sensitivity.45 Despite this attempt, we did not observe noticeable improvement in the read-out of the ECFP-RL7-Venus sensor expressed in the ER environment of CHO cells (data not shown). Accordingly, all data on changes in glutathione redox potentials within the ER were collected with the FRET sensor erCY-RL7. The main alteration in FRET signal was achieved with application of DTT at 1 mmol/L, providing additional confirmation of the highly oxidative state of the sensor in the ER (Figure 3h). It should again be noted that the dynamic range of the sensor in the ER was twice of that found in cytosol (Figures 1c and 3h).
Figure 3.
Visual confirmation of spatial localization and monitoring of the sensor erCY-RL7 responsiveness to diamide and DTT treatments. Images of CHO cells were taken for (a) the sensor localization with 488 nm excitation and 500–590 nm emission; (b) the ER-Tracker Red dye with 594 nm excitation and emission over 600 nm; (c) merge (overlay a , b); and (d) co-localized pixels from data volume. Time-lapse FRET response of CY-RL7 targeted to the ER of CHO cells treated with diamide (e, g) and DTT (f, h). Each trace within panels (g) and (h) indicates a separate cell. Diamide (vertical solid line) and DTT (vertical dashed line) were added to cells at concentrations of 1 mmol/L. The data are representative of four independent experiments using a minimum of four ROIs. CHO, Chinese hamster ovary; DTT, dithiothreitol; FRET, Förster resonance energy transfer; ER, endoplasmic reticulum
FRET sensor targeting ER responds to GSH synthesis in both non-transformed and transformed cells
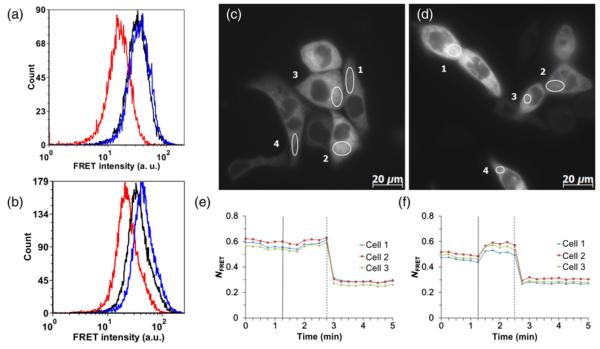
To test the specificity of the sensor to the glutathione couple, stably transfected cells were treated with NAC, a substrate for cytosolic synthesis of GSH. To obtain quantitative data, we employed flow cytometry that allows the rapid collection of data from thousands of individual cells, providing a large data-set, which confers a statistical advantage over microscopy with its cell-to-cell variability. Changes in FRET signal of untreated cells versus cells pretreated with 5 mmol/L NAC were measured under oxidative and reductive insults provided by 2 mmol/L diamide and 1 mmol/L DTT. Data collected on NAC-free cells show that the sensor at steady state (black line) exhibits only a minor increase in FRET signal after treatment with diamide (blue line) and a large decline in FRET after reduction (red line) with DTT (Figure 4a). On the other hand, in NAC-pretreated cells, the sensor became less oxidized by shifting its FRET signal at a basal state (black line peak) from the fully oxidized state (blue line peak) towards the fully reduced state (red line peak) (Figure 4b). The increase in degree of sensor reduction in response to NAC treatment was approximately two-fold and it was consistently observed for all experiments (Table 1). At the same time, the oxidation level of erCY-RL7 was not changed after cell incubation with BSO, an inhibitor of GSH biosynthesis (data not shown).
Figure 4.
Representative histograms of flow cytometric data for the CY-RL7 probe targeted to the ER of CHO cells non-treated (a) and pretreated (b) with 5 mmol/L NAC for 24 h. The black line corresponds to basal level and red and blue lines correspond to treatment with 1 mmol/L DTT or 2 mmol/L diamide, respectively. The ordinate (‘Count’) is the number of cells counted, and the abscissa (‘FRET intensity’) is the fluorescence intensity excited at 405 nm and collected through a 525 nm filter with a halfwidth of 25 nm. Time-lapse response of CY-RL7 targeted to the ER of HCT116 cells cultured without (c, e) and with (d, f) 5 mmol/L NAC to a sequential treatment with an exogenous oxidant and reductant. Each trace within panels (e) and (f) indicates a separate cell. Diamide (vertical solid line) and DTT (vertical dashed line) were added to cells at concentrations of 1 mmol/L. The data are representative of six independent experiments using a minimum of three ROIs. ER, endoplasmic reticulum; CHO, Chinese hamster ovary; NAC, N-acetylcysteine; DTT, dithiothreitol; FRET, Förster resonance energy transfer
Table 1.
The erCY-RL7 sensor expressed in CHO cells responds to NAC treatment by increase in reduced form
| Treatment | Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | Average | SD |
|---|---|---|---|---|---|---|
| Basal | 12.5 | 13.4 | 10.9 | 11.4 | 12.1 | 1.1 |
| NAC | 31.6 | 31.3 | 20.9 | 28.9 | 28.2 | 5.0 |
| Δ | 19.1 | 17.9 | 10 | 17.5 | 16.1 | 4.1 |
CHO, Chinese hamster ovary; NAC, N-acetylcysteine
Fractions of erCY-RL7 in reduced form (%) in CHO cells before (basal) and after treatment with 5 mmol/L NAC for 24 hr were determined as described in Materials and methods. Δ Denotes increase in the probe’s reduced form after NAC treatment. Data for each experiment were collected by flow cytometry and represent an average of three replicates collected on 10,000 cells±SD
To demonstrate the efficacy of the erCY-RL7 in application to tumor cells and the sensor’s broad base response to a NAC-modulated increase of the cellular GSH pool, HCT116 cells were transiently transfected with the probe. In control cells, the sensor at steady state was observed to be in a highly oxidized state (Figures 4c and e). The increase in the FRET signal was modest, but present in all HCT116 cells in response to treatment with diamide at 1 mmol/L. Alternatively, in NAC-pretreated cells, an increase in the FRET signal was much stronger in response to diamide (Figures 4d and f). The larger change of FRET signal in response to an oxidative insult indicates the more reductive state of the probe at steady state. Thus, the redox state of the erCY-RL7 probe is responsive to an elevated cellular GSH pool in both transformed and non-transformed mammalian cells. The dynamic range of the probe was similar in ER environments of both CHO and HCT116 cells. The midpoint potential of −143 mV previously determined for the CY-RL7 sensor enabled a quantitative estimate of the redox state of the ER.34,37 Based on the assumptions that the steady state equilibrium of the probe expressed in ER is oxidized at ≈88% and is specifically managed by the GSH/GSSG couple, the ER redox state of mammalian cells is estimated to be around −118 mV at pH 7.0.
Analysis of the ER redox environment with the roGFP1-iL probe
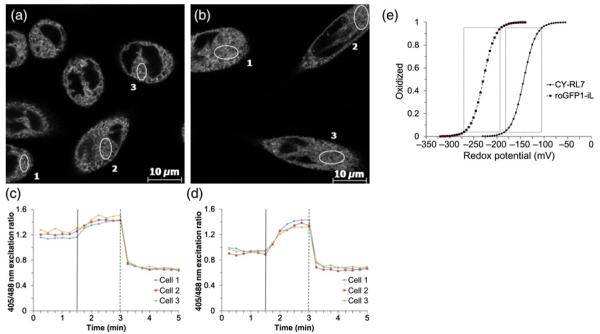
Recently developed roGFP1-iX probes with lowered thermodynamic stability have exhibited moderately high midpoint potentials suitable for relatively oxidizing subcellular environments.34 It was demonstrated that roGFP1-iE and roGFP1-iL probes were only 50–55% reduced in the ER of live yeast and mammalian cells.29,35 As a result, the ER redox potential was estimated in the range of −230 to −240 mV. Knowing that both the CY-RL7 and roGFP1-iX probes were developed for detecting changes in the redox potentials of the GSH/GSSG couple in live cells, a considerable difference in read-outs of FRET-based and roGFP1-iX probes motivated us to test the specificity of ERroGFP1-iL to alterations in intracellular GSH content. Initial data collected by flow cytometry on CHO cells stably expressing roGFP1-iL in ER (ERroGFP1-iL) revealed that the redox status of the probe at steady state is close to recently reported values (Table 2).29,35 Temporal response of ERroGFP1-iL to redox changes in live cells was acquired on a laser scanning confocal microscope. The typical profiles of the ERroGFP1-iL at steady state and its response to sequential acute insults with a reductant and an oxidant are shown in Figures 5c and d. The steady-state 405/488 nm ratio of fluorescence, which reflects the redox equilibrium between the sensor and the ER, often varied between samples of the same experiment (Figures 5c and d). Variability observed in excitation ratios taken at steady state between experiments may be explained by changes to the redox status during the cell cycle, differences in cell density or light-induced changes.29,46
Table 2.
Comparison of the redox states of ERroGFP1 expressed in CHO cells cultured with and without alteration of intracellular GSH content
| Treatment | Exp. 1 | Exp. 2 | Exp. 3 | Exp.4 | Exp. 5 | Exp. 6 | Average | SD |
|---|---|---|---|---|---|---|---|---|
| Basal | 47.6 | 54.4 | 47.5 | 50.4 | 38.6 | 46 | 47.4 | 5.2 |
| NAC | 64.6 | N/A | 38.4 | 51.5 | 38 | 39.5 | 46.4 | 10.4 |
| BSO | N/A | 47 | 41.4 | 43.9 | 39.9 | 36.5 | 41.7 | 3.6 |
CHO, Chinese hamster ovary; GSH, glutathione; NAC, N-acetylcysteine; BSO, l-buthionine-sulfoximine
Fractions of ERroGFP1-iL in reduced form (%) in cells before (basal) and after treatment with either 5 mmol/L NAC or 0.1 mmol/L BSO (BSO) were determined as described in Materials and methods. Data for each experiment were collected by flow cytometry and represent an average of three replicates collected on 10,000 cells±SD
Figure 5.
Representative images of the roGFP1-iL sensor targeted to the ER of CHO cells. (c, d) Corresponding time-lapse responses of 405/488 nm ratio to sequential treatment with diamide (vertical solid line) and DTT (vertical dashed line) added to cells at concentrations of 1 and 10 mmol/L, respectively. The data are representative of six independent experiments using a minimum of three ROIs. (e) Relationship between the redox potentials in mV and the fractions of CY-RL7 (solid line) and roGFP1-iL (dashed line) in oxidized form. Boxes denote changes in probe fluorescence from 5 to 95% oxidation, which translates to an effective measuring range equal to a given probe midpoint potential ± 40 mV. ER, endoplasmic reticulum; CHO, Chinese hamster ovary; DTT, dithiothreitol
According to data acquired by flow cytometry, the ER redox state is very close to the middle point potential of ERroGFP1-iL where the probe has the highest sensitivity (Table 2). Therefore, ERroGFP1-iL was expected to be more sensitive to alterations in the ER redox state than erCY-RL7, which is approximately 88% oxidized at a basal state. Next, to examine responsiveness of ERroGFP1-iL to endogenous perturbations in GSH status, CHO cells were treated with 5 mmol/L NAC and 0.1 mmol/L BSO for 24 h. The flow cytometry data revealed large alterations in the GSH levels of cells pretreated with NAC and BSO (Supplementary Figure 2). Yet, significant differences were not observed in the redox level of the probe at steady state in response to reductive or oxidative insults with NAC and BSO (Table 2). Nevertheless, CDNB treatment at 1 mmol/L provided an oxidative insult similar to diamide (Supplementary Figure 3). Hydrogen peroxide at 1 mmol/L was as effective as the oxidants CDNB and diamide in spite of a relatively small decrease in GSH level (Supplementary Figures 2 and 3). Thus, the ERroGFP1-iL response to H2O2 does not follow alterations in GSH content.
Discussion
In this work, we exploited a recently developed FRET-based redox probe for live cell imaging in two subcellular environments, the cytosol and ER. The data obtained by this non-invasive redox-sensitive probe provide unambiguous evidence that redox potentials within these two intracellular compartments are at a non-equilibrium steady state with respect to each other. In the cytosol, the redox sensor is fully reduced at its steady state, reflecting the inherently reductive environment of living cells. Furthermore, the CY-RL7 probe responds to intracellular GSH deprivation with CDNB, but not with BSO. The absence of the sensor’s response to BSO-mediated GSH depletion may reflect the cells’ ability to maintain a cytosolic potential in response to acute oxidative insults. It is of interest to note that the probe equilibrates with the glutathione redox potential and not intracellular GSH concentrations. On the other hand, the midpoint potential of approximately −143 mV implies that the FRET probe becomes saturated below −190 mV in the cytosolic reductive environment, which ranged from −280 to −320 mV.11,15
Because of its strongly oxidizing potential, the CY-RL7 probe provides an ideal tool for monitoring intraorganellar glutathione potentials in more oxidative environments such as the ER, Golgi apparatus and lysosomes. Only 12% of the FRET-based probe targeting the ER of CHO cells was in a reduced form, which implies approximately −118 mV for the ER glutathione potential at pH 7.0. Furthermore, we observed a significant alteration in the level of probe reduction after induction of glutathione biosynthesis with NAC (Table 1). Specifically, NAC-mediated increase in the degree of probe reduction from 12% to 28% reflects changes in the ER redox state of the GSH/GSSG couple from −118 to −131 mV at pH 7.0. However, the biosensor’s quantitative read-out of redox potential in mV is based on a number of assumptions, such as pH, and therefore cannot be taken as an absolute value.
Besides the pH-dependent nature of redox potential values, the most intriguing question relates to the specificity of the current redox probe within the ER. In contrast to the roGFPs, the CY-RL7 probe was not sensitive to H2O2 either in vitro or within cultured cells.37 Interestingly, the fluorescence signal of the FRET sensor was not altered by H2O2 even in CHO cells with a low glutathione content mediated by BSO (Figure 2). That the CY-RL7 probe is specific to GSH/GSSG pair in vitro, however, does not necessarily indicate the same mode of specificity in living cells, specifically in the ER where thiol/disulfide exchange is different from the cytosol. Therefore, the observation of a two-fold increase of the reduced form of the erCY-RL7 probe in CHO cells after NAC-modulated GSH increase may be considered as key evidence of probe specificity for the glutathione redox potential. Real-time visualization of redox changes of erCY-RL7 in the transformed HCT116 cell line revealed the same pattern in response to NAC treatment (Figures 4e and f).
Additionally, the dynamic range of erCY-RL7 is twice as high as CY-RL7. Moreover, the dynamic range of the erCY-RL7 was not altered in cells treated with BSO, also in contrast to the cytosolic probe. As such, the CY-RL7 probe exhibits a similar dynamic range in the ER and cytosol for CHO cells pretreated with BSO. This observation may indicate a lower buffering capacity of the GSH/GSSG couple in the ER versus the cytosol.
Real-time monitoring of redox changes in the ER of yeast and mammalian cells was reported recently with roGFP1-iX probes.29,35 The probes at steady state are near equilibrium between oxidized and reduced forms and are suited for monitoring subtle changes in the ER redox status in response to both reductive and oxidative insults. Moreover, the reduction potential of the ER of eukaryotic cells was determined to be in the range of −230 to −240 mV.29,35 Thus, FRET-based erCY-RL7 and ERroGFP1-iL sensors provide differences of more than 100 mV in measurement of the ER redox potential at steady state. Although the exact reason(s) behind this difference is not known, it may highlight a differential ability of the two probes to respond to subtle changes in redox couples other than the glutathione buffer.
We also monitored redox changes with the roGFP1-iL sensor targeting the ER of live cells to better understand the differential read-outs of erCY-RL7 and ERroGFP1-iL. It was expected that ERroGFP1-iL, being equilibrated within the ER near its midpoint redox potential, would be equally sensitive to oxidative or reductive insults. This consideration provides a simple assay to test probe specificity in the ER. Surprisingly, neither BSO-mediated depletion nor a NAC-stimulated increase of the total cellular GSH content affected the redox status of the probe (Table 2). In contrast to ERroGFP1-iL, the reduced form of erCY-RL7 was doubled in response to a NAC-mediated increase of the GSH pool. The same signal peptide and retention signals were used to target CY-RL7 and roGFP1-iL to the ER and both of their ER localizations were confirmed, indicating that their differential read-outs were not likely a result of dissimilar compartmentalization (Figures 3a-d and Supplementary Figure 4). Additionally, the responsiveness of ERroGFP1-iL was not specific to alterations in GSH content. For instance, ERroGFP1-iL was fully oxidized in response to H2O2 despite only a modest decrease of intracellular GSH. In contrast, the probe was not sensitive to an oxidative insult with BSO, a strong GSH depletor that decreases cellular glutathione levels to 10% of control values. Nonetheless, both erCY-RL7 and ERroGFP1-iL exhibited similar responses to an oxidative insult induced with CDNB, which depletes cellular GSH by conjugation in a manner of minutes. Defining the biochemical basis for the differential sensitivity of erCY-RL7 and ERroGFP1-iL may reveal novel mechanisms mediating redox homeostasis in the ER.
In spite of the insensitivity to H2O2, the erCY-RL7 probe is more oxidized at its steady state than the H2O2-sensitive ERroGFP1-iL. This is consistent with recent observations that H2O2 produced in the ER is efficiently metabolized by the ER-localized enzyme peroxiredoxin IV.47 Furthermore, the CY-RL7 probe with its more oxidized midpoint potential would be assumed to be fully reduced in an environment close to the midpoint potential of the roGFP-iL probe (Figure 5e). Thus, the large difference in read-outs of the ER redox state between the two redox sensors cannot be explained by their distinct sensitivity to H2O2 or their differences in midpoint redox potentials. Together, these findings indicate that while the CY-RL7 and roGFP1-iL probes can measure redox potentials of the ER, their specificity may extend beyond the GSH/GSSG couple to other pathways that contribute to ER thiol/disulfide exchange. The recent observation that BSO-mediated depletion of cellular GSH in HEK293 cells does not alter the redox state of certain members of the PDI family indeed indicates that mechanism(s) other than ER export of disulfides or import of thiols underlie ER redox balance.26
In summary, the present data demonstrate that the FRET-based erCY-RL7 probe can be utilized for monitoring the redox status of the very oxidizing environment of the ER. The CY-RL7 probe targeted to the ER of both non-transformed and transformed cells responded to moderate increases in GSH, which were mediated by NAC, a substrate for GSH synthesis. Additionally, we estimated the ER glutathione potential to be approximately −118 mV at pH 7.0 or approximately −141 mV assuming pH 7.4.18,48,49 Finally, after demonstrating that both erCY-RL7 and ERroGFP1-iL respond to glutathione depletion by CDNB, but exhibit a differential response to H2O2, it became evident that yet another redox pathway(s) may contribute to the distinct read-out of the ER redox status by these two probes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Barbara Pilas and Ben Montez at the Flow Cytometry Facility of the University of Illinois Biotechnology Center as well as Dr Shiv Sivaguru at the Institute for Genomic Biology for assistance with fluorescence microscopy.
This work was supported by NIH grant R33-CA137719 to HRG and PJAK.
Footnotes
Author contributions: All authors participated in the interpretation of the studies, analysis of the data and review of the manuscript. VLK, PJAK and HRG participated in the experimental design; VLK, MTL, AC and BMS conducted the experiments; and VLK and HRG wrote the manuscript.
REFERENCES
- 1.Palumaa P. Biological Redox Switches. Antioxid Redox Signal. 2009;11:981–3. doi: 10.1089/ars.2009.2468. [DOI] [PubMed] [Google Scholar]
- 2.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 3.Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–79. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 4.Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/ disulfide redox systems: a perspective on redox systems biology. Free Radic Biol Med. 2008;44:921–37. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones DP. Redox sensing: orthogonal control in cell cycle and apoptosis signalling. J Inter Med. 2010;268:432–48. doi: 10.1111/j.1365-2796.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banhegyi G, Benedetti A, Csala M, Mandl J. Stress on redox. FEBS Lett. 2007;581:3634–40. doi: 10.1016/j.febslet.2007.04.028. [DOI] [PubMed] [Google Scholar]
- 7.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic-reticulum. Science. 1992;257:1496–502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 8.Bass R, Ruddock LW, Klappa P, Freedman RB. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J Biol Chem. 2004;279:5257–62. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- 9.Dixon BM, Heath SHD, Kim R, Suh JH, Hagen TM. Assessment of endoplasmic reticulum glutathione redox status is confounded by extensive ex vivo oxidation. Antioxid Redox Signal. 2008;10:963–72. doi: 10.1089/ars.2007.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjornberg O, Ostergaard H, Winther JR. Measuring intracellular redox conditions using GFP-based sensors. Antioxid Redox Signal. 2006;8:354–61. doi: 10.1089/ars.2006.8.354. [DOI] [PubMed] [Google Scholar]
- 11.Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal. 2010;13:621–50. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 12.Oku M, Sakai Y. Assessment of physiological redox state with novel FRET protein probes. Antioxid Redox Signal. 2012;16:698–704. doi: 10.1089/ars.2011.4251. [DOI] [PubMed] [Google Scholar]
- 13.Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–62. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–53. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 15.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279:22284–93. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 16.Jiang K, Schwarzer C, Lally E, Zhang S, Ruzin S, Machen T, Remington SJ, Feldman L. Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 2006;141:397–403. doi: 10.1104/pp.106.078246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007;52:973–86. doi: 10.1111/j.1365-313X.2007.03280.x. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzer C, Illek B, Suh JH, Remington SJ, Fischer H, Machen TE. Organelle redox of CF and CFTR-corrected airway epithelia. Free Radic Biol Med. 2007;43:300–16. doi: 10.1016/j.freeradbiomed.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yano T, Oku M, Akeyama N, Itoyama A, Yurimoto H, Kuge S, Fujiki Y, Sakai Y. A novel fluorescent sensor protein for visualization of redox states in the cytoplasm and in peroxisomes. Mol Cell Biol. 2010;30:3758–66. doi: 10.1128/MCB.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivashchenko O, Van Veldhoven PP, Brees C, Ho YS, Terlecky SR, Fransen M. Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Mol Biol Cell. 2011;22:1440–51. doi: 10.1091/mbc.E10-11-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–47. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–4. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- 23.Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–5. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csala M, Margittai E, Banhegyi G. Redox control of endoplasmic reticulum function. Antioxid Redox Signal. 2010;13:77–108. doi: 10.1089/ars.2009.2529. [DOI] [PubMed] [Google Scholar]
- 25.Appenzeller-Herzog C. Glutathione-and non-glutathione-based oxidant control in the endoplasmic reticulum. J Cell Sci. 2011;124:847–55. doi: 10.1242/jcs.080895. [DOI] [PubMed] [Google Scholar]
- 26.Appenzeller-Herzog C, Riemer J, Zito E, Chin KT, Ron D, Spiess M, Ellgaard L. Disulphide production by Ero1 alpha-PDI relay is rapid and effectively regulated. EMBO J. 2010;29:3318–29. doi: 10.1038/emboj.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lappi AK, Ruddock LW. Reexamination of the role of interplay between glutathione and protein disulfide isomerase. J Mol Biol. 2011;409:238–49. doi: 10.1016/j.jmb.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 28.Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–4. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- 29.van Lith M, Tiwari S, Pediani J, Milligan G, Bulleid NJ. Real-time monitoring of redox changes in the mammalian endoplasmic reticulum. J Cell Sci. 2011;124:2349–56. doi: 10.1242/jcs.085530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salminen A, Kaarniranta K. ER stress and hormetic regulation of the aging process. Ageing Res Rev. 2010;9:211–7. doi: 10.1016/j.arr.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–92. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–58. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 33.Matus S, Glimcher LH, Hetz C. Protein folding stress in neurodegenerative diseases: a glimpse into the ER. Curr Opin Cell Biol. 2011;23:239–52. doi: 10.1016/j.ceb.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Lohman JR, Remington SJ. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry. 2008;47:8678–88. doi: 10.1021/bi800498g. [DOI] [PubMed] [Google Scholar]
- 35.Delic M, Mattanovich D, Gasser B. Monitoring intracellular redox conditions in the endoplasmic reticulum of living yeasts. FEMS Microbiol Lett. 2010;306:61–6. doi: 10.1111/j.1574-6968.2010.01935.x. [DOI] [PubMed] [Google Scholar]
- 36.Kolossov VL, Spring BQ, Sokolowski A, Conour JE, Clegg RM, Kenis PJA, Gaskins HR. Engineering redox-sensitive linkers for genetically encoded FRET-based biosensors. Exp Biol Med. 2008;233:238–48. doi: 10.3181/0707-RM-192. [DOI] [PubMed] [Google Scholar]
- 37.Kolossov VL, Spring BQ, Clegg RM, Henry JJ, Sokolowski A, Kenis PJA, Gaskins HR. Development of a high-dynamic range, GFP-based FRET probe sensitive to oxidative microenvironments. Exp Biol Med. 2011;236:681–91. doi: 10.1258/ebm.2011.011009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci USA. 2008;105:19264–9. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osibow K, Malli R, Kostner GM, Graier WF. A new type of non-Ca2+-buffering Apo(a)-based fluorescent indicator for intraluminal Ca2+ in the endoplasmic reticulum. J Biol Chem. 2006;281:5017–25. doi: 10.1074/jbc.M508583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia ZP, Liu YH. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys J. 2001;81:2395–402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C, Kolossov VL, Tsvid G, Trump L, Henry JJ, Henderson JL, Rund LA, Kenis PJA, Schook LB, Gaskins HR, Timp G. Imaging in real-time with FRET the redox responses of tumorigenic cells to glutathione petrubations in a microscale flow. Integr Biol. 2011;3:208–17. doi: 10.1039/c0ib00071j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice GC, Bump EA, Shrieve DC, Lee W, Kovacs M. Quantitative analysis of cellular glutathione by flow cytometry utilizing monochlorobimane – some applications to radiation and drug resistance in vitro and in vivo. Cancer Res. 1986;46:6105–10. [PubMed] [Google Scholar]
- 43.Fernandezcheca JC, Kaplowitz N. The use of monochlorobimane to determine hepatic GSh levels and synthesis. Anal Biochem. 1990;190:212–9. doi: 10.1016/0003-2697(90)90183-a. [DOI] [PubMed] [Google Scholar]
- 44.Bertsche U, Schorn H. Glutathione depletion by DL_Buthionine-SR-Sulfoximine (BSO) potentiates x-ray induced chromosome lesions after liquid holding recovery. Radiat Res. 1986;105:351–69. [PubMed] [Google Scholar]
- 45.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotech. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 46.Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–9. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 47.Tavender TJ, Springate JJ, Bulleid NJ. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 2010;29:4185–97. doi: 10.1038/emboj.2010.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paroutis P, Touret N, Grinstein S. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology. 2004;19:207–15. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- 49.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.