Abstract
Estrogen signaling is a critical pathway that plays a key role in the pathogenesis of breast cancer. In a previous transcriptional profiling study, we identified a novel panel of estrogen-induced genes in breast cancer. One of these genes is solute carrier family 22 member 5 (SLC22A5), which encodes a polyspecific organic cation transporter (also called OCTN2). In this study, we found that estrogen stimulates SLC22A5 expression robustly in an estrogen receptor (ER)-dependent manner and that SLC22A5 expression is associated with ER status in breast cancer cell lines and tissue specimens. Although the SLC22A5 proximal promoter is not responsive to estrogen, a downstream intronic enhancer confers estrogen inducibility. This intronic enhancer contains a newly identified estrogen-responsive element (ERE) (GGTCA-CTG-TGACT) and other transcription factor binding sites, such as a half ERE and a nuclear receptor related 1 (NR4A2/Nurr1) site. Estrogen induction of the luciferase reporter was dependent upon both the ERE and the NR4A2 site within the intronic enhancer. Small interfering RNA against either ER or Nurr1 inhibited estrogen induction of SLC22A5 expression, and chromatin immunoprecipitation assays confirmed the recruitment of both ER and Nurr1 to this enhancer. In functional assays, knockdown of SLC22A5 inhibited L-carnitine intake, resulted in lipid droplet accumulation, and suppressed the proliferation of breast cancer cells. These results demonstrate that SLC22A5 is an estrogen-dependent gene regulated via a newly identified intronic ERE. Since SLC22A5 is a critical regulator of carnitine homeostasis, lipid metabolism, and cell proliferation, SLC22A5 may serve as a potential therapeutic target for breast cancer in the future.
Keywords: SLC22A5/OCTN2, Breast cancer, Estrogen, Estrogen receptor, ERE
Introduction
Breast cancer is the most frequently diagnosed cancer and the second leading cause of cancer-related deaths in women [1]. Among many factors that contribute to the development and progression of breast cancer, estrogen signaling is regarded as the most critical one, playing a key role in the development, progression, prevention, and treatment of the disease [2, 3]. The effects of estrogen on breast cancer are primarily mediated by estrogen receptor alpha (ER), a member of the nuclear receptor superfamily [3–5]. Previous studies have demonstrated that estrogen and ER regulate target gene expression via several different mechanisms [6–9], including the classical genomic pathway [10], transcription factor crosstalk, also known as the tethering pathway [11, 12], and the non-genomic (cytoplasmic) pathway [13–15]. Recently, genome-wide ER binding sites have been identified [16, 17]. These studies demonstrate that many estrogen-regulated genes do not have consensus estrogen-response elements (EREs) in their promoters. In addition, this data identifies the positioning of ER binding sites in distal regions not previously annotated as cis-regulatory elements rather than in the proximal promoter regions.
To further investigate the molecular mechanism of estrogenic effects on breast cancer, we have performed transcriptional profiling experiments and identified a novel panel of estrogen-induced genes in breast cancer [18]. Many identified genes are critical regulators of cell cycle, cell growth, and metabolism. While the effects of estrogen on breast cancer cell cycle and growth have been well documented in our previous studies [11, 12], estrogenic effects on metabolism remain understudied. Among the estrogen-induced genes we identified, solute carrier family 22 member 5 (SLC22A5) is of particular interest as it encodes a polyspecific organic cation transporter (OCTN2) that carries a high affinity for carnitine [19, 20]. Carnitine has been implicated in lipid metabolism, because it is essential for the transfer of fatty acids across the inner mitochondrial membrane before β-oxidation [21–23], and OCTN2 is primarily responsible for carnitine-reabsorption in the kidney once it has been filtered. Mutations in SLC22A5 gene and polymorphisms in its promoter have been associated with many diseases, including type 1 diabetes (T1D) [24], chronic inflammatory disease of the gastrointestinal tract (Crohn’s disease) [25], and systemic primary carnitine deficiency (hypoketotic hypoglycemia and skeletal/cardio-myopathy) [26–29]. However, little information is available regarding the role of the SLC22A5 gene in cancer.
In this study, we show that SLC22A5 is an ER-dependent gene and that its expression is associated with ER status in both breast cancer cells and tumor specimens. We also define the molecular mechanism by which estrogen regulates SLC22A5 expression in breast cancer cells. Specifically, the SLC22A5 proximal promoter is not sensitive to estrogen. However, within the +9 kb region downstream of the transcriptional start site of the SLC22A5 gene is an estrogen-activated intronic enhancer element containing a novel ERE. Furthermore, we found that SLC22A5 is critical for carnitine intake, lipid metabolism, and cell proliferation. This study therefore identifies SLC22A5 as a newly discovered intronic ERE-dependent gene implicated in carnitine homeostasis, lipid metabolism, and breast cancer cell proliferation.
Materials and methods
Cell culture
The breast cancer cell lines BT474, BT483, HCC1428, HCC1500, HS578T, MCF-7, MDA-MB-231, MDA-MB-361, MDA-MB-415, MDA-MB-453, MDA-MB-468, SKBr3, SUM-159PT, T47D, and ZR75-1 were purchased from American Type Culture Collection (ATCC, Manassas, VA).
Chemicals and treatment
17-β-estradiol (E2), 12-O-tetradecanoyl-phorbol-13-acetate (TPA), forskolin (FSK), ICI 182.780 (ICI), actinomycin D (ActD), and cycloheximide (CHX) were purchased from Sigma-Aldrich (St. Louis, MO) and dissolved in ethanol or dimethyl sulfoxide (DMSO). The final concentrations of E2, TPA, FSK, ICI, ActD, and CHX were 1 nmol/l, 100 nmol/l, 10 μmol/l, 1 μmol/l, 1 μg/ml, and 10 μg/ml, respectively.
Constructs
The CMV-Nurr1 plasmid was purchased from OriGene (Rockville, MD). The −5.7 kb L and −5.0 kb LΔE SLC22A5 promoter plasmids were generously provided by Dr. Bing Ren [30]. The SLC22A5 proximal promoter region (−527 to +39 bp) was amplified by PCR and inserted into the pGL3basic plasmid. The +9 kb downstream enhancer region (1,063 bp) was cloned by PCR from human genomic DNA. Primer sequences are listed in Supplemental Table 1. For site-directed mutagenesis, the half ERE (hERE) GGTCA was mutated to GagCA and the hERE TGACC/T was mutated to TGctC/T; the cyclic adenosine monophosphate (cAMP)-response element (CRE) TGACATCA was mutated to TtctATCA; the nuclear receptor subfamily 4, group A, member 2 (NR4A2) site AtGTCA was mutated to AtagaA. All constructs were verified by sequencing.
Luciferase assay
Hormone-depleted MCF-7 cells were transfected with plasmids using FuGENE 6 Transfection Reagent (Roche, Basel, Switzerland). The next day, cells were treated with E2 for 6 h. Luciferase activity was measured using a commercially available kit (Promega, Madison, WI).
RNA extraction and quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from cells using the RNeasy Mini Kit (Qiagen, Valencia, CA), then reverse transcribed. qRT-PCR was carried out on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems/Life Technologies, Carlsbad, CA). The relative gene expression was determined using the comparative CT method as previously described [31]. Primers and probes are listed in Supplemental Table 1.
Small interfering RNA (siRNA) experiment
Cells were transfected with chemically synthesized siRNAs (Sigma, St. Louis, MO) using RNAiMAX Transfection Reagent (Invitrogen, Carlsbad, CA). siRNA for the luciferase gene (Sigma) was used as a control.
Western blot
Total protein was extracted from cells using RIPA buffer containing protease inhibitors (Roche). 30 μg protein per lane was resolved by SDS-PAGE and transferred to a nitrocellulose membrane. After incubating sequentially with primary and secondary antibodies, blots were developed using the SuperSignal West Pico Kit (Pierce, Rockford, IL) and exposed to film.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed according to the published protocols with minor modifications [11, 12, 18, 32]. Primers and probes are listed in Supplemental Table 1. The recruitment was presented as the ratio of the q-PCR result of immunoprecipitated fragments normalized to the total input DNA.
Breast cancer tissue datasets
Clinical microarray datasets consisting of primary breast cancer tissue specimens were used to analyze the association between ER and SLC22A5 expression in breast cancer specimens [33–47]. ER and SLC22A5 probe set IDs, corresponding annotations, and clinical data were extracted from these datasets.
Carnitine transport assay
Prior to carnitine transport assays, cells were washed three times with PBS. Carnitine hydrochloride, L-[N-methyl-3H] (100 μmol/l final concentration, 89.5 Ci/mmol, PerkinElmer, Waltham, MA) were added to the cells. At the indicated times, cells were washed three times with cold PBS and harvested. Radioactivity was quantified using a liquid scintillation counter (Beckman Coulter LS-6500).
Lipid droplets assay
Cells were seeded in 96-well plates, transfected with SLC22A5 siRNA for 2 days, and then treated with estrogen or vehicle for another 2 days. Afterward, cells were washed with PBS and fixed in 0.4% paraformaldehyde overnight. Neutral lipids were stained using HCS Lipid-TOX™ Green neutral lipid stains and nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes/Invitrogen, Carlsbad, CA) for 30 min at room temperature. Cell images were acquired using ImageXpress Micro Widefield High Content Screening System and analyzed using MetaXpress software version 3.1.0.79 (Molecular Devices, Inc., Sunnyvale, CA).
Cell proliferation assays
Cell proliferation assays were performed using 3-(4,5-dimethyl-thiazol-2-yl)-5-(3-carboxymethoxy-phenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assays in tandem with cell counting. After the indicated number of days, MTS assays were performed using the CellTiter 96 kit (Promega, Madison, WI). Cell numbers were determined using the Countess Automated Cell Counter (Invitrogen, Carlsbad, CA).
Results
SLC22A5 is expressed in breast cancer
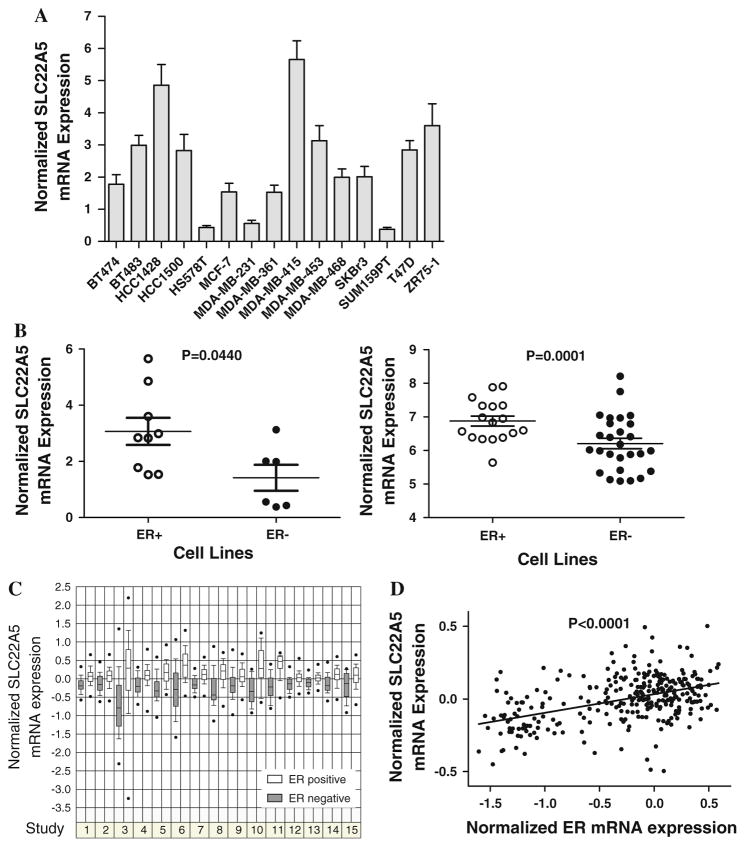
In our previous transcriptional profiling study, we observed that SLC22A5 is an estrogen-induced gene in breast cancer cells [18]. However, no information is available regarding SLC22A5 in breast cancer. To gain a better understanding of the regulation and function of the SLC22A5 gene in breast cancer, we first examined SLC22A5 expression in a panel of breast cancer cell lines. As shown in Fig. 1a, SLC22A5 is expressed in all the breast cancer cell lines examined. Of special interest, SLC22A5 expression is significantly overexpressed in ER-positive cell lines compared with ER-negative cell lines (P < 0.001; Fig. 1b, left panel). Similar results were obtained upon analysis of the cDNA microarray dataset of breast cancer cell lines [48] (P < 0.001; Fig. 1b, right panel). Examination of SLC22A5 expression in breast tumor tissue specimens through clinical microarray dataset [33–47] analysis further supported SLC22A5 expression being significantly over-expressed in ER-positive tumor tissue specimens compared with ER-negative tumor tissue specimens. In fact, this held true in all 15 independent datasets studied, including more than 2,000 patients (Fig. 1c). We also found that SLC22A5 expression correlates significantly with ER expression in breast tumor tissue specimens using data from these clinical microarray datasets (P < 0.0001, Fig. 1d).
Fig. 1.
SLC22A5 expression in breast cancer. a SLC22A5 expression in breast cancer cell lines as determined by qRT-PCR. b SLC22A5 expression in ER-positive and ER-negative cell lines. Left panel qRT-PCR data of this study; right panel cDNA microarray dataset [48]. c SLC22A5 expression in breast tumor tissue specimens from 15 studies including a total of more than 2,000 patients. Studies 1–15 are from the clinical microarray datasets [33–47]. d The correlation between SLC22A5 and ER expression in breast tumor tissue specimens. Shown is the correlation data from the largest dataset, comprised of 295 patients [42]. Analyses of other datasets also generated similar results (data not shown)
SLC22A5 expression is upregulated by estrogen
Due to the correlative relationship between SLC22A5 and ER mRNA levels, our next step was to examine the effect of estrogen treatment on SLC22A5 expression in different cell lines. As shown in Fig. 2a, E2 induced SLC22A5 expression in ER-positive cell lines, but had no effect on its expression in ER-negative cell lines. To investigate the molecular mechanism by which estrogen induces SLC22A5 gene expression, we chose MCF-7 as a model system for further studies, as this cell line displayed the highest E2-regulated induction of SLC22A5 expression. We first examined SLC22A5 expression in response to estrogen at different time points. As shown in Fig. 2b, E2 induces SLC22A5 mRNA expression very rapidly. This induction could, in fact, be detected within 0.5 h, peaking at 2 h, and remaining elevated thereafter (Fig. 2b). In order to determine whether E2-induced SLC22A5 expression is through a primary transcriptional regulatory mechanism, as suggested by the expression-over-time analysis, we tested the effect of E2 on SLC22A5 expression in MCF-7 cells pretreated individually with inhibitors of RNA synthesis, ActD, and protein synthesis, CHX, versus that observed with the vehicle, DMSO. As shown in Fig. 2c, ActD, but not CHX, was able to block E2 induction of SLC22A5 expression, indicating that estrogen-regulated induction of SLC22A5 expression requires RNA, as opposed to protein, synthesis. Next, we investigated whether ER is required for estrogen induction of SLC22A5 expression. ICI 182.780 (ICI), a pure ER antagonist, completely blocked E2-induced expression of SLC22A5 (Fig. 2d). Moreover, siRNA to ER similarly abolished E2 induction of SLC22A5 expression (Fig. 2e). These results indicate that E2 induction of SLC22A5 expression is through a primary transcriptional regulatory mechanism, and further, that this regulation is absolutely dependent on ER.
Fig. 2.
SLC22A5 expression is regulated by estrogen. a E2 regulation of SLC22A5 expression in a panel of breast cancer cell lines. Hormone-depleted cells were treated with vehicle/E2 for 6 h, followed by mRNA expression analysis by qRT-PCR. b Time course of E2 induction of SLC22A5 expression in MCF-7 cells. c Effects of ActD and CHX on E2 induction of SLC22A5 expression. Hormone-depleted MCF-7 cells were treated with ActD (1 μg/ml) or CHX (10 μg/ml) for 1 h before the addition of vehicle/E2. RNA was harvested 2 h after treatment with vehicle/E2. d Effect of ICI 182.780 (ICI) on E2 induction of SLC22A5 expression. Hormone-depleted MCF-7 cells were treated with ICI (1 μmol/l) for 1 h before the addition of vehicle/E2. RNA was harvested 2 h after treatment with vehicle/E2. e Effects of siRNA against ER induction of SLC22A5 expression. Hormone-depleted MCF-7 cells were transfected with siRNA against luciferase (Luc) or estrogen receptor (ER) for 36 h before the addition of vehicle/E2. RNA was harvested 2 h after treatment with vehicle/E2. Inset verification of ER knockdown by Western blot
The SLC22A5 proximal promoter is not responsive to estrogen
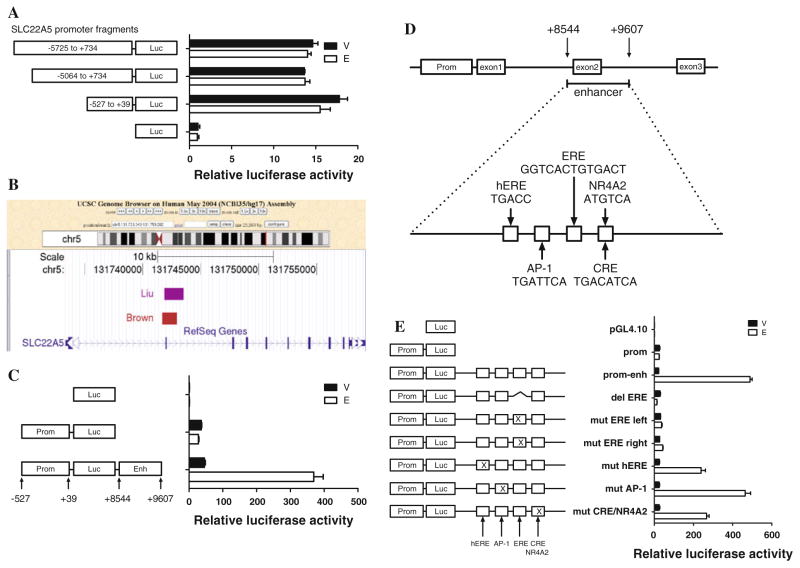
Since SLC22A5 is an ER/E2-dependent gene, we wished to further define the physical nature of the interaction and chose to investigate whether the SLC22A5 proximal promoter was responsible for the gene’s estrogen inducibility. As estrogen may regulate gene expression by binding to an ERE or by tethering other transcription factors at gene promoter regions, this seemed a logical location for ER/E2 interaction with SLC22A5. We tested three SLC22A5 promoter luciferase reporter plasmids with different promoter fragments lengths and found that none of the constructs were responsive to estrogen treatment (Fig. 3a). The −527 to +39 bp fragment retains strong basal activity and is regarded as the core promoter. This portion of the proximal promoter was used for the following experiments.
Fig. 3.
Luciferase assay of SLC22A5 enhancer/promoter luciferase constructs. a Effect of E2 on the reporter activity of different lengths of SLC22A5 promoter luciferase constructs. Hormone-depleted MCF-7 cells were transfected with different lengths of SLC22A5 promoter luciferase plasmids for 16 h. Cells were then treated with vehicle/E2 for 6 h and lysed for luciferase analysis. b Schematic representation of SLC22A5 gene structure in the genomic context, showing the ER binding site (chr5:131741886-131742949, +8544 to +9607 from the transcriptional start site), visualized using the UCSC genome browser (http://genome.ucsc.edu). c Effect of E2 on the reporter activity of the SLC22A5 promoter luciferase construct with or without the intronic +9 kb enhancer. Luciferase analysis was performed as described above. d Potential transcription factor binding sites within the intronic +9 kb enhancer, showing the predicted half ERE, AP-1 site, full ERE, and CRE/NR4A2 site. e Effect of E2 on the reporter activity of SLC22A5 promoter-enhancer luciferase constructs, each carrying a deletion or mutation within one of the predicted binding sites. Luciferase analysis was performed as described before
A downstream enhancer confers SLC22A5 estrogen responsiveness
Since the SLC22A5 proximal promoter was unresponsive to estrogen, we examined SLC22A5 gene structure in a global genomic context and found a potential ER binding region located approximately 9 kb downstream of the transcriptional start site (Fig. 3b). This region has been identified in two independent genome-wide studies and may therefore contain a functional ER binding site [16, 17]. The potential ER binding region (1,063 bp) encompasses a portion of introns 1 and 2, as well as exon 2 in its entirety. We cloned this DNA fragment and inserted it downstream of the SLC22A5 promoter–luciferase gene cassette in a manner resembling native SLC22A5 genomic structure. This SLC22A5 promoter–luciferase gene-intronic fragment construct is dramatically induced by estrogen, indicating that the ER binding region included, the 1,063 bp fragment, functions as an enhancer, thereby contributing to the estrogen responsiveness of SLC22A5 (Fig. 3c).
The enhancer contains an ERE and multiple transcription factor binding sites
Since this intronic enhancer enables estrogen-regulated induction of the SLC22A5 promoter, we next sought to determine which cis-acting elements and their cognate trans-acting factors are required for estrogen induction. We analyzed this intronic enhancer region and identified many potential transcription factor binding sites, of which those previously shown to be related with the ER signaling pathway are presented in Fig. 3d. Interestingly, four potential transcription factor binding sites are juxtapositioned in the middle of this intronic enhancer: sequentially, a hERE, an activator protein-1 (AP-1) site, a full ERE, and CRE. These elements are located within a small stretch of DNA only 113 bp in size. We cloned this 113 bp fragment containing the four elements in addition to the 3 bp flanking protective sequences on either side of the string of transcription factor binding sites and inserted the full 119 bp DNA fragment into the SLC22A5 promoter–luciferase construct. This small fragment is highly responsive to estrogen, retaining at least 75% of the estrogen inducibility of the 1,063 bp full-length enhancer (data not shown). Therefore, the estrogen-responsive region must be located within this 119 bp strand of DNA.
Since an ERE-like sequence, GGTCA-CTG-TGACT, is located in this enhancer, we hypothesized that it would act as a functional ERE. Therefore, we generated plasmids with deletions or mutations within this ERE-like element. Indeed, deletion of the entire 13 bp ERE-like sequence, as well as a mutation of 2 bp in either the left or the right half of the ERE site abolished estrogen responsiveness (Fig. 3e). These results clearly identify this 13 bp ERE-like sequence (GGTCA-CTG-TGACT) as a new intronically located ERE, which is only 1 bp different from the classical vitellogenin ERE (vit-ERE) consensus sequence (GGTCA-NNN-TGACC) [49].
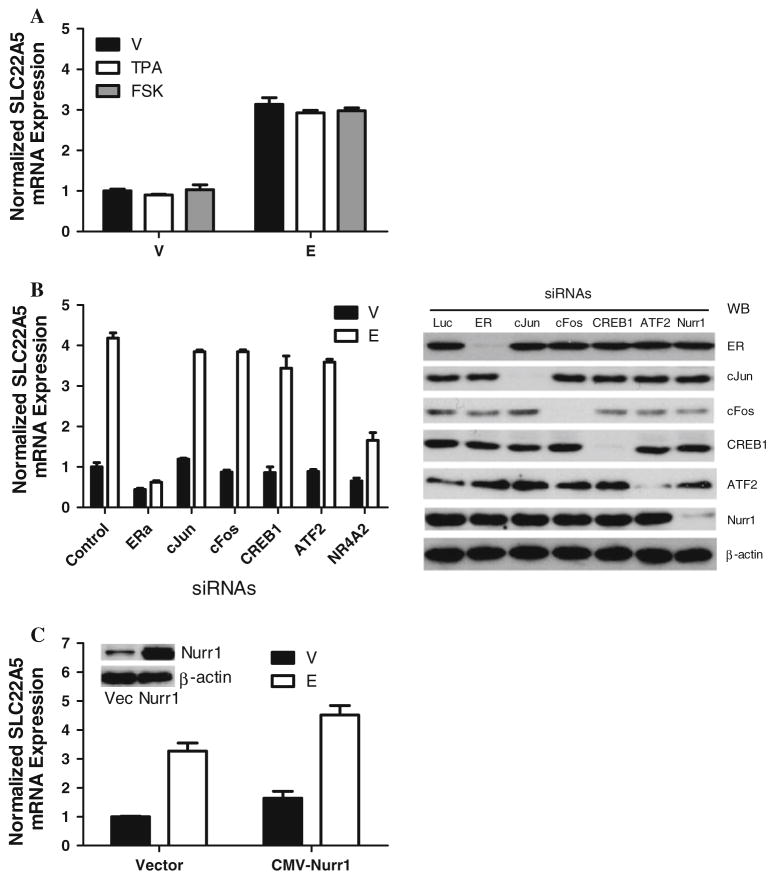
To determine whether other specific elements within the enhancer region are also required for estrogen responsiveness, we generated several additional constructs carrying a mutation within the hERE, AP-1, or CRE sites and tested whether any affected estrogen responsiveness. Manipulation of either the hERE (TGACC) or the potential CRE site (TGACATCA) significantly decreased estrogen-induced luciferase activity by about 50%, whereas mutation of the potential AP-1 site (TGATTCA) had little effect on estrogen-induced luciferase activity (Fig. 3e). These results suggest that the cAMP/CRE binding (CREB) pathway rather than AP-1 pathway may be involved in estrogen regulation of SLC22A5 expression. To test this hypothesis, we used a dual approach: (1) stimulating the cAMP pathway or AP-1 pathway using the associated activating chemicals (FSK, an activator of adenylyl cyclase, or TPA, an activator of AP-1, respectively) (see Fig. 4a), and (2) abrogating the pathways using siRNAs against CREB1/ATF2 or AP-1 proteins (see Fig. 4b). Intriguingly, none of these approaches affected basal or estrogen-stimulated SLC22A5 gene expression (Fig. 4a, b).
Fig. 4.
Effects of the AP-1, cAMP, and Nurr1 pathways on SLC22A5 expression. a Effect of small molecule chemicals for AP-1 and cAMP pathways on estrogen-induced SLC22A5 expression in MCF-7 cells. TPA or FSK was added with vehicle/E2 for 2 h. RNA was harvested and analyzed by qRT-PCR. b siRNA knockdown of ER, AP-1, CREB1, ATF2, or Nurr1 on estrogen-induced SLC22A5 expression. Hormone-depleted MCF-7 cells were transfected with siRNA against each gene, as indicated, for 36 h. The cells were then treated with vehicle/E2 for 2 h. The data for siLuc transfected and vehicle-treated samples were arbitrarily set as 1. The efficiency of the siRNAs has been validated to suppress the expression of target genes as shown in the Western blot. c Effect of Nurr1 overexpression on estrogen-induced SLC22A5 expression. Hormone-depleted MCF-7 cells were transfected with vector (vec) or CMV-Nurr1 plasmid for 36 h and then treated with vehicle/E2 for 2 h. RNA was harvested and analyzed by qRT-PCR. Inset verification of Nurr1 expression by Western blot
An NR4A2/Nurr1 site overlapping with the potential CRE site is critical for estrogen induction of SLC22A5 expression
Based on the above findings that mutation of the potential CRE site significantly decreased estrogen inducibility while activation or abrogation of cAMP pathway did not affect either the endogenous or estrogen-induced SLC22A5 expression, we hypothesized that this potential CRE site is not functional but that mutation of the site may simultaneously affect another cis-acting element. If this is the case, then binding of a trans-acting factor contributing to estrogen inducibility specific for the alternate cis-acting element could be affected by the mutation. We analyzed this enhancer region and found a potential NR4A2/Nurr1 binding site (ATGTCA, minus strand) that overlaps with the potential CRE site (TGACATCA, plus strand).
NR4A2 gene is a member of the steroid–thyroid hormone–retinoid receptor superfamily [50, 51]. The encoded protein, Nurr1, is an orphan nuclear receptor that acts as a transcription factor [52]. We used a dual method approach to test our hypothesis of NR4A2/Nurr1 involvement in the estrogen induction of SLC22A5 expression. First, we abrogated the NR4A2/Nurr1 pathway using NR4A2 siRNAs. siRNA knockdown of NR4A2 partially blocked E2-induced SLC22A5 expression without affecting the basal level of SLC22A5 expression (Fig. 4b). Second, we overexpressed the Nurr1 transcription factor by transfection of the cytomegalovirus-nuclear receptor related 1 (CMV-Nurr1) plasmid. Overexpression of Nurr1 not only stimulated SLC22A5 expression, but also cooperated with E2 to induce SLC22A5 expression to a higher extent (Fig. 4c). These results demonstrate that NR4A2/Nurr1 is involved in estrogen induction of SLC22A5 gene expression.
ER and Nurr1 proteins are recruited to the intronic enhancer
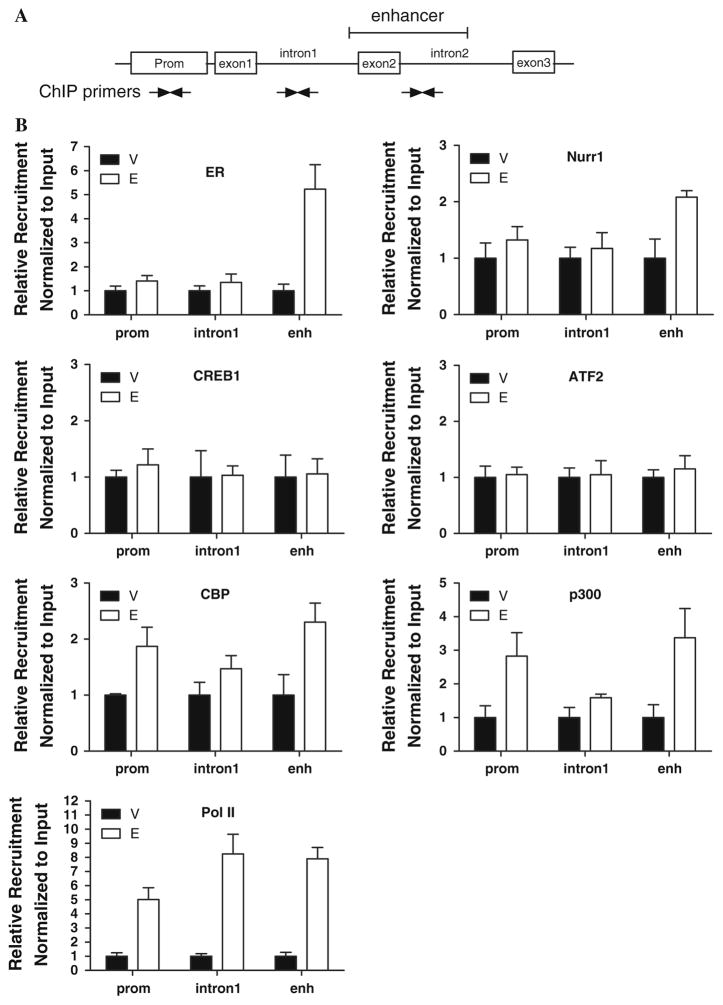
In order to validate the recruitment of transcription factors to the SLC22A5 intronic enhancer and proximal promoter elements, we performed ChIP experiments. Immunoprecipitated DNA was analyzed by q-PCR using primers for both the SLC22A5 intronic enhancer and proximal promoter (Fig. 5a). Primers for the pS2 gene promoter and primers for intron 1 of the SLC22A5 gene were used as positive or negative controls, respectively (data for positive control not shown) (Fig. 5a). Upon treatment with E2, both ER and Nurr1 were recruited to the intronic enhancer within intron 2 as opposed to the proximal promoter region (Fig. 5b). By contrast, neither CREB1 nor ATF2 were recruited to either the SLC22A5 intronic enhancer or the proximal promoter regions. In addition to the observed Nurr1 localization, integrator proteins CREB binding protein (CBP) and E1A binding protein p300 (p300) were also recruited to the intronic enhancer, as well as to the promoter. RNA polymerase II (Pol II), on the other hand, was recruited to the proximal promoter and introns 1 and 2 upon E2 treatment.
Fig. 5.
ChIP assay of the recruitment of transcription factors to the SLC22A5 gene locus. a Schematic representation of the primers used to amplify the SLC22A5 promoter, intron 1, or intronic enhancer (within intron 2) regions. b Recruitment of ER, Nurr1, CREB1, ATF2, p300, CBP, and Pol II to the SLC22A5 promoter, intron 1, or intronic enhancer (within intron 2) regions. Data were presented as the relative amount of immunoprecipitated DNA normalized to input as measured by q-PCR assay. The data for vehicle-treated samples were arbitrarily set as 1
The physiological relevance of SLC22A5 expression in breast cancer
SLC22A5 gene encodes a polyspecific organic cation transporter with sodium-dependent high affinity to carnitine, which is an essential nutrient involved in fatty acid transport into mitochondria and oxidation to produce energy. To investigate whether SLC22A5 is involved in carnitine transportation and lipid metabolism in breast cancer, we performed carnitine transport assays examining carnitine intake in breast cancer cells in response to estrogen treatment. As shown in Fig. 6a, E2 treatment resulted in increased carnitine intake into MCF-7 breast cancer cells. We also tested the effect of downregulation of SLC22A5 expression on carnitine intake. In this case, carnitine intake was inhibited by pretreatment with siRNA against SLC22A5 (Fig. 6b). Further examination of carnitine intake in another cell line (T47D) revealed similar results (data not shown). Collectively, these results demonstrate that SLC22A5 functions as a carnitine transporter in breast cancer cells.
Fig. 6.
Carnitine intake in MCF-7 breast cancer cells. a Effect of E2 on carnitine intake. Right panel verification of upregulation of SLC22A5 expression in response to E2. b Effect of siRNA against SLC22A5 on carnitine intake. Right panel verification of knockdown of SLC22A5 expression by its siRNA
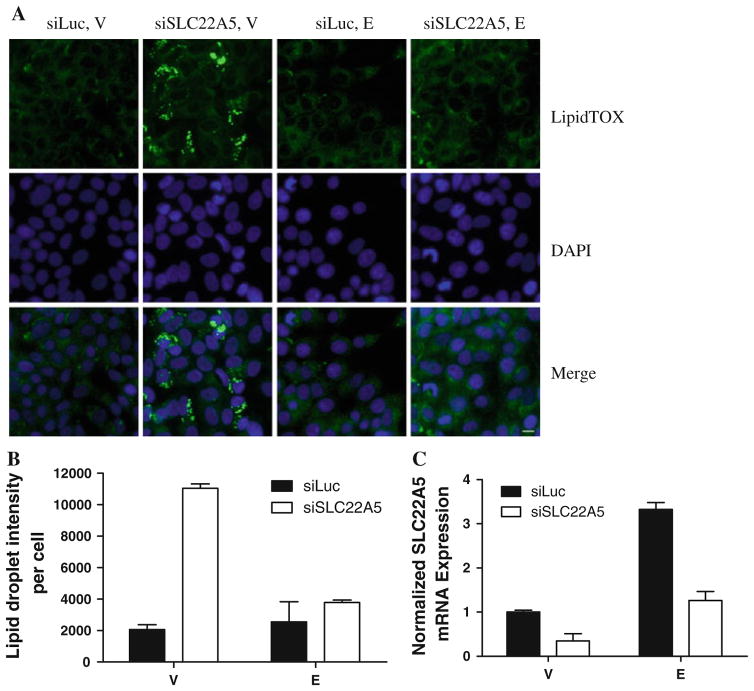
Since carnitine is a critical regulator of fatty acid transport and lipid metabolism, we investigated the possible effects of SLC22A5 on lipid content. As shown in Fig. 7, SLC22A5 knockdown resulted in increased lipid droplet accumulation in vehicle-treated cells. Estrogen treatment was able to prevent the accumulation of lipid when SLC22A5 was knocked down. This result is consistent with our studies showing that estrogen upregulates SLC22A5 expression and rescued the effect of SLC22A5 knockdown (Fig. 7).
Fig. 7.
Lipid droplet accumulation in MCF-7 breast cancer cells. a Fluorescent images of cells stained with HCS LipidTOX (green fluorescence) and DAPI (blue fluorescence). b Quantification of lipid content. c SLC22A5 expression in cells transfected with siRNA against Luc/SLC22A5 and treated with vehicle/E2
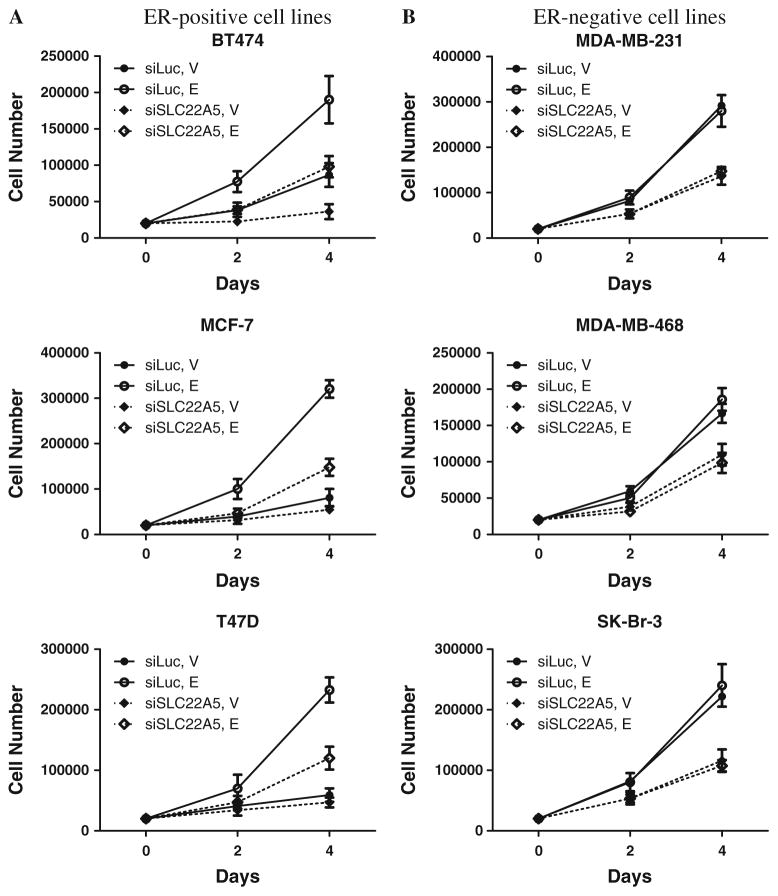
Since previous studies have shown that increased cellular lipid content is associated with both a less malignant state and a reduction in cell growth rate [53], we assessed the proliferation of breast cancer cells after knockdown of SLC22A5 expression. siRNA against SLC22A5 significantly inhibited growth of three ER-positive and three ER-negative breast cancer cell lines, indicating the functional importance of SLC22A5 in cell proliferation at a fundamental level (Fig. 8). Confirmation of these results was achieved through MTS assay, the results of which were in accordance with our initial findings (data not shown). Taken together, these results demonstrate that SLC22A5 is a critical gene for carnitine intake, lipid metabolism, and breast cancer cell proliferation.
Fig. 8.
Cell proliferation as determined using the Countess Automated Cell Counter. a ER-positive cell lines, b ER-negative cell lines
Discussion
Estrogen and its receptors regulate many genes implicated in breast cancer. Indeed, the ER/E2 signaling pathways play an important role in the pathogenesis of breast cancer. Although transcriptional profiling studies have shown that thousands of genes are regulated by ER/E2 in breast cancer, only a small number of genes have ER binding sites that have been experimentally verified. Therefore, the molecular mechanisms of ER/E2 regulation of target gene expression and biological function are not completely understood. Previous studies by us and others have shown that ER/E2 regulate genes via different pathways, including the classical genomic pathway [54] and transcription factor crosstalk (the tethering pathway) [11, 12]. However, more complex mechanisms may also be involved in ER/E2 signaling in breast cancer. Recent genome-wide studies have revealed that many ER binding sites are located in distal regions rather than in proximal promoter regions [16, 17]. It has also been demonstrated that the regulatory elements located within the intronic region are critical for the regulation of many important genes in breast cancer, including ER [55], IGFBP-6 [56], and ERBB2 [57].
In a previous genome-wide transcriptional profiling study, we identified SLC22A5 as a novel estrogen-induced gene [18]. In the present study, we show that estrogen stimulates SLC22A5 expression through an ER-dependent transcriptional regulatory mechanism via a distal enhancer located in the second intron. We characterized a half ERE, a critical newly identified full ERE, and an NR4A2/Nurr1 site, juxtapositioned within the intronic enhancer, as important elements for estrogen induction of SLC22A5 expression. Finally, we found that crosstalk between ER and Nurr1 is involved in the estrogen-regulated induction of SLC22A5 expression.
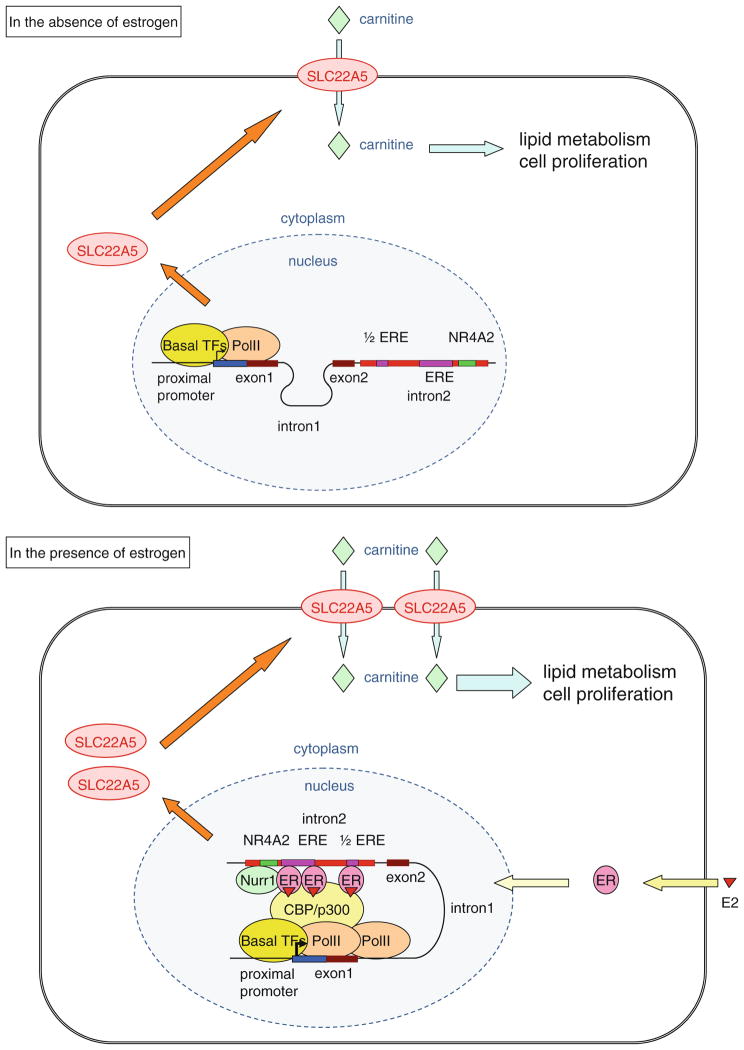
Based on our results, we propose a novel molecular mechanism by which ER and Nurr1 cooperate to regulate SLC22A5 expression (Fig. 9). In this model, ER and Nurr1 are not bound to the distal intronic enhancer region in the absence of estrogen, and SLC22A5 is expressed at a low level. When estrogen is added to the cells, estrogen-liganded ER translocates to the nucleus and the dimeric ER complex binds to the full ERE. This complex is stabilized by the interaction of ER with Nurr1 binding to the NR4A2 site as well as another ER binding to the half ERE. The ER–Nurr1 complex then functions as a transactivator, interacting with RNA polymerase II bound to the SLC22A5 promoter region, and ultimately upregulating SLC22A5 gene expression, and subsequently carnitine intake and lipid metabolism, in breast cancer cells.
Fig. 9.
Schematic representation of the molecular mechanism by which estrogen regulates SLC22A5 gene expression
Previous studies have shown that SLC22A5 is highly expressed in the kidney and functions primarily as a transporter of carnitine [19, 20], the level of which has been implicated in lipid metabolism and energy production [21–23]. There are only a few studies of the regulation of carnitine levels by sex hormones. For example, plasma carnitine concentration decreases upon estrogen treatment in rats [58] and serum-free carnitine concentration is inversely correlated with estrogen levels in women [59]. Estrogen has also been shown to upregulate carnitine palmitoyltransferase I gene expression in rat skeletal muscle [60, 61]. These studies have demonstrated a potential role of estrogen in regulating carnitine homeostasis, lipid metabolism, and cellular energy. However, no information is available to date regarding the regulation and function of the SLC22A5 gene in breast cancer, or in any other cancers. In this study, we demonstrate that estrogen treatment results in increased carnitine intake into MCF-7 breast cancer cells. We also show that knockdown of SLC22A5 expression dramatically inhibits carnitine intake, increases lipid droplet accumulation, and suppresses proliferation of breast cancer cells. The underlying mechanism in breast cancer could be an interruption of carnitine transport due to SLC22A5 inhibition, thereby leading to aberrant lipid metabolism, reduced cellular energy, and decreased cell proliferation. Since estrogen is a mitogen, ER-positive cells require more carnitine intake for lipid metabolism and growth, which may be the evolutionary reason why an ERE is present in the SLC22A5 regulatory region.
In summary, the study presented here demonstrates that SLC22A5 expression is regulated by estrogen via a novel intronic ERE and indicates that SLC22A5 is required for carnitine intake, lipid metabolism, and proliferation of breast cancer cells. This study provides novel insights into the underlying molecular mechanism by which estrogen regulates metabolism and proliferation, and also raises the potential for specifically targeting SLC22A5 for the treatment of breast cancer in the future.
Supplementary Material
Acknowledgments
This study was supported by the NIH grant R01-CA123246. The authors thank Michelle I. Savage for proofreading the manuscript.
Abbreviations
- ER
Estrogen receptor
- ERE
Estrogen-response element
- ActD
Actinomycin D
- CHX
Cycloheximide
- ChIP
Chromatin immunoprecipitation
- siRNA
Small interfering RNA
- SLC22A5
Solute carrier family 22 member 5
- OCTN2
Organic cation transporter
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-011-1925-0) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Russo J, Russo IH. The role of estrogen in the initiation of breast cancer. J Steroid Biochem Mol Biol. 2006;102(1–5):89–96. doi: 10.1016/j.jsbmb.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sommer S, Fuqua SA. Estrogen receptor and breast cancer. Semin Cancer Biol. 2001;11(5):339–352. doi: 10.1006/scbi.2001.0389. [DOI] [PubMed] [Google Scholar]
- 4.Shao W, Brown M. Advances in estrogen receptor biology: prospects for improvements in targeted breast cancer therapy. Breast Cancer Res. 2004;6(1):39–52. doi: 10.1186/bcr742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali S, Coombes RC. Estrogen receptor alpha in human breast cancer: occurrence and significance. J Mammary Gland Biol Neoplasia. 2000;5(3):271–281. doi: 10.1023/a:1009594727358. [DOI] [PubMed] [Google Scholar]
- 6.Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276(40):36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- 7.Moggs JG, Orphanides G. Estrogen receptors: orchestrators of pleiotropic cellular responses. EMBO Rep. 2001;2(9):775–781. doi: 10.1093/embo-reports/kve185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87(3):905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 9.Marino M, Galluzzo P, Ascenzi P. Estrogen signaling multiple pathways to impact gene transcription. Curr Genomics. 2006;7(8):497–508. doi: 10.2174/138920206779315737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29(14):2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Mayer JA, Mazumdar A, Fertuck K, Kim H, Brown M, Brown PH. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Mol Endocrinol. 2011;25(9):1527–1538. doi: 10.1210/me.2011-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Mayer JA, Mazumdar A, Brown PH. The rearranged during transfection/papillary thyroid carcinoma tyrosine kinase is an estrogen-dependent gene required for the growth of estrogen receptor positive breast cancer cells. Breast Cancer Res Treat. 2012 doi: 10.1007/s10549-011-1775-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly MJ, Levin ER. Rapid actions of plasma membrane estrogen receptors. Trends Endocrinol Metab. 2001;12(4):152–156. doi: 10.1016/s1043-2760(01)00377-0. [DOI] [PubMed] [Google Scholar]
- 14.Moriarty K, Kim KH, Bender JR. Minireview: estrogen receptor-mediated rapid signaling. Endocrinology. 2006;147(12):5557–5563. doi: 10.1210/en.2006-0729. [DOI] [PubMed] [Google Scholar]
- 15.Kampa M, Pelekanou V, Castanas E. Membrane-initiated steroid action in breast and prostate cancer. Steroids. 2008;73(9–10):953–960. doi: 10.1016/j.steroids.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 17.Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei CL, Liu ET. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3(6):e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeNardo DG, Kim HT, Hilsenbeck S, Cuba V, Tsimelzon A, Brown PH. Global gene expression analysis of estrogen receptor transcription factor cross talk in breast cancer: identification of estrogen-induced/activator protein-1-dependent genes. Mol Endocrinol. 2005;19(2):362–378. doi: 10.1210/me.2004-0267. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Prasad PD, Leibach FH, Ganapathy V. cDNA sequence, transport function, and genomic organization of human OCTN2, a new member of the organic cation transporter family. Biochem Biophys Res Commun. 1998;246(3):589–595. doi: 10.1006/bbrc.1998.8669. [DOI] [PubMed] [Google Scholar]
- 20.Tamai I, Ohashi R, Nezu J, Yabuuchi H, Oku A, Shimane M, Sai Y, Tsuji A. Molecular and functional identification of sodium ion-dependent, high affinity human carnitine transporter OCTN2. J Biol Chem. 1998;273(32):20378–20382. doi: 10.1074/jbc.273.32.20378. [DOI] [PubMed] [Google Scholar]
- 21.Ramsay RR. The role of the carnitine system in peroxisomal fatty acid oxidation. Am J Med Sci. 1999;318(1):28–35. doi: 10.1097/00000441-199907000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Hoppel C. The role of carnitine in normal and altered fatty acid metabolism. Am J Kidney Dis. 2003;41(4 Suppl 4):S4–S12. doi: 10.1016/s0272-6386(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 23.Longo N, Amat di San Filippo C, Pasquali M. Disorders of carnitine transport and the carnitine cycle. Am J Med Genet C Semin Med Genet. 2006;142C(2):77–85. doi: 10.1002/ajmg.c.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santiago JL, Martinez A, de la Calle H, Fernandez-Arquero M, Figueredo MA, de la Concha EG, Urcelay E. Evidence for the association of the SLC22A4 and SLC22A5 genes with type 1 diabetes: a case control study. BMC Med Genet. 2006;7:54. doi: 10.1186/1471-2350-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36(5):471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 26.Shoji Y, Koizumi A, Kayo T, Ohata T, Takahashi T, Harada K, Takada G. Evidence for linkage of human primary systemic carnitine deficiency with D5S436: a novel gene locus on chromosome 5q. Am J Hum Genet. 1998;63(1):101–108. doi: 10.1086/301911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nezu J, Tamai I, Oku A, Ohashi R, Yabuuchi H, Hashimoto N, Nikaido H, Sai Y, Koizumi A, Shoji Y, Takada G, Matsuishi T, Yoshino M, Kato H, Ohura T, Tsujimoto G, Hayakawa J, Shimane M, Tsuji A. Primary systemic carnitine deficiency is caused by mutations in a gene encoding sodium ion-dependent carnitine transporter. Nat Genet. 1999;21(1):91–94. doi: 10.1038/5030. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Ye J, Ganapathy V, Longo N. Mutations in the organic cation/carnitine transporter OCTN2 in primary carnitine deficiency. Proc Natl Acad Sci USA. 1999;96(5):2356–2360. doi: 10.1073/pnas.96.5.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang NL, Ganapathy V, Wu X, Hui J, Seth P, Yuen PM, Wanders RJ, Fok TF, Hjelm NM. Mutations of OCTN2, an organic cation/carnitine transporter, lead to deficient cellular carnitine uptake in primary carnitine deficiency. Hum Mol Genet. 1999;8(4):655–660. doi: 10.1093/hmg/8.4.655. [DOI] [PubMed] [Google Scholar]
- 30.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 31.Strecker TE, Shen Q, Zhang Y, Hill JL, Li Y, Wang C, Kim HT, Gilmer TM, Sexton KR, Hilsenbeck SG, Osborne CK, Brown PH. Effect of lapatinib on the development of estrogen receptor-negative mammary tumors in mice. J Natl Cancer Inst. 2009;101(2):107–113. doi: 10.1093/jnci/djn436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103(6):843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 33.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA, Jr, Marks JR, Dressman HK, West M, Nevins JR. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 34.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10(6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d’Assignies MS, Bergh J, Lidereau R, Ellis P, Harris AL, Klijn JG, Foekens JA, Cardoso F, Piccart MJ, Buyse M, Sotiriou C. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 36.Ginestier C, Cervera N, Finetti P, Esteyries S, Esterni B, Adelaide J, Xerri L, Viens P, Jacquemier J, Charafe-Jauffret E, Chaffanet M, Birnbaum D, Bertucci F. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12(15):4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 37.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, Rouzier R, Sneige N, Ross JS, Vidaurre T, Gomez HL, Hortobagyi GN, Pusztai L. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 38.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, Wong JE, Liu ET, Bergh J, Kuznetsov VA, Miller LD. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 39.Miller WR, Larionov AA, Renshaw L, Anderson TJ, White S, Murray J, Murray E, Hampton G, Walker JR, Ho S, Krause A, Evans DB, Dixon JM. Changes in breast cancer transcriptional profiles after treatment with the aromatase inhibitor, letrozole. Pharmacogenet Genomics. 2007;17(10):813–826. doi: 10.1097/FPC.0b013e32820b853a. [DOI] [PubMed] [Google Scholar]
- 40.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 42.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 43.van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijervan Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 45.Yu K, Ganesan K, Miller LD, Tan P. A modular analysis of breast cancer reveals a novel low-grade molecular signature in estrogen receptor-positive tumors. Clin Cancer Res. 2006;12(11 Pt 1):3288–3296. doi: 10.1158/1078-0432.CCR-05-1530. [DOI] [PubMed] [Google Scholar]
- 46.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA, Malmstrom P, Memeo L, Isola J, Bendahl PO, Rosen N, Hibshoosh H, Ringner M, Borg A, Parsons R. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA. 2007;104(18):7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, Van de Vijver MJ, Bergh J, Piccart M, Delorenzi M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 48.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein-Hitpass L, Schorpp M, Wagner U, Ryffel GU. An estrogen-responsive element derived from the 5′ flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986;46(7):1053–1061. doi: 10.1016/0092-8674(86)90705-1. [DOI] [PubMed] [Google Scholar]
- 50.Law SW, Conneely OM, DeMayo FJ, O’Malley BW. Identification of a new brain-specific transcription factor, NURR1. Mol Endocrinol. 1992;6(12):2129–2135. doi: 10.1210/mend.6.12.1491694. [DOI] [PubMed] [Google Scholar]
- 51.Mages HW, Rilke O, Bravo R, Senger G, Kroczek RA. NOT, a human immediate-early response gene closely related to the steroid/thyroid hormone receptor NAK1/TR3. Mol Endocrinol. 1994;8(11):1583–1591. doi: 10.1210/mend.8.11.7877627. [DOI] [PubMed] [Google Scholar]
- 52.Murphy EP, Dobson AD, Keller C, Conneely OM. Differential regulation of transcription by the NURR1/NUR77 subfamily of nuclear transcription factors. Gene Expr. 1996;5(3):169–179. [PMC free article] [PubMed] [Google Scholar]
- 53.Mueller E, Sarraf P, Tontonoz P, Evans RM, Martin KJ, Zhang M, Fletcher C, Singer S, Spiegelman BM. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1(3):465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 54.Barkhem T, Haldosen LA, Gustafsson JA, Nilsson S. pS2 Gene expression in HepG2 cells: complex regulation through crosstalk between the estrogen receptor alpha, an estrogen-responsive element, and the activator protein 1 response element. Mol Pharmacol. 2002;61(6):1273–1283. doi: 10.1124/mol.61.6.1273. [DOI] [PubMed] [Google Scholar]
- 55.Schausi D, Tiffoche C, Thieulant ML. Regulation of the intronic promoter of rat estrogen receptor alpha gene, responsible for truncated estrogen receptor product-1 expression. Endocrinology. 2003;144(7):2845–2855. doi: 10.1210/en.2003-0024. [DOI] [PubMed] [Google Scholar]
- 56.Uray IP, Shen Q, Seo HS, Kim H, Lamph WW, Bissonnette RP, Brown PH. Rexinoid-induced expression of IGFBP-6 requires RARbeta-dependent permissive cooperation of retinoid receptors and AP-1. J Biol Chem. 2009;284(1):345–353. doi: 10.1074/jbc.M804721200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, Jiang J, Howat WJ, Ali S, Carroll JS. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456(7222):663–666. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borum PR. Regulation of the carnitine concentration in plasma. In: Frenkel RA, McGarry JD, editors. Carnitine biosynthesis, metabolism, and function. Academic Press; New York: 1980. pp. 115–126. [Google Scholar]
- 59.Takiyama N, Matsumoto K. Age-and sex-related differences of serum carnitine in a Japanese population. J Am Coll Nutr. 1998;17(1):71–74. doi: 10.1080/07315724.1998.10720458. [DOI] [PubMed] [Google Scholar]
- 60.Campbell SE, Febbraio MA. Effect of ovarian hormones on mitochondrial enzyme activity in the fat oxidation pathway of skeletal muscle. Am J Physiol Endocrinol Metab. 2001;281(4):E803–E808. doi: 10.1152/ajpendo.2001.281.4.E803. [DOI] [PubMed] [Google Scholar]
- 61.Campbell SE, Mehan KA, Tunstall RJ, Febbraio MA, Cameron-Smith D. 17beta-estradiol upregulates the expression of peroxisome proliferator-activated receptor alpha and lipid oxidative genes in skeletal muscle. J Mol Endocrinol. 2003;31(1):37–45. doi: 10.1677/jme.0.0310037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.