Abstract
We present a new haplotype-based approach for inferring local genetic ancestry of individuals in an admixed population. Most existing approaches for local ancestry estimation ignore the latent genetic relatedness between ancestral populations and treat them as independent. In this article, we exploit such information by building an inheritance model that describes both the ancestral populations and the admixed population jointly in a unified framework. Based on an assumption that the common hypothetical founder haplotypes give rise to both the ancestral and the admixed population haplotypes, we employ an infinite hidden Markov model to characterize each ancestral population and further extend it to generate the admixed population. Through an effective utilization of the population structural information under a principled nonparametric Bayesian framework, the resulting model is significantly less sensitive to the choice and the amount of training data for ancestral populations than state-of-the-art algorithms. We also improve the robustness under deviation from common modeling assumptions by incorporating population-specific scale parameters that allow variable recombination rates in different populations. Our method is applicable to an admixed population from an arbitrary number of ancestral populations and also performs competitively in terms of spurious ancestry proportions under a general multiway admixture assumption. We validate the proposed method by simulation under various admixing scenarios and present empirical analysis results from a worldwide-distributed dataset from the Human Genome Diversity Project.
Keywords: local ancestry, admixture, infinite hidden Markov model (iHMM), Dirichlet process (DP), hierarchical Dirichlet process
THE problem of inferring genetic ancestries in a population has been widely investigated for various applications such as disease gene mapping and population history inference. For example, the inferred ancestry information has been used in correcting the confounding effect by population stratification in association studies (Price et al. 2006; Wang et al. 2010). The examination of loci that have elevated probabilities of a specific ancestry has also given critical clues in selecting out potential causal variants of certain diseases in admixture mapping (Cheng et al. 2009, 2010; Zhu et al. 2011). Broadly, two different problem settings have been commonly considered for ancestral structure analysis (Alexander et al. 2009), one on the “global ancestry” that considers the average proportion of each contributing population across the genome in an “unsupervised” way (i.e., ancestral labeling of the study population is unknown) (Falush et al. 2003; Patterson et al. 2006; Alexander et al. 2009) and the other on the “local ancestry” that is more concerned with a locus-by-locus ancestry given reference population data (Tang et al. 2006; Pasaniuc et al. 2009; Price et al. 2009). In this article, we consider the problem of estimating the local ancestry in an admixed population. As a common scenario of this problem, consider the decomposition of chromosomes of modern African-Americans into blocks that have either African or European ancestry given the reference population data close to ancient African and European populations, which we call ancestral populations. The populations of Europeans (CEU) and Africans (YRI) are the most typical choices for such ancestral population data when an admixed population of African-Americans is considered. We present a new haplotype-based method for local ancestry estimation that can deal with an arbitrary number of ancestral populations in a nonparametric Bayesian framework.
A natural approach to this problem involves a hidden Markov model (HMM) that traces the ancestry of each individual along the markers on a chromosome. Most previous approaches using the HMM can be largely categorized into two families, depending on how they encode the ancestral population. The first family of methods uses a population-specific allele-frequency profile to characterize each ancestral population. Such an allele-frequency profile has been typically used as the latent component that generates population data in traditional admixture studies for global ancestry estimation (Pritchard et al. 2000; Falush et al. 2003; Huelsenbeck and Andolfatto 2007; Alexander et al. 2009). When adopted in the local ancestry estimation problem as in LAMP (Sankararaman et al. 2008a,b; Pasaniuc et al. 2009), it has the general advantages of low computational cost and availability of such frequency profiles in representative data sets. However, the correlations between loci are reflected only by the variation in such allele frequencies and not by the actual recombination events at the chromosome level, so it is rather unnatural to model linkage disequilibrium (LD) structure between tightly linked SNPs. Therefore, either a subset of markers in low LD has to be selected in a preprocessing step or a recombination process needs to be indirectly embedded to utilize a denser set of markers (Patterson et al. 2004; Tang et al. 2006; Pasaniuc et al. 2009). The representation power of this family of methods thus tends to diminish when the correlations between markers are not carefully considered (Price et al. 2008).
Another family of methods is based on haplotype data that may contain richer information. These methods utilize representative haplotypes taken from each of the ancestral population data as reference information for the local ancestry estimation (Sundquist et al. 2008; Price et al. 2009). Each haplotype in an ancestral population, which we call an ancestral haplotype, constitutes a hidden state in an HMM and the basic transition mechanism involves traversing among these ancestral haplotypes. Therefore, these approaches provide a more natural way to reflect the underlying admixing process by simulating recombinations at a real chromosome level. However, the inference result can be rather sensitive to the size and the choice of such ancestral haplotype data because the admixed haplotype is directly compared with the ancestral haplotypes. Moreover, few existing methods make use of the genetic relatedness between ancestral populations resultant from ancient population history and therefore the populations have been typically treated as independent. To improve the robustness and the accuracy in light of these issues, HAPMIX (Price et al. 2009) introduces a “miscopying” parameter that allows a small possibility for an allele to be copied from population 2 even when it is assumed to be originated from an ancestral haplotype in population 1. In this way, it prevents unnecessary transitions among ancestral populations during inference and the allelic information in one population can be naturally borrowed by another population. However, this method is limited to two-way admixture that involves only two ancestral populations, and it is not trivial to generalize this model to consider more general demographic scenarios.
We propose a new Bayesian approach for local ancestry estimation that uses the multipopulation haplotype data in a more systematic way. Our method is built on the assumption of a common pool of hypothetical founder haplotypes from which the ancestral haplotypes in multiple ancestral populations are to be inherited and from which in turn the individuals in an admixed population are generated as well by the admixing process between ancestral populations. Motivated by the population model called SPECTRUM in Sohn and Xing (2007), we model the ancestral population data by an infinite hidden Markov model in which the hidden states correspond to the unknown number of hypothetical founder haplotypes. The recombination and mutation events are then modeled with respect to these founders as transition and emission processes. For an individual in an admixed population, we extend the hidden state space to a joint space of founder haplotypes and ancestral populations. That is, we incorporate a hidden state variable consisting of two indicator variables at each marker, one for selecting the ancestral population and the other for selecting the hypothetical founder haplotype. The hidden state variable corresponding to the ancestral population determines the local admixing status and hence defines the local ancestry along the markers. Furthermore, population-specific scale parameters are incorporated to allow variable recombination probabilities in different populations. These scale parameters can be interpreted as being proportional to two major factors that affect the recombination probabilities in the corresponding populations: the effective population size and the hypothetical time since the hypothetical era of founder haplotypes. We observe that this parameterization also enhances the robustness of our model under scenarios that deviate from the common modeling assumption that all the populations participate in the admixture simultaneously.
A subtle issue in the proposed representation is how to choose the number of founders and how to construct them efficiently across multiple populations. Naively, we may assume K founders per ancestral population, but under this setting, not only does one have to employ a nontrivial model selection process to determine K, but also there is in general no correspondence between the K founders in one population and another set of K founders in a different population. This problem not only would result in a serious identifiability and multimodality issue that can severely slow down inference, but also will restrict the information sharing across populations and hence compromise the accuracy of ancestry estimation as well. On the other hand, if we are to use one shared set of K founders, the representational power of the population-specific HMM can also be limited. A nonparametric Bayesian framework using an infinite hidden Markov model gives a natural solution for this (Beal et al. 2002; Teh et al. 2010). Under an infinite HMM, an unbounded number of founder haplotypes can be systematically handled to describe a study population. If we employ multiple such infinite HMMs defined over the same set of founders, one infinite HMM per population, then it allows the founders to be shared between populations, while different populations do not have to include all these founders and can have a unique set of founders with its own frequency and recombination patterns among them. The number and the haplotypes of the founders are recovered as a result of posterior inference from data. Under a Dirichlet process prior, the posterior typically yields a parsimonious set of founders. This nonparametric Bayesian framework allows us to exploit the genetic relatedness between populations in a principled way by describing the ancestral populations in terms of a common set of founder haplotypes. In Sohn and Xing (2009), a similar approach using a hierarchical Dirichlet process has been successfully used for the problem of haplotype inference from multipopulation data. However, the recombination process was not explicitly modeled in that work and a rather heuristic approach was employed to handle the linkage disequilbrium structure.
In our comparative study with two state-of-the-art methods of LAMP (Sankararaman et al. 2008a,b; Pasaniuc et al. 2009) and HAPMIX (Price et al. 2009), we show that the proposed method, which we call the admixture model based on multiple SPECTRUM representations (mSpectrum), enjoys enhanced robustness and accuracy, evidenced by its substantially reduced sensitivity to the choice and the amount of ancestral population data. In particular, our method shows very competitive performance even when the sample size of the ancestral population data is very small. This highlights the potential usefulness of this method in the analysis involving underrepresented populations of limited data availability. In addition, the compact population characterization by an infinite hidden Markov model improves the model flexibility over that of existing haplotype-based approaches so that it can naturally handle an arbitrary number of ancestral populations instead of only two in HAPMIX. It is also robust even under deviation from the common modeling assumption that multiple populations participate in the admixture at the same time as in Pasaniuc et al. (2009). The performance of our model is superior in terms of the proportion of spuriously estimated ancestries under general multiway admixture assumption as well.
In the remainder of this article, we first describe the statistical model and the inference method. Then we validate the proposed method through simulation study and show the empirical analysis result using Human Genome Diversity Panel data (Jakobsson et al. 2008). A discussion follows and concludes the article.
Methods
Problem setting
We consider an admixed population in which J ancestral populations have mixed since G generations ago. For example, if we are to recover the local ancestry of individuals in a Latino population (admixed population), we can incorporate J = 3 populations of ancient Africans, Europeans, and Native Americans as our ancestral populations. In our problem setting, we assume that the haplotypes composed of single-nucleotide polymorphisms are given for the ancestral populations and the admixed population. We will recover the pool of hypothetical founder haplotypes and their associations to individuals by statistical inference. The association of admixed individuals to the ancestral populations will be recovered along with their association to the founders, which would lead to the estimation of local ancestry.
Overview of admixture model based on founder haplotypes
The choice of representation about how to characterize a population is the crucial starting point in admixture modeling. Unlike most previous approaches that use allele-frequency profiles (Sankararaman et al. 2008a,b; Pasaniuc et al. 2009) or representative ancestral haplotypes in their raw forms (Sundquist et al. 2008; Price et al. 2009), we employ a new haplotype-based method that builds on an assumption of hypothetical founder haplotypes of unknown cardinality. The founder-based population model with explicit recombination modeling has been introduced in Sohn and Xing (2007) with the application to population structure and recombination analysis. Under this approach, each individual in a population is generated from the hypothetical pool of founders via a series of recombination and mutation. An individual haplotype can then be viewed as a mosaic of the founders whose pattern is determined by the association with founders. This mosaic process could be modeled as a hidden Markov model in which the founders correspond to the hidden states, the individual haplotypes correspond to the observation sequences, the transition process is modeled by the recombination process, and the emission process is modeled by the mutation from founders to the individuals. By employing an infinite hidden Markov model, the number and the haplotypes of the founders can be recovered through posterior inference rather than being prespecified, and the local inheritance association between the founders and the study individuals can also be derived.
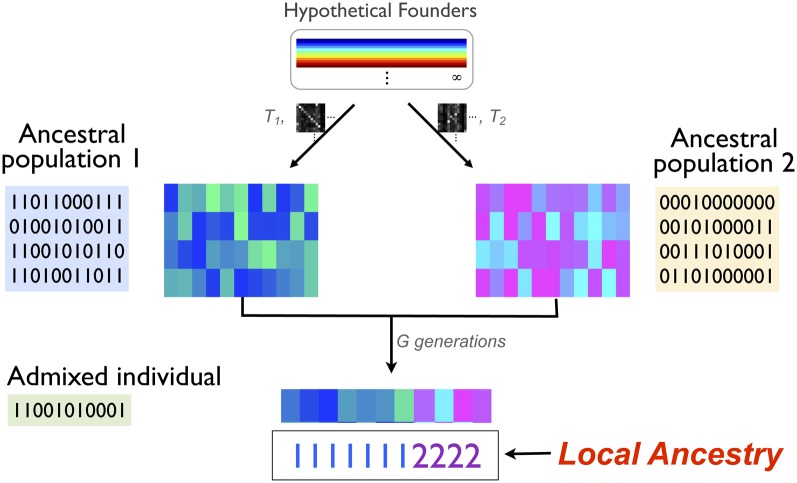
Now we further extend this population model to describe admixture events from an arbitrary number of ancestral populations. When the ancestral populations start to mix and form an admixed population, each individual haplotype in the admixed population can be decomposed into blocks with distinct ancestry. For each of these blocks, we can trace back the source of the genetic materials to a haplotype in the corresponding ancestral population. Now, recall that this “ancestral haplotype” is modeled as a mosaic of its founders. This means that each ancestry block in an admixed individual is further dissected into a finer-grained mosaic of founders. Therefore, the admixed inheritance process is a composite process with two different resolutions, one from the founders to ancestral haplotypes and the other from the ancestral haplotypes to the admixed individuals. A graphical illustration of the proposed model is shown in Figure 1. A variant of the infinite hidden Markov model is employed to make the choice of founders and the ancestral populations at the same time along the chromosome.
Figure 1 .
Graphical illustration of the proposed model.
Statistical model for generating ancestral and admixed population data
We now describe in detail the admixed inheritance model as a generative process of the individual haplotypes in ancestral populations and an admixed population with respect to a set of hypothetical founders.
Transition and emission probabilities:
For ease of description, we assume that the individuals are haploids. Let individual haplotypes in an admixed population be indexed by i, ancestral populations by j, and the markers by t. And let Hit ε {0, 1} and Fkt ε {0, 1} represent the alleles of individual i and founder k at marker t, respectively. We introduce a set of hidden state variables Sit = (Cit, Zit), where Cit ε {1, 2, …} and Zit ε {1, … , J} represent the indicator variables that select a founder haplotype and an ancestral population, respectively, on an ith admixed haplotype at marker t. For each ancestral population j, let νjk be the initial and background probability of founder k, and let be the transition probability that determines the probability of switching from copying founder k′ to founder k. We also introduce a set of scale parameters Tj ε (0, ∞) that scale the recombination rate in each population j by Tj. The role of these parameters is to take into account the difference in the hypothetical time since the founder population and also the effective population sizes of different ancestral populations. Let η = (η1, … , ηJ) denote the global admixing proportion such that ηj is the expected proportion of ancestral population j in the admixed population, let G ε [0, ∞) represent the time since admixture in the admixed population, and let r = (r1, r2, … , rT) and d = (d1, … , dT) represent the recombination rate and the physical distance between each neighboring marker, respectively. The final transition probabilities and the emission probabilities are defined as
| (1) |
| (2) |
where I(⋅) represents an indicator function such that I(i = j) = 1 if i = j and 0 otherwise. We assume a founder-specific mutation parameter δk that determines the probability of mutation during the inheritance from a founder k to individuals.
The overall idea underlying this representation is the two-layered inheritance framework, one from the time of hypothetical founders to ancestral populations and the other from those ancestral populations to the admixed population. If we set G = 0 in Equation 1, this two-layered framework is reduced to the model of the first layer that characterizes the ancestral populations with respect to the founder haplotypes. Under the reduced model, each population is associated with its own hidden Markov model parameters and the recombination rate scaled by Tj. Suppose (Ci,t−1, Zi,t−1) = (k′, j′), which means the ith haplotype has inherited from founder k′ at marker t − 1 in ancestral population j′. At the next marker t, either it selects a new founder k with probability and assigns Cit = k, or no recombination takes place with the remaining probability and Cit = Ci,t−1. If we trace the values of Cit across all the t, it will decompose the haplotype i into blocks with distinct associated founders. Therefore, each chromosome can be thought of as a mosaic of such founders.
Now, at the second layer that involves the admixture, this sequential process for selecting founders Cit occurs within the same ancestral population with probability so that Zit = Zi,t−1. Or with probability , a new population j as well as a new founder k is chosen jointly with a probability proportional to the product of admixing proportion ηj and the background probability νjk. Therefore, haplotypes both in the ancestral populations and in the admixed population are modeled as mosaics of founders determined by the sequence of Cit. In addition, each admixed individual i is associated with another resolution of mosaic determined by the sequence of Zit across t. The estimation of local ancestry can be done by tracing the posterior probability of Zit along the markers.
Note that even when no admixing is assumed, we still have the flexibility of choosing a different founder haplotype. This feature helps to control the number of transitions among populations effectively so that the hidden state does not need to change excessively. Moreover, although we assume the J populations participate in the admixture simultaneously, the population-specific scale parameters would explicitly allow heterogenous resolution of the genetic mosaics in different ancestral populations to be generated. This greatly improves robustness of the model against the violations of such modeling assumption as well as the accuracy of the ancestry estimation.
The cardinality of the founder space:
Instead of fixing the number of hypothetical founders by doing statistical model selection, we adapt a more flexible nonparametric approach using an infinite hidden Markov model (iHMM) (Beal et al. 2002; Teh et al. 2010). Recall that if we consider finite, say K, hidden states, the transition probabilities will be represented as a K × K matrix. Each row k of this matrix sums to one and defines the probabilities of switching from a source state k to all the target states.
Now, if we consider an infinite hidden state space, each row of the transition matrix would be an infinite dimensional vector that sums to one. Dirichlet process (DP) (Blackwell and MacQueen 1973; Ferguson 1973) has been effectively used to describe such probability distribution. A DP is defined by two parameters: the base measure (“mean” of the DP) and the scale parameter that controls the concentration around the mean. To ensure all the row-specific DPs are built on the same state space, another Dirichlet process is shared as a common base measure at a top level. This model for the hidden Markov transition probabilities actually corresponds to a hierarchical Dirichlet process (Teh et al. 2010). We omit the statistical details of an infinite hidden Markov model formulation in terms of a hierarchical Dirichlet process here (see Beal et al. 2002, Sohn and Xing 2007, and Teh et al. 2010 for more details). Basically, the (k, k′) element of the transition matrix πj defines the transition probability from state k to state k′ in population j, and for a given source state k, the target state index k′ can increase as large as needed by the given data. Infinite-dimensional vector of initial probabilities νj can be defined in a similar way under the same hierarchical Dirichlet process framework. Since we consider multiple such infinite HMMs for multiple populations, we let the same base measure be shared across all the populations. This infinite HMM-based framework leads to a very simple solution to how many founders to consider and how to construct the founder space across multiple populations. The iHMM parameters of our admixture model thus can be summarized as
where α0 and γ define the scale parameters for the population-specific DPs and the top-level DP, respectively.
Other parameter description:
We assume a Dirichlet distribution prior for the population proportion parameter η ∼ Dirichlet(ξ1, … , ξJ) and a Beta prior for each of the mutation parameters δk.
For simplicity of inference, we transform the variables such that rt and Tj are combined as . Similarly, we use the notation . We assume these variables are i.i.d. under a Gamma prior. Then Equation 1 is transformed as follows:
| (3) |
In summary, infinite hidden Markov model parameters combined with population genetics parameters are used to capture different characteristics in populations and to describe an admixture event from an arbitrary number of ancestral populations. While we assume an infinite number of founders a priori, the posterior inference usually produces a small number of founders and this leads to a compact representation of a population for the admixture analysis.
Posterior inference
To overcome the drawbacks of slow convergence in traditional Gibbs sampling, we employ a variant of beam sampling proposed for an infinite HMM (Van Gael et al. 2008). Basically, it extends the well-known dynamic programming technique of a forward–backward algorithm in a finite-state HMM to an infinite-state space case. It exploits the property that in an observation sequence of finite length, the number of actually realized hidden states is finite at each iteration step. Therefore, the number of states to be considered in a forward–backward algorithm can be adaptively changed over iterations. More specifically, a set of auxiliary variables u is sampled conditional on S such that given u1, … , uT, the number of states K having positive forward probabilities is finite. More details of the beam sampling scheme for the proposed model are described in supporting information, File S1.
Since the entire inheritance process from founders to ancestral populations and then to the admixed population is modeled in a single Bayesian framework, it allows the exact posterior inference by putting the ancestral and admixed population data together in a single series of beam sampling iterations (see File S1). However, this is not optimal in terms of time complexity as we often favor running multiple test sets after we extract reference information about the ancestral populations. Therefore, we split the whole inference process into two phases: (1) the training phase where the model parameters about ancestral populations are learned and (2) the ancestry estimation phase that actually recovers the ancestry of admixed individuals.
One caveat of this decomposition is that we may not fully take advantage of the flexibility of the infinite model. This is because we need to constrain the hidden state space somehow as a finite space when the output from the training phase is returned. As an nth posterior sample from Bayesian inference of the training phase, we get a finite number K(n) of founder haplotypes and the related HMM parameters of π(n) and ν(n) with for each j. Averaging these results as one training output is not straightforward as K(n) can be different across different n. A plausible approach would be to keep multiple, say N posterior samples □ = {F(n), π(n), ν(n)}n = 1,…,N and run the ancestry estimation routine N times using each of these parameters in □. Then the N posterior distributions of the ancestry indicator variable Z can be easily averaged to form the final posterior distribution since Z is defined over a fixed number of populations J unlike C or other parameters that depend on K. Note that does not depend on K, so we can use the posterior mean of as the final estimate for it. Another practical approach would be to select a single output from the training phase such as a MAP solution and estimate the local ancestry based on the single set of parameters. Empirically, we observe that the performance degradation by this MAP solution with respect to the first approach is relatively small.
Training phase:
For an individual in an ancestral population j, we can set the time since admixture G to be zero and the population indicator variables Z to be observed as constant. Then the hidden state variable Sit = (Cit, Zit) can be replaced with a Cit indicating the founder and Equation 3 is reduced to the following:
We infer the variable C through the beam sampling algorithm described in Equation A2 in File S1 and the other variables through the standard Gibbs sampling.
Note that the contribution of transition at neighboring loci t − 1 and t to the parameter π and is not all equal because of the self-transition probability forced by the recombination model in Equation 3. We handle this by sampling auxiliary binary variables to indicate whether the jump occurs in the transition or not. The transition probability can be decomposed as follows:
Then we sample Mit given Cit and Ci,t−1 backward in a forward–backward process from
Now, π can be sampled as in Van Gael et al. (2008), but conditional on M, which involves the transitions with Mit = 1 only. can also be sampled conditional on M, using . The overall sampling procedure is summarized in Algorithm 1.
Algorithm 1 Procedure for training iHMMs in reference populations
Input: Haplotype data H for ancestral populations
Output: N posterior samples of founders and the related HMM parameters {F(n), π(n), ν(n), gr(n)} for n = 1, … , N
-
1: repeat
-
2: for each individual chromosome i do
-
3: Sample the auxiliary variables uit for t = 0, … , T − 1.
-
4: Sample Cit | u, H, F using the beam sampling algorithm
-
5: Sample Fk,t and δk
-
6: Sample parameters ν, π, β, and gr.
-
7: end for
-
8: until convergence.
Ancestry estimation phase:
As the variables F, gr, ν, and π are returned in the training stage, the unknown variables now are the global admixing proportion η, the generations since admixture G, the mutation rate δk of founders, and S = (C, Z) for the admixed individuals. We resample δk in the ancestry estimation phase instead of getting it from the training step because δk can reflect additional information about the admixed population by describing it in terms of the discrepancy between founders and the population. As we now deal with a finite number of hidden states obtained from the training phase, it is not necessary to incorporate the auxiliary variable u to sample S in the ancestry estimation phase. The variables Sit thus are sampled through a standard forward–backward algorithm. As in the training stage, the transition probability at each marker can be decomposed into two parts, depending on whether the jump process for admixture occurs or not. We use a similar technique to sample Gr by introducing an auxiliary variable . The overall sampling scheme is summarized in Algorithm 2.
If the time since admixture G, admixing proportion η, and the recombination rate r are assumed to be known as is often the case in admixture analysis, we can omit the second step of parameter sampling (line 5 in Algorithm 2) and reuse δk that can be returned from the training stage. Then it is also possible to get an approximate solution by use of a posterior decoding from forward–backward steps in a finite dimensional HMM.
Algorithm 2 Procedure for estimating local ancestry in an admixed individual
Input: Haplotype data H for an admixed population, estimated parameters {F(n), π(n), ν(n), gr(n)}
Output: Posterior distribution of Z = (Zit).
-
1: for n = 1, … , N do
-
2: repeat
-
3: for each individual chromosome i do
-
4: Sample Sit = (Cit,Zit) | H, F using the forward–backward algorithm
-
5: Sample δk, η, and Gr .
-
6: end for
-
7: until convergence
-
8: Keep S posterior samples of Z
-
9: end for
10: Average N ⋅ S posterior samples and return the final posterior distribution of Z.
Results
Simulation design
To validate the proposed method, we simulated admixed haplotypes using the Human Genome Diversity Project (HGDP) data genotyped on Illumina Infinium HumanHap550 BeadChips (Jakobsson et al. 2008). Considering previous results that have revealed distinct genetic characteristics across different continents, we selected the following reference populations that would serve as putative ancestral populations: YRI for African, CEU for European, JPT and CHB for East Asian, and Maya for Native American ancestry. Each of the resulting ancestral populations contained 30, 30, 28, and 13 individuals, respectively. In the simulation study, we first focus on chromosome 22 for computational efficiency under diverse types of simulation scenarios.
To take into account the discrepancy between real ancestral populations and those used in training, we generated admixed individuals using populations that are similar but not identical to those used as ancestral populations. For example, individuals in Russian and BantuKenya populations are mixed to simulate an admixed population and then the local ancestries of these individuals are estimated with respect to CEU and YRI populations. A simulation scheme similar to that in Price et al. (2009) was used to generate admixed haplotypes as follows. For each haplotype in an admixed population, we first sample the ancestry j ε {1, … , J} at the first marker according to the probabilities η = (η1, … , ηJ) and randomly select an ancestral haplotype in the corresponding population j to copy the allele at the first marker. For the following markers, either we assign the same ancestry as that of the previous marker with probability exp(−rtdtG) and copy the allele of the same ancestral haplotype at the corresponding marker or, with probability 1 − exp(−rtdtG), we resample the ancestry j′ among the J possible populations on the basis of the probabilities η and randomly reselect the ancestral haplotype for the allele copy within the selected population j′. We use a constant recombination rate of rt = 10−8 per base pair per generation as in previous studies (Sankararaman et al. 2008a,b). Note that our simulation data are not generated under our modeling assumption based on founder haplotypes, but in a more general setting that is commonly considered in previous admixture studies. For each simulation scenario below, we generate 30 admixed individuals per data set.
The performance is measured as the mean squared error rate of ancestry probabilities along the loci. Specifically, let pijt denote the probability of ancestry j at a locus t in an individual i. The average error rate of across all the individuals is reported. We compare our results with the two state-of-the-art methods: LAMP (Sankararaman et al. 2008a,b; Pasaniuc et al. 2009), the method based on allele-frequency profiles as reference information, and HAPMIX (Price et al. 2009) that uses representative ancestral haplotypes. These methods appear to outperform other methods such as HAPPA (Sundquist et al. 2008), SABER (Tang et al. 2006), or ANCESTRYMAP (Patterson et al. 2004) in previous studies (Price et al. 2009). Since the benchmark algorithms require the parameters for recombination r, the admixture time G, and the population proportion η to be specified as input, we provided the true values of these parameters to all the algorithms in the simulation study. Additionally, each set of haplotype data for ancestral populations was converted to allele-frequency profiles and then LAMP was run with these frequency data as input. For the analysis below, we used the MAP solution as our parameter estimation from the training phase.
Performance on two-way admixture
The first simulation scenario considers two-way admixture of ancient European and African populations. We generate admixed individuals using BantuKenya and Russian population data with the admixing proportion of η = (0.5, 0.5) and then the local ancestries of the admixed individuals are estimated with respect to YRI and CEU. In Figure 2, we first display the true and the estimated local ancestry probabilities of two sample individuals in an admixed population. The yellow color corresponds to YRI ancestry, and the dark green corresponds to CEU ancestry. The length of the vertical color bar at each chromosomal location along the x-axis is proportional to the corresponding ancestry probability. While all the algorithms produce reasonable results in general, the proposed method denoted by mSpectrum is especially effective in picking out fine details of ancestry changes as can be seen in the example.
Figure 2 .
True and estimated local ancestries of two sample individuals in an admixed population from African and European populations. The x-axis corresponds to chromosomal position and the y-axis corresponds to the ancestry probability (yellow, African; dark grean, European).
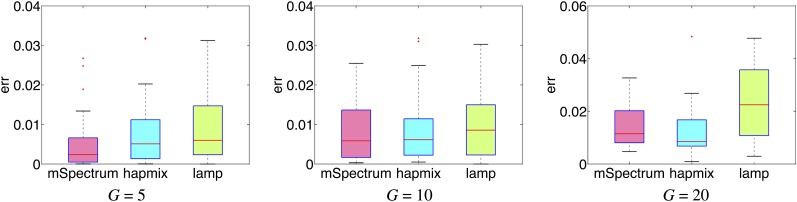
The overall performance of each algorithm across all the generated samples is shown in Figure 3. Roughly, we can see that mSpectrum and HAPMIX perform comparably to each other and tend to outperform LAMP in the case of two-way admixture. Still, all three algorithms perform reasonably well as can be seen in the small overall error rates. For example, the average error rates for G = 10 were 0.0077, 0.0086, and 0.0116 in mSpectrum, HAPMIX, and LAMP, respectively.
Figure 3 .
Boxplot for mean squared error rates of ancestry estimation for two-way admixture of African and European populations since G generations ago.
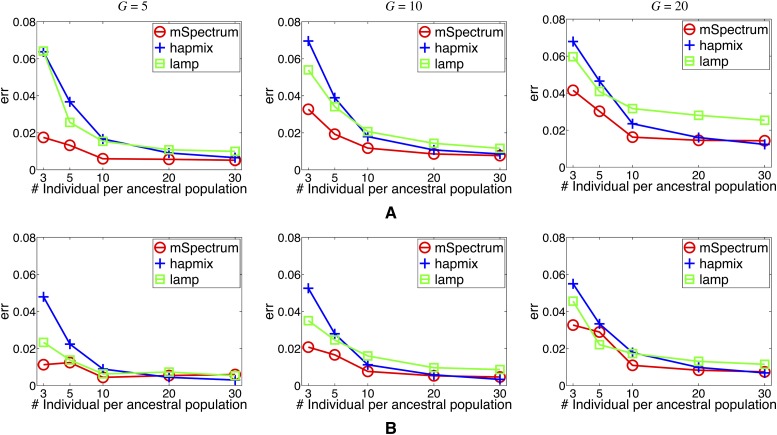
Performance as a function of data size in the training set
To further evaluate each method in terms of its performance with respect to the training data size, we varied the number of available individual samples per ancestral population. We trained the model using 3, 5, 10, 20, and 30 individuals, hence 6, 10, 20 40, and 60 haplotypes, per ancestral population and estimated the ancestries on the basis of each of the trained models. The performance of each algorithm is presented as a function of training data size in Figure 4 for two scenarios: (a) the two-way admixture scenario from BantuKenya and Russian populations of which the result on the full data set is shown in Figure 3 and (b) the admixture of YRI and CEU populations where the individuals not contained in the training data are used to generate the admixed individuals. It is clearly seen that the proposed method substantially outperforms the other benchmark algorithms in both cases, especially when the data size is small. Even when only a few ancestral haplotypes are available, it still gives very good estimates of the local ancestries compared to the others. Therefore, our method can be especially useful in the analysis of admixture effect involving nontraditional populations where the amount of available genotypes is still limited. In addition, our method shows greater performance gain over the other two methods when the discrepancy between the training population and the one used in the simulation is large. This implies that the hierarchical structure put on top of the ancestral population data allows a more general description of the ancestral populations and hence enhances the accuracy of the ancestry estimation even when the ancestral population used for reference has diverged from the true ancestral populations.
Figure 4 .
Error rate as a function of the number of individuals per training population. (A and B) Two-way admixture of African and European populations since G generations ago using (A) Russian and BantuKenya populations and (B) CEU and YRI populations.
Performance on three-way admixture
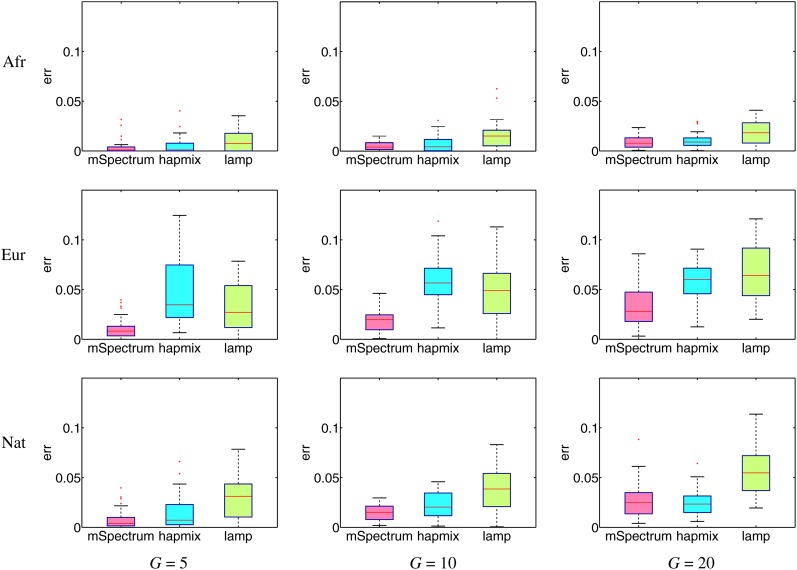
We now consider the admixture involving more than two ancestral populations. Analogous to the formation of the Puerto Rican population (Tang et al. 2007), we included CEU, YRI, and Maya as ancestral populations for African, European, and Native American ancestry and generated an admixed population using Russian, BantuKenya, and Pima with admixing proportions of 0.66, 0.18, and 0.16, respectively.
Figure 5 shows the resulting error rates across different values of G. Since HAPMIX cannot handle more than two ancestral populations directly, we ran it in three different modes such that each run tries to estimate the targeted ancestry vs. the other two ancestries as was done in its original procedure (Price et al. 2009). For this reason, we compare the performance on each ancestry separately. Overall, our method performs significantly better than the other two in most of the analyzed cases.
Figure 5 .
Boxplot for mean squared error rates of ancestry estimation. Shown is three-way admixture of African, European, and Native American populations since G generations ago. Since HAPMIX is applicable to only the two-way admixture case and was run to estimate each ancestry vs. the other two, we report the error rate on each ancestry separately.
Robustness under deviation from admixture assumption
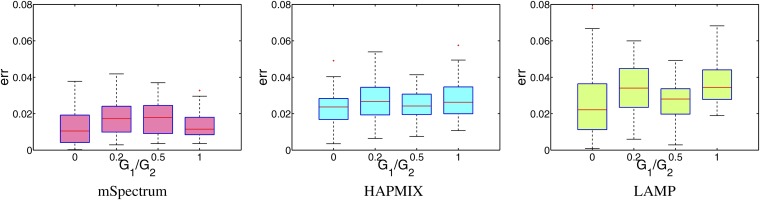
The modeling assumption that all the ancestral populations participate in the admixing simultaneously does not hold in reality, especially in the case of multiway admixture involving multiple ancestral populations. We test the robustness of each method under deviation from such a modeling assumption by generating admixed haplotypes from three ancestral populations that started to mix at two different time points. More specifically, Russian and BantuKenya populations are mixed for G1 generations with a 50%/50% proportion. Then this admixed population is mixed with the third population of Pima for G2 generations with 50%/50%, resulting in the overall proportion of 0.5, 0.25, 0.25. We fixed G2 to be 10 and varied G1 to be 0, 2, 5, and 10, where G1 = 0 corresponds to the case in which the modeling assumption holds. The result is summarized in Figure 6. In each plot, the x-axis corresponds to the values of G1/G2 and the y-axis shows the error rates. The proposed method resulted in not only the lowest error rates, but also the most stable performance across different values of G1/G2. For more quantitative comparison of robustness across different algorithms, we calculated the linear regression coefficient of G1/G2 vs. the error rates. The resulting slopes were −0.0011, 0.0029, and 0.0074 for mSpectrum, HAPMIX, and LAMP, which again supports the superior robustness of the proposed method.
Figure 6 .
Robustness under deviation from the modeling assumption. The x-axis represents the ratio G1/G2, where G1 denotes the number of generations for which the first two populations mixed and G2 means the additional number of generations since the third population joined and they have further mixed together.
Performance under four-way admixture assumption
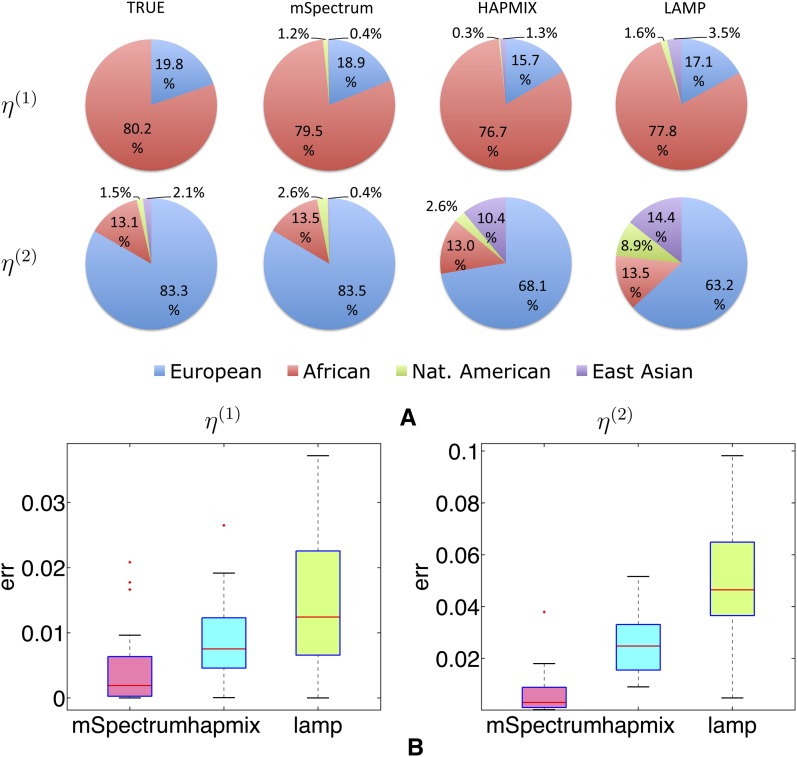
When it is unclear how many or which ancestral populations have contributed to the given admixed population due to unknown population history, one needs to run the local ancestry estimation under the general assumption of multiway admixture involving all the candidate ancestral populations. In this case, the proportion of spurious association to the noncontributing population is also an important measure for performance comparison in addition to the mean squared error rates for local ancestry estimation. Or when the contribution of a certain population is extremely small, we can test how sensitively each algorithm detects such a small portion of ancestries. To examine the behavior of each algorithm under such cases, we let each algorithm assume four ancestral populations of CEU, YRI, Maya, and JPT+CHB and then estimate the ancestry of admixed haplotypes generated from Russian, BantuKenya, Pima, and Yi populations with admixing proportions of η(1) = (0.2, 0.8, 0, 0) and η(2) = (0.8, 0.15, 0.03, 0.02).
We first illustrate the true and the estimated local ancestry probabilities of two sample individuals in each of the admixed populations generated using η(1) and η(2) in Figure 7, A and B. The red color corresponds to YRI ancestry, the black corresponds to CEU, and the yellow and the white correspond to Maya and JPT+CHB ancestries, respectively. We find that mSpectrum shows the most accurate and stable result with the least amount of spurious association in both cases.
Figure 7 .
True and estimated local ancestries of two sample individuals in an admixed population from African and European populations when the ancestry is estimated with respect to four ancestral populations of YRI (red), CEU (black), Maya (yellow), and JPT+CHB (white). The x-axis corresponds to chromosomal position and the length of each colored vertical bar is proportional to the corresponding ancestry probability.
The global admixing proportion computed as the average local ancestry proportion across all the markers and all the individuals in the admixed population is summarized in Figure 8A. For the first scenario using η(1) that involves only European and African ancestries, the mean proportions of spuriously estimated ancestries are 0.016, 0.016, and 0.051 for mSpectrum, HAPMIX, and LAMP, respectively. (For HAPMIX, since each ancestry proportion is estimated under a two-way admixture assumption of one ancestry vs. all the others, the ancestry proportions across all the populations do not necessarily sum to one. While the pie charts and the illustration in Figure 8 show the normalized results, we report the numbers before normalization on the pie charts because we find this estimation is more accurate than that after normalization.) In the case of the second scenario using η(2) where the true combined proportion of Maya and JPT+CHB populations is 0.036, the estimated proportions of these ancestries in each algorithm are 0.03, 0.13, and 0.23, for mSpectrum, HAPMIX, and LAMP. This result shows that our method is indeed effective in preventing excessive transitions between ancestral populations and hence reducing the proportion of spurious estimations. Figure 8B shows that mSpectrum significantly outperforms HAPMIX or LAMP in terms of the mean squared error rates for the local ancestry estimation as well.
Figure 8 .
Performance under the four-way admixture assumption when the admixed population is generated with admixing proportions of η(1) = (0.2, 0.8, 0, 0) and η(2) = (0.8, 0.15, 0.03, 0.02) using Russian, BantuKenya, Pima, and Yi populations. We show two performance measures of (A) the true and the empirical η estimated as the average local ancestry proportions across individuals and markers and (B) the mean squared error rates of local ancestry estimation.
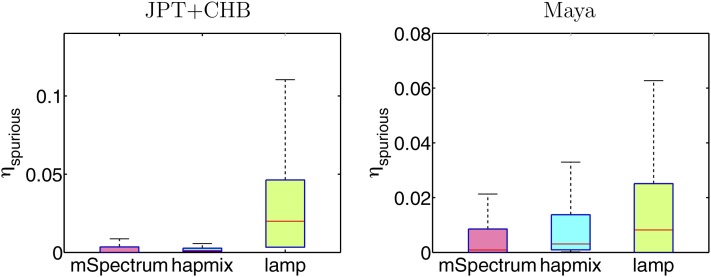
For more detailed comparison of the proportion of spurious ancestries in different methods, in Figure 9, we show the overall distribution of the spuriously estimated ancestry measured over 50 data sets simulated by η(1) . We find that mSpectrum and HAPMIX estimate similar proportions of spurious JPT+CHB ancestry, which is substantially less than that from LAMP. On the other hand, mSpectrum is the most accurate in preventing spurious Maya ancestry.
Figure 9 .
Spuriously estimated ancestry proportions under the four-way admixture assumption when the admixed population is generated using Russian and BantuKenya populations only, computed over 50 data sets, each of which contains 30 individuals.
Sensitivity analysis on model parameters
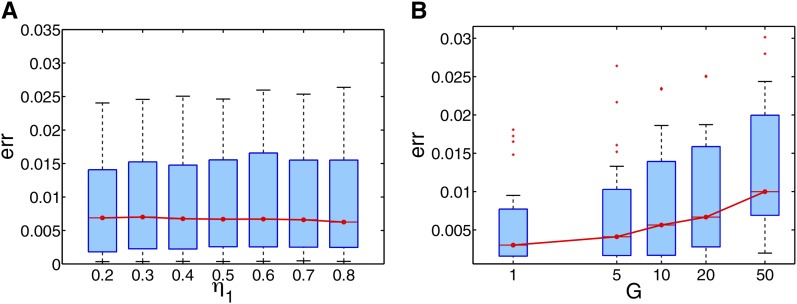
Since the parameters of η and G were assumed to be known in our simulation study in parallel with other methods, we also examine how the performance of mSpectrum is affected by incorrectly specifying these parameters. The performance is shown for the data set simulated with G = 10 and η = (0.5, 0.5) with respect to YRI and CEU ancestries in Figure 10. In each plot, the x-axis shows the specified parameters where the values are shown in log scale in the case of G. We could see that there was almost no effect when η was incorrectly set in the range from 0.2 to 0.8. When we examined the result on G, the algorithm had the general tendency to favor a specified value G smaller than the true value. The effect of a misspecified value of G was minimal when the discrepancy was within a factor of 2. Even in the extreme case such as G varied by a factor of 5, the error still remained within twice the error rates when the true value was given.
Figure 10 .
Sensitivity analysis: boxplot for error rates as a function of specified parameter values (A) η1 and (B) G when the true values are ηtrue = (0.5, 0.5) and Gtrue = 10.
Empirical analysis of HGDP data
To illustrate our method on real data, we applied it to 22 autosomes of the HGDP data set (Jakobsson et al. 2008). Four ancestral populations of YRI, CEU, JPT+CHB, and Maya were chosen as in the simulation study to represent African, European, East Asian, and Native American ancestries. We then recovered the local ancestries in the remaining 28 populations. Since the time since admixture is not available for real data, we let our program estimate the parameters by posterior inference.
The mean ancestry proportion of each population estimated from our algorithm is summarized in Table 1. Overall, the ancestry vector agrees very well with their geographical locations or known history. For example, populations such as Yoruba, Mandenka, BiakaPygmy, or BantuSouthAfrica recovered pure African ancestries; Druze, Basque, Russian, and Adygei populations had dominant European ancestries (≥0.978); and Pima or Colombian populations resulted in almost pure Native American ancestries (≥0.983).
Table 1 . Estimated ancestry proportions of populations in the HGDP data set.
| African | European | East Asian | Native American | |
|---|---|---|---|---|
| Yoruba | 1.000 | 0.000 | 0.000 | 0.000 |
| Mandenka | 1.000 | 0.000 | 0.000 | 0.000 |
| BiakaPygmy | 1.000 | 0.000 | 0.000 | 0.000 |
| BantuSouthAfrica | 1.000 | 0.000 | 0.000 | 0.000 |
| San | 0.999 | 0.001 | 0.000 | 0.000 |
| MbutiPygmy | 0.999 | 0.000 | 0.000 | 0.001 |
| BantuKenya | 0.998 | 0.001 | 0.000 | 0.000 |
| Mozabite | 0.141 | 0.818 | 0.013 | 0.028 |
| Bedouin | 0.035 | 0.941 | 0.006 | 0.018 |
| Palestinian | 0.013 | 0.966 | 0.006 | 0.015 |
| Basque | 0.000 | 0.998 | 0.000 | 0.001 |
| Russian | 0.000 | 0.990 | 0.003 | 0.007 |
| Druze | 0.002 | 0.989 | 0.002 | 0.006 |
| Adygei | 0.000 | 0.978 | 0.008 | 0.014 |
| Kalash | 0.000 | 0.930 | 0.027 | 0.043 |
| Balochi | 0.015 | 0.888 | 0.031 | 0.066 |
| Burusho | 0.000 | 0.741 | 0.088 | 0.170 |
| Uyghur | 0.000 | 0.348 | 0.414 | 0.239 |
| Yakut | 0.000 | 0.045 | 0.848 | 0.106 |
| Mongola | 0.000 | 0.006 | 0.960 | 0.034 |
| Daur | 0.000 | 0.004 | 0.972 | 0.024 |
| Cambodian | 0.000 | 0.004 | 0.977 | 0.019 |
| Lahu | 0.000 | 0.000 | 0.987 | 0.013 |
| Yi | 0.000 | 0.001 | 0.991 | 0.009 |
| Melanesian | 0.001 | 0.039 | 0.821 | 0.140 |
| Papuan | 0.002 | 0.081 | 0.733 | 0.185 |
| Pima | 0.001 | 0.012 | 0.004 | 0.983 |
| Colombian | 0.002 | 0.001 | 0.001 | 0.996 |
More interestingly, the result also identifies the populations that have strong evidence of admixing effect among multiple ancestries. For instance, the proportion of European ancestry in the Uyghur population was 0.35, that of East Asian ancestry was 0.41, and the remaining proportion of 0.24 was of Native American ancestry. Although only one or two populations are selected to serve as each putative ancestral population in our study and hence the interpretation of this result needs to be done carefully, our result largely agrees with the previously reported ancestry proportion in this population. For example, the analysis in Xu et al. (2008; Xu and Jin 2008) claimed that Uyghur had ∼50–60% of European ancestry and 40–50% of East Asian ancestry from the analysis based on two-way admixture. More recent study in Li et al. (2009) showed evidence that the estimation of European ancestry in these studies appears to be biased and suggested a newly estimated proportion of ∼30%. Our estimation of East Asian ancestry (41%) is similar to that in Xu et al. (2008) and in addition the estimation of European ancestry (35%) is closer to the more recent result in Li et al. (2009) than that in Xu et al. (2008). Considering its geographical location and the resulting population history, our result suggests that the Uyghur population has ∼35% of European ancestry, 41% of East Asian ancestry, and the remaining proportions of ancestries in other contributing populations that have greater similarity to the Native American population.
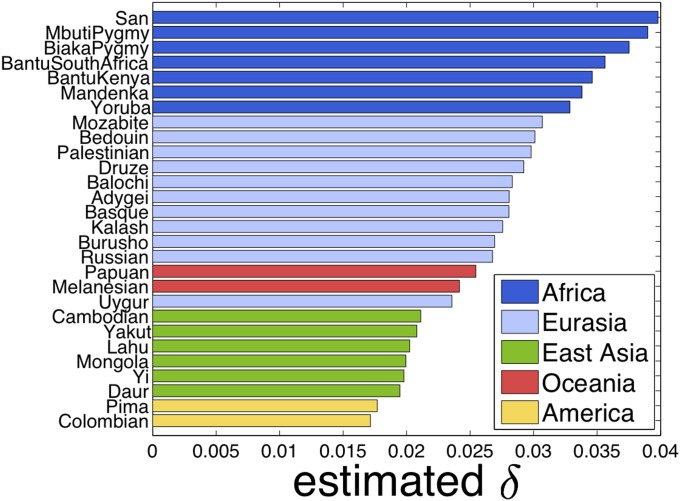
To further analyze each set of population data and the behavior of the proposed method, we examined the empirical mutation parameter of each study population computed as an average discrepancy between individuals and corresponding founders within each of the populations. Therefore, can be viewed as reflecting the level of divergence from the founder population. The result is displayed in Figure 11 where the colors of the bars are based on the geographic location of the corresponding population. The ordering of populations by their parameter values almost exactly agrees with the geographic locations out of Africa. That is, all the populations in the African continent had the largest values of δ, populations in Eurasia came next, and Oceanian populations were the third. Populations in the East Asian region formed the fourth cluster and then Pima and Colombian populations showed the smallest values of δ. It is noteworthy that Yoruba, which appears to be the closest to the training population of YRI, recovers a much larger value of mutation rate δ than all the populations in geographic locations other than the African continent. This comes from the nice property of our model that we do not directly use the training haplotype data as our reference; we rather infer the corresponding common founders across all the population data together and then work in a framework dealing with founders and admixed individuals. Otherwise, it would be impossible to obtain such a result because the discrepancy of Yoruba and its reference data would be much smaller than that of most of the other populations.
Figure 11 .
The empirical mutation rate δ of each HGDP population computed as the average discrepancy between individuals and their founders.
Discussion
Previous admixture studies have suggested that the world populations are not independent of each other, but rather are structured through population admixing history and the resulting gene flow. Most existing approaches for local ancestry estimation have ignored such relatedness and treated the populations as unrelated. We explore this dependency among populations and efficiently utilize it by building a unified model that covers all the ancestral populations and the admixed population together. As shown in Results, this modeling strategy is especially helpful when only a limited amount of data are available to represent the ancestral populations. Since genetic information in one population can be naturally shared by another population in such a framework, it effectively enhances the robustness of the proposed model regarding the choice of the ancestral population data.
In our comparative study, HAPMIX appears to perform very well when enough data for ancestral populations are given and also for older admixture events. However, this method does not allow one to analyze the admixing effect from more than two ancestral populations. Instead, one ancestry vs. all the other ancestries should be estimated. While this setting may be fine for some applications, this constraint limits its applicability to complex admixture scenarios and may compromise its ability to deal with older admixtures.
LAMP has a slightly different focus: while its performance was shown to be worse than the other two in general in our simulation study, it can deal with multiple ancestral populations as does our model. And computationally this method was significantly faster than the other two haplotype-based methods. LAMP seems to be more suited for the very recent admixture case, and its performance tends to drop quite sharply as we consider more ancient admixture events. On the other hand, in a very recent admixture case, LAMP tends to be less sensitive to the amount of training data than HAPMIX as shown in Figure 4. Our approach is more general and of more practical utility in that it can incorporate an arbitrary number of ancestral populations with comparable or superior performance to that in HAPMIX under various scenarios. In comparison of computation time with HAPMIX, our method requires additional, but off-line computation time for model training, which is linear in the number of individuals and the number of markers. For the ancestry estimation phase, we would additionally need a series of MCMC iteration time if we want to estimate the parameters of interest such as admixture time or mutation rates. As an of example running time of our algorithm, it took ∼5 min to run on a data set with 30 admixed individuals on chromosome 22.
In the proposed model, we adopted population-specific recombination rates by using a scaling parameter of Tj that explains the different effect of population size and the time since the founder population. Although it makes sense to scale the mutation rate by Tj as well in each of the ancestral populations, we found that the performance for the local ancestry estimation did not improve in our experiments. This might be due to a statistical reason. During inference, it is observed that the algorithm tends to favor the ancestral population with the smallest mutation rate excessively, so this might have created excessive bias toward such an ancestral population instead of selecting the correct ancestry.
Although our method allows us to estimate the admixture time parameter G instead of requiring it as an input when inferring the local ancestry, the parameter estimation result was not very accurate in general. Still, the local ancestry estimation performance was not significantly affected by incorrect estimation of the parameter as implied from our sensitivity analysis in Figure 10B. It appears that the likelihood surface from our statistical model is relatively flat over the space of model parameters, so the single optimal point on the model parameter space could not be achieved stably. When we let our program estimate G instead of fixing it in the same scenario considered in Figure 10B, the estimate of G averaged over 50 repetitions was ∼14 when the true value was G = 10. The ancestry estimation accuracy was comparable to the case when we fixed G as 10.
It is worth mentioning some of the previous approaches for global ancestry analysis as well to position our method in context. STRUCTURE (Pritchard et al. 2000) has been one of the most widely used softwares for admixture analysis, and more recently, other softwares such as EIGENSTRAT (Patterson et al. 2006) and ADMIXTURE (Alexander et al. 2009) have also gained great popularity especially for their computational efficiency. In global ancestry estimation problems, typically no prior information is provided for the ancestral populations and the ancestries of given individuals are recovered as mean proportions of each possible ancestry. Therefore, it can be considered as an unsupervised problem. In contrast, local ancestries are mostly estimated on the basis of the given reference information such as allele frequencies or genotypes of putative ancestral populations. There has been more recent work that bridges the gap between these two approaches. For example, LAMP can also run in an “unsupervised mode” such that it recovers the allele-frequency profiles of ancestral populations as well as the local ancestries. Also, ADMIXTURE, which is for global ancestry estimation, recently added a new feature that the known ancestries of some reference individuals can be exploited (Alexander and Lange 2011). For haplotype-based approaches, this extension is not straightforward in general because one needs to deal with a set of hidden haplotypes that results in a large number of parameters. Regarding this aspect, our model for the local ancestry has the desirable property that it integrates out the ancestral population data during the inference and works with the hypothetical founders and the admixed population data. Therefore, we expect that the extension of the model to an unsupervised case would also be a promising direction to pursue.
In this article, we assumed that phased haplotype data are given. In practice, a number of softwares are available for haplotype phasing (Scheet and Stephens 2006; Browning and Browning 2009; Li et al. 2010), so the phase information can be readily available in a processing step. It is also possible to extend our model to deal with unphased genotypes. For example, we may assume that the haplotypes of ancestral populations are given, and then we allow unphased genotypes for admixed individuals, as in the setting considered in Price et al. (2009). The only additional computation then would be one more step in our posterior sampling to recover the phasing of genotypes as well as the hidden states in the ancestry estimation phase.
Supplementary Material
Acknowledgments
This material is based upon work supported by a National Science Foundation Career Award to E.P.X. under grant DBI-0546594 and National Institutes of Health grant 1R01GM087694.
Footnotes
Communicating editor: A. M. Beaumont
Literature Cited
- Alexander D. H., Lange K., 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J., Lange K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal M. J., Ghahramani Z., Rasmussen C. E., 2002. The infinite hidden Markov model, pp. 577\x{2013}585 in Advances in Neural Information Processing Systems 14, edited by M. J. Beal, Z. Ghahramani, and C. E. Rasmussen. MIT Press, Cambridge, MA [Google Scholar]
- Blackwell D., MacQueen J. B., 1973. Ferguson distributions via Polya urn schemes. Ann. Stat. 1: 353–355 [Google Scholar]
- Browning B. L., Browning S. R., 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84: 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-Y., Kao W. H. L., Patterson N., Tandon A., Haiman C. A., et al. , 2009. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet. 5: e1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-Y., Reich D., Wong T. Y., Klein R., Klein B. E. K., et al. , 2010. Admixture mapping scans identify a locus affecting retinal vascular caliber in hypertensive African Americans: the Atherosclerosis Risk in Communities (ARIC) study. PLoS Genet. 6: e1200308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson T. S., 1973. A Bayesian analysis of some nonparametric problems. Ann. Stat. 1: 209–230 [Google Scholar]
- Huelsenbeck J. P., Andolfatto P., 2007. Inference of population structure under a Dirichlet process model. Genetics 175: 1787–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M., Scholz S. W., Scheet P., Gibbs J. R., VanLiere J. M., et al. , 2008. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451: 998–1003 [DOI] [PubMed] [Google Scholar]
- Li H., Cho K., Kidd J. R., Kidd K. K., 2009. Genetic landscape of Eurasia and ”admixture” in Uyghurs. Am. J. Hum. Genet. 85: 934–937, author reply 937–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Willer C. J., Ding J., Scheet P., Abecasis G. R., 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34: 816–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B., Sankararaman S., Kimmel G., Halperin E., 2009. Inference of locus-specific ancestry in closely related populations. Bioinformatics 25: i213–i221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Hattangadi N., Lane B., Lohmueller K. E., Hafler D. A., et al. , 2004. Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 74: 979–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909 [DOI] [PubMed] [Google Scholar]
- Price A. L., Weale M. E., Patterson N., Myers S. R., Need A. C., et al. , 2008. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 83: 132–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Tandon A., Patterson N., Barnes K. C., Rafaels N., et al. , 2009. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5: e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S., Kimmel G., Halperin E., Jordan M. I., 2008a On the inference of ancestries in admixed populations, pp. 424–433 in RECOMB’08: Proceedings of the 12th Annual International Conference on Research in Computational Molecular Biology, edited by M. Vingron and L. Wong. Springer-Verlag, Berlin/Heidelberg, Germany/New York
- Sankararaman S., Sridhar S., Kimmel G., Halperin E., 2008b Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 82: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K.-A., Xing E. P., 2007. Spectrum: joint Bayesian inference of population structure and recombination events. Bioinformatics 23: i479–i489 [DOI] [PubMed] [Google Scholar]
- Sohn K.-A., Xing E. P., 2009. A hierarchical Dirichlet process mixture model for haplotype reconstruction from multi-population data. Ann. Appl. Stat. 3: 791–821 [Google Scholar]
- Sundquist A., Fratkin E., Do C. B., Batzoglou S., 2008. Effect of genetic divergence in identifying ancestral origin using HAPAA. Genome Res. 18: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Coram M., Wang P., Zhu X., Risch N., 2006. Reconstructing genetic ancestry blocks in admixed individuals. Am. J. Hum. Genet. 79: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Choudhry S., Mei R., Morgan M., Rodriguez-Cintron W., et al. , 2007. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am. J. Hum. Genet. 81: 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh Y. W., Jordan M. I., Beal M. J., Blei D. M., 2010. Hierarchical Dirichlet processes. J. Am. Stat. Assoc. 101: 1566–1581 [Google Scholar]
- Van Gael J., Saatci Y., Teh Y. W., Ghahramani Z., 2008 Beam sampling for the infinite hidden Markov model, pp. 1088–1095 in ICML ’08: Proceedings of the 25th International Conference on Machine Learning. ACM, New York
- Wang X., Zhu X., Qin H., Cooper R. S., Ewens W. J., et al. , 2010. Adjustment for local ancestry in genetic association analysis of admixed populations. Bioinformatics 27: 670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Jin L., 2008. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am. J. Hum. Genet. 83: 322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Huang W., Qian J., Jin L., 2008. Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am. J. Hum. Genet. 82: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Young J. H., Fox E., Keating B. J., Franceschini N., et al. , 2011. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum. Mol. Genet. 20: 2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.