Abstract
The use of narrow bore LC capillaries operated at ultralow flow rates coupled with mass spectrometry provides a desirable convergence of figures of merit to support high performance LC-MS/MS analysis. This configuration provides a viable means to achieve in-depth protein sequence coverage while maintaining a high rate of data production. Here we explore potential performance improvements afforded by use of 25 μm × 100 cm columns fabricated with 5 μm diameter reversed phase particles and integrated electrospray emitter tips. These columns achieve a separation peak capacity of ≈750 in a 600 minute gradient, with average chromatographic peak widths of less than one minute. At room temperature a pressure drop of only ≈1500 psi is sufficient to maintain an effluent flow rate of ≤ 10 nL/min. Using mouse embryonic stem cells as a model for complex mammalian proteomes we reproducibly identify over 4000 proteins across duplicate 600 min LC-MS/MS analyses.
Keywords: proteome profiling, nanoflow HPLC, reversed phase, LC-MS/MS, proteomics, high throughput, narrow bore column, capillary HPLC
INTRODUCTION
Improvements in chromatographic performance are an important component of efforts to achieve systematic and in depth proteome sequence analysis.1-5 Pioneering work from multiple labs demonstrated the benefits of small particles (< 2 μm),6-8 ultra-narrow bore columns (≤ 25 μm I.D.),9-11 and high pressure solvent delivery (up to 70,000 psi).6 Various combinations of these technologies have been used to attain single dimension separation peak capacity values of 700 1500.2, 6-7, 9, 12,18 As demonstrated originally by Novotny19-20 and Jorgenson,21 overall chromatographic performance improves with smaller diameter columns; in fact several variables converge to the benefit of electrospray performance under these circumstances. First, the volumetric flow at which optimal plate height is achieved varies with column inner diameter. Secondly, the effective analyte concentration, and hence signal intensity observed for LC-MS, increases approximately with the inverse square root of column inner diameter.9, 22 Finally, for aspect ratios (e.g., column diameter: particle diameter, < 10) obtained with small inner diameter (≤25 μm) capillaries and readily available reversed phase resins (3-5 μm dia.), the column cross section is dominated by the loosely packed “wall region,” creating a more homogeneous packing structure.19-21, 23-24
More recently the commercial availability of columns packed with particles smaller than 2 μm and ultra-high pressure pump systems (UHPLC) have been widely used for mass spectrometry-based proteomics, typically with capillary columns of ≥ 75 μm inner diameter.5, 17 The use of smaller particles at a fixed column I.D. maintains chromatographic resolution at increased flow rates, enabling so-called fast separations.6, 25-27 However, recent work from our lab,28 along with related studies,10-11, 29-31 has provided compelling evidence that the gains in electrospray ionization efficiency achieved at ultra-low effluent flow rates more than compensate for diminished chromatographic performance. Moreover, multiple studies have suggested that the use of large particles packed in long beds is the best route to achieve maximum peak capacity for separation of complex mixtures.32-35
Collectively these data and observations suggest that a focus on smaller diameter capillaries packed with larger particles and operated in flow regimes below Van Deemter minima represents a promising path for improved LC-MS performance. Towards this end we fabricated 25 μm × 100 cm columns with integrated electrospray emitters based on our previously described protocol.28 Using mouse embryonic stem cells as a model for complex mammalian proteomes we observed significant improvements in multiple analytical figures of merit for these extended length columns. Our data suggest that the use of narrow bore capillaries packed with larger particles in extended bed lengths, and operated at ultra-low flow rates provides a useful convergence of high peak capacity separation, high ionization efficiency, improved protein sequence analysis, and increased data production rate.
EXPERIMENTAL SECTION
Due to space considerations experimental methods related to cell culture, sample preparation, and general mass spectrometry acquisition parameters are provided in Supplementary Materials.
Construction of 25 μm × 100 cm fused silica analytical columns with integrated emitter tips
The column packing procedure is similar to that described previously.28 In brief, silicate based frits were cast in situ as follows: A 2.5 cm section of polyimide was removed approximately 3 cm from one end of the fused silica tubing. A silicate solution was allowed to migrate via capillary action to four fifths the length of the exposed window. Next, polymerization was induced using a soldering iron, with care taken to form frits of 1 - 2 mm in length. After ejection of excess silicate solution the frits were re heated with the soldering iron at 400°C for several seconds. Columns were slurry packed as previously described,28 with 5 Hm diameter, 120 Å pore size Monitor C18 beads (Column Engineering, Ontario, CA) suspended in acetonitrile. Bed lengths of 100 cm were obtained after 48 hours of continuous packing at ≈1500 psi in a stainless steel vessel pressurized with helium. Next, the columns were dried with helium for 30 minutes. Finally, an integrated emitter tip of 0.75 – 1.5 μm diameter was formed 2 – 4 mm beyond the frit using a laser-based pipette puller (P-2000, Sutter Instruments, Novato, CA). A 150 μm × 6 cm column packed with POROS 10R2 resin was prepared as previously described28 and used as a precolumn (PC).
Our nanoflow LC platform was based on a Waters NanoACQUITY UHPLC system as described previously,36 and equipped with a 6-port, 2-position valve (VICI Valco, Houston, TX). We removed the first dimension column of the original 2D RP-RP configuration to create a one dimension LC-MS/MS platform. The autosampler was used to load samples. True nanoflow rates in the analytical column were achieved through use of a passive flow split located prior to the reversed phase PC; the pre- and analytical-columns were configured in a vented geometry37-38 as previously described.28 We used two independent means to measure column flow rates. When the spray voltage was set >5kV bubbles form periodically in the column and we estimated flow rate based on the time required to traverse a predefined segment of the capillary. The bubbles purge quickly once the spray voltage was reduced to normal operating levels (<2.5kV). Alternatively we measured the displaced effluent volume by collection from the tip into an empty piece of 25 mm I.D. fused silica capillary, held in place perpendicular to the emitter by a secondary clamp on the ESI source platform. Linear velocity was calculated based on parameters previously reported 9 for similar geometry LC assemblies.
Data Analysis
Our API-based multiplierz39-40 software framework were used to extract and format MS/MS data for subsequent search against an IPI mouse database v3.68. MS/MS data was searched using Protein Pilot V3.0 (AB Sciex, Foster City, CA) with the following parameters: instrument =“Orbi/FT MS (sub ppm), LTQ MS/MS” for data acquired from Orbitrap XL and Velos, or “QSTAR Elite” for the 5600 Triple TOF. A fixed modification of +46 corresponding to methylthioylation of cysteine was also included. Only those peptides with scores at or above a FDR threshold of 1% were further considered. Multiplierz scripts were used to systematically extract XICs and calculate corresponding peak widths for all identified peptides.
Data Availability
Lists of proteins and peptides associated with Figures 1, 3, and 4 are provided in Supplementary Files S1 and S2. In addition, all native mass spectrometry files are available for download from our web site: http://blaispathways.dfci.harvard.edu/mz/?id=doi:10.1021/ac2031404
Figure 1.
(A) Total ion chromatogram (TIC) for a 600 minute gradient LC-MS/MS acquisition at a flow rate of 5 nL/min. (B-E) Representative extracted ion chromatograms (XIC) for peptides detected throughout the gradient: (B) IFAPNHVVAK; (C) LVGEIMQETGTR; (D) LNVDFALIHK; and (E) AEYLASIFGTEK.
Figure 3.
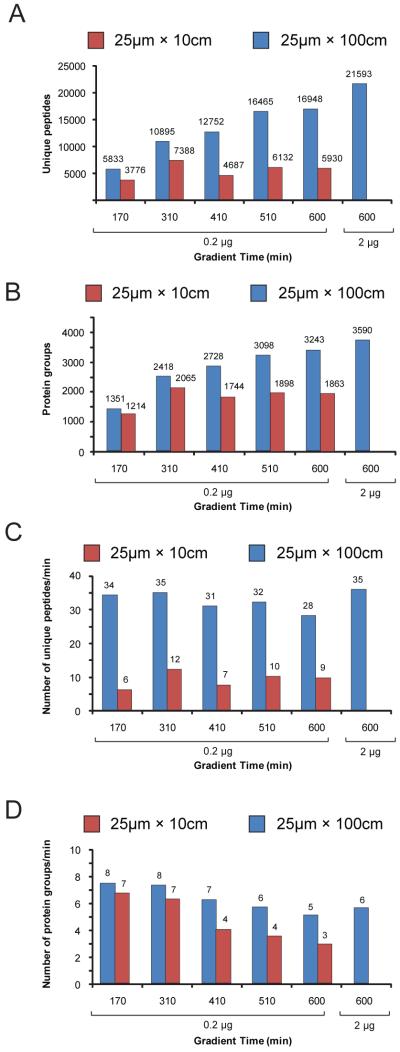
Number of (A) peptide and (B) protein identifications as a function of gradient length and sample quantity for (red) 25 μm × 10 cm and (blue) 25 μm × 100 cm columns. Rate of (C) peptide and (D) protein identifications per minute of LC-MS/MS acquisition time for (red) 25 μm × 10 cm and (blue) 25 μm × 100 cm columns.
Figure 4.
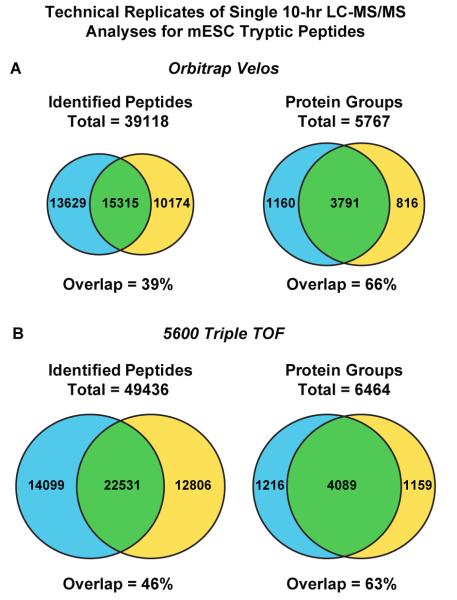
Venn diagrams representing the overlap of identified (left) peptides and (right) proteins for technical replicate 600 min. gradient LC-MS/MS analyses of tryptic peptides derived from mouse embryonic stem cells. (A) Data acquired from Thermo Fisher Velos Orbitrap. (B) Data acquired from AB Sciex 5600 TripleTOF.
SAFETY CONSIDERATIONS
Safety glasses should be worn at all times during construction and use of fused silica based capillaries. In addition, a lab coat and gloves should be worn when manipulating cell cultures in biosafety cabinets or when handling neat TFA and other volatile organics in a chemical fume hood.
RESULTS AND DISCUSSION
We recently demonstrated the use of 25 μm × 10 cm analytical columns28 operated at true nanoflow regimes in online, automated 2D RP-RP36 and 3D RP-SAX-RP43-44 configurations for the analysis of complex proteomes. The latter system provided highly orthogonal separation between all three dimensions; based on these results we asked whether improved peak capacity in the final reversed phase stage would provide higher LC-MS/MS performance. Towards this end we extended the bed length of our analytical columns to 100 cm. In order to facilitate column packing and minimize the operating backpressure we used 5 μm diameter C18 resin. As noted by Ehlert et al.,23 columns with a column to particle diameter ratio () below ≈10 exhibit more homogenous cross sections dominated by loosely packed particles. Serendipitously, this configuration provides somewhat greater inter particle porosity, and hence lower backpressure as compared to columns with closely packed ( > 10) core regions.23 We observed that a pressure drop of approximately 1400 psi across this column provided for an effluent flow rate of ≈5nL/min. at room temperature. This volumetric flow rate corresponds to a reduced velocity of ≈0.021 cm/sec., or nearly an order of magnitude less than that determined for optimal chromatographic peak width in similar slurry packed, microcapillary configurations.9
We first asked whether our 25 μm × 100 cm columns would provide high peak capacity separation in the context of extended gradients. Towards this end we analyzed tryptic peptides derived from murine embryonic stem cells (mESC). We chose these cells as they are reported to exhibit a greater degree of genomic stability as compared to typical immortalized tumor lines,45 and hence may represent a suitable model system for platform performance comparisons across time and technologies. An aliquot corresponding to 200 ng of solubilized protein lysate was subjected to data dependent LC-MS/MS on an Orbitrap XL in the context of a 10 hour organic gradient. Figure 1A shows the total ion chromatogram (TIC) along with several extracted ion chromatograms (XIC) for peptides (panels B, C, D, and E) detected at various points throughout the gradient. Peak widths of less than 1 min. were readily observed even near the end of the gradient (panel E). Next we repeated this analysis on both 25 μm × 10 cm and 25 μm × 100 cm columns at various gradient lengths ranging from ≈3 to 10 hours. For each LC-MS/MS acquisition we calculated an average XIC peak width for all detected peptides within our multiplierz software environment39-40 and plotted these as a function of gradient length (Figure 2A). We observed that the average peak width broadened rapidly as a function of gradient length for the 10 cm column while that for the 100 cm column remained less than 60 s, even for the longest gradient tested. Based on these data we next calculated separation peak capacity and observed that the 25 μm × 100 cm columns provided superior performance, reaching a maximum at ≈750 for a 10 hour gradient (Figure 2B). Importantly the observed peak capacity for the 10 cm length columns was less than 50% of that for the extended length column and in fact diminished somewhat beyond a gradient length of ≈300 minutes. These results are consistent with previous reports suggesting that larger particles packed in extended length beds maximize separation peak capacity.32-35
Figure 2.
(A) Chromatographic peak width (4σ, where 2σ is defined as FWHM of the corresponding XIC) and (B) separation peak capacity for (red) 25 μm × 10 cm and (blue) 25 μm × 100 cm columns as a function of LC gradient length. Peak capacity was estimated by dividing the time over which peptides were detected by the median peptide XIC peak width.
To test whether the increased peak capacity translated into improved sequence coverage, we next plotted the number of unique peptides and proteins identified in these analyses (Figure 3 A and B). In each case the number of identifications mirrored the observed peak capacity, suggesting that separation quality was a major determinant of proteome sequence coverage as opposed to simply providing more time for exhaustive data-dependent MS/MS acquisition. In fact, at a gradient length of 600 min. the 25 μm × 100 cm analytical column provided a nearly 3-fold increase in the number of unique peptides identified as compared to the same analysis performed on the 10 cm column. We next increased the quantity of sample loaded to 2 μg, near the estimated upper limit of the 150 μm × 4 cm reversed phase pre column assembly28 and repeated the 10-hr. LC-MS/MS analysis; consistent with a previous report that systematically collated LC-MS/MS analytical figures of merit,46 we observed an approximate 25% and 10% increase in the number of peptide and protein groups identifications, respectively (Figure 3A and B, rightmost bars). The rate of data production is also a useful metric to evaluate overall system efficiency. Towards this end we plotted the numbers of peptides (Figure 3C) and proteins (Figure 3D) identified per minute of LC-MS/MS acquisition time in the above experiments. The 100 cm long column exhibited remarkably stable efficiency across all conditions tested. Consistent with the reduced peak capacity observed as a function of gradient length with the 25 μm × 10 cm column, we also observed a concomitant drop in the rate of data production at longer gradients.
To explore further improvements afforded by a higher performance mass spectrometer, we next coupled the 25 μm × 100 cm column to a LTQ Velos Orbitrap instrument. In replicate, 10 hr gradient acquisitions performed with 2 μg mESC whole cell lysate digests, we identified 4951 and 4607 protein groups, respectively (Figure 4A) and 5767 protein groups in total. The average data production rate across these two LC-MS/MS acquisitions was 45 and 9 peptide and protein identifications per minute, respectively. These data represent an approximate ≈25% improvement relative to those acquired on the Orbitrap XL platform. Given that we observed a 39% overlap in peptide sequences between replicate LC-MS/MS analyses (Figure 4A), we next systematically compared the relative retention times of the 200 most abundant peptides and found that they varied on average by 6% relative to the total acquisition time of 600 minutes. In addition we observed that some 80% of the corresponding precursor intensity ratios were within ± 2 fold across replicate runs. It is conceivable that the convolution of peptide complexity in whole proteome digests, along with variation in retention time and the associated spray suppression effects, results in identification of different sequences across replicate LC-MS/MS acquisitions. To explore the possible contribution of stochastic precursor sampling during MS/MS, we repeated the 600 minute gradient LC-MS experiments on an AB Sciex 5600 Triple TOF mass spectrometer, an instrument with improved MS/MS acquisition rates. The reproducibility of sequence identification across replicate analyses increased to 46% (Figure 4B), while the average number of unique peptide and protein identifications increased by 32% and 10%, respectively, as compared to the Orbitrap Velos data. The average data production rate across these two LC-MS/MS acquisitions was 60 peptide and 9 protein identifications per minute, respectively. While the continued evolution of mass spectrometry instrumentation will undoubtedly provide broader proteome coverage independent of developments in LC technology,47-49 detailed analyses have indicated that the incremental improvement provided by commercial advances in mass spectrometry hardware is somewhat offset by higher peak capacity separation.48
In general our data compare favorably in terms of sequence coverage and total acquisition time with a number of recent studies50-53 that reported deep proteome analysis on the same mass spectrometry platform as described herein. For example, both the Mann54 and Zeng55 laboratories used an Orbitrap XL mass spectrometer in conjunction with SDS-PAGE gel and “Ying-Yang” prefractionation techniques, respectively, to analyze the mESC proteome. Results from Köcher and colleagues,56 based on LC-MS/MS analysis on an Orbitrap Velos of proteins derived from human cells, revealed similar trends as reported herein with respect to column and gradient length. In contrast to our column configuration (25 μm I.D. packed with 5 μm dia. beads) they opted for 75 μm I.D. capillaries packed with 2 μm dia. beads (25 cm and 50 cm bed lengths). Here again direct comparisons with our data are tempered by the experimental differences; however the large discrepancy between reported protein identifications, 2516 I.D.s (average value across triplicate measurements)56 vs 4171 I.D.s (reproduced across replicate measurements, Figure 4A) is consistent with the notion that smaller diameter columns packed with larger particles and operated at ultra-low effluent flow rates maximizes LC-MS/MS data output.
Consistent with other recent reports,5, 9, 14, 30, 47, 56-59 our data support the idea that high-performance, single-dimension chromatography may provide a reasonable alternative to multi-dimensional fractionation in terms of the depth of proteome coverage and the total required acquisition time. In many cases these systems are operated at ultra-high pressure or elevated temperature. Open tubular11, 29, 31, 60-62 or monolithic stationary phases10, 57, 60, 63 have been successfully utilized as alternatives to slurry packed columns, although the overall binding capacity is often substantially less as compared to porous or semi-porous64 silica- or polymer-based particles. Similar to some of these recent reports, we also opted to use smaller diameter capillaries, but chose somewhat larger diameter reversed phase beads in order to simultaneously maintain high resolving power at low effluent flow rate, and hence optimal electrospray ionization efficiency,28 along with a relatively low backpressure requirement of only ≈1500 psi. Indeed our 25 μm I.D. column assemblies provided a peak capacity of ≈750 at a flow rate of ≈5nL/min. (Figure 2B). This is approximately 50% of the total peak capacity reported by Shen, et al., for a 200 cm long column packed with 1.8 μm particles and operated at a backpressure of nearly 20,000 psi.16 Similarly the peak capacity obtained on our 25 μm × 100 cm columns compares favorably to that obtained with 100 μm × 400 cm long monolithic C18 columns operated at ≈6000 psi.63 Finally, as previously described28 our fabrication protocol is economical and requires no specialized equipment; in addition the integrated electrospray emitters minimize deleterious band broadening often encountered when mechanical fittings are used to couple spray emitters with microcapillary columns. As with our 25 μm × 10 cm analytical columns, the extended length versions have proven very reliable and work is ongoing to integrate these with our fully automated 3D RP SAX-RP43-44 separation platform. Other interesting extensions of this work include using somewhat smaller particles (e.g., ~3 μm dia.) in (i) the current 25 μm I.D. capillaries to achieve higher throughput while maintaining relatively low backpressure (e.g., < 5000 psi.), or (ii) 15 μm I.D. columns in order to maintain the same aspect ratio but utilize still lower effluent flow rates for potentially higher ionization efficiency during LC-MS/MS.
CONCLUSIONS
Despite significant improvements in mass spectrometry performance,48-49, 65 obtaining in-depth sequence coverage for complex proteomes remains a difficult challenge. Moreover, the inherent limitations for parallel acquisition in mass spectrometry-based proteomics require that the depth of proteome coverage be considered within the context of data production rate and total instrument time. In this work we describe the fabrication and relative performance improvements afforded by the use of 25 μm × 100 cm analytical columns packed with 5 μm diameter reversed phase particles. These assemblies provide multiple figures of merit towards an optimal coupling of high peak capacity separation and high efficiency electrospray ionization in LC-MS/MS. When used in conjunction with state-of-the-art mass spectrometry instrumentation, this 1-D LC configuration simultaneously provides in-depth sequence coverage (average = 5277 protein groups across replicate analyses) and high data production rate (60 and 9 peptide and protein identifications per minute, respectively).
Supplementary Material
ACKNOWLEDGMENTS
Generous support for this work was provided by the Dana Farber Cancer Institute and the National Institutes of Health, NHGRI (P50HG004233), and NINDS (P01NS047572). The authors thank Karl Mechtler (IMP and IMBA, Vienna, Austria) for helpful suggestions and critial reading of our manuscript.
REFERENCES
- (1).Sandra K, Moshir M, D’Hondt F, Tuytten R, Verleysen K, Kas K, Francois I, Sandra P. J. Chromatogr. B. 2009;877:1019–1039. doi: 10.1016/j.jchromb.2009.02.050. [DOI] [PubMed] [Google Scholar]
- (2).De Villiers A, Cabooter D, Lynen F, Desmet G, Sandra P. J. Chromatogr. A. 2009;1216:3270–3279. doi: 10.1016/j.chroma.2009.02.038. [DOI] [PubMed] [Google Scholar]
- (3).Slaughter C, High A, Galea C, Thomas P, Mishra A, Gebler J, Zhou M, Stine M, Finkelstein D, Obenauer J, Doherty P, Kriwacki R. Mol. Cell. Proteomics. 2007;6:46–46. [Google Scholar]
- (4).Michalski A, Cox J, Mann M. J. Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- (5).Thakur SS, Geiger T, Chatterjee B, Bandilla P, Frohlich F, Cox J, Mann M. Molecular & cellular proteomics: MCP. 2011;10:M110–003699. doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).MacNair JE, Patel KD, Jorgenson JW. Analytical Chemistry. 1999;71:700–708. doi: 10.1021/ac9807013. [DOI] [PubMed] [Google Scholar]
- (7).Patel KD, Jerkovich AD, Link JC, Jorgenson JW. Analytical Chemistry. 2004;76:5777–5786. doi: 10.1021/ac049756x. [DOI] [PubMed] [Google Scholar]
- (8).Mellors JS, Jorgenson JW. Anal Chem. 2004;76:5441–50. doi: 10.1021/ac049643d. [DOI] [PubMed] [Google Scholar]
- (9).Shen YF, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. Analytical Chemistry. 2002;74:4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- (10).Wang DD, Hincapie M, Rejtar T, Karger BL. Analytical Chemistry. 2011;83:2029–2037. doi: 10.1021/ac102825g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yue GH, Luo QZ, Zhang J, Wu SL, Karger BL. Analytical Chemistry. 2007;79:938–946. doi: 10.1021/ac061411m. [DOI] [PubMed] [Google Scholar]
- (12).MacNair JE, Lewis KC, Jorgenson JW. Analytical Chemistry. 1997;69:983–989. doi: 10.1021/ac961094r. [DOI] [PubMed] [Google Scholar]
- (13).Shen YF, Tolic N, Zhao R, Pasa Tolic L, Li LJ, Berger SJ, Harkewicz R, Anderson GA, Belov ME, Smith RD. Analytical Chemistry. 2001;73:3011–3021. doi: 10.1021/ac001393n. [DOI] [PubMed] [Google Scholar]
- (14).Shen Y, Tolic N, Masselon C, Pasa Tolic L, Camp DG, Hixson KK, Zhao R, Anderson GA, Smith RD. Analytical Chemistry. 2004;76:144–154. doi: 10.1021/ac030096q. [DOI] [PubMed] [Google Scholar]
- (15).Mazzeo JR, Neue UD, Kele M, Plumb RS. Analytical Chemistry. 2005;77:460A–467A. [Google Scholar]
- (16).Shen YF, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EA, Udseth HR, Smith RD. Analytical Chemistry. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- (17).Liu HJ, Finch JW, Lavallee MJ, Collamati RA, Benevides CC, Gebler JC. J. Chromatogr. A. 2007;1147:30–36. doi: 10.1016/j.chroma.2007.02.016. [DOI] [PubMed] [Google Scholar]
- (18).Min HK, Hyung SW, Shin JW, Nam HS, Ahn SH, Jung HJ, Lee SW. Electrophoresis. 2007;28:1012–1021. doi: 10.1002/elps.200600501. [DOI] [PubMed] [Google Scholar]
- (19).Karlsson KE, Novotny M. Analytical Chemistry. 1988;60:1662–1665. doi: 10.1021/ac00168a006. [DOI] [PubMed] [Google Scholar]
- (20).Kennedy RT, Jorgenson JW. Analytical Chemistry. 1989;61:1128–1135. doi: 10.1021/ac00180a012. [DOI] [PubMed] [Google Scholar]
- (21).Hsieh SC, Jorgenson JW. Analytical Chemistry. 1996;68:1212–1217. doi: 10.1021/ac950682m. [DOI] [PubMed] [Google Scholar]
- (22).Banks JF. J. Chromatogr. A. 1996;743:99–104. [Google Scholar]
- (23).Ehlert S, Roesler T, Tallarek U. J. Sep. Sci. 2008;31:1719–1728. doi: 10.1002/jssc.200800018. [DOI] [PubMed] [Google Scholar]
- (24).Knox JH, Parcher JF. Analytical Chemistry. 1969;41:1599. [Google Scholar]
- (25).Castro Perez J, Plumb R, Granger JH, Beattie I, Joncour K, Wright A. Rapid communications in mass spectrometry: RCM. 2005;19:843–8. doi: 10.1002/rcm.1859. [DOI] [PubMed] [Google Scholar]
- (26).MacNair JE, Opiteck GJ, Jorgenson JW, Moseley MA., 3rd. Rapid communications in mass spectrometry: RCM. 1997;11:1279–85. doi: 10.1002/(SICI)1097-0231(199708)11:12<1279::AID-RCM18>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- (27).Tolley L, Jorgenson JW, Moseley MA. Analytical Chemistry. 2001;73:2985–91. doi: 10.1021/ac0010835. [DOI] [PubMed] [Google Scholar]
- (28).Ficarro SB, Zhang Y, Lu Y, Moghimi AR, Askenazi M, Hyatt E, Smith ED, Boyer L, Schlaeger TM, Luckey CJ, Marto JA. Analytical Chemistry. 2009;81:3440–3447. doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
- (29).Ivanov AR, Zang L, Karger BL. Analytical Chemistry. 2003;75:5306–5316. doi: 10.1021/ac030163g. [DOI] [PubMed] [Google Scholar]
- (30).Shen YF, Moore RJ, Zhao R, Blonder J, Auberry DL, Masselon C, Pasa Tolic L, Hixson KK, Auberry KJ, Smith RD. Analytical Chemistry. 2003;75:3596–3605. doi: 10.1021/ac0300690. [DOI] [PubMed] [Google Scholar]
- (31).Luo QZ, Tang KQ, Yang F, Elias A, Shen YF, Moore RJ, Zhao R, Hixson KK, Rossie SS, Smith RD. J. Proteome Res. 2006;5:1091–1097. doi: 10.1021/pr050424y. [DOI] [PubMed] [Google Scholar]
- (32).Wang XL, Barber WE, Carr PW. J. Chromatogr. A. 2006;1107:139–151. doi: 10.1016/j.chroma.2005.12.050. [DOI] [PubMed] [Google Scholar]
- (33).Poppe H. J. Chromatogr. A. 1997;778:3–21. [Google Scholar]
- (34).Scott RPW, Kucera P. JCh. 1979;169:51–72. [Google Scholar]
- (35).Wang XL, Stoll DR, Schellinger AP, Carr PW. Analytical Chemistry. 2006;78:3406–3416. doi: 10.1021/ac0600149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zhou F, Cardoza JD, Ficarro SB, Adelmant GO, Lazaro JB, Marto JA. J. Proteome Res. 2010;9:6242–6255. doi: 10.1021/pr1004696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).van der Heeft E, ten Hove GJ, Herberts CA, Meiring HD, van Els C, de Jong A. Analytical Chemistry. 1998;70:3742–3751. doi: 10.1021/ac9801014. [DOI] [PubMed] [Google Scholar]
- (38).Licklider LJ, Thoreen CC, Peng JM, Gygi SP. Analytical Chemistry. 2002;74:3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- (39).Askenazi M, Parikh JR, Marto JA. Nat. Methods. 2009;6:240–242. doi: 10.1038/nmeth0409-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Parikh JR, Askenazi M, Ficarro SB, Cashorali T, Webber JT, Blank NC, Zhang Y, Marto JA. BMC Bioinformatics. 2009;10:364. doi: 10.1186/1471-2105-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Grobei MA, Qeli E, Brunner E, Rehrauer H, Zhang RX, Roschitzki B, Basler K, Ahrens CH, Grossniklaus U. Genome Res. 2009;19:1786–1800. doi: 10.1101/gr.089060.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Qeli E, Ahrens CH. Nat. Biotechnol. 2010;28:647–650. doi: 10.1038/nbt0710-647. [DOI] [PubMed] [Google Scholar]
- (43).Ficarro SB, Zhang Y, Carrasco Alfonso MJ, Garg B, Adelmant GO, Webber JT, Luckey CJ, Marto JA. Mol. Cell. Proteomics. 2011;10:M111.011064. doi: 10.1074/mcp.O111.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhou F, Sikorski TW, Ficarro SB, Webber JT, Marto JA. Analytical Chemistry. 2011;83:6996–7005. doi: 10.1021/ac200639v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, Wersto RP, Ko MSH. Nature. 2010;464:858–U66. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Rudnick PA, Clauser KR, Kilpatrick LE, Tchekhovskoi DV, Neta P, Blonder N, Billheimer DD, Blackman RK, et al. Mol. Cell. Proteomics. 2010;9:225–241. doi: 10.1074/mcp.M900223-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Nagaraj N, Alexander Kulak N, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Analytical Chemistry. 2009;81:7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Andrews GL, Simons BL, Young JB, Hawkridge AM, Muddiman DC. Analytical Chemistry. 2011;83:5442–5446. doi: 10.1021/ac200812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Nature. 2008;455:1251–U60. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- (51).Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. Science. 2008;320:938. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- (52).Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Graumann J, Hubner NC, Kim JB, Ko K, Moser M, Kumar C, Cox J, Scholer H, Mann M. Mol. Cell. Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- (55).Li QR, Xing XB, Chen TT, Li RX, Dai J, Sheng QH, Xin SM, Zhu LL, Jin Y, Pei G, Kang JH, Li YX, Zeng R. Mol. Cell. Proteomics. 2011;10:14. doi: 10.1074/mcp.M110.001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Kocher T, Swart R, Mechtler K. Anal Chem. 2011;83:2699–704. doi: 10.1021/ac103243t. [DOI] [PubMed] [Google Scholar]
- (57).Iwasaki M, Miwa S, Ikegami T, Tomita M, Tanaka N, Ishihama Y. Analytical Chemistry. 2010;82:2616–2620. doi: 10.1021/ac100343q. [DOI] [PubMed] [Google Scholar]
- (58).Shen YF, Jacobs JM, Camp DG, Fang RH, Moore RJ, Smith RD, Xiao WZ, Davis RW, Tompkins RG. Analytical Chemistry. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- (59).Iwasaki M, Sugiyama N, Tanaka N, Ishihama Y. J Chromatogr A. 2012;1228:292–7. doi: 10.1016/j.chroma.2011.10.059. [DOI] [PubMed] [Google Scholar]
- (60).Luo QZ, Shen YF, Hixson KK, Zhao R, Yang F, Moore RJ, Mottaz HM, Smith RD. Analytical Chemistry. 2005;77:5028–5035. doi: 10.1021/ac050454k. [DOI] [PubMed] [Google Scholar]
- (61).Luo Q, Page JS, Tang KQ, Smith RD. Analytical Chemistry. 2007;79:540–545. doi: 10.1021/ac061603h. [DOI] [PubMed] [Google Scholar]
- (62).Luo Q, Yue G, Valaskovic GA, Gu Y, Wu SL, Karger BL. Analytical Chemistry. 2007;79:6174–6181. doi: 10.1021/ac070583w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Eghbali H, Sandra K, Detobel F, Lynen F, Nakanishi K, Sandra P, Desmet GJ. Chromatogr. A. 2011;1218:3360–3366. doi: 10.1016/j.chroma.2010.10.045. [DOI] [PubMed] [Google Scholar]
- (64).Gritti F, Cavazzini A, Marchetti N, Guiochon G. J Chromatogr A. 2007;1157:289–303. doi: 10.1016/j.chroma.2007.05.030. [DOI] [PubMed] [Google Scholar]
- (65).Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mol. Cell. Proteomics. 2011;10:11. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Lists of proteins and peptides associated with Figures 1, 3, and 4 are provided in Supplementary Files S1 and S2. In addition, all native mass spectrometry files are available for download from our web site: http://blaispathways.dfci.harvard.edu/mz/?id=doi:10.1021/ac2031404
Figure 1.
(A) Total ion chromatogram (TIC) for a 600 minute gradient LC-MS/MS acquisition at a flow rate of 5 nL/min. (B-E) Representative extracted ion chromatograms (XIC) for peptides detected throughout the gradient: (B) IFAPNHVVAK; (C) LVGEIMQETGTR; (D) LNVDFALIHK; and (E) AEYLASIFGTEK.
Figure 3.
Number of (A) peptide and (B) protein identifications as a function of gradient length and sample quantity for (red) 25 μm × 10 cm and (blue) 25 μm × 100 cm columns. Rate of (C) peptide and (D) protein identifications per minute of LC-MS/MS acquisition time for (red) 25 μm × 10 cm and (blue) 25 μm × 100 cm columns.
Figure 4.
Venn diagrams representing the overlap of identified (left) peptides and (right) proteins for technical replicate 600 min. gradient LC-MS/MS analyses of tryptic peptides derived from mouse embryonic stem cells. (A) Data acquired from Thermo Fisher Velos Orbitrap. (B) Data acquired from AB Sciex 5600 TripleTOF.