Abstract
Saccharomyces cerevisiae Actin-Binding Protein 1 (Abp1p) is a member of the Abp1 family of proteins, which are in diverse organisms including fungi, nematodes, flies, and mammals. All proteins in this family possess an N-terminal Actin Depolymerizing Factor Homology (ADF-H) domain, a central Proline-Rich Region (PRR), and a C-terminal SH3 domain. In this study, we employed sequence analysis to identify additional conserved features of the family, including sequences rich in proline, glutamic acid, serine, and threonine amino acids (PEST), which are found in all family members examined, and two motifs, Conserved Fungal Motifs 1 and 2 (CFM1 and CFM2), that are conserved in fungi. We also discovered that, similar to its mammalian homologs, Abp1p is phosphorylated in its PRR. This phosphorylation is mediated by the Cdc28p and Pho85p kinases, and it protects Abp1p from proteolysis mediated by the conserved PEST sequences. We provide evidence for an intramolecular interaction between the PRR region and SH3 domain that may be affected by phosphorylation. Although deletion of CFM1 alone caused no detectable phenotype in any genetic backgrounds or conditions tested, deletion of this motif resulted in a significant reduction of growth when it was combined with a deletion of the ADF-H domain. Importantly, this result demonstrates that deletion of highly conserved domains on its own may produce no phenotype unless the domains are assayed in conjunction with deletions of other functionally important elements within the same protein. Detection of this type of intragenic synthetic lethality provides an important approach for understanding the function of individual protein domains or motifs.
Keywords: Abp1p, conserved motif, intragenic synthetic lethality, phosphorylation
SACCHAROMYCES cerevisiae Acting-Binding Protein 1 (Abp1p) was the first described member of a highly conserved family of actin-binding proteins (Drubin et al. 1988) found in diverse organisms including fungi, worms, flies, and humans. The common features of these proteins are an N-terminal Actin Depolymerizing Factor Homology (ADF-H) domain (Lappalainen et al. 1998), followed by a large, mainly unstructured central region including a Pro-Rich Region (PRR) and a C-terminal SH3 domain (Figure 1). The conservation among the SH3 domains of these proteins is particularly high (e.g., the human and yeast domains are 45% identical), and they recognize very similar target peptide sequences (Stollar et al. 2009). Given the high conservation and ubiquitous occurrence of Abp1 family members, these proteins undoubtedly fulfill a critical function, and investigating these functions is an important objective. In this work, we have used yeast Abp1p as a model to gain further insight into this family.
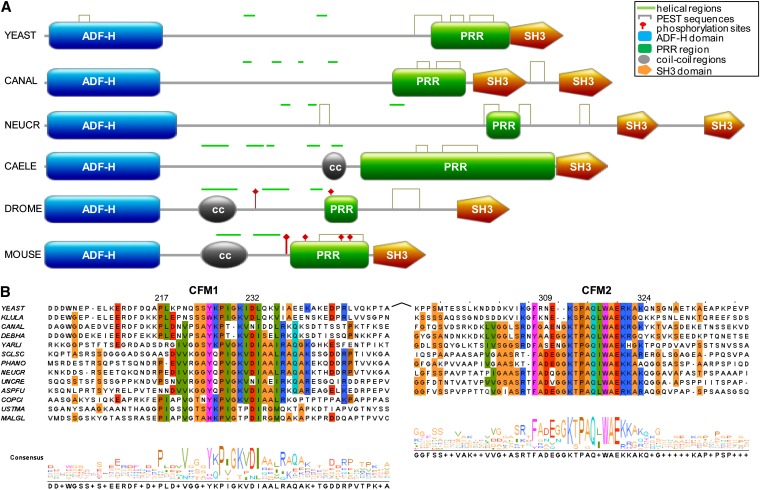
Figure 1 .
Conserved features of Abp1 family members. (A) Analysis of the domain structure of Saccharomyces cerevisiae Abp1p (YEAST) and other Abp1p homologs from different species: Candida albicans (CANAL), Neurospora crassa (NEUCR), Caenorhabditis elegans (CAELE), Drosophila melanogaster (DROME), and Mus musculus (MOUSE). Regions predicted with high probability to be PEST sequences or to be helical are indicated by gray brackets or green horizontal bars, respectively. Confirmed phosphorylation sites in the DROME and MOUSE Abp1 homologs are also indicated. PRRs and the boundaries of ADFH and SH3 domains were defined by using Scanprosite (http://prosite.expasy.org/scanprosite). (B) Alignment of Conserved Fungal Motifs (CFM1 and CFM2) from Abp1p homologs from diverse fungal species. The following groups of residues were shaded with the same colors: (C, I, L, V, M), (F, Y, W, H), (D, E), (N, Q), (R, K), (G, A, S, T), and P. Numbering is according to S. cerevisiae (YEAST) Abp1p. Other species included in the analysis are Kluyveromyces lactis (KLULA), Candida albicans (CANAL), Debaryomyces hansenii (DEBHA), Yarrowia lipolytica (YARLI), Sclerotinia sclerotiorum (SCLSC), Phaeosphaeria nodorum (PHANO), Neurospora crassa (NEUCR), Uncinocarpus reesii (UNCRE), Aspergillus fumigatus (ASPFU), Coprinopsis cinerea (COPCI), Ustilago maydis (USTMA), and Malassezia globosa (MALGL). A sequence logo, which quantifies conservation at each position in the alignment, is also shown.
Abp1p was originally identified as an actin-binding protein by actin-affinity chromatography (Drubin et al. 1988), and it has been shown to localize to cortical actin patches. Abp1p plays important roles in actin organization and endocytosis. It binds to actin filaments, but not actin monomers, mainly through the ADF-H domain (Lappalainen et al. 1998, Goode et al. 2001), and also possesses two acidic motifs that are required for binding and activation of the Arp2/3 complex (Goode et al. 2001). The SH3 domain mediates biologically relevant interactions with several other proteins involved in endocytosis, such as Ark1p, Scp1p, and Sjl2p (Lila and Drubin 1997; Fazi et al. 2002; Stefan et al. 2005; Haynes et al. 2007; Stollar et al. 2009). The mammalian homolog of Abp1p (mAbp1), similar to the yeast Abp1p, also binds F-actin with its N-terminal actin-binding domain and is involved in receptor-mediated endocytosis (Kessels et al. 2001; Mise-Omata et al. 2003). The SH3 domain mediates protein–protein interactions with proteins involved in synaptogenesis, endocytosis, and cell motility (Kessels et al. 2001; Fenster et al. 2003; Han et al. 2003; Cortesio et al. 2010). mAbp1p is recruited to dynamic actin structures (Kessels et al. 2000), and this localization is reminiscent of the localization of the yeast protein, which is found in cortical actin patches accumulating in the yeast bud but not at actin cables (Drubin et al. 1988).
Although deletion of the yeast ABP1 gene does not result in slower growth, this deletion is synthetically lethal with deletions of SAC6, SLA1, or SLA2, which also encode actin-associated components involved in endocytosis (Holtzman et al. 1993). In addition, combined deletion of ABP1 and PRK1, which encodes an actin patch-associated protein kinase, results in a temperature-sensitive phenotype (Cope et al. 1999). An interesting aspect of Abp1p function is that the in vivo requirements for its domains differ depending on the genetic background in which the assay is carried out. For example, although the SH3 domain is required in all known ABP1-dependent genetic backgrounds (Lila and Drubin 1997; Haynes et al. 2007), certain amino acid substitutions that partially decrease the affinity of this domain for its targets cause a marked reduction in viability only in the abp1Δsac6Δ and abp1Δsla2Δ backgrounds (Haynes et al. 2007). Surprisingly, deletion of the conserved ADF-H domain resulted in loss of viability in these same two backgrounds, but not in the abp1Δsla1Δ background (Quintero-Monzon et al. 2005), but the functional roles of the other regions of Abp1p under varying conditions remain unknown. Another uninvestigated aspect of Abp1p is its regulation through post-translation modification. Since mAbp1 is known to be phosphorylated by Src- and Syk-family kinases (Larbolette et al. 1999; Han et al. 2003; Larive et al. 2009; Boateng et al. 2012) and phosphorylated peptides from Abp1p have been identified in several global studies of yeast protein phosphorylation (Ficarro et al. 2002; Ubersax et al. 2003; Chi et al. 2007; Holt et al. 2009), a role for phosphorylation in the function of Abp1p might be expected.
In this study, we have analyzed the sequences of diverse Abp1 family members and identified previously unrecognized conserved motifs. By analyzing the effects of a variety of ABP1 deletion mutants assayed in several different genetic backgrounds, we have revealed functional roles for these conserved elements of Abp1p. We have also discovered that Abp1p is phosphorylated in the same region as its mammalian counterparts. Our results highlight the importance of conservation analysis and assessment of phenotypes under a variety of conditions for the elucidation of protein function.
Materials and Methods
Yeast strains and growth conditions
Yeast strains used in this study are listed in Table 1. ABP1 gene disruption to create the abp1Δsla2Δcoil1 strain was carried out by homologous recombination at the chromosomal locus by standard PCR-based methods (Longtine et al. 1998). Yeast cells were grown either in liquid YPD complete medium (1% yeast extract, 2% bactopeptone, supplemented with 2% glucose) or in yeast nitrogen base (YNB) minimal medium supplemented with 2% glucose (SD), unless otherwise stated. Tetrads were dissected using a Singer Instruments MSM manual micromanipulator. Yeast cells were transformed by using the lithium-acetate/SS carrier/PEG method (Gietz and Woods 2002). Yeast cells expressing the analog-sensitive (as) alleles were grown in YPD media at 30° to exponential growth phase (0.5 OD600) and then split into two equal volumes, one-half of which was allowed to grow without treatment and the second half of which was treated with CDK inhibitor 1NM-PP1 at 25 μM final concentration.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| Y263 | MATa trp1Δ63 ura3-52 lys2-801 ade2-107 his3Δ200 leu2-Δ1 | Measday et al. (1994) |
| BY1009a | BY263 abp1Δ::Kan | Haynes et al. (2007) |
| BY3002b | a/α abp1Δ::Nat sla1Δ::Kan | B. Andrew lab |
| abp1Δprk1Δ | MATa abp1Δ::Nat prk1Δ::Kan | Cross of BY1689 and BY2985 Haynes et al. (2007) |
| RH3395 | MATa lys2 leu2 ura3 his3 bar1 sla2/end4::HIS3 end4Δ376-501::TRP1 | Wesp et al. (1997) |
| abp1Δsla2Δcoil1 | MATa lys2 leu2 ura3 his3 bar1 sla2/end4::HIS3 end4Δ376-501::TRP1 abp1::Nat | This study |
| BY4068b | MATa cdc28-as::Nat ura3 leu2 his3 met15 | B. Andrew lab |
| BY4131b | MATα pho85-as::Hph ura3 leu2 his3 met15 | B. Andrew lab |
| BY4129b | MATa cdc28-as::Nat pho85-as::Hph ura3 leu2 his3 met15 CAN1+LYP1+ | B. Andrew lab |
Strains are isogenic to the parent strain, BY263, an S288C derivative.
Strains from the deletion consortium are isogenic to the parent strain, BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0), which is also derived from S288C (Brachmann et al. 1998).
To recover slow-growing double mutants that expressed mutant versions of Abp1p in combination with sla1 deletions, we used a similar strategy to the one described in our previous work (Haynes et al. 2007), in which double mutants were isolated from the heterozygous diploid abp1Δsla1Δ strain by tetrad dissection and allowed to grow at room temperature. In the cases of abp1Δprk1Δ and abp1Δsla2Δcoil1 strains, the double-mutant haploid strains were transformed directly with the different genetic constructs carrying mutant forms of Abp1p.
Recombinant DNA manipulations
Plasmids used in this study are listed in Table 2. Plasmid p6-1 contains a 3.5-kb EcoRI fragment carrying the ABP1 gene with 1.5 kb upstream from the initiator ATG codon and 180 bp downstream from the stop codon subcloned into pRS316ΔSalI (Haynes et al. 2007). This plasmid was used to generate yeast expression plasmids for wild-type (WT) and mutant versions of Abp1p using standard cloning techniques. All generated plasmids were sequenced to verify the integrity of the constructs.
Table 2. Plasmids used in this study.
| Plasmid | Construction | Source |
|---|---|---|
| ABP1WT | p6-1 contains a 3.5-kb EcoRI fragment carrying the ABP1 gene with 1.5 kb upstream from the initiator ATG codon and 180 bp downstream from the stop codon subcloned into pRS316ΔSalI | Haynes et al. (2007) |
| abp1ΔADH-F | p6-1 derivative containing ABP1 with the ADF-H domain deletion (47–200 amino acids) | This study |
| abp1Δ200-440 | p6-1 derivative containing ABP1 with a 200- to 440-amino-acid deletion | This study |
| abp1ΔPR\R | p6-1 derivative containing ABP1 with the PRR deletion (440–530 amino acids) | This study |
| abp1ΔSH3 | p6-1 derivative containing ABP1 with the SH3 domain deletion | Haynes et al. (2007) |
| abp1ΔCFM1 | p6-1 derivative containing ABP1 with a 217- to 232-amino-acid deletion | This study |
| abp1ΔCFM2 | p6-1 derivative containing ABP1 with a 309- to 324-amino-acid deletion | This study |
| abp1ΔCFM1ΔADH-F | p6-1 derivative containing ABP1 with the deletion of the ADF-H domain and the region comprising amino acids 217–232 | This study |
| abp1ΔCFM2ΔADH-F | p6-1 derivative containing ABP1 with the deletion of the ADF-H domain and the region comprising amino acids 309–324 | This study |
| Abp1S*T*Pro | p6-1 derivative containing ABP1 with substitutions of all Ser and Thr residues within the PRR to Ala | This study |
| Abp1T526Pro | p6-1 derivative containing ABP1 with the Thr526-to-Ala substitution | This study |
| Abp1S*Pro | p6-1 derivative containing ABP1 with substitutions of all Ser residues within the PRR to Ala | This study |
| pRS426-ABP1 | ABP1 overexpression plasmid, contains a 3.5-kb EcoRI fragment from p6-1 and subcloned into pRS426 | This study |
| EP1-E11 (pCDC28) | CDC28 with its own promoter and terminator subcloned in a MAGIC plasmid | C. Boone lab |
Protein immunoprecipitation and Western blot analysis
Yeast-cell protein extracts were prepared from exponentially growing cells expressing WT or mutant Abp1p by lysing cells with glass beads (Lee et al. 1998). Protein concentrations were measured using the BCA Protein Assay Reagent kit (Pierce Chemical, Rockford, IL). Protein extracts were separated on denaturing 10% SDS-PAGE using the Mini-Protean III system (BioRad). After electrophoresis, the proteins were transferred to nitrocellulose filters and detected by Western blotting with a polyclonal chicken anti-Abp1p antibody (supplied by B. Goode) using enhanced chemiluminescence.
For Abp1p immunoprecipitation, 1 mg of total protein extracted from the BY263 yeast was incubated with 1 µl of Abp1SH3 mouse monoclonal antibody (18-A, produced by the University of Toronto monoclonal antibody facility) in a total volume of 100 µl of lysis buffer [50 mM Tris–HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.1% NP-40, 50 mM NaF, and protease inhibitor cocktail (Sigma)]. After incubation at 4° for 1 hr, 15 µl of a 50% (v/v) Protein A-Sepharose slurry in lysis buffer was added and further incubated overnight at 4°. The resin was washed three times with 1 ml of wash buffer containing 50 mM Tris–HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 1 mM DTT, and 0.1% NP-40 and then resuspended in SDS-gel loading buffer or phosphatase reaction buffer [10 mM Tris–HCl (pH 7.5), 1 M NaCl, 10 mM MgCl2, 1 mM DTT, and 0.1 mM 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) (Sigma)]. Immune complexes were visualized by Western blotting with anti-Abp1p antibody. In some experiments, membranes were stripped of anti-Abp1p and reprobed with anti-tubulin antibody (Sigma).
In vivo labeling experiments
For overexpression of Abp1p, plasmid pRS426-ABP1 was transformed into strain BY263. Transformants were grown to mid-log phase in SD-URA, transferred to YPD for two generations, and then shifted to YPD minus Pi and allowed to duplicate once. The cells were pelleted and concentrated 20-fold to a final volume of 5 ml in YPD-Pi, and 1 mCi of [32P]orthophosphate (Dupont/NEN) was then added. After 1 hr of incubation with shaking, cells were collected by centrifugation, washed with 50 mM NaF, and flash-frozen. 32P-labeled Abp1p was immunoprecipitated, subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and detected by autoradiography. The nitrocellulose membrane was further analyzed by Western blotting.
Phosphatase treatment
Whole-cell extracts were obtained following the procedure described above using a modified lysis buffer [100 mM Tris (pH 8.0), 100 mM NaCl, 2 mM MnCl2, 10% glycerol, 1 mM DTT, and protease inhibitor cocktail (Sigma)]. For phosphatase treatment (Ho et al. 1999), 100 μg of total proteins were used and incubated for 30 min at 30° in a total volume of 20 μl of lysis buffer in the presence of 2 mM MnCl2 and 1600 U of λ-phosphatase (New England Biolabs). Where indicated, the protease/phosphatase inhibitor (250 mM NaF, 10 mM EDTA, 4 mM sodium orthovanadate, 2 mM AEBSF) was added. The phosphatase reaction was stopped by addition of SDS sample buffer, and lysates were analyzed by Western blotting.
Spot dilution growth assays
To assay growth on solid media, yeast cells were grown overnight to saturation in selective media supplemented with 2% dextrose and the appropriate amino acids. Five microliters of 10-fold serial dilutions of equivalent cell concentrations (0.1 OD600nm) from each strain expressing WT or mutant Abp1p variants was spotted onto minimal media lacking uracil (SD-URA). Growth differences were recorded following incubation of the plates for 48 hr at the indicated temperature. In some cases, 5 mM caffeine (Sigma) was added to emphasize growth defects. All growth assays were repeated at least three times, and one representative experiment is shown in Figure 2.
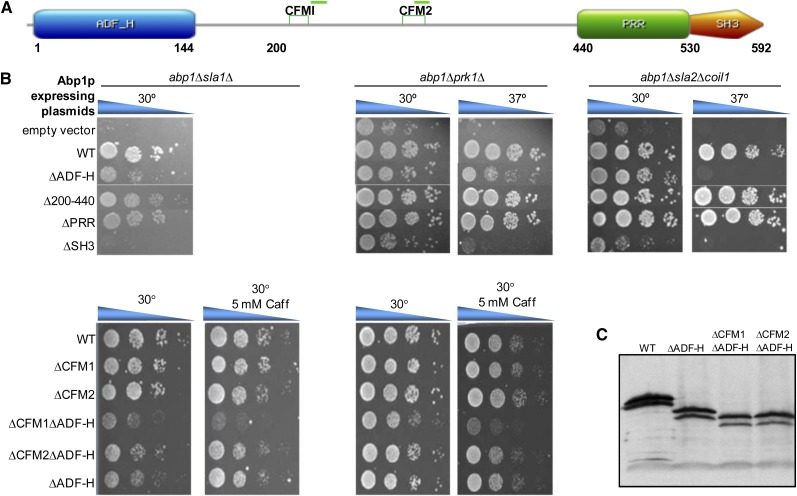
Figure 2 .
The novel fungal conserved motif CFM1 is required in the absence of the ADF-H domain. (A) Domain structure of Abp1p showing CFM1 and CFM2 and predicted helical regions are indicated by green horizontal bar. Relevant amino acid coordinates for Abp1p are shown beneath the diagram. (B) Yeast strains bearing the indicated double- gene deletions were complemented by plasmids expressing either WT Abp1p or Abp1p mutants with the indicated deleted regions on the left. Cultures were spotted in serial 10-fold dilutions and incubated at the indicated temperatures for 48 hr. Caffeine was added in some cases to emphasize growth defects. (C) Yeast cells bearing plasmids expressing mutant forms of Abp1p, as indicated at the top, were analyzed by Western blotting using anti-Abp1p antibody.
Protein stability assays using cycloheximide
For analyses of protein stability, yeast cultures were grown in SD-URA to exponential growth phase (0.5 OD600nm) and divided in two halves. One-half was allowed to continue growth, and 35 μg/ml of cycloheximide (Sigma) was added to the other. Subsequently, equal numbers of cells were collected after 4 hr by centrifugation and immediately frozen; degradation was stopped by the addition of 1 mM sodium azide. Proteins were extracted from whole-cell lysates, as described earlier, and analyzed by Western blotting using anti-Abp1p and antitubulin probes. Three independent experiments were carried out, and one representative experiment is shown in Figure 4D.
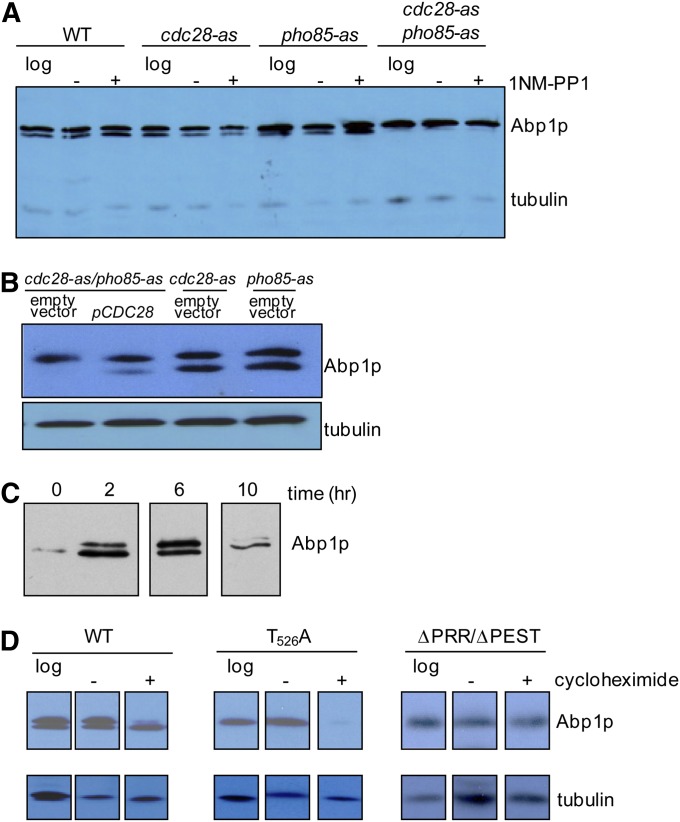
Figure 4.
Investigation of the role of Cdc28p and Pho85p in the phosphorylation of Abp1p using analog sensitive (as) mutants. (A) Wild-type (WT), cdc28-as, pho85-as, and cdc28-as/pho85-as yeast strains were grown to mid-log phase (log), and then inhibitor suspended in DMSO [1NM-PP1 (+)] or just DMSO (−) was added. In A–D, cell extracts were analyzed by Western blotting using anti-Abp1p antibodies. Loading controls were performed using antitubulin antibody. (B) Extracts from cdc28-as, pho85-as, and cdc28-aspho85-as yeast cells bearing empty vector or a plasmid expressing wild-type Cdc28p (pCDC28) from its own promoter were subjected to Western blot analysis. (C) Yeast cells in stationary phase were diluted into fresh YPD broth (time 0), and the cells were then grown at 30°. Samples were removed at the indicated times, and cell lysates were analyzed by Western blotting with anti-Abp1p antibody. (D) Yeast cells bearing plasmids expressing wild-type Abp1p (WT), Abp1p with a Thr526-to-Ala substitution (T526A), or a mutant (ΔPRR/ΔPEST)—in which the PRR, including Thr526 and the PEST sequences, was deleted—were grown in the absence (−) or presence (+) of cycloheximide for 4 hr. The images were assembled from one gel with extraneous lanes removed. In C and D, lanes were extracted from the same exposure of the same gel. However, lanes containing extraneous data were excised. Lanes have been placed in boxes to indicate lanes that were not adjacent to each other in the original gel.
NMR spectroscopy
The extended Abp1p SH3 domain construct, which included the SH3 domain and the 28 residues lying N-terminal to the domain, was expressed as a thioredoxin fusion with a tobacco etch virus protease cleavage site and was purified as described previously (Stollar et al. 2009). The 1H-15N HSQC spectrum of the extended construct was collected using standard methods (Kay et al. 1995) at 30° on a Varian 500 MHz spectrometer equipped with pulsed field gradients and a triple resonance probe with actively shielded z-gradients. Data were processed and analyzed using NMRPipe/NMRDraw (Delaglio et al. 1995) and NMRView (Johnson and Blevins 1994). Spectra were recorded in 50 mM sodium phosphate, 100 mM NaCl, and 1 mM EDTA (pH 7.0). Using the established assignments of Ark1p peptide-bound and free Abp1p SH3 domain (E. J. Stollar H. Lin, A. R. Davidson, and J. D. Forman-Kay, unpublished results), peptide titrations were used to monitor the movement of peaks and assign the final position of the Abp1p SH3 domain:PRR peptide complex resonances. Almost all of the peptide complexes were exchanging in the fast timescale regime facilitating this method of assignment. The 1H-15N HSQC spectrum of the extended PRR-SH3 domain construct was essentially superimposable on that of the SH3 domain:PRR peptide complex; thus, further experiments to assign the peaks within this spectrum were unnecessary. Amide hydrogen chemical shift differences were calculated as previously described (Stollar et al. 2009).
Results
Conserved sequence features are found in the Abp1 family
To gain more insight into the functionally important regions of the Abp1 protein family, we compared the sequence features of these proteins from diverse fungal and higher eukaryotic species. As seen in Figure 1A, all members of the family possess an N-terminal ADF-H domain and at least one C-terminal SH3 domain. In addition, a PRR containing at least 10% Pro is found upstream of the SH3 domains in all cases. Using the algorithm Epestfind (http://bioweb2.pasteur.fr/docs/EMBOSS/epestfind.html), we were also able to detect sequences predicted with high PEST scores within the PRR regions of all Abp1p homologs examined. The prevalence of PEST sequences, which have been shown to mediate protein degradation (Rogers et al. 1986; Rechsteiner and Rogers 1996), within the whole Abp1 family has not been previously reported.
Since regions C-terminal to the ADF-H domain in mouse Abp1 were predicted with high confidence to form helices, we searched for predicted helices in other homologs using the Jpred3 algorithm (Cole et al. 2008). We found that all members of the Abp1 family displayed predicted helices within the same region (Figure 1A). Furthermore, examination of the predicted helical regions of the fungal homologs revealed two highly conserved sequence motifs, Conserved Fungal Motifs 1 and 2 (CFM1 and CFM2) (Figure 1B). CFM1 lies just amino terminal to the first conserved predicted helix, and CFM2 lies within a predicted conserved helix (Figure 2A). These motifs are very conspicuous because they lie within an area of these proteins that is generally highly diverse in sequence despite the conserved presence of predicted helices. The conservation of CFM1 (Figure 1B) is particularly striking since it is found even in very distantly related Abp1p orthologs from Basidiomycete species (Ustilago, Coprinopsis, and Malassezia), which diverged from S. cerevisiae at least 500 million years ago (Taylor and Berbee 2006).
Deletion of the ADF-H domain reveals a functional role for CFM1
To assess the biological roles of the conserved features identified in Abp1p, we constructed plasmids expressing mutant versions of Abp1p with various regions deleted. The biological activity of these plasmid-expressed proteins was tested in several yeast genetic backgrounds in which Abp1p is required for viability under some conditions. These backgrounds included abp1Δsla1Δ, which is unable to grow at 30°, but can be propagated at room temperature (Haynes et al. 2007), and abp1Δprk1Δ, which is unable to grow at 37° (Cope et al. 1999). We also utilized a strain combining abp1Δ with a partial deletion of SLA2 (sla2Δcoil1), which displays impaired growth at 37° (Wesp et al. 1997). Transformation of these mutant strains with the plasmid expressing wild-type ABP1 resulted in robust growth while cells containing an empty vector did not grow under the restrictive conditions (Figure 2B), confirming the requirement for Abp1p in these strain backgrounds and growth conditions.
As has been previously observed (Haynes et al. 2007), expression of Abp1p lacking its SH3 domain resulted in growth that was no better than vector alone in all conditions and in genetic backgrounds in which Abp1p was required. By contrast, deletion of the central region (residues 200–440) or PRR region (residues 440–530) caused no growth defects as compared to WT under any conditions. Deletion of residues 47–200 (abp1ΔADF-H), which includes the conserved ADF-H domain (residues 1–144), caused only a partial reduction in viability in the abp1Δsla1Δ and abp1Δprk1Δ backgrounds, yet no growth was observed in abp1Δsla2Δcoil1 at 37°. These results are consistent with another study in which the ADF-H domain was found to be essential for viability in an abp1Δsla2Δ strain, but not in an abp1Δsla1Δ strain (Quintero-Monzon et al. 2005); however, the abp1Δprk1Δ and abp1Δsla2Δcoil1 were not tested in this previous study. We also tested a deletion in which both the ADF-H domain and central region were deleted (Δ47–440), but this mutant was very poorly expressed and meaningful growth data could not be obtained.
The presence of an ADF-H domain, which binds to actin, is a conserved feature of all Abp1p homologs; thus, it was surprising that this domain was not fully required for growth under all conditions. Since a second actin-binding activity was identified in the putative helical region of mouse Abp1, we investigated the possibility that the CFM1 or CFM2 motifs (Figure 1), which are within the same region of yeast Abp1p, might play a functional role when the ADF-H domain is deleted. As shown in Figure 2B, combining a deletion of CFM1 (Δ217–232) with a deletion of a fragment including the ADF-H (Δ47–200) domain resulted in a much greater reduction in growth than deletion of either region alone under the conditions where the ADF-H domain was not essential for growth. Importantly, the expression levels of all three of these Abp1p deletion mutants was very similar (Figure 2C). It should be noted that different stress conditions were used in these assays as compared to those described above because these conditions were found to best emphasize the effect of the CFM1 deletion. We did not observe any reduction in viability when a deletion of the CFM2 motif (Δ309–324) was combined with an ADF-H domain deletion, suggesting that this region possesses a function distinct from that of the CFM1 region.
Yeast Abp1p is phosphorylated within the PRR
Since mAbp1 is phosphorylated (Larbolette et al. 1999; Han et al. 2003; Larive et al. 2009; Boateng et al. 2012) and several phosphorylated peptides from yeast Abp1p have been identified in large-scale studies (Ficarro et al. 2002; Ubersax et al. 2003; Holt et al. 2009), we sought to further investigate the possible phosphorylation of Abp1p. Interestingly, Abp1p migrates as a double-protein band on SDS-PAGE gels (Drubin et al. 1988). To determine whether the existence of this double band is due to phosphorylation, whole-yeast extracts were treated with λ-phosphatase, and the electrophoretic mobility of the treated protein was evaluated. As seen in Figure 3A, phosphatase treatment caused the disappearance of the band of faster mobility, implying that this band represents a phosphorylated form of Abp1p. When phosphatase and phosphatase inhibitor were added to the reactions, the faster band remained. To confirm that this band contained phosphate, cells were grown in 32P-orthophosphate, and immunoprecipitated Abp1p that had incorporated 32P was visualized on SDS-PAGE gels by exposure to film. It can be seen that a labeled band was observed in a WT extract, but not in an extract from an abp1Δ strain (Figure 3B). Furthermore, the labeled band was more intense in extracts of a strain carrying a plasmid that overexpressed Abp1p. Western blotting of the same gels demonstrated that the 32P-labeled bands corresponded to the faster-migrating band of Abp1p, confirming that this band is phosphorylated.
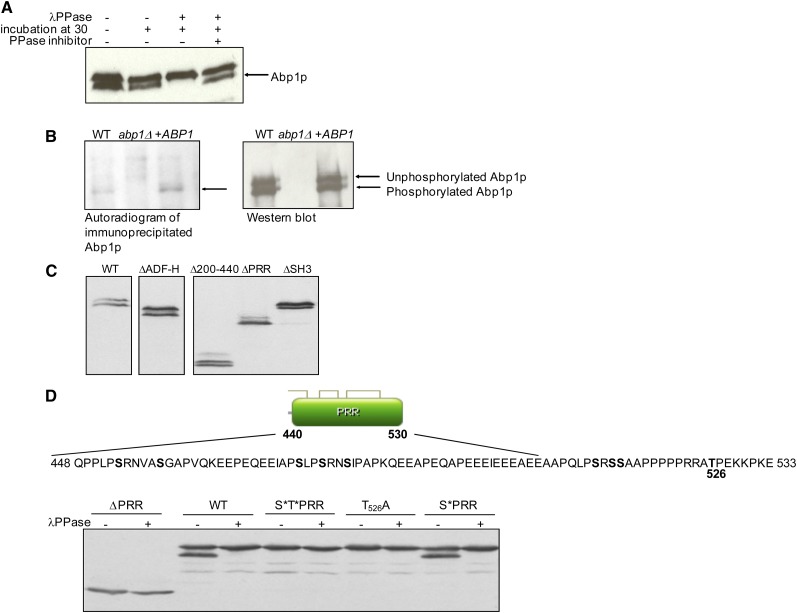
Figure 3.
Yeast Abp1p is phosphorylated in vivo. (A) Wild-type yeast extracts were treated with λ-phosphatase in the presence or absence of phosphatase inhibitor as indicated. Abp1p was detected by Western blot analysis using a polyclonal anti-Abp1p antibody. (B, left) 32P-labeled Abp1p was detected after in vivo labeling and immunoprecipitation of Abp1p from WT yeast cells or WT cells bearing a plasmid overexpressing ABP1 (+ABP1). The abp1Δ strain was included as a negative control. Proteins separated in SDS-PAGE gels were transferred to a nitrocellulose membrane and analyzed by autoradiography. (Right) Western blot of the same membrane shown in the left panel probed with an anti-Abp1p antibody. The positions of phosphorylated and unphosphorylated Abp1p are indicated. (C) Yeast cells bearing plasmids expressing mutant forms of Abp1p, as indicated, were analyzed by Western blotting using anti-Abp1p antibody. This figure is composed of lanes extracted from the same exposure of the same gel. However, lanes containing extraneous data were excised. Lanes have been place in boxes to indicate lanes that were not adjacent to each other in the original gel. (D) Phosphorylation of PRR mutants. The sequence of the Abp1p PRR is shown (residues 448–533) with potential sites of phosphorylation (Ser and Thr residues) in boldface type. Thr residue in position 526 is indicated. Yeast protein extracts from cells bearing plasmids expressing wild-type Abp1p (WT), Abp1p lacking the PRR (ΔPRR), Ala substitutions in all potential phosphosites within the PRR (S*T*PRR), all Ser residues mutated to Ala (S*PRR), or Thr526Ala substitution (T526A) were treated with (+) or without (−) λ-phosphatase and analyzed by Western blotting using anti-Abp1p antibody.
To identify which region of Abp1p was phosphorylated, Western blots were carried out on extracts of an abp1Δ strain carrying the ABP1 mutant plasmids described above. Bands of similar intensity were detected in each of these extracts, indicating that the various deletions did not have a large effect on Abp1p expression or stability. In addition, a double band was detected in all extracts except when the PRR was deleted (Figure 3C). The absence of phosphorylation of this mutant was confirmed by phosphatase treatment (Figure 3D, bottom). Since this result indicated that the phosphorylation site was located within the PRR, we mutagenized all Ser and Thr residues within this region (no Tyr residues were found in the PRR) as a means to identify the specific site of phosphorylation. As expected, simultaneous replacement of all nine Ser and Thr residues within the PRR with Ala (S*T*PRR) abrogated phosphorylation as detected by the absence of the faster-migrating Abp1p band. The remaining band, which comigrated with the upper band in the WT Abp1p extract, was not affected by addition of phosphatase. By substituting each Ser and Thr residue in the PRR individually, we found that only substitution of Thr526 (T526A) resulted in a loss of phosphorylation. Thus, we conclude that phosphorylation of Thr526 causes the change in mobility of Abp1p observed in SDS-PAGE. Although this site is clearly phosphorylated, the Thr526Ala substitution did not cause a detectable growth phenotype in any of the ABP1-dependent strain backgrounds described above (data not shown). In addition, testing of a phosphomimetic Thr526Glu substitution in the abp1Δ/prk1Δ and abp1Δ/sla2Δcoil1 backgrounds did not reveal a growth phenotype (data not shown).
Cdc28p and Pho85p phosphorylate Abp1p
The sequence surrounding the Thr526 residue (TPEK) exactly matches the CDK consensus phosphorylation site [(S/T)-P-X-(K/R)], and this region was predicted with very high probability to be phosphorylated by the yeast CDKs, Cdc28p, and Pho85p using the Predikin software (Saunders et al. 2008; Saunders and Kobe 2008). To determine whether Abp1p is a target of these CDKs, we assessed the level of Abp1p phosphorylation in yeast strains carrying inhibitor-sensitive alleles of CDC28 (cdc28-as) and PHO85 (pho85-as). In these strains, kinase activity can be specifically inhibited by the addition of specific nucleotide analogs (Carroll et al. 2001; Ubersax et al. 2003), such as 1NM-PP1, which is the compound that we used here. As can be seen in Figure 4A, phosphorylation could still be detected in strains carrying the cdc28-as and pho85-as alleles on their own, but the cdc28-as/pho85-as double-mutant strain displayed no detectable phosphoylated Abp1p. This result implies that both of these CDKs are able to phosphorylate Abp1p so that Abp1p phosphorylation persists in the single-mutant strains. It was somewhat surprising that the cdc28-as/pho85-as double-mutant strain displayed no Abp1p phosphorylation even in the absence of inhibitor. Since the cdc28-as mutant is reduced by sixfold in overall in vitro activity in the absence of inhibitor (Bishop et al. 2000), we surmise that the loss of Abp1p phosphorylation in the double-mutant strain is the result of simultaneous partial reduction in the activity of both kinases. We show here that the pho85-as mutant is definitely inhibited further by addition of 1NM-PP1 (Supporting Information, Figure S1), and the specific inhibition of the cdc28-as mutant by this compound has been previously demonstrated (Bishop et al. 2000; Zimmermann et al. 2011).
To directly test the ability of Cdc28p to phosphorylate Abp1p, a plasmid producing wild-type Cdc28p was introduced into the cdc28-as/pho85-as strain. In this case, phosphorylation of Abp1p was partially restored (Figure 4B), proving that the presence of Cdc28p can mediate phosphorylation of Abp1p. The incomplete restoration of Abp1p phosphorylation in this strain background may be due to perturbed regulation of the plasmid-borne CDC28 gene or others factors specific to the double-mutant strain. Since many Cdc28p and Pho85p substrates are phosphorylated in a cell-cycle-dependent manner (Huang et al. 2007; Enserink and Kolodner 2010), we assessed the cell cycle dependence of Abp1p phosphorylation by α-factor growth synchronization experiments. Although the levels of Abp1p phosphorylation varied with time after release of α-factor arrest, these changes did not correlate with cell cycle progression as monitored by the changes in cellular DNA content (Figure S2).
To determine whether any other yeast protein kinases are involved in Abp1p phosphorylation, strains bearing single deletions of known nonessential kinase-encoding genes were examined. We found that none of these 80 strains displayed any alteration in Abp1p phosphorylation as assessed by Western blotting of lysates of exponentially growing cells. Similarly, overexpression of 27 different essential kinases caused no change in the degree of Abp1p phosphorylation (Figure S3).
Novel role for the PRR and phosphorylation in Abp1p stabilization
Since substitution of the Thr526 phosphorylation site with Ala did not cause a detectable growth phenotype, we sought to uncover other possible biological roles for this phosphorylation. Monitoring of the relative levels of phosphorylated and unphosphorylated Abp1p during exponential growth showed that these levels changed with a relatively greater amount of the unphosphorylated form in the early stages of growth followed by accumulation of mostly the phosphorylated form when stationary phase was reached (Figure 4C). These data suggested that the phosphorylated form may be more stable. To directly test this idea, we monitored the degradation of Abp1p over time after inhibition of protein translation by the addition of cycloheximide. Strikingly, the unphosphorylated form of Abp1p was almost completely degraded after 4 hr of incubation in cycloheximide while little change in the level of the phosphorylated form was seen (Figure 4D, left). Consistent with this result, the Thr526Ala mutant, which cannot be phosphorylated, was also mostly degraded after 4 hr in cycloheximide (Figure 4D, center). These data demonstrate that the phosphorylation of Thr526 leads to stabilization of Abp1p.
As described above, the presence of PEST sequences, which mediate protein degradation (Rogers et al. 1986; Rechsteiner and Rogers 1996), is a conserved feature of the PRRs of both fungal and higher eukaryotic Abp1p orthologs. To determine whether these PEST regions are required for the rapid degradation of unphosphorylated Abp1p, the effect of deletion of the PRR (ΔPRR) on Abp1p stability after inhibition of protein synthesis was analyzed. As is shown in Figure 4D (right), the ΔPRR mutant is resistant to proteolysis after addition of cycloheximide, supporting the importance of the PEST sequences in mediating this degradation. It should be noted that the Abp1p phosphorylation site is within the region deleted in the ΔPRR mutant; thus, the increased stability of this mutant occurs in spite of its being unphosphorylated. In summary, these data indicate that protein degradation mediated by PEST sequences within the PRR is inhibited by phosphorylation of Abp1p at Thr526.
Abp1p SH3 domain can potentially form an intramolecular interaction with the PRR
Since the Abp1p SH3 domain lies immediately adjacent to the PRR region, we hypothesized that an intramolecular interaction between these two regions could play a role in regulating Abp1p activity. To test this idea, we expressed and purified a version of the Abp1p SH3 domain that included 28 residues N-terminal to the start of the SH3 domain. This region includes several PXXP motifs and the phosphorylation site at Thr526. To assess the ability of this region to interact with the SH3 domain, we collected an 1H-15N HSQC NMR spectrum for this construct and compared it to spectra previously collected for free and peptide-bound forms of the Abp1p SH3 domain (Stollar et al. 2009). In this type of spectrum, a cross-peak can be observed corresponding to each amide group in the protein. The positions of these cross-peaks (i.e., their chemical shifts) are very sensitive to the chemical environment; thus, amide groups in residues close to the peptide-binding surface display significant chemical shift changes upon peptide binding. It can be seen in Figure 5A that similar changes in chemical shifts are seen when comparing the free Abp1p SH3 domain to the Ark1p peptide-bound domain (top) or to the extended Abp1p SH3 domain construct (bottom). These data indicate that the PRR of the extended construct interacts with the SH3 domain in a manner similar to a bound target peptide. We also found that a synthesized Pro-rich peptide derived from the Abp1p PRR was able to bind the Abp1p SH3 domain intermolecularly when added to the NMR sample (Figure 5A, middle). The dissociation constant of this interaction as measured by isothermal titration calorimetry was 64.3 µM (E. J. Stollar, H. Lin, A. R. Davidson, and J. D. Forman-Kay, unpublished results). Comparison of the NMR spectrum of this intermolecular complex with the spectrum of the extended SH3 domain construct revealed additional residues in the extended construct with significant chemical shift changes (Figure 5A, bottom). A plot of these residues on the structure of the Abp1p SH3 domain:peptide complex shows that these residues lie on one surface of the domain (Figure 5B) and likely are positioned on the path that the PRR region would have to follow to engage in an intramolecular interaction with the peptide-binding surface of the domain.
Figure 5.
The PRR binds to the Abp1p SH3 domain. (A) The backbone amide chemical shift differences between the unbound Abp1p SH3 domain and the Abp1p SH3 domain bound to a high-affinity target peptide derived from Ark1p (top) (Stollar et al. 2009) or the PRR-derived peptide (middle) are shown. The sequence of the PRR peptide used is shown in the middle. The bottom shows the chemical shift differences between the unbound Abp1p SH3 domain and the extended SH3 domain construct. The sequence of the PRR region attached to the SH3 domain in the extended construct is shown. The residues highlighted in blue are those that significantly differ between the extended construct and the Abp1p SH3 domain:PRR peptide intermolecular complex. (B) Amide groups that show significant chemical shift differences between the PRR peptide-bound Abp1p SH3 domain intermolecular complex and the extended SH3 domain are indicated by blue spheres on the structure of the Abp1p SH3 domain:Ark1p peptide complex. The dotted line shows a possible path connecting the domain to the peptide as it could be within an intramolecular complex and indicates that Thr526 would lie within this region. The structure on the right has been rotated 90° relative to the one on the left. The Abp1p SH3 domain is shown in gray and the Ark1p peptide in green.
Discussion
Members of the Abp1 family of proteins have been found in diverse fungal and metazoan organisms ranging from yeast to humans. The high degree of conservation seen among these proteins indicates a crucial function in all of these organisms. It is surprising, therefore, that deletion of the ABP1 gene in yeast causes no detectable phenotype and that measurable effects of ABP1 mutations are seen only when they are combined with other gene deletions (Holtzman et al. 1993; Lila and Drubin 1997; Cope et al. 1999; Haynes et al. 2007). These findings emphasize that deletion of functionally important proteins or regions of proteins will not always cause a detectable phenotype when tested under laboratory conditions. Thus, uncovering the roles of these conserved regions may require phenotypic assays to be carried out under a wide variety of growth conditions and genetic backgrounds. The results presented in this work clearly demonstrate the difficulties associated with assigning functions to the conserved features of yeast Abp1p, yet also show that a thorough investigation can reveal requirements for regions that at first may appear to be dispensable.
Through sequence alignment analysis, we discovered two highly conserved motifs, CFM1 and CFM2, within the fungal Abp1p orthologs. Given the high conservation of these motifs, it was striking that deletion of the whole central portion of Abp1p (Δ200–440), which includes both motifs, resulted in absolutely no reduction in viability in any strain backgrounds or growth conditions tested (Figure 2). We were, however, able to uncover a function for CFM1 by deleting this motif within the context of Abp1p lacking the ADF-H domain. This discovery implies that CFM1 is able to fulfill part of the function of the ADF-H domain. One possibility is that CFM1 binds actin, similarly to a charged helical sequence found in mAbp1 (Larbolette et al. 1999; Kessels et al. 2000). It might also interact with another protein that is associated with actin. We expect that any potential direct or indirect association between CFM1 and actin would be weak because deletion of the ADF-H domain was observed to abrogate localization of Abp1p to cortical actin patches as assessed by GFP localization experiments (Quintero-Monzon et al. 2005). An important conclusion from our studies on CFM1 is that, even when a motif is highly conserved, its function may not become apparent unless the protein is sensitized by other perturbations, such as deletion of the ADF-H domain in this case. This type of “intragenic synthetic lethality” has been recognized in a handful of other studies (Teixeira et al. 1997; Kou and Pugh 2004; Murphy et al. 2004; Dias et al. 2008; Garrey and Mackie 2011), but systematic searches for intragenic synthetic lethality have not been carried out. Such experiments could provide a generally applicable approach for uncovering the roles of conserved regions within multi-domain proteins.
Our work has also illuminated other potentially important features of Abp1p. We have demonstrated that Abp1p is phosphorylated at Thr526 by the Cdc28p and Pho85p kinases (Figure 4), which recognize the same consensus phosphorylation sites and are functionally interchangeable under some conditions (Nishizawa et al. 1998; Ptacek et al. 2005; Huang et al. 2007, 2009; Sopko et al. 2007; Enserink and Kolodner 2010). While other sites of Cdc28p-mediated phosphorylation have been detected in Abp1p through large-scale phosphoproteomic studies (Ficarro et al. 2002; Ubersax et al. 2003; Holt et al. 2009), Thr526 is the first phosphorylation site within Abp1p to be experimentally verified through mutagenesis. We cannot exclude the possibility that other residues within Abp1p are phosphorylated because some phosphorylation events may be insensitive to phosphatase or may not affect electrophoretic mobility. Our studies on Abp1p proteolysis indicate a role for Abp1p phosphorylation since the phosphorylated form of Abp1p is considerably more resistant to proteolysis than the unphosphorylated form (Figure 4D). Consistent with an increased stability for phosphorylated Abp1p, we observed that the relative level of unphosphorylated Abp1p peaks in mid-log phase when de novo synthesis is highest (Figure 4C). At later growth stages, Abp1p is either phosphorylated or degraded, so that most Abp1p present in the cell at stationary phase is phosphorylated. We also showed that Abp1p proteolysis results from the presence of PEST sequences in the PRR that are located near the site of phosphorylation (Figure 1). Thus, phosphorylation at Thr526 appears to provide a means of regulating proteolysis mediated by the nearby PEST sequences. Since PEST sequences are a conserved feature of Abp1p homologs in fungal and mammalian species and phosphorylation within the PRR is seen in mouse (Larbolette et al. 1999; Han et al. 2003; Zanivan et al. 2008; Larive et al. 2009; Boateng et al. 2012) and human Abp1 (Olsen et al. 2006; Molina et al. 2007), we propose that this regulation of Abp1p stability may be a conserved feature of this family. It is possible that the putative intramolecular interaction between the SH3 domain and the PRR, which is supported by our NMR data, is affected by phosphorylation at Thr526 since this site lies between the SH3 domain and the PRR (Figure 5B). This intramolecular interaction may play a role in regulating Abp1p stability through modulating the accessibility of the PRR to other binding partners. In mammalian cortactin, a protein with a similar domain layout and function as Abp1p (Olazabal and Machesky 2001), phosphorylation has also been suggested to modulate the intramolecular interaction between a C-terminal SH3 domain and an upstream PRR (Lua and Low 2005).
The investigation of Abp1p provides an instructive example of a protein that is highly conserved in evolution, yet deletion of the gene encoding it in yeast results in no detectable growth defects under laboratory conditions. The presence of Abp1p orthologs in divergent species implies that organisms lacking this protein would certainly display a decreased evolutionary fitness. Clearly, the requirements for survival in the natural environment over hundreds of millions of years are much more stringent than those found in the laboratory. For this reason, detection of phenotypes for ABP1 deletions in the laboratory requires additional manipulation of genetic backgrounds and growth conditions (Lila and Drubin 1997; Fazi et al. 2002; Stefan et al. 2005; Haynes et al. 2007; Stollar et al. 2009). In this work, we show that deletions of conserved domains and motifs of Abp1p, which also must be functionally important, do not always lead to detectable growth phenotypes, yet further manipulation of conditions can reveal these phenotypes. In some cases, multiple conserved regions within a protein may have to be deleted before a phenotype emerges as we observed here in the case of the CFM1 motif. Detection of this type of intragenic synthetic lethality could be an important new addition to the repertoire of systematic approaches that are generally employed for uncovering protein functions. Due to the frequent difficulty in establishing conditions in the laboratory that can reveal phenotypes for highly conserved protein regions, we suggest that the detection of conservation is a more reliable approach for identifying functionally important regions in a protein than is the identification of phenotypes resulting from mutations. The future challenge is to detect phenotypes associated with protein regions that are strongly indicated to be important through conservation analysis. High-throughput genetics studies on strains containing deletions of one or more conserved regions of a protein may provide the means to achieve this goal.
Supplementary Material
Acknowledgments
We thank Brenda Andrews and Charlie Boone for plasmids and yeast strains; Bruce Goode for anti-Abp1p antibody; and Karen Maxwell for critical reading of the manuscript. This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR ARD grant MOP-13609 to A.R.D.). E.J.S. was supported by grants from the National Center for Research Resources (5P20RR016480-12) and the National Institute of General Medical Sciences (8P20GM103451-12) of the National Institutes of Health.
Footnotes
Communicating editor: D. Lew
Literature Cited
- Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., et al. , 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 [DOI] [PubMed] [Google Scholar]
- Boateng L. R., Cortesio C. L., Huttenlocher A., 2012. Src-mediated phosphorylation of mammalian Abp1 (DBNL) regulates podosome rosette formation in transformed fibroblasts. J. Cell Sci. 125: 1329–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Carroll A. S., Bishop A. C., DeRisi J. L., Shokat K. M., O’Shea E. K., 2001. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. USA 98: 12578–12583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., et al. , 2007. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA 104: 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Barber J. D., Barton G. J., 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36: W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope M. J., Yang S., Shang C., Drubin D. G., 1999. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J. Cell Biol. 144: 1203–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesio C. L., Perrin B. J., Bennin D. A., Huttenlocher A., 2010. Actin-binding protein-1 interacts with WASp-interacting protein to regulate growth factor-induced dorsal ruffle formation. Mol. Biol. Cell 21: 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., et al. , 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Dias C. A., Cano V. S., Rangel S. M., Apponi L. H., Frigieri M. C., et al. , 2008. Structural modeling and mutational analysis of yeast eukaryotic translation initiation factor 5A reveal new critical residues and reinforce its involvement in protein synthesis. FEBS J. 275: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G., Miller K. G., Botstein D., 1988. Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107: 2551–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J. M., Kolodner R. D., 2010. An overview of Cdk1-controlled targets and processes. Cell Div. 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi B., Cope M. J., Douangamath A., Ferracuti S., Schirwitz K., et al. , 2002. Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: structural and functional analysis. J. Biol. Chem. 277: 5290–5298 [DOI] [PubMed] [Google Scholar]
- Fenster S. D., Kessels M. M., Qualmann B., Chung W. J., Nash J., et al. , 2003. Interactions between Piccolo and the actin/dynamin-binding protein Abp1 link vesicle endocytosis to presynaptic active zones. J. Biol. Chem. 278: 20268–20277 [DOI] [PubMed] [Google Scholar]
- Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., et al. , 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20: 301–305 [DOI] [PubMed] [Google Scholar]
- Garrey S. M., Mackie G. A., 2011. Roles of the 5′-phosphate sensor domain in RNase E. Mol. Microbiol. 80: 1613–1624 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A., 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Goode B. L., Rodal A. A., Barnes G., Drubin D. G., 2001. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J. Cell Biol. 153: 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Kori R., Shui J. W., Chen Y. R., Yao Z., et al. , 2003. The SH3 domain-containing adaptor HIP-55 mediates c-Jun N-terminal kinase activation in T cell receptor signaling. J. Biol. Chem. 278: 52195–52202 [DOI] [PubMed] [Google Scholar]
- Haynes J., Garcia B., Stollar E. J., Rath A., Andrews B. J., et al. , 2007. The biologically relevant targets and binding affinity requirements for the function of the yeast actin-binding protein 1 Src-homology 3 domain vary with genetic context. Genetics 176: 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Costanzo M., Moore L., Kobayashi R., Andrews B. J., 1999. Regulation of transcription at the Saccharomyces cerevisiae start transition by Stb1, a Swi6-binding protein. Mol. Cell. Biol. 19: 5267–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt L. J., Tuch B. B., Villen J., Johnson A. D., Gygi S. P., et al. , 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D. A., Yang S., Drubin D. G., 1993. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122: 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Friesen H., Andrews B., 2007. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol. Microbiol. 66: 303–314 [DOI] [PubMed] [Google Scholar]
- Huang D., Kaluarachchi S., van Dyk D., Friesen H., Sopko R., et al. , 2009. Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast. PLoS Biol. 7: e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. A., Blevins R. A., 1994. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4: 603–614 [DOI] [PubMed] [Google Scholar]
- Kay L. E., Keifer P., Saarinen T., 1995. Pure absorption gradient enhanced hetero-nuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114: 10663–10665 [Google Scholar]
- Kessels M. M., Engqvist-Goldstein A. E., Drubin D. G., 2000. Association of mouse actin-binding protein 1 (mAbp1/SH3P7), an Src kinase target, with dynamic regions of the cortical actin cytoskeleton in response to Rac1 activation. Mol. Biol. Cell 11: 393–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels M. M., Engqvist-Goldstein A. E. Y., Drubin D. G., Qualmann B., 2001. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J. Cell Biol. 153: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou H., Pugh B. F., 2004. Engineering dimer-stabilizing mutations in the TATA-binding protein. J. Biol. Chem. 279: 20966–20973 [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Kessels M. M., Cope M. J., Drubin D. G., 1998. The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol. Biol. Cell 9: 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larbolette O., Wollscheid B., Schweikert J., Nielsen P. J., Wienands J., 1999. SH3P7 is a cytoskeleton adapter protein and is coupled to signal transduction from lymphocyte antigen receptors. Mol. Cell. Biol. 19: 1539–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larive R. M., Urbach S., Poncet J., Jouin P., Mascre G., et al. , 2009. Phosphoproteomic analysis of Syk kinase signaling in human cancer cells reveals its role in cell-cell adhesion. Oncogene 28: 2337–2347 [DOI] [PubMed] [Google Scholar]
- Lee J., Colwill K., Aneliunas V., Tennyson C., Moore L., et al. , 1998. Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr. Biol. 8: 1310–1321 [DOI] [PubMed] [Google Scholar]
- Lila T., Drubin D. G., 1997. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell 8: 367–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lua B. L., Low B. C., 2005. Cortactin phosphorylation as a switch for actin cytoskeletal network and cell dynamics control. FEBS Lett. 579: 577–585 [DOI] [PubMed] [Google Scholar]
- Measday V., Moore L., Ogas J., Tyers M., Andrews B., 1994. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science 266: 1391–1395 [DOI] [PubMed] [Google Scholar]
- Mise-Omata S., Montagne B., Deckert M., Wienands J., Acuto O., 2003. Mammalian actin binding protein 1 is essential for endocytosis but not lamellipodia formation: functional analysis by RNA interference. Biochem. Biophys. Res. Commun. 301: 704–710 [DOI] [PubMed] [Google Scholar]
- Molina H., Horn D. M., Tang N., Mathivanan S., Pandey A., 2007. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 104: 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. W., Olson B. L., Siliciano P. G., 2004. The yeast splicing factor Prp40p contains functional leucine-rich nuclear export signals that are essential for splicing. Genetics 166: 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Kawasumi M., Fujino M., Toh-e A., 1998. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol. Biol. Cell 9: 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olazabal I. M., Machesky L. M., 2001. Abp1p and cortactin, new “hand-holds” for actin. J. Cell Biol. 154: 679–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., et al. , 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., et al. , 2005. Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- Quintero-Monzon O., Rodal A. A., Strokopytov B., Almo S. C., Goode B. L., 2005. Structural and functional dissection of the Abp1 ADFH actin-binding domain reveals versatile in vivo adapter functions. Mol. Biol. Cell 16: 3128–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M., Rogers S. W., 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21: 267–271 [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M., 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234: 364–368 [DOI] [PubMed] [Google Scholar]
- Saunders N. F., Kobe B., 2008. The Predikin webserver: improved prediction of protein kinase peptide specificity using structural information. Nucleic Acids Res. 36: W286–W290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders N. F., Brinkworth R. I., Huber T., Kemp B. E., Kobe B., 2008. Predikin and PredikinDB: a computational framework for the prediction of protein kinase peptide specificity and an associated database of phosphorylation sites. BMC Bioinformatics 9: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Smith J. C., Figeys D., Andrews B. J., 2007. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 26: 4487–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C. J., Padilla S. M., Audhya A., Emr S. D., 2005. The phosphoinositide phosphatase Sjl2 is recruited to cortical actin patches in the control of vesicle formation and fission during endocytosis. Mol. Cell. Biol. 25: 2910–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollar E. J., Garcia B., Chong P. A., Rath A., Lin H., et al. , 2009. Structural, functional, and bioinformatic studies demonstrate the crucial role of an extended peptide binding site for the SH3 domain of yeast Abp1p. J. Biol. Chem. 284: 26918–26927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Berbee M. L., 2006. Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia 98: 838–849 [DOI] [PubMed] [Google Scholar]
- Teixeira M. T., Siniossoglou S., Podtelejnikov S., Benichou J. C., Mann M., et al. , 1997. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 16: 5086–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., et al. , 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Wesp A., Hicke L., Palecek J., Lombardi R., Aust T., et al. , 1997. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8: 2291–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanivan S., Gnad F., Wickstrom S. A., Geiger T., Macek B., et al. , 2008. Solid tumor proteome and phosphoproteome analysis by high resolution mass spectrometry. J. Proteome Res. 7: 5314–5326 [DOI] [PubMed] [Google Scholar]
- Zimmermann C., Chymkowitch P., Eldholm V., Putnam C. D., Lindvall J. M., et al. , 2011. A chemical-genetic screen to unravel the genetic network of CDC28/CDK1 links ubiquitin and Rad6-Bre1 to cell cycle progression. Proc. Natl. Acad. Sci. USA 108: 18748–18753 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.