Abstract
Glycolytic enzymes are cytosolic proteins, but they also play important extracellular roles in cell-cell communication and infection. We used Saccharomyces cerevisiae to analyze the secretory pathway of some of these enzymes, including enolase, phosphoglucose isomerase, triose phosphate isomerase, and fructose 1,6-bisphosphate aldolase. Enolase, phosphoglucose isomerase, and an N-terminal 28-amino-acid-long fragment of enolase were secreted in a sec23-independent manner. The enhanced green fluorescent protein (EGFP)-conjugated enolase fragment formed cellular foci, some of which were found at the cell periphery. Therefore, we speculated that an overview of the secretory pathway could be gained by investigating the colocalization of the enolase fragment with intracellular proteins. The DsRed-conjugated enolase fragment colocalized with membrane proteins at the cis-Golgi complex, nucleus, endosome, and plasma membrane, but not the mitochondria. In addition, the secretion of full-length enolase was inhibited in a knockout mutant of the intracellular SNARE protein-coding gene TLG2. Our results suggest that enolase is secreted via a SNARE-dependent secretory pathway in S. cerevisiae.
INTRODUCTION
Glycolytic enzymes play various roles inside and outside the cell (53). Although they are cytosolic proteins, numerous large-scale analyses have revealed their extracellular existence, in both unicellular and multicellular organisms (9, 28, 37, 39, 40, 46). Many glycolytic enzymes have been reported to play roles in important cellular processes, such as signal transduction and surface binding (16, 30, 42, 48). For example, extracellular enolase, which is a glycolytic enzyme, is a virulence factor in Candida albicans and some parasites (2, 24). Enolase has been found in small vesicles outside the cell (39, 40) and in the cell wall (13). In addition, enolase is secreted in a sequence-dependent manner (29, 56) and is present in the cell wall with no enzymatic activity, but it binds to plasminogen and helps the pathogen invade (49). Enolase is also found in viral particles (3, 8, 45) and is required for transcription of Sendai virus (38). Therefore, enolase is a therapeutic target for many diseases, including candidiasis (7, 55). Another extracellular glycolytic enzyme, phosphoglucose isomerase, enhances the motility of tumor cells (11) and performs like a cytokine (52), although it possesses no enzymatic activity outside the cell (54). However, the secretory pathway of glycolytic enzymes such as enolase and phosphoglucose isomerase remains to be revealed. This pathway appears to be unconventional, because glycolytic enzymes have no known secretion signals. Therefore, in this study we analyzed the secretory pathway of glycolytic enzymes.
A number of secreted proteins without known secretion signals have been found (25), and several unconventional secretory pathways have been discovered and suggested (12, 31, 36). Recently, Duran and coworkers identified the novel unconventional secretory pathway of the Acb1 protein (12). The budding yeast Saccharomyces cerevisiae is a useful organism to identify previously unknown secretory pathways, because it is a commonly used and well-understood model for studying cellular processes (43).
Two popular methods can be used to detect cellular secretion, namely, secretome analysis and the glucoamylase assay (23). Although these methods are highly informative and convenient, 3 major problems arise when using them to detect unknown secretory pathways. First, because proteome analysis targets naturally produced proteins, the proportion of each protein varies. Therefore, the secretory abilities of different proteins are incommensurable, and detecting leakage is inevitable. Second, the glucoamylase assay cannot detect changes in the sizes of proteins. Therefore, this method can miss the processing of proteins during secretion, which is important for the prediction of the secretory pathway. Third, neither method can visually trace the intracellular secretory pathway. Therefore, it is important to be cautious with the information obtained by these methods and to investigate all the possible pathways.
We previously developed a novel two-dimensional high-performance liquid chromatography (2D-HPLC)-based method that detects proteins on the living cell surface (34). Using this method, we gained an overview of the proteins on the outside of the cell and select glycolytic enzymes suitable for secretion analyses. In this study, we utilized enhanced green fluorescence protein (EGFP) conjugated with FLAG tag (EGFP-FLAG-tagged) glycolytic enzymes to analyze the secretory pathway of glycolytic enzymes. Western blot analysis enabled detection of the secreted proteins in the culture medium. Moreover, the use of plasmid-based protein expression facilitated uniform protein levels and analysis of the secreted proteins. Moreover, the secretory pathway was visualized and assessed with the aid of the conjugated fluorescent proteins (19, 22).
MATERIALS AND METHODS
Strains and media.
Escherichia coli strain DH5α [F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1] was used for the host cells in the cloning experiments. The temperature-sensitive sec23-1 strain RSY282 (MATa leu2Δ ura3Δ sec23-1) was kindly provided by Randy Schekman (Department of Molecular and Cell Biology and Howard Hughes Medical Institute, University of California at Berkeley). The yeast strain BY4741 (MATa his3Δ1 leu2Δ met15Δ ura3Δ) and the derived deletion strains SED1 (sed1Δ), SSO1 (sso1Δ), SSO2 (sso2Δ), SEC22 (sec22Δ), SNC2 (snc2Δ), TLG2 (tlg2Δ), BTN2 (btn2Δ), PEP12 (pep12Δ), VPS51 (vps51Δ), GOS1 (gos1Δ), ATG1 (atg1Δ), ATG8 (atg8Δ), ATG11 (atg11Δ), ATG17 (atg17Δ), ATG20 (atg20Δ), VAM3 (vam3Δ), and GRH1 (grh1Δ) were purchased from Euroscarf (Frankfurt, Germany). The yeast GFP clones (Invitrogen, Carlsbad, CA) with GFP-tagged endogenous proteins (Pma1p, Nup84p, Mae1p, Chs5p, Snf7p, Vrg4p, Pex11p, and Sec13p) and HIS3 marker in the parent BY4741 strain were used to determine the localization of proteins. E. coli was grown in lysogeny broth (LB; 1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] sodium chloride, and 100 μg/ml ampicillin). The yeast cells were grown in yeast extract-peptone-dextrose (YPD) medium (1% [wt/vol] yeast extract, 2% [wt/vol] polypeptone, and 2% [wt/vol] glucose), SD+HM medium (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] glucose, 0.002% l-histidine-HCl, and 0.003% l-methionine), SDC+HM medium (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% (wt/vol) glucose, 0.002% l-histidine-HCl,0.003% l-methionine, 2% Casamino Acids (BD, Franklin Lakes, NJ), and 2% (wt/vol) agar), SC+ML medium (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] glucose, 0.003% l-methionine,0.003% l-leucine, 0.13% SD multiple drop out [-Ade, -His, -Leu, -Lys, -Trp, and -Ura; Funakoshi Co., Ltd., Tokyo, Japan], 2% [wt/vol] agar), SD+ML medium (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] glucose, 0.003% l-methionine,0.003% l-leucine), or SDC+ML medium (SD+ML supplemented with 2% Casamino Acids).
Construction of S. cerevisiae expression plasmids.
The S. cerevisiae expression plasmids were constructed using a conventional PCR-based method and our novel PCR-free method (one-step construction method for plasmids [OSCoM-P]) (see Fig. S1 in the supplemental material). In addition, iProof DNA polymerase (Bio-Rad, Richmond, CA), KOD-plus DNA polymerase (Toyobo, Osaka, Japan), KOD-plus-Neo DNA polymerase (Toyobo), Ligation High (Toyobo), and synthetic oligonucleotides (Japan Bio Services, Saitama, Japan) were used. All primers used in this study are shown in Table S2 of the supplemental material. The plasmids for the internal production of the recombinant proteins were constructed from pULSG1 (32). The primers coding for the ATG codon were mixed with the pULSG1 digest and inserted using the EcoRI and XhoI sites by OSCoM-P (see Fig. S1); the resulting plasmid was named pUL-ATG-EGFP. The section of pUL-ATG-EGFP, including the GAPDH promoter, the terminator, the FLAG tag (DYKDDDDK [21]), and the EGFP sequence were amplified and added to the BamHI and NotI sites by PCR using the primers GAPDH promoter-F and GAPDH terminator-R and then inserted into the BamHI-NotI section of pRS423 (ATCC 77104) (47); the resulting plasmid was named pRS423-ATG-EGFP. For constructing the plasmid pULGI2, OSCoM-P was also performed. Oligonucleotide fragments with several restriction sites were inserted into pULSG1 by using the EcoRI and XhoI sites. The plasmids for the internal expression of the glycolytic enzymes conjugated with EGFP-FLAG were constructed as follows. The yeast genomic DNA was extracted and purified from the S. cerevisiae BY4741 strain, and each gene coding for a glycolytic enzyme was cloned using the appropriate primer set (see Table S2 in the supplemental material). The fragments were digested and inserted into pULGI2 by using the BamHI and XhoI sites or the BamHI and SacI sites. The internal expression vector without EGFP was constructed from pULSG1C (32) and pWGP3 (50). The multicloning site followed by the GAPDH terminator sequence was amplified from pWGP3 and inserted into pULSG1C by using the SacI and KpnI sites; the resulting plasmid was named pULI1. For the construction of the plasmid for the intercellular production of enolase-EGFP-FLAG with the N-terminal peptide sequence (hemagglutinin [HA] tag), the HA tag sequence was inserted into pULGI2 by using OSCoM; the resultant plasmid was named pULGI2-HA. The ENO2 coding sequence from pULGI2-ENO2 was inserted into pULGI2-HA, and the resultant plasmid was named pULGI2-HA-ENO2. For the construction of plasmids to produce red fluorescent proteins, the Discosoma sp. red fluorescent protein (DsRed) monomer with EcoRI and XhoI sites at the N terminus and SalI at the C terminus was cloned from pKRD4 (26). The amplified fragments were digested with EcoRI and SalI and then inserted into the same site of pULSG1; the resultant plasmid was named pUL-ATG-DsRed. For production of the enolase fragments fused with EGFP or DsRed, amplified fragments were digested and inserted into pULSG1 or pUL-ATG-DsRed by using the EcoRI and XhoI sites (see Table S2). Plasmids for production of Tlg2p (p413-ADH-TLG2) were constructed as follows: TLG2 coding sequence was cloned from the genomic DNA extracted from S. cerevisiae BY4741 and inserted into multiple cloning site of p413-ADH (ATCC 87370) by using EcoRI and XhoI sites. The plasmid construction was confirmed by DNA sequencing performed using a BigDye Terminator v3.1 cycle sequencing kit and an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA).
Production of recombinant proteins in yeast cells.
The yeast cells were transformed with plasmids by using a Frozen-EZ yeast transformation II kit (Zymo Research, Orange, CA) and grown on SDC+HM agar plates. The transformants were selected as single colonies and inoculated into 10 ml of SD+HM medium for precultivation at 25°C with shaking. At the late log phase, the preculture was subcultured into 10 ml (for secretion analysis of the sec23-1 strains) or 25 ml of the same medium to obtain an optical density at 600 nm (OD600) of 0.01 (for secretion analysis of the glycolytic enzymes) or 0.3. The cells were cultivated at 25°C for 26 h (for secretion analysis of the glycolytic enzymes) or 4 h with shaking until they reached an OD600 of 0.9 to 1.1 or 0.5. Genotypes of knockout strains were checked by colony PCR. For secretion analysis of plasmid-cotransformed strains, plasmids for internal overexpression of EGFP-FLAG-conjugated proteins (pUL-X) were cotransformed with p413-plasmids into BY4741wt and ΔTLG2 strains. Transformants were grown on SC+ML agar medium. Single colonies were picked up and recultivated on the same medium and then used for secretion analysis. Transformants were precultivated in 10 ml of SDC+ML mediun at 25°C for 26 h with shaking. Cells were washed with fresh SD+ML medium and inoculated into 25 ml of SD+ML medium. The cells were cultivated at 25°C for 4 h with shaking until they reached an OD600 of 0.5.
Identification of noncovalently bound cell surface proteins of living cells.
The budding yeast strain BY4741 sed1Δ transformed with the plasmids pRS423-ATG-EGFP and pKRD4 (26) was grown in SDC+HM medium. The noncovalently bound cell surface proteins extracted from living yeast cells by using 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate were separated by a 2D-HPLC system optimized for protein separation (34). The fractionated proteins were lyophilized using a vacuum centrifuge (Labconco, MO) and solubilized in 50 mM ammonium hydrogen carbonate. The collected proteins were reduced with 50 mM dithiothreitol for 30 min at 60°C and alkylated with 500 mM iodoacetamide for 45 min at room temperature. The alkylated proteins were digested by trypsin (sequencing-grade, modified trypsin; Promega Corp., Madison, WI) for 12 h at 37°C for protein identification by mass spectrometry using a Prominence nanoflow system (Shimadzu, Kyoto, Japan) and an LTQ Velos linear ion trap mass spectrometer (Thermo Scientific Inc., Bremen, Germany). The proteolytic digests were separated by reversed-phase chromatography using a packed tip column (NTCC-360; 150 mm by 100 μm inner diameter; Nikyo Technos, Tokyo, Japan) at a flow rate of 500 nl/min. The gradient was provided by changing the mixing ratio of the 2 eluents (A, with 0.1% [vol/vol] formic acid and B, acetonitrile containing 0.1% [vol/vol] formic acid). The gradient was started with 5% B, increased to 45% B for 60 min, further increased to 95% B to wash the column, and then returned to the initial condition and held for reequilibration. For data-dependent acquisition of mass spectrometry detection, the method was set to automatically analyze the top 3 most intense ions observed in the mass spectrometry scan. An electrospray ionization voltage of 1.9 kV was directly applied to the flow using a microtee. The ion transfer tube temperature on the LTQ Velos ion trap was set to 300°C. The experiments were independently repeated twice. Protein identification was performed using the combined tandem mass spectrometry data and the Protein Discoverer software (Thermo Scientific). The results were compared to the information in the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org) and filtered at a q value of ≤0.05, corresponding to a 5% false discovery rate (FDR) on a spectral level, and the identified proteins contained >2 peptide fragments.
Preparation of extracellular and intracellular proteins.
The detection of extracellular and intracellular proteins was performed according to the previously described method (33) with minor modifications. The culture medium (25 ml) was centrifuged at 900 × g for 10 min at 4°C to remove cells. Following this, the culture medium was filtered through a 0.2-μm Acrodisc syringe filter (PALL Corporation, MI) and concentrated by ultrafiltration [M-10 filter for pULI1, pULSG1C, pUL-ATG-EGFP, pUL-eno(1–17), pUL-eno(1–28), and pUL-eno(1–30), and a YM-30 filter for the others; Amicon; Millipore, Millford, MA]. After washing thrice with 4 volumes of 20 mM Tris-HCl (pH 7.8), the concentrated proteins were frozen and lyophilized. The proteins were then suspended with 15 μl (for secretion analysis of the glycolytic enzymes) or 30 μl of loading buffer and analyzed by SDS-PAGE. The cells were suspended with 500 μl of 20 mM Tris-HCl (pH 7.8) containing 0.1% SDS. After homogenization at 4,000 rpm for 20 s using glass beads (GB-05; diameter 0.5 mm; TOMY, Tokyo, Japan) and Bead Smash 12 (Wakenyaku, Kyoto, Japan), the sample solutions were centrifuged at 9,700 × g for 5 min. Aliquots (5 μl) of the supernatants were suspended with 5 μl of 2× loading buffer and analyzed by SDS-PAGE.
Inhibition of conventional secretion by the sec23-1 strain.
After preculture in SD+HM medium, the cells were washed with fresh medium and inoculated into a fresh 10 ml of the same medium to an OD600 of 0.3 and incubated at 25 or 37°C with shaking. After 4 h, the culture medium was filtered and concentrated. Following measurement of the protein concentration, the solution was lyophilized and suspended in 10 μl of loading buffer.
SDS-PAGE.
SDS-PAGE was conducted according to the previously described method (27) by using a continuous polyacrylamide gel (5% to 20%; 120 by 100 mm; e-PAGEL; Atto, Tokyo, Japan). The samples were heated in the loading buffer at 100°C for 3 min, centrifuged at 21,900 × g at 4°C for 5 min to remove the debris, and loaded. As an external standard, the FLAG protein (48-kDa cleavage control protein; Novagen, Inc., WI) was used.
Western blotting.
After transfer to a nitrocellulose membrane (0.45 μm pore size) by using trans-blot transfer medium (Bio-Rad), Western blot analysis was performed using an anti-FLAG M2 antibody conjugated with horseradish peroxidase (HRP; Sigma). The loading control, Pgi1p, was detected using the rabbit anti-baker's yeast Pgi1p antibody (Acris Antibodies GmbH, Hiddenhausen, Germany) and an anti-rabbit antibody conjugated to HRP (GE Healthcare, UK, Ltd., Buckinghamshire, United Kingdom). The detection was enhanced by using the Can Get signal immunoreaction enhancer solution (Toyobo). After detection, the antibodies were removed using a stripping agent (WB stripping solution; Nacalai), and the membranes were blocked and reprobed using an anti-FLAG M2 antibody conjugated to HRP (Sigma). The chemiluminescence was detected using ECL Plus Western blotting detection reagents (GE Healthcare). The membranes treated with the detection reagent were exposed to Amersham Hyperfilm ECL (GE Healthcare) and developed using Rendol and Renfix (Fujifilm, Kanagawa, Japan) to detect the secreted glycolytic enzymes and enolase fragments. Other data were obtained by using the ImageQuant LAS 4000 minisystem (GE Healthcare). Gained signals from extracellular Pgi1p and Eno2p conjugated with EGFP-FLAG tag using anti-Pgi1p and anti-FLAG were processed by setting signals obtained from 0.4 ng/lane of FLAG protein as 1. Relative amounts of Eno2p-EGFP-FLAG tag were calculated as follows: (signal intensities after anti-FLAG treatment)/(signal intensities after anti-Pgi1p treatment). One-tailed t tests were performed to detect significant differences.
Fluorescence microscopy.
For confocal microscopy, the cells were grown to the mid-log phase and fixed with phosphate-buffered saline (pH 7.4) containing 3.7% paraformaldehyde. The cells were then fixed to the bottom of a 35-mm glass-base dish (Synapse fine view dish SF-G-D27; FPI Inc., Kyoto, Japan) by using the same buffer. The fluorescence images were obtained at room temperature with a 60× objective (oil immersion; numerical aperture [NA], 1.35) by using a laser-scanning confocal microscope (FluoView FV1000; Olympus) and FV10-ASW software (Olympus). The efficiency of colocalization was analyzed using ImageJ software (http://rsb.info.nih.gov/ij/). To observe the cells producing recombinant proteins and to perform time course observations, an epifluorescence IX71 microscope (Olympus) with a 100× objective (oil immersion; NA, 1.40) and Aquacosmos software (Hamamatsu Photonics, Hamamatsu, Japan) were used.
Time course observations of living cells on an agarose pad.
To observe the time-dependent localization changes of the foci, an agarose pad was prepared using a slightly modified method (51). Briefly, 2% agarose was added to the SD+HM medium, heated, and dissolved. The solution at 60°C was spotted onto a glass slide with vinyl tape wrapped on each side. Immediately following this, the coverglass was overlaid and left at room temperature. Before observation, the yeast culture in mid-log phase at an OD600 of 0.4 was spotted onto the agarose pad and covered with the coverglass. Time-lapse observations were conducted by fitting a small incubator (MI-IBC-IF; Olympus) onto the microscopy system and by manually photographing every 5 min for 30 min. The incubator was prewarmed to 30°C, and the excitation light source was turned on only during image recording.
RESULTS
Detection of unconventional secretion of glycolytic enzymes.
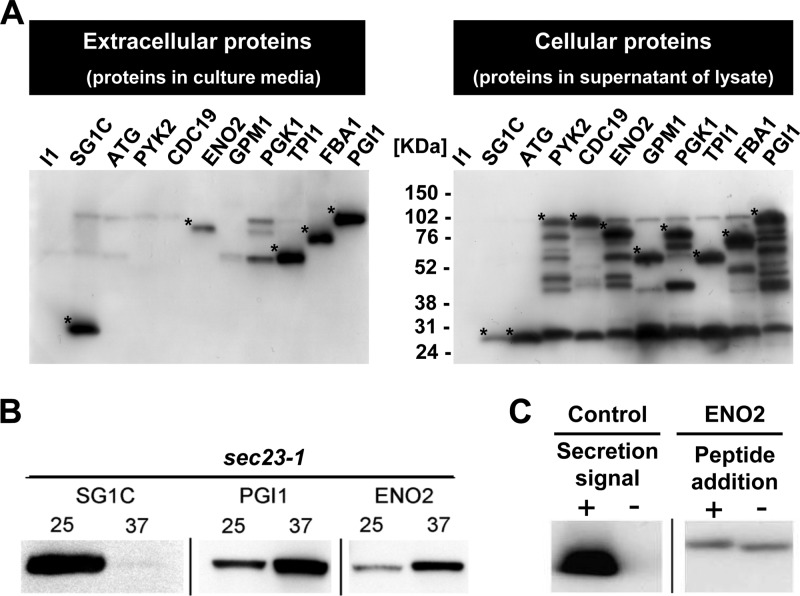
We performed 2D-HPLC-based cell surface proteome analysis to detect the extracellular presence of 11 glycolytic enzymes, namely, enolases (Eno1p and Eno2p), glyceraldehyde-3-phosphate dehydrogenases (Tdh1p, Tdh2p, and Tdh3p), 3-phosphoglycerate kinase (Pgk1p), fructose 1,6-bisphosphate aldolase (Fba1p), phosphoglucose isomerase (Pgi1p), triose phosphate isomerase (Tpi1p), phosphoglycerate mutase (Gpm1p), and pyruvate kinase (Cdc19p) (Table 1; see also Table S1 in the supplemental material). To shortlist the candidate proteins for analysis of the secretory pathway, the detected glycolytic enzymes were produced as recombinant proteins fused to EGFP-FLAG. Although all the glycolytic enzymes were successfully produced in the cell, only 4 (Eno2p, Pgi1p, Tpi1p, and Fba1p) were reproducibly detected in the culture medium (Fig. 1A, left). Among these, Eno2p and Pgi1p were both thought to be important molecules when secreted and thus were used for further investigation. To examine whether Eno2p and Pgi1p were secreted via the conventional pathway, a sec23-1 temperature-sensitive mutant strain was used. The secretion of EGFP fused with the conventional glucoamylase secretion signal sequence was successfully inhibited at 37°C. Comparatively, under the same conditions and using the same strain, both Eno2p and Pgi1p were detected in the culture medium (Fig. 1B). To examine whether a cleavable peptide sequence existed at the N terminus of Eno2p, which is typical for conventionally secreted proteins, an extra N-terminal peptide sequence (in this case, HA tag) was added to Eno2p. In the wild-type cells, Eno2p with the extra N-terminal peptide sequence was detected in the culture medium, as was Eno2p without the extra sequence (Fig. 1C). These results suggest that glycolytic enzymes, at least Eno2p, can be secreted via an unconventional pathway in S. cerevisiae. Among the glycolytic enzymes detected in the culture medium, Pgi1p-EGFP-FLAG gave the clearest bands. Moreover, the secretion of endogenous Pgi1p was detected in a previous study (40). Therefore, we used endogenous Pgi1p as a control in the following experiments.
Table 1.
Identified noncovalently bound cell surface proteins
| Function group | Cellular process | No. of identified proteins |
|---|---|---|
| Metabolism | Glycolysis | 11 |
| Amino acid biosynthesis | 6 | |
| TCAa cycle | 1 | |
| Pentose phosphate pathway | 1 | |
| Alcoholic fermentation | 1 | |
| Fatty acid metabolism | 1 | |
| Protein binding | 6 | |
| Homeostasis | 5 | |
| Translation | 4 | |
| Signaling | 1 | |
| Folding | 1 | |
| Traffic | 1 | |
| Unknown | 3 | |
| Total | 42 |
TCA, tricarboxylic acid.
Fig 1.
Detection of unconventional secretion of glycolytic enzymes. Anti-FLAG antibody was used for detection. (A) Secretion of glycolytic enzymes. (Left) Secreted proteins; (right) cellular proteins. I1, pULI1; SG1C, pULSG1C; ATG, pUL-ATG-EGFP; PYK2, pULGI2-PYK2; CDC19, pULGI2-CDC19; ENO2, pULGI2-ENO2; GPM1, pULGI2-GPM1; PGK1, pULGI2-PGK1; TPI1, pULGI2-TPI1; FBA1, pULGI2-FBA1; PGI1, pULGI2-PGI1. (B) The effect of inhibition of the conventional pathway on the secretion of the glycolytic enzymes. Secretion of recombinant proteins in sec23-1 strains at 25°C or 37°C is shown. (C) The effect of the N-terminal peptide (HA tag) on the secretion of enolase. Control (secretion signal +, secretion of EGFP-FLAG protein with conventional secretion signal sequence), pULSG1C; control (secretion signal -, secretion of EGFP-FLAG protein without secretion signal sequence), pUL-ATG-EGFP; ENO2 (peptide addition +, secretion of N-terminal HA peptide-tagged Eno2p-EGFP-FLAG), pULGI2-ATG-HA-ENO2; ENO2 (peptide addition -, secretion of Eno2p-EGFP-FLAG without peptide addition), pULGI2-ENO2. Similar results were obtained from 3 independent experiments. *, glycolytic enzymes conjugated to EGFP and FLAG. Additional bands are either nonspecific binding of antibody or degradation products of target proteins.
Search for the enolase sequence responsible for secretion.
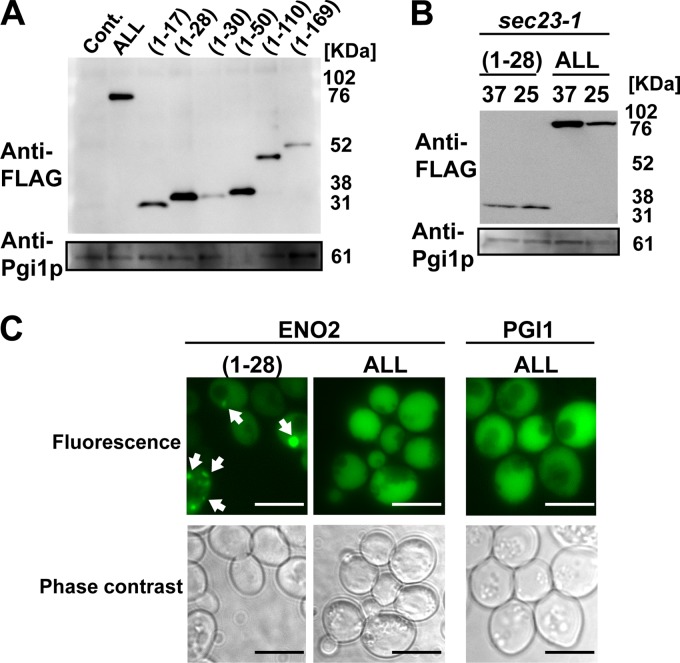
N-terminal fragments of Eno2p containing 169 amino acids (fragment 1–169) were prepared as described in a previous report (29). In addition, N-terminal fragments of Eno2p containing 17, 28, 30, 50, and 110 amino acids (fragments 1–17, 1–28, 1–30, 1–50, and 1–110) were prepared. Subsequently, the (1–28), (1–50), and (1–169) fragments were reproducibly detected in the culture medium (Fig. 2A). In addition, secretion of the (1–28) Eno2p fragment [eno(1–28)] conjugated to EGFP and FLAG in the sec23-1 strain was not inhibited at 37°C (Fig. 2B). These results suggest that eno(1–28), as well as Eno2p, is secreted via an unconventional pathway.
Fig 2.
Detection and monitoring of the secretion of the N-terminal fragment of enolase conjugated to EGFP and FLAG. (A) Detection of secreted enolase fragments. Cont., pUL-ATG-EGFP; ALL, pULGI2-ENO2; (1–17), pUL-eno(1–17); (1–28), pUL-eno(1–28); (1–30), pUL-eno(1–30); (1–50), pUL-eno(1–50); (1–110), pUL-eno(1–110); (1–169), pUL-eno(1–169). (B) SEC23-independent secretion of the enolase fragment. Secretion levels of recombinant proteins in sec23-1 strains under 25°C or 37°C are shown. (1–28), pUL-eno(1–28); ALL, pULGI2-ENO2; 37, cultivated at 37°C; 25, cultivated at 25°C. (C) Fluorescence microscopy of cells transformed with pUL-eno(1–28), pULGI2-ENO2, and pULGI2-PGI1. Bar, 10 μm. Similar results were obtained from 3 independent experiments.
Focus formation and intracellular translocation of eno(1–28).
To monitor the secretion of the eno(1–28) fragment, cells producing eno(1–28) conjugated with EGFP and FLAG were observed by fluorescence microscopy (Fig. 2C). The green fluorescence from EGFP was detected as a dot, suggesting that eno(1–28) formed foci. In addition, some of the foci changed location when observed at 30°C on the agarose pad (see Fig. S2 in the supplemental material).
Colocalization of eno(1–28) with organelle marker proteins.
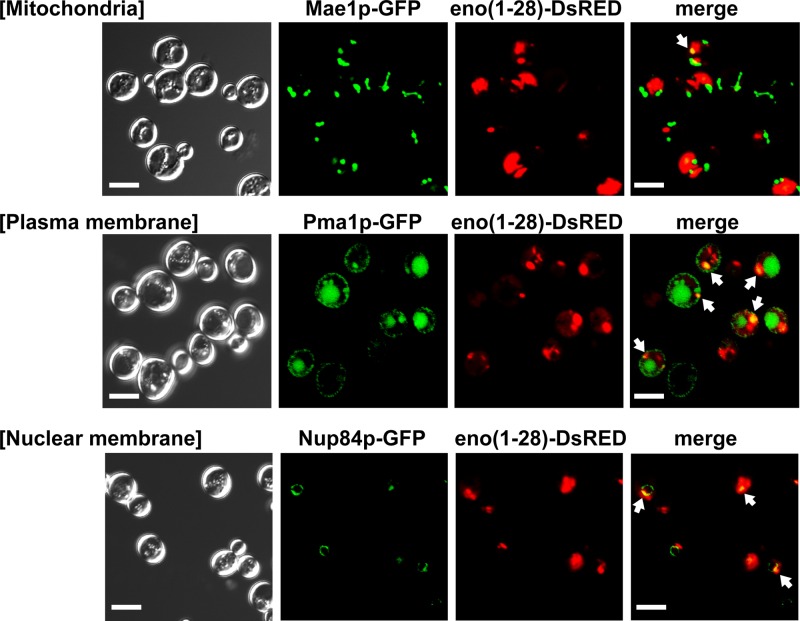
To examine the localization of the foci formed by eno(1–28) in the cells, eno(1–28) conjugated with the DsRed monomer was produced in the GFP clones carrying organelle marker protein-coding genes (see Table S3 in the supplemental material) (PMA1 [plasma membrane], NUP84 [nuclear membrane], MAE1 [mitochondria], CHS5 [exomer], SNF7 [endosome vesicle and multivesicular body, or MVB], VRG4 [cis-Golgi complex], PEX11 [peroxisome], and SEC13 [endoplasmic reticulum-to-Golgi complex transport vesicle]), fused with the GFP-coding sequence at the 3′ end. Our results showed that eno(1–28)-DsRed colocalized with the plasma membrane, nuclear membrane, exomer, endosome/MVB vesicle, Golgi complex, and peroxisome, but not with the mitochondria (Fig. 3 and Table 2; see also Fig. S3 in the supplemental material).
Fig 3.
Colocalization of enolase fragments conjugated to DsRed- and GFP-tagged organelle markers. The fixed cells were observed at room temperature. The numbers of cells forming foci are shown in Table 2. White arrow, foci that colocalized with organelle markers. Bar, 5 μm.
Table 2.
Localization of enolase fragments conjugated to DsRed- and GFP-tagged organelle markersa
| Organelle marker | No. of cells forming foci with marker, based on fluorescence with: |
No. of cells with colocalized foci | (No. of cells with colocalized foci)/(no. of cells forming DsRed foci) (%) | |
|---|---|---|---|---|
| DsRed | GFP | |||
| Nup84p | 28 | 72 | 19 | 68 |
| Pma1p | 21 | 224 | 14 | 67 |
| Snf7p | 28 | 77 | 15 | 54 |
| Chs5p | 20 | 80 | 9 | 45 |
| Sec13p | 20 | 159 | 8 | 40 |
| Pex11p | 48 | 40 | 16 | 33 |
| Vrg4p | 41 | 198 | 15 | 27 |
| Mae1p | 72 | 27 | 1 | 3.7 |
These findings are illustrated in Fig. 3 (see also Fig. S3 in the supplemental material).
Inhibition of enolase secretion by knockout strains.
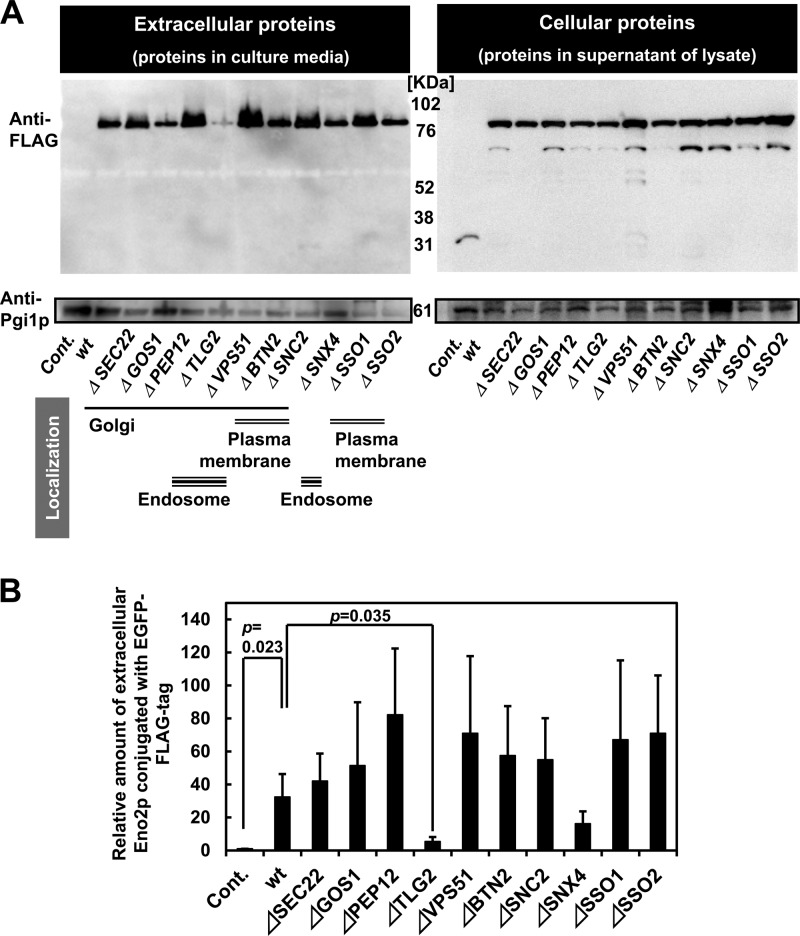
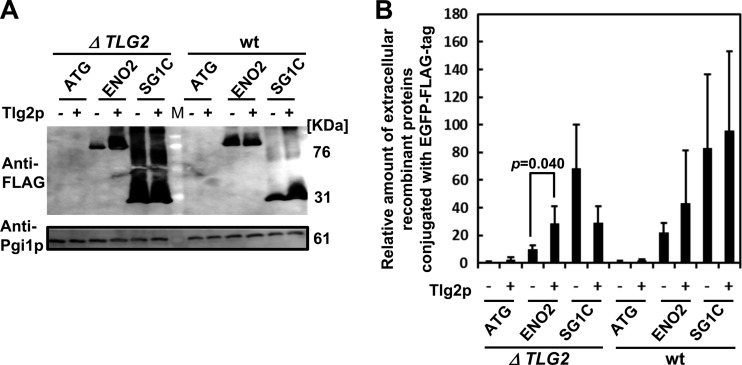
Knockout strains of SNAREs (see Table S3 in the supplemental material [SEC22, GOS1, PEP12, TLG2, VPS51, BTN2, SNC2, SNX4, SSO1, and SSO2]), which play a major role in intracellular protein transportation, were utilized to examine their effects on the secretion of Eno2p conjugated with EGFP and endogenous Pgi1p (Fig. 4A). Pgi1p was detected in the culture medium of all strains, while the levels of Eno2p-EGFP-FLAG were lower in the culture medium of the TLG2 knockout strain. Inhibition of the translocation of Eno2p-EGFP-FLAG to the cell surface was also tested using immunostaining (Fig. 4B). The ATG1, ATG8, ATG11, ATG17, ATG20, VAM3, and GRH1 knockout strains (see Table S3) as well as wild-type BY4741 were further tested to investigate the role of autophagy-related genes in secretion (see Fig. S4 in the supplemental material). The secretion of Eno2p was not inhibited in the knockout strains of the autophagy-related genes, demonstrating that of all the SNARE and autophagy-related genes analyzed, only the TLG2 knockout strain inhibited the secretion of Eno2p (Fig. 4A). TLG2 dependency of Eno2p-EGFP-FLAG secretion was further confirmed by complementation of the mutation with a wild-type plasmid (Fig. 5). Therefore, we concluded that Eno2p is secreted by an unknown TLG2-dependent pathway.
Fig 4.
Intracellular SNARE dependence of Eno2p and Pgi1p secretion. (A) Secretion in strains transformed with Eno2-expressing plasmids. Cont., wild-type BY4741 cells transformed with pUL-ATG-EGFP; wt, wild-type BY4741 cells transformed with pULGI2-ENO2; Δ, knockout strains transformed with pULGI2-ENO2. (B) Calculated amounts of secreted Eno2p compared to the levels of Pgi1p secreted. Values are means ± standard errors of the means of ≥3 independent experiments.
Fig 5.
TLG2 dependency of Eno2p secretion. (A) Secretion in strains transformed with Eno2-expressing plasmids. (B) Calculated amounts of secreted proteins, compared to the levels of Pgi1p secreted. ATG, cells transformed with pUL-ATG-EGFP; ENO2, cells transformed with pULGI2-ENO2; SG1C, cells transformed with pULSG1C; ΔTLG2, TLG2 knockout strains; wt, wild-type BY4741 strains; M, marker; Tlg2p -, cells transformed with p413-ADH (control vector); Tlg2p +, cells transformed with p413-ADH-TLG2 (plasmid for producing Tlg2p). Values are means ± standard errors of the means of ≥3 independent experiments.
DISCUSSION
The 11 glycolytic enzymes detected in our cell surface proteome analysis, including the 4 detected in the secretion analysis, have all been previously reported to be secreted or present in the cell wall in S. cerevisiae (37, 40). These results demonstrate both the different detection capacities of proteome analysis and Western blotting and the efficacy of our detection method (see Table S1 in the supplemental material). The lower number of glycolytic enzymes detected in the secretion analysis compared to those detected by proteome analysis suggests that the secretion of some of the proteins was undetectable because the tendency to secrete was too low. However, it is also possible that the conjugated extra amino acid sequence inhibited the secretion of the glycolytic enzymes. We utilized Eno2p to analyze the secretory pathway, because enolase secretion is thought to be important for many diseases and the secretory pathway has not yet been investigated. We utilized Pgi1p as a control because it was detected in large amounts in the culture medium. Conjugation of an extra peptide sequence to the N terminus of Eno2p slightly increased its molecular weight (Fig. 1C), suggesting that Eno2p does not possess a conventional secretion signal sequence that is cleaved during secretion. In addition, Eno2p and Pgi1p were secreted in the sec23-1 mutant at 37°C. These results provide persuasive evidence that the secretion of glycolytic enzymes, at least Eno2p and Pgi1p, is not dependent on the conventional secretory pathway.
Although full-length Eno2p conjugated with EGFP had a broad subcellular localization, the Eno2p fragment formed foci in the cell, and some of the foci changed location from the center of the cell to the cell periphery (Fig. 2C; see also Fig. S2 in the supplemental material). Therefore, we assumed that the short amino acid sequence of enolase that can be secreted from the cell exemplifies the secretory pathway of enolase. In a previous report, Lopez-Villar and colleagues demonstrated that eno(1–46) and (1–101) do not exist in the cell wall, whereas the eno(1–169) fragment, conjugated with glucoamylase, does (29). Recently, Yang et al. identified the hydrophobic domain required for enolase secretion (56). The domain includes the 96- to 132-amino acid (aa) region of S. cerevisiae enolase that is a conserved membrane-embedded (EM) domain (56), and the domain is not identical to the (1–28) region of enolase. Although the EM domain is a bacterial domain, eukaryotic S. cerevisiae may have similar mechanisms for secretion of enolase. Because the eno(1–28) region has similarity with the 96- to 132-aa region and the EM domain of yeast enolase to some extent, respectively (see Fig. S5 in the supplemental material), the secretion of eno(1–28) may depend on the same secretion mechanism as that of the 96- to 132-aa region. There may be a sequence in the N-terminal (29–96) region that inhibits secretion, because in contrast to eno(1–28), eno(1–30) was hardly detected in the culture medium. It will be important to complete further investigations and to determine the precise signal sequence required for enolase secretion.
The eno(1–28) fragment conjugated with EGFP and FLAG formed foci in the cell and changed location over time (see Fig. S2 in the supplemental material). The fragment localized to various cellular membranes, but not to the mitochondria (Fig. 3 and Table 2; see also Table S3 in the supplemental material). It has been previously reported that glycolytic enzymes, including enolase, associate with the post-Golgi complex vesicles (15), and that yeast enolase takes part in a macromolecular complex associated with the mitochondria (5) and assists in the transport of tRNA (14). However, the localization of enolase to the mitochondria seems to be regulated by a different region of enolase.
It is reasonable that not all DsRed colocalized with a particular marker, considering that enolase colocalized with several markers (Fig. 3 and Table 2; see also Fig. S3). The difference in the number of DsRed- versus GFP-tagged cells reflects the difference in the way each fluorescent protein is incorporated into cells; the GFP-encoding gene is integrated in the genome of yeast cells after each open reading frame encoding the organelle marker protein, while DsRed is produced by plasmids. The numbers of GFP-conjugated organelle marker proteins in the cell is dependent on endogenous promoters for each organelle marker protein. Although GFP should be produced in all the cells, some organelle marker proteins are weakly translated and, therefore, in some cases the fluorescence is undetectable. The number of eno(1–28)-DsRed-tagged molecules seems to be dependent on the transfection efficiency of plasmids, and the plasmid uses the strong GAPDH promoter.
We assumed that the major elements that participate in intracellular trafficking also play a role in the secretion of Eno2p. SNAREs govern the translocation of proteins, and although many SNARE-coding genes are lethal when deleted, some nonlethal deletion mutants are available. We used the SNAREs from the S. cerevisiae genome database (http://www.yeastgenome.org/) that participate in the translocation of proteins between the Golgi complex, endosome, and plasma membrane (see Table S3 in the supplemental material). Analyses of the deletion mutants revealed that knocking out TLG2 inhibited enolase secretion. However, we propose other proteins may be involved, because the inhibition was not complete. In contrast to the previously reported unconventional secretion of the Acb1 protein (12, 31), secretion levels of Eno2p and Pgi1p were not inhibited in the GRH1, SSO1, and BTN2 knockout strains (Fig. 4). Therefore, it is probable that regulation of the secretion of these glycolytic enzymes differs from that of the Acb1 protein. There was also no incorporation of the Golgi complex reassembly and stacking protein (GRASP) Vps51p; this was surprising, as GRASPs have been reported to participate in several unconventional secretory pathways (17, 25, 31, 44). Therefore, the secretion of glycolytic enzymes in S. cerevisiae seems to be independent of the GRASP-regulated pathway. Moreover, Gos1p, which has a role in the cytoplasm-to-vacuole (Cvt) pathway (4), had no influence on the secretion of Eno2p. In addition to its involvement in the Cvt pathway, Tlg2p is a syntaxin-like t-SNARE that participates in vesicle fusion, endocytosis, Golgi complex-to-vacuole transport, endosomal protein sorting, and protein release from the endoplasmic reticulum (1, 10, 18, 20, 35, 41). Mousley and colleagues previously showed that Tlg2p has a role in protein secretion in combination with Sec14p (35), and our results suggest that the participation of Tlg2p in the secretion of Eno2p is plausible. Therefore, we conclude that the secretory pathway of glycolytic enzymes is regulated in a different manner from the Cvt pathway. S. cerevisiae is reported to have autophagosome-mediated membrane compartments for the unconventional secretion of proteins (6), and it is, therefore, possible that the secretion of Eno2p entails an autophagy-related pathway. However, our results using the knockout strains of autophagy-related proteins (see Fig. S4) suggest the absence of active participation of autophagy-related genes in Eno2p secretion.
In conclusion, by using the above methods, we detected secreted glycolytic enzymes and traced the translocation changes of the fragments of Eno2p. Our results contribute to identification of the unknown machinery, which is dependent on TLG2, involved in the translocation of glycolytic enzymes inside the cell.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Randy Schekman at the University of California, Berkeley, for providing the RSY282 (sec23-1) strain.
This work was supported by a grant-in-aid for JSPS fellows.
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Abeliovich H, Darsow T, Emr SD. 1999. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 18: 6005–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avilan L, et al. 2011. Enolase: a key player in the metabolism and a probable virulence factor of trypanosomatid parasites: perspectives for its use as a therapeutic target. Enzyme Res. 2011: 932549.doi:10.4061/2011/932549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bechtel JT, Winant RC, Ganem D. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79: 4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bensen ES, Yeung BG, Payne GS. 2001. Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol. Biol. Cell 12: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brandina I, et al. 2006. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim. Biophys. Acta 1757: 1217–1228 [DOI] [PubMed] [Google Scholar]
- 6. Bruns C, McCaffery JM, Curwin AJ, Duran JM, Malhotra V. 2011. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J. Cell Biol. 195: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capello M, Ferri-Borgogno S, Cappello P, Novelli F. 2011. α-Enolase: a promising therapeutic and diagnostic tumor target. FEBS J. 278: 1064–1074 [DOI] [PubMed] [Google Scholar]
- 8. Chertova E, et al. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80: 9039–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiellini C, et al. 2008. Characterization of human mesenchymal stem cell secretome at early steps of adipocyte and osteoblast differentiation. BMC Mol. Biol. 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coe JG, Lim AC, Xu J, Hong W. 1999. A role for Tlg1p in the transport of proteins within the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell 10: 2407–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dobashi Y, et al. 2006. Autocrine motility factor/glucose-6-phosphate isomerase is a possible predictor of metastasis in bone and soft tissue tumours. J. Pathol. 208: 44–53 [DOI] [PubMed] [Google Scholar]
- 12. Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edwards SR, Braley R, Chaffin WL. 1999. Enolase is present in the cell wall of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 177: 211–216 [DOI] [PubMed] [Google Scholar]
- 14. Entelis N, et al. 2006. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 20: 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forsmark A, et al. 2011. Quantitative proteomics of yeast post-Golgi vesicles reveals a discriminating role for Sro7p in protein secretion. Traffic 12: 740–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghosh AK, Jacobs-Lorena M. 2011. Surface-expressed enolases of Plasmodium and other pathogens. Mem. Inst. Oswaldo Cruz 106(Suppl. 1): 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giuliani F, Grieve A, Rabouille C. 2011. Unconventional secretion: a stress on GRASP. Curr. Opin. Cell Biol. 23: 498–504 [DOI] [PubMed] [Google Scholar]
- 18. Gurunathan S, Marash M, Weinberger A, Gerst JE. 2002. t-SNARE phosphorylation regulates endocytosis in yeast. Mol. Biol. Cell 13: 1594–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirschberg K, Lippincott-Schwartz J. 1999. Secretory pathway kinetics and in vivo analysis of protein traffic from the Golgi complex to the cell surface. FASEB J. 13(Suppl. 2): S251–S256 [DOI] [PubMed] [Google Scholar]
- 20. Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. 1998. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 17: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hopp TP, et al. 1988. A short polypeptide marker sequence useful for recombinant protein identification and purification. Nat. Biotechnol. 6: 1204–1210 [Google Scholar]
- 22. Huang D, Shusta EV. 2005. Secretion and surface display of green fluorescent protein using the yeast Saccharomyces cerevisiae. Biotechnol. Prog. 21: 349–357 [DOI] [PubMed] [Google Scholar]
- 23. Innis MA, et al. 1985. Expression, glycosylation, and secretion of an Aspergillus glucoamylase by Saccharomyces cerevisiae. Science 228: 21–26 [DOI] [PubMed] [Google Scholar]
- 24. Jong AY, et al. 2003. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J. Med. Microbiol. 52: 615–622 [DOI] [PubMed] [Google Scholar]
- 25. Kinseth MA, et al. 2007. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell 130: 524–534 [DOI] [PubMed] [Google Scholar]
- 26. Kuroda K, et al. 2009. Enhancement of display efficiency in yeast display system by vector engineering and gene disruption. Appl. Microbiol. Biotechnol. 82: 713–719 [DOI] [PubMed] [Google Scholar]
- 27. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 28. Lamonica JM, et al. 2005. Comparative secretome analyses of three Bacillus anthracis strains with variant plasmid contents. Infect. Immun. 73: 3646–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez-Villar E, et al. 2006. Genetic and proteomic evidences support the localization of yeast enolase in the cell surface. Proteomics 6(Suppl. 1): S107–S118 [DOI] [PubMed] [Google Scholar]
- 30. Makhina T, et al. 2009. Extracellular GAPDH binds to L1 and enhances neurite outgrowth. Mol. Cell Neurosci. 41: 206–218 [DOI] [PubMed] [Google Scholar]
- 31. Manjithaya R, Anjard C, Loomis WF, Subramani S. 2010. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 188: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matsui K, Kuroda K, Ueda M. 2009. Creation of a novel peptide endowing yeasts with acid tolerance using yeast cell-surface engineering. Appl. Microbiol. Biotechnol. 82: 105–113 [DOI] [PubMed] [Google Scholar]
- 33. Miura N, Aoki W, Tokumoto N, Kuroda K, Ueda M. 2009. Cell-surface modification of non-GMO without chemical treatment by novel GMO-coupled and -separated cocultivation method. Appl. Microbiol. Biotechnol. 82: 293–301 [DOI] [PubMed] [Google Scholar]
- 34. Morisaka H, Kirino A, Kobayashi K, Ueda M. 2012. Two-dimensional protein separation by the HPLC system with a monolithic column. Biosci. Biotechnol. Biochem. 76: 585–588 [DOI] [PubMed] [Google Scholar]
- 35. Mousley CJ, et al. 2008. trans-Golgi network and endosome dynamics connect ceramide homeostasis with regulation of the unfolded protein response and TOR signaling in yeast. Mol. Biol. Cell 19: 4785–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nickel W, Rabouille C. 2009. Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10: 148–155 [DOI] [PubMed] [Google Scholar]
- 37. Nombela C, Gil C, Chaffin WL. 2006. Non-conventional protein secretion in yeast. Trends Microbiol. 14: 15–21 [DOI] [PubMed] [Google Scholar]
- 38. Ogino T, Yamadera T, Nonaka T, Imajoh-Ohmi S, Mizumoto K. 2001. Enolase, a cellular glycolytic enzyme, is required for efficient transcription of Sendai virus genome. Biochem. Biophys. Res. Commun. 285: 447–455 [DOI] [PubMed] [Google Scholar]
- 39. Oliveira DL, et al. 2010. Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infect. Immun. 78: 1601–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oliveira DL, et al. 2010. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One 6: e11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paumet F, et al. 2001. A t-SNARE of the endocytic pathway must be activated for fusion. J. Cell Biol. 155: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Renigunta A, et al. 2011. The glycolytic enzymes glyceraldehyde 3-phosphate dehydrogenase and enolase interact with the renal epithelial K channel ROMK2 and regulate its function. Cell. Physiol. Biochem. 28: 663–672 [DOI] [PubMed] [Google Scholar]
- 43. Schekman R. 2010. Charting the secretory pathway in a simple eukaryote. Mol. Biol. Cell 21: 3781–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schotman H, Karhinen L, Rabouille C. 2008. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev. Cell 14: 171–182 [DOI] [PubMed] [Google Scholar]
- 45. Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. 2008. Cellular proteins in influenza virus particles. PLoS Pathog. 6: e1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shinya R, Morisaka H, Takeuchi Y, Ueda M, Futai K. 2010. Comparison of the surface coat proteins of the pine wood nematode appeared during host pine infection and in vitro culture by a proteomic approach. Phytopathology 100: 1289–1297 [DOI] [PubMed] [Google Scholar]
- 47. Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sriram G, Martinez JA, McCabe ER, Liao JC, Dipple KM. 2005. Single-gene disorders: what role could moonlighting enzymes play? Am. J. Hum. Genet. 76: 911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Swenerton RK, et al. 2011. The oligopeptidase B of Leishmania regulates parasite enolase and immune evasion. J. Biol. Chem. 286: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takahashi S, Ueda M, Tanaka A. 2001. Function of the prosequence for in vivo folding and secretion of active Rhizopus oryzae lipase in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 55: 454–462 [DOI] [PubMed] [Google Scholar]
- 51. Tanaka K, Kitamura E, Tanaka TU. 2010. Live-cell analysis of kinetochore-microtubule interaction in budding yeast. Methods 51: 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Torimura T, et al. 2001. Autocrine motility factor enhances hepatoma cell invasion across the basement membrane through activation of beta1 integrins. Hepatology 34: 62–71 [DOI] [PubMed] [Google Scholar]
- 53. Tristan C, Shahani N, Sedlak TW, Sawa A. 2011. The diverse functions of GAPDH: views from different subcellular compartments. Cell. Signal. 23: 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsutsumi S, et al. 2003. The enzymatic activity of phosphoglucose isomerase is not required for its cytokine function. FEBS Lett. 534: 49–53 [DOI] [PubMed] [Google Scholar]
- 55. van Deventer HJ, Goessens WH, van Vliet AJ, Verbrugh HA. 1996. Anti-enolase antibodies partially protective against systemic candidiasis in mice. Clin. Microbiol. Infect. 2: 36–43 [DOI] [PubMed] [Google Scholar]
- 56. Yang CK, et al. 2011. Nonclassical protein secretion by Bacillus subtilis in the stationary phase is not due to cell lysis. J. Bacteriol. 193: 5607–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.