Abstract
C. Uhlenhaut, J.I. Cohen, S. Pavletic, G. Illei, J.C. Gea‐Banacloche, M. Abu‐Asab, T. Krogmann, L. Gubareva, S. McClenahan, P.R. Krause. Use of a novel virus detection assay to identify coronavirus HKU1 in the lungs of a hematopoietic stem cell transplant recipient with fatal pneumonia. Transpl Infect Dis 2011. All rights reserved
Abstract: A 38‐year‐old female patient with systemic lupus erythematosus presented with pulmonary infiltrates and hypoxemia for several months following immunodepleting autologous hematopoietic stem cell transplantation. She was treated for influenza, which was isolated repeatedly from oropharynx and bronchoalveolar lavage (BAL) fluids, and later empirically for lupus pneumonitis, but died 6 months after transplant. Autopsy findings failed to show influenza in the lungs or lupus pneumonitis. A novel generic polymerase chain reaction (PCR)‐based assay using degenerate primers identified human coronavirus (CoV) HKU1 RNA in BAL fluid at autopsy. CoV was confirmed by virus‐specific PCRs of lung tissue at autopsy. Electron microscopy showed viral particles consistent with CoV HKU1 in lung tissue both at autopsy and from a previous biopsy. Although human CoV HKU1 infection is not usually severe, in highly immunocompromised patients, it can be associated with fatal pneumonia.
Keywords: viral infections, generic virus detection, virus discovery, immunocompromised, coronavirus, HKU1
We report a 38‐year‐old woman who underwent an autologous hematopoietic stem cell transplant (HSCT) for severe systemic lupus erythematosus. She subsequently developed fatal pneumonia of unknown origin. Here we describe her clinical course, including development of oseltamivir‐resistant H3N2 influenza infection, and the use of a previously described (1) generic virus detection assay to detect human coronavirus (CoV) HKU1 (CoV HKU1) in lung biopsy tissue and in lung tissue obtained at autopsy from the patient.
Case report
The patient had a past history of diffuse proliferative lupus nephritis (World Health Organization Class IV) that was resistant to treatment with high‐dose corticosteroids, cyclophosphamide, and mycophenolate mofetil. She was enrolled in an approved study of autologous HSCT for patients with refractory systemic lupus erythematosus (http://Clinicaltrials.gov Identifier NCT00076752). After receiving a conditioning regimen consisting of fludarabine, cyclophosphamide, and rituximab, she received a T‐cell depleted autologous HSCT.
The patient was admitted to the Clinical Center at the National Institutes of Health 11 weeks after transplant with dyspnea and cough (see Fig. 1 for a summary of her clinical course). A computed tomography (CT) scan showed no pulmonary disease and she was discharged 6 days later after improvement on bronchodilators.
Figure 1.
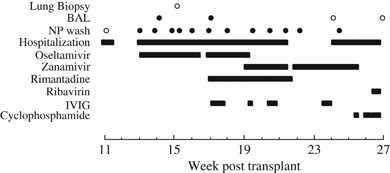
Hospital course. Critical events during the patient's hospitalizations are indicated, including use of antiviral and cytotoxic drugs. Cultures obtained at lung biopsy, bronchoalveolar lavage (BAL), and nasopharyngeal (NP) wash are denoted by circles. Closed circles represent positive cultures for influenza A virus, while open circles represent negative cultures. IVIG, intravenous immune globulin.
The patient was readmitted to the National Institutes of Health Clinical Center 13 weeks after transplant with dry cough and dyspnea on exertion, nasal congestion, rhinorrhea, and postnasal drip. Her 9‐year‐old daughter and 5‐year‐old son had had ‘a cold’ one after another in the previous week, and cases of influenza were reported at their school.
Physical examination showed a mildly cushingoid young woman with tachypnea and a constant cough. Her temperature was 36°C, blood pressure 105/55, respiratory rate 24/min, pulse 82/min. Pulmonary examination showed disseminated wheezing and rhonchi and the remainder of the examination was unremarkable. The white blood cell count was 6950/mL with 75% neutrophils and 19% lymphocytes, the hemoglobin was 11.1 gm/dL, and the platelet count was 107,000/mL. Creatinine was 1.4 mg/dL, total protein was 4.9 g/dL, and albumin was 3.1 g/dL. The arterial blood gas on 35% oxygen showed hypoxemia, with a pO2 of 62 mm Hg, a pCO2 of 26 mm Hg, and a pH of 7.35. A CT scan of the lungs performed at the time of this admission showed bilateral pleural effusions and patchy areas of nodular infiltration of the lungs, involving all of the lobes.
A nasopharyngeal wash grew influenza A H3N2 (later shown to be oseltamivir sensitive), and was negative for influenza B, respiratory syncytial virus (RSV), parainfluenza viruses 1‐3, and adenovirus (AdV). Induced sputum showed a few neutrophils and gram‐positive cocci in pairs and clusters and gram‐negative rods. Fungal and mycobacterial cultures were negative.
Because the differential diagnosis included influenza infection and lupus pneumonitis, she was begun on oseltamivir 75 mg twice daily and her prednisone was increased to 20 mg daily. A repeat CT scan after 8 days of treatment showed no improvement. Bronchoalveolar lavage (BAL) fluid grew influenza A virus, and was negative for bacteria, fungi, mycobacteria, Pneumocystis, mycoplasma, Chlamydia, Legionella, cytomegalovirus, and herpes simplex virus.
A thoracentesis showed an exudate with 395 white blood cells/mm3 with 26% neutrophils, 4% lymphocytes, 140 red blood cells/mm3, and 70% other cells. The albumin was 1.9 g/dL (serum albumin 2.5 g/dL), lactic acid dehydrogenase was 136 U/L (serum lactic acid dehydrogenase 242 U/L), and glucose was 73 mg/dL. No organisms were identified by microscopy or by culture. Because of worsening pulmonary infiltrates, open lung biopsies were performed on day 15 of the hospitalization. The right lung showed diffuse alveolar damage with hyaline membranes (Fig. 2). Stains for fungus, acid‐fast bacilli, Pneumocystis, and cytomegalovirus were negative. Cultures for viruses, including influenza, were negative. Immunohistochemistry of the lung tissue was negative for influenza and AdV.
Figure 2.
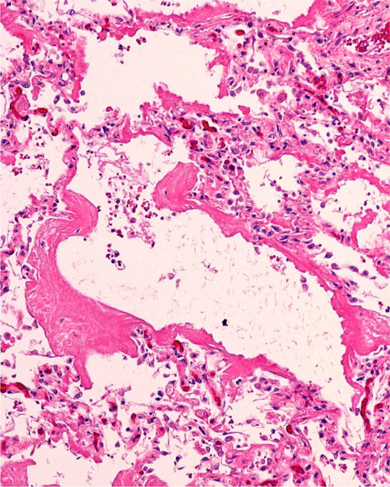
Lung biopsy. Hematoxylin and eosin staining of right lung biopsy obtained at week 15 post transplant.
Because of continued concerns about potential lupus pneumonitis, based on the non‐specific results of the lung biopsy, the dose of steroids was increased to methylprednisolone 60 mg daily. Oseltamivir was discontinued after 21 days (16 weeks after transplant). Initially, her oxygenation improved and the dose of methylprednisolone was decreased; however, her hypoxemia subsequently worsened. As repeat nasopharyngeal washes and BAL fluid continued to grow influenza, oseltamivir was reinstituted at 150 mg twice daily on hospital day 29 (17 weeks after transplant). Intravenous immune globulin was administered and steroids were continued; because an antiviral sensitivity test subsequently showed high level (IC50=770 nM, 14,000‐fold increase) oseltamivir resistance, oseltamivir was discontinued and inhaled zanamivir was begun. A CD4 count obtained 20 weeks after transplant was 135 (normal range 385–1259).
The patient was discharged from the hospital on zanamivir and a tapering dose of steroids at week 21 after transplant. The steroids were reinstituted when her infiltrates worsened and her pulmonary effusions returned. After improvement of her pulmonary status, steroids were again tapered. Sequencing of the oseltamivir‐resistant H3N2 influenza virus isolate A/Bethesda/956/2006 showed an arginine to lysine mutation in the neuraminidase at amino acid 292, a mutation previously shown to be associated with oseltamivir resistance (2) and with diminished viral fitness in animal experiments and instability in vitro (3).
The patient was readmitted 3 weeks later (24 weeks after transplant) with worsening pulmonary infiltrates. Zanamivir was continued because nasopharyngeal washes continued to grow influenza virus, although BAL fluid culture was negative for respiratory pathogens including influenza virus. Ribavirin was added in view of her increasing pulmonary infiltrates and persistent shedding of influenza virus from her nasopharynx (which was suggestive of reduced sensitivity of the influenza virus to zanamivir). Cyclophosphamide was added in the final week of life, because of persistent concerns about lupus pneumonitis and worsening of her pulmonary status on tapering doses of corticosteroids. Empiric antibacterials and antifungals were added. The patient died 27 weeks after transplant of hypotension and cardiac arrest.
At autopsy, the lungs showed bronchopneumonia, organizing pneumonitis, and diffuse alveolar damage. BAL fluid and lung tissue both obtained at autopsy were negative by culture for influenza, parainfluenza viruses 1‐3, AdV, RSV, bacteria, fungi, mycobacteria, and Nocardia. Immunohistochemistry of lung tissue was negative for influenza and AdV.
Methods
Because standard tests did not provide an explanation for the progressive clinical course leading to death of this HSCT recipient, we employed a degenerate oligonucleotide primer (DOP) polymerase chain reaction (PCR) assay to detect potential viruses in the lungs of this patient. The DOP‐PCR assay combines digesting cellular DNA in the sample, enriching for virus nucleocapsids, and performing degenerate PCR and was previously used to identify human metapneumovirus RNA in BAL fluid (4).
We obtained BAL fluid and lung tissue at autopsy and performed DOP‐PCR. To remove cellular DNA from the sample, and purify viral DNA and RNA protected in nucleocapsids, 400 μL of the BAL sample was treated with DNase I (Sigma, St Louis, Missouri, USA) at a final concentration of 80 U/mL DNase I. The DNase digestion was stopped by adding EDTA to a final concentration of 41.5 mM. Viral capsids were further purified by ultracentrifugation through 2 mL of a 1 M NaCl, 10 mM Tris/HCl pH 7.5 solution for 1.5 h, 4°C at 64,000 g in a SW60 rotor (Beckman Coulter Inc., Fullerton, California, USA). The pellet was resuspended in RLT+buffer (AllPrep DNA/RNA kit, Qiagen, Valencia, California, USA) with 143 mM β‐mercaptoethanol. DNA and RNA were isolated according to the manufacturer's instructions. DNA was eluted in 100 μL elution buffer; RNA was eluted in 50 μL of water. RNA was reverse transcribed into cDNA. The cDNA synthesis was carried out using a first strand kit and random hexamer primers (Invitrogen, Carlsbad, California, USA) according to the manufacturer's instructions.
Nucleic acids obtained from these preparations were then subjected to non‐specific DOP‐PCR. DOP‐PCR was carried out using 2.5 U low DNA Taq (Applied Biosystems, Foster City, California, USA) with the PCR buffer provided by the manufacturer, 0.01% Brij‐35, 200 μM dNTP, and 2.4 μM DOP primer (5′‐CCGACTCGAGINNNNNNTGTGG‐3′ with N representing an equimolar distribution of all 4 dNTPs, I represents inosine). The following cycling conditions were used: initial denaturation for 5 min at 95°C, followed by 5 cycles of 1 min at 94°C, 5 min at 25°C ramping at 0.1°C/s to 30°C, 4 min at 30°C, ramping at 0.1°C/s to 37°C, 3 min at 37°C, ramping at 0.1°C/s to 42°C, 2 min at 42°C, ramping at 0.1°C/s to 55°C, 1 min at 55°C and 2 min at 72°C; and 35 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min extension at 72°C, with the addition of 14 s per cycle to each extension step; and a final extension step of 10 min at 72°C. These conditions were modified from those previously published (1, 4), because additional validation studies demonstrated that they provided increased sensitivity for non‐specific amplification of virus‐sized genomes (data not shown).
Samples were prepared for electron microscopy (EM) by removing lung tissue from paraffin blocks by incubation overnight in xylene, rehydration in PBS, post‐fixation in osmium tetroxide (0.5%), dehydration, and embedding into Maraglas epoxy resin. Ultrathin sections (90 nm) were prepared and double‐stained with uranyl acetate and lead citrate, and viewed by EM with a Philips CM10 transmission electron microscope (Amsterdam, The Netherlands) .
Viral respiratory cultures for influenza, parainfluenza viruses 1‐3, AdV, and RSV were performed using the shell vial method with A549 cells and Madin‐Darby Canine Kidney cells (R‐Mix Too™, Diagnostic Hybrids, Athens, Ohio, USA) and the D3 Ultra Respiratory Staining Kit (Diagnostic Hybrids).
Results
DOP‐PCR products obtained from cDNA and DNA extracted from BAL fluid at autopsy were analyzed and purified by gel electrophoresis. All visible bands (Fig. 3) were purified and cloned. A total of 63 sequence reads were obtained from DOP‐PCR. Sequences from the clones obtained were compared with the non‐redundant database in GenBank using TBLASTX (National Center for Biotechnology Information, Bethesda, Maryland, USA). Of these 63 sequences, 53 represented human sequences, 8 represented bacterial or yeast sequences, and 1 showed greatest homology to an insect sequence. The bacterial and fungal sequences detected in DOP‐PCR were judged likely to be unrelated to her illness, because bacterial and fungal cultures at autopsy were negative. The 63rd sequence matched human CoV HKU1 genotype A with 410 of 412 identical nucleotides, overlapping the M and N genes. There were 2 nucleotide changes (G28236A; A28384G) from the reference sequence (accession number DQ415914). All CoV HKU1 genotype A sequences available in GenBank have the same sequence in this genome region; none has the nucleotide changes found in the CoV from the HSCT recipient.
Figure 3.
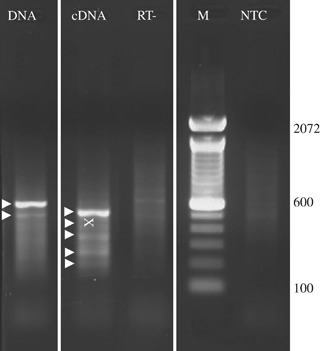
Degenerate oligonucleotide primer (DOP)‐polymerase chain reaction (PCR) results. Results of DOP‐PCR on lung biopsy and lung tissue at autopsy are shown. Arrowheads indicate bands that were excised and cloned. The ‘X’ marks the location of the band that yielded human coronavirus HKU1 sequence. RT− is the DOP‐PCR reaction performed without a prior reverse transcriptase reaction. M, molecular weight marker; NTC, no template control.
The results obtained with generic DOP‐PCR were confirmed in both BAL fluid and autopsy samples with 3 independent CoV PCRs using primers published elsewhere (Fig. 4) (5). The quality of RNA, as assessed by attempted PCR amplification of a cellular housekeeping gene (porphobilinogen deaminase) was insufficient to confirm the presence of CoV HKU1 by reverse transcription (RT)‐PCR of RNA extracted from the lung biopsy obtained during life.
Figure 4.
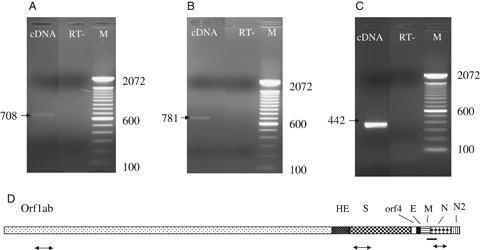
Coronavirus (CoV) HKU1 polymerase chain reaction (PCR). Results of PCR on lung tissue obtained at autopsy to detect RNA sequences in the human CoV HKU1 ORF1ab gene (A), spike gene (B), or nucleocapsid gene (C), including reverse transcriptase‐negative (RT−) controls, are shown. (D) Genome organization and localization of PCR products. Locations relative to the human CoV HKU1 genome of DOP‐PCR product (solid line) and of confirmatory PCR products (arrows) are shown. ORF1ab, open reading frame 1a and1b; HE, hemagglutinin‐esterase; S, spike; E, envelope; M, membrane; N, nucleocapsid proteins of CoV HKU1; DOP, degenerate oligonucleotide primer.
EM at autopsy showed virus‐like particles with a diameter of approximately 80 nm (Fig. 5B). Examination of the previously obtained biopsy material also confirmed presence of these particles (Fig. 5A). The diameter of the particles was consistent with CoV, although at the low end of the typical range of 80–160 nm (6).
Figure 5.
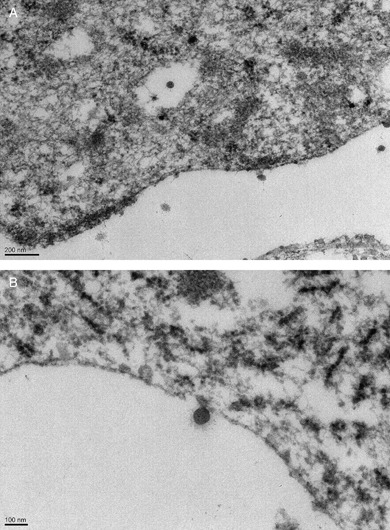
Electron microscopy showing viral particles. Electron micrographs were performed on tissue obtained at lung biopsy at week 15 (A) (× 11,500) and at autopsy at week 27 (B) (× 15,500).
Discussion
We detected sequences of human CoV HKU1 in lung tissue and in BAL fluid at autopsy from this HSCT recipient who had a previously treated influenza virus infection resistant to oseltamivir, and died with pneumonia. In addition, viral particles consistent with CoV HKU1 were detected by EM in biopsy and autopsy lung tissue. The patient was also intermittently suspected to have lupus pneumonitis. This case illustrates the difficulty of making a diagnosis in an HSCT patient with multiple possible etiologies for lung disease, and shows the value of an unbiased virus detection technique in expanding the differential diagnosis and identifying a pathogen in this individual.
Our patient had CoV HKU1 genotype A virus and sequences in the lungs. HKU1 (7) is a group 2 CoV, most closely related to human CoV OC43 (8), and is distinguished from group 1 CoVs 229E and NL63 (9) and from SARS CoV, which is distantly related to group 2 CoVs (10, 11). There are 3 known genotypes of CoV HKU1 (A, B, and C). Genotypes A and B were more prevalent than C in studies in Australia (12) and Italy (13), while a retrospective study of respiratory specimens in the US described similar prevalence rates (14). Other studies suggest that this virus is distributed worldwide, although the reported number of cases remains very limited (10).
Four lower respiratory syndromes potentially caused by human CoVs have been described: community‐acquired pneumonia, acute tracheobronchitis, acute exacerbations of chronic obstructive pulmonary disease, and acute asthma exacerbation (6). CoV HKU1 usually causes relatively mild community‐acquired infections in children, characterized by rhinorrhea, cough, and fever (12, 13, 14, 15, 16, 17, 18); infection can result in bronchiolitis and pneumonia; occasionally, hypoxia and abnormal chest x‐rays are described (12, 14). CoV HKU1 has also been related to a high incidence of febrile seizures (15). Another study implies that CoV HKU1 could be associated with enteric symptoms (5). Pneumonia related to CoV HKU1 infection occurs mostly in older people with underlying diseases, in particular severe respiratory and cardiovascular diseases, and in transplant recipients, although the virus was not identified in lung tissue in these prior studies (16). Two fatal cases of CoV HKU1‐associated community‐acquired pneumonia were reported, in which virus was detected in nasopharyngeal aspirates by RT‐PCR (16). Both patients had lymphopenia and were suffering from diabetes mellitus; 1 patient was 66 years old and had gastric lymphoma, while the other was 72 years old and had prostate carcinoma. Our patient received a lymphodepleting conditioning regimen for autologous HSCT, which results in prolonged T and B cell lymphopenia, and she was severely immunocompromised from both her HSCT and from the immunosuppressive therapy used to treat possible lupus pneumonitis.
Although influenza was suspected as a cause of the patient's pneumonitis and was repeatedly isolated from nasopharyngeal washes and BAL fluids, immunohistochemistry of lung tissue obtained at biopsy and at autopsy failed to show influenza virus. While inhaled zanamivir may have reduced the sensitivity of culture, immunohistochemistry would be expected to show evidence of influenza infection if the virus were a significant cause of disease in this patient (19) Although influenza virus infection may have been a contributor to the patient's symptoms early after HSCT, the continued progression of bronchopneumonia while on antivirals for influenza, along with failure to identify influenza in the lower respiratory tract in any clinical sample after the initial positive BALs at weeks 14 and 17 after transplantation, or at autopsy, strongly suggested that influenza was not the cause of her fatal illness. Highly immunocompromised patients are known to shed influenza in the upper respiratory tract for months (20) and the influenza virus in the BAL fluids may have been due to shedding of virus from the upper respiratory tract.
Another possible diagnosis in our patient was idiopathic pneumonia syndrome (IPS), which presents as diffuse lung injury without evidence of infection. Our patient was in the intermediate period after transplant (days 30–100), which is the median time for onset of IPS (between 21 and 87 days post transplant) (21). IPS is typically described after allogeneic HSCT in which the level of immunosuppression is usually more profound than after standard autologous HSCT. The reported frequency of IPS ranges between 2% and 17%, averaging about 10% (21). CoV and other respiratory viruses may be responsible for some cases of IPS, if reliable means of detection are not employed. An increasing arsenal of diagnostic tools for detection of respiratory viruses is developing, including rapid antigen detection, monoclonal antibody testing, cell culture, and various nucleic acid amplification techniques. However, detection of some respiratory viruses including CoVs remains a challenge.
The DOP‐PCR protocol, used to identify CoV HKU1 in this case, has several significant advantages over other methods. This universal virus detection tool can provide sequence information of the pathogen present in a given sample without any prior information about the genome. DOP‐PCR is unbiased and has a detection limit that is close to that of nucleic acid testing techniques that use virus‐specific primers (1). Subtractive techniques that might be used in order to identify a pathogen require an uninfected but otherwise genetically identical control, which may be difficult to obtain for clinical samples. We previously showed that the detection limit of the DOP‐PCR is several orders of magnitude more sensitive than that of other generic detection techniques (1). Isolation of nucleic acids from purified nucleocapsids in the DOP‐PCR protocol is more likely to detect intact infectious particles than PCR of the whole sample, which has large amounts of contaminating cellular nucleic acids. DOP‐PCR, in which viral nucleic acid sequences are highly enriched before PCR is performed, may be especially useful for finding pathogens that cannot be readily identified by standard methods. Nonetheless, results from non‐specific screening techniques like the DOP‐PCR must be independently verified, as exemplified by the detection by DOP‐PCR of non‐viral sequences in the autopsy specimen.
Conclusion
This is the first report, to our knowledge, of CoV HKU1 detected directly in lung tissue from an individual with fatal pneumonia. Although there may have been other co‐factors, CoV HKU1 infection likely contributed to this patient's fatal outcome. The patient shed influenza virus from the respiratory tract for several months, and this virus was thought to be a cause of pneumonia during much of her hospital course after transplantation. When influenza infection was considered less likely, and lupus pneumonitis was considered a possibility, she received immunosuppressive therapy that may have exacerbated her CoV HKU1 infection. While we cannot rule out the possibility that the transient improvements observed after increases in immunosuppressive therapy were due to reduction in the inflammatory response to CoV HKU1 infection, this case illustrates the importance of obtaining appropriate specimens from patients with unexplained pneumonitis and the value of unbiased approaches such as the universal virus detection assay employed here to identify other causes of pulmonary infiltrates and hypoxemia.
Acknowledgements:
The authors thank David Kleiner, who interpreted histological results at biopsy and autopsy; Sherif Zaki of the Centers for Disease Control and Prevention, who performed immunohistochemistry on the lung tissue for influenza and AdV; Kening Wang, who performed additional experiments on material obtained at autopsy; and Susanne Duwe, Robert Koch‐Institute, Berlin, who provided helpful comments.
This work was supported by the intramural research programs of the Center for Biologics Evaluation and Research, the National Institute of Allergy and Infectious Diseases, the National Institute of Dental and Craniofacial Research, the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the National Cancer Institute, and the Centers for Disease Control and Prevention.
Disclosure and funding: The authors of this manuscript have no conflicts of interest to disclose. This research was supported by the intramural research programs of the US Food and Drug Administration, the National Institute of Allergy and Infectious Diseases, the National Cancer Institute, and the National Institute of Dental and Craniofacial Research.
References
- 1. Nanda S, Jayan G, Voulgaropoulou F, et al Universal virus detection by degenerate‐oligonucleotide primed polymerase chain reaction of purified viral nucleic acids. J Virol Methods 2008; 152 (1–2): 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moscona A. Oseltamivir resistance–disabling our influenza defenses. N Engl J Med 2005; 353 (25): 2633–2636. [DOI] [PubMed] [Google Scholar]
- 3. Carr J, Ives J, Kelly L, et al Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo . Antiviral Res 2002; 54 (2): 79–88. [DOI] [PubMed] [Google Scholar]
- 4. Uhlenhaut C, Cohen JI, Fedorko D, Nanda S, Krause PR. Use of a universal virus detection assay to identify human metapneumovirus in a hematopoietic stem cell transplant recipient with pneumonia of unknown origin. J Clin Virol 2009; 44 (4): 337–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vabret A, Dina J, Gouarin S, Petitjean J, Corbet S, Freymuth F. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin Infect Dis 2006; 42 (5): 634–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rohde G, Drosten C, Borg I, et al [Detection of respiratory viruses – how, when, where and why?] Pneumologie 2009; 63 (1): 14–22. [DOI] [PubMed] [Google Scholar]
- 7. Woo PC, Lau SK, Chu CM, et al Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 2005; 79 (2): 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci USA 1967; 57 (4): 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van der Hoek L, Pyrc K, Jebbink MF, et al Identification of a new human coronavirus. Nat Med 2004; 10 (4): 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sloots TP, Whiley DM, Lambert SB, Nissen MD. Emerging respiratory agents: new viruses for old diseases? J Clin Virol 2008; 42 (3): 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woo PC, Lau SK, Huang Y, Tsoi HW, Chan KH, Yuen KY. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia. Arch Virol 2005; 150 (11): 2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol 2006; 35 (1): 99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerna G, Percivalle E, Sarasini A, et al Human respiratory coronavirus HKU1 versus other coronavirus infections in Italian hospitalised patients. J Clin Virol 2007; 38 (3): 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. Coronavirus HKU1 infection in the United States. Emerg Infect Dis 2006; 12 (5): 775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau SK, Woo PC, Yip CC, et al Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol 2006; 44 (6): 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woo PC, Lau SK, Tsoi HW, et al Clinical and molecular epidemiological features of coronavirus HKU1‐associated community‐acquired pneumonia. J Infect Dis 2005; 192 (11): 1898–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cilla G, Onate E, Perez‐Yarza EG, Montes M, Vicente D, Perez‐Trallero E. Viruses in community‐acquired pneumonia in children aged less than 3 years old: high rate of viral coinfection. J Med Virol 2008; 80 (10): 1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garbino J, Crespo S, Aubert JD, et al A prospective hospital‐based study of the clinical impact of non‐severe acute respiratory syndrome (non‐SARS)‐related human coronavirus infection. Clin Infect Dis 2006; 43 (8): 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guarner J, Shieh WJ, Dawson J, et al Immunohistochemical and in situ hybridization studies of influenza A virus infection in human lungs. Am J Clin Pathol 2000; 114 (2): 227–233. [DOI] [PubMed] [Google Scholar]
- 20. Weinstock DM, Gubareva LV, Zuccotti G. Prolonged shedding of multidrug‐resistant influenza A virus in an immunocompromised patient. N Engl J Med 2003; 348 (9): 867–868. [DOI] [PubMed] [Google Scholar]
- 21. Afessa B, Peters SG. Major complications following hematopoietic stem cell transplantation. Semin Respir Crit Care Med 2006; 27 (3): 297–309. [DOI] [PubMed] [Google Scholar]


