Abstract
The consequences of deprivation of the molecular chaperone Hsp104 in the fungal pathogen Candida albicans were investigated. Mutants lacking HSP104 became hypersusceptible to lethally high temperatures, similarly to the corresponding mutants of Saccharomyces cerevisiae, whereas normal susceptibility was restored upon reintroduction of the gene. By use of a strain whose only copy of HSP104 is an ectopic gene under the control of a tetracycline-regulated promoter, expression of Hsp104 prior to the administration of heat shock could be demonstrated to be sufficient to confer protection from the subsequent temperature increase. This result points to a key role for Hsp104 in orchestrating the cell response to elevated temperatures. Despite their not showing evident growth or morphological defects, biofilm formation by cells lacking HSP104 proved to be defective in two established in vitro models that use polystyrene and polyurethane as the substrates. Biofilms formed by the wild-type and HSP104-reconstituted strains showed patterns of intertwined hyphae in the extracellular matrix. In contrast, biofilm formed by the hsp104Δ/hsp104Δ mutant showed structural defects and appeared patchy and loose. Decreased virulence of the hsp104Δ/hsp104Δ mutant was observed in the Caenorhabditis elegans infection model, in which high in vivo temperature does not play a role. In agreement with the view that stress responses in fungal pathogens may have evolved to provide niche-specific adaptation to environmental conditions, these results provide an indication of a temperature-independent role for Hsp104 in support of Candida albicans virulence, in addition to its key role in governing thermotolerance.
INTRODUCTION
Candida albicans is a frequent component of the microbial community in healthy humans which can, under certain conditions, turn into a pathogen causing cutaneous infections, vulvovaginitis, esophagitis, and oral thrush. People undergoing wide-spectrum antibiotic therapy, chemotherapy, and organ transplantation, as well as neutropenic and diabetic patients, are at risk of more severe problems of invasive candidiasis (9, 21). These are frequently initiated by entry of C. albicans into the bloodstream, a condition known as candidemia, followed by the dissemination of fungal cells in target organs, causing acute or chronic states. In order to thrive in the human host body, C. albicans must, especially as a pathogen, adapt to several microenvironments and challenges, from different nutrient availability to different pH to the host immune defense. Genome-wide studies of C. albicans gene expression in infected tissues have contributed to form the general idea that adaptation to stress contributes to the virulence potential of this fungus (4). On the other hand, several observations about pathways involved in the stress response in C. albicans suggest that such responses, while evolutionarily related to those of nonpathogenic yeasts, may have specialized to provide niche-specific adaptation (20).
The presence of an ortholog of Hsp104 in C. albicans, a species that has been uniquely found in nature in association with warm-blooded animals and which therefore is not expected to be exposed to dramatic temperature changes during its life cycle, raises the question whether additional features of this protein that contribute to the general fitness of the fungus or to its virulence may exist.
Hsp104 is a yeast molecular chaperone that was discovered in Saccharomyces cerevisiae as a heat shock-induced protein required for thermotolerance (29) and later further described as also required for resistance to several different types of stress (30). Hsp104 appears to be capable of resolving protein aggregates, allowing protein refolding (15), in cooperation with Hsp70, Hsp40, and trehalose (34). Interestingly, while orthologs of Hsp104 have been identified in other fungi (33, 40) and in bacteria and plants (31), and while a functional homolog (known as Hsp78) has also been found in yeast mitochondria (32), no homologs of Hsp104 have been found in animal cells. Expression of yeast Hsp104, however, proved to be able to counteract protein aggregation in a transgenic mouse model of Huntington's disease (38).
Both overexpression and absence of Hsp104 have been described to lead to defects in the propagation of [PSI+], the prion form of the yeast translational terminator factor Sup35 (6–8, 37). In addition, Hsp104 also plays an important role in the propagation of several other known yeast prions, such as [URE3] and [PIN+]. Substitution of S. cerevisiae Hsp104 for its C. albicans ortholog led to the conclusion that the C. albicans protein can propagate the [PSI+] prion in yeast (40), suggesting that the machinery for prion propagation also exists in C. albicans. Although a recent report claimed that the Ure2 protein of C. albicans can be a prion in S. cerevisiae (10), the existence of prions in C. albicans is still an open question that will have to wait for the development of appropriate molecular tools.
In this study, we investigated the consequences of deprivation of Hsp104 for C. albicans physiology and its virulence potential. While the absence of Hsp104 does not affect cell growth under normal conditions, it results in decreased resistance to exposure to high temperatures. The decreased proficiency of hsp104Δ/hsp104Δ mutants in biofilm formation in vitro in two different model systems and their attenuated virulence in a room temperature worm infection model suggest that Hsp104 may be a subtle regulator of cell fitness independently from its role in maintaining thermotolerance.
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Table 1. They were routinely refreshed from frozen stocks at −80°C and maintained on YPD plates (1% yeast extract, 2% peptone, 2% dextrose). Cells were grown in minimal medium (1.7 g/liter Difco yeast nitrogen base without ammonium sulfate, 5 g/liter ammonium sulfate) supplemented with 2% glucose (SD). For experiments using acetate medium, glucose was replaced by 1% potassium acetate. Ten percent serum, Spider, and sucrose media were prepared as described in reference 39. Media were solidified with 2% agar.
Table 1.
C. albicans strains used in this study
| Strain | Genotype | Comment | Source or reference |
|---|---|---|---|
| SC5314 | Wild type | 14a | |
| AFA46b | SC5314 hsp104Δ∷FRT/HSP104 | Heterozygous mutant | This study |
| AFA48b | SC5314 hsp104Δ∷FRT/hsp104Δ∷FRT | Homozygous mutant | This study |
| AFA54a | AFA48B hsp104Δ∷FRT/hsp104Δ∷HSP104 | Reconstituted strain | This study |
| AFA58a | AFA48B adh1Δ∷Ptet-HSP104/ADH1 | Strain with doxycycline-inducible HSP104 as the only HSP104 allele | This study |
Plasmid constructions.
A plasmid containing the HSP104 disruption cassette was constructed as follows. The 3′ untranslated region of HSP104 (position +2676 to +3178) was amplified from genomic DNA of strain SC5314 using oligonucleotides HSP104-3 and HSP104-4 (oligonucleotides used in this study are listed in Table 2) and cloned as a SacI-SacII fragment into pSFS2 (26). The 5′ untranslated region of HSP104 (position −422 to +82), amplified using oligonucleotides HSP104-1 and HSP104-2, was subsequently cloned in the generated plasmid as an ApaI-XhoI fragment, generating pAFC78a.
Table 2.
Primers used in this study
| Name | Sequence |
|---|---|
| HSP104-1 | TATTAATCCGGGCCCTGGACAAGTAGTTG |
| HSP104-2 | CCCGCTCGAGACAATTGCGAATTGGCCTGTT |
| HSP104-3 | ATTCACTTCACCGCGGCTTGACTAAACTAA |
| HSP104-4 | TATTTCAAAATAGAGCTCCAGAATATTTCTAC |
| HSP104-5 | CCCGCTCGAGTTAGTTTAGTCAAGTCCAGGTG |
| CaHSP104-s | ACGCGTCGACATGGAAGATTTTACAGATAACGC |
| CaHSP104-as | GGAAGATCTTTAGTCAAGTCCAGGTGAAGTG |
| HSP104-upstream | GTTCTACCGCATACAAGTGAC |
| HSP104-downstream2 | GGGCAATTTAACGTTTGTTTGA |
| SAT1flp-check1 | GTGTAATGGTACAGATGGTACTAG |
| SAT1flp-check2 | GGTTCTCGGGAGCACAGGATGA |
| tetO7_check | GGAGCTCGACTTTCACTTTTCT |
| CaADH1_check | CAACTGGTGTCCAATACGTATC |
For construction of the HSP104 reintegration plasmid, a DNA fragment spanning from nucleotide −422 to nucleotide +2720 of HSP104 was PCR amplified from genomic DNA of SC5314 using AccuPrime Taq high-fidelity DNA polymerase (Invitrogen) with primers HSP104-1 and HSP104-5. This fragment was cloned as an ApaI-XhoI fragment in the HSP104 3′-containing pSFS2 plasmid described above, generating pAFC82. The HSP104 open reading frame contained in this plasmid was sequence verified (it contains the silent mutation C2211T).
For construction of the tetracycline-regulated HSP104-containing plasmid, the HSP104 open reading frame was PCR amplified from genomic DNA of SC5314 with primer pair CaHSP104-s and CaHSP104-as using AccuPrime Taq DNA polymerase and cloned as a SalI-BglII fragment into plasmid pNIM1 (22), generating pAFC80a. The cloned insert was sequence verified.
Strain constructions.
DNA fragments containing the HSP104 disruption cassette were recovered as ApaI-SacI digests of plasmid pAFC78a and transformed into C. albicans SC5314 using the lithium acetate transformation procedure to generate the hsp104Δ∷FRT/HSP104 heterozygous strain AFA46b. After 4 h of outgrowth in YPD medium, transformation mixes were plated onto YPD plates containing 200 mg/liter nourseothricin (Werner BioAgents, Jena, Germany). Transformants were identified as resistant colonies after 1 to 2 days of incubation at 30°C and restreaked on YPD plates with nourseothricin for confirmation of resistance. Insertion of the disruption cassette at the HSP104 genomic locus was verified via PCR on both sides of the HSP104 gene, using oligonucleotides annealing on the promoter and terminator of HSP104 external to the disruption cassette coupled to primers internal to the SAT1 marker. Primer pairs were HSP104-upstream with SAT1flp-check1 and HSP104-downstream2 with SAT1flp-check2, respectively. hsp104Δ∷FRT∷SAT1∷FRT/HSP104 heterozygous strains were cultivated overnight in liquid YPD to obtain FLP-mediated excision of the SAT1 marker, and strains in which excision had occurred were initially identified from their weak growth on plates containing 25 mg/liter nourseothricin. Correct pop-out of the SAT1 marker was verified via PCR using oligonucleotides HSP104-upstream and HSP104-downstream2, which produced DNA fragments of 3.7 kb and 1.2 kb for the wild-type gene and for hsp104Δ∷FRT, respectively. Cells of the heterozygous strain were retransformed with the HSP104 disruption cassette, and strains were again PCR analyzed for gene disruption as described above. A PCR with the primer pair HSP104-upstream and HSP104-downstream2 was also performed on these transformants to discriminate between those in which the disruption cassette had integrated at the locus of the previously deleted gene from those in which integration had replaced the remaining wild-type allele. Only strains for which this PCR still produced a 1.2-kb DNA fragment were considered for further SAT1 excision and characterization. This procedure produced the hsp104Δ∷FRT/hsp104Δ∷FRT homozygous strain AFA48b.
The hsp104Δ∷FRT/hsp104Δ∷HSP104-reconstituted strain AFA54a was generated by transforming AFA48b with the transformation cassette obtained by restriction of plasmid pAFC82 with ApaI and SacI. Transformants were selected on the basis of their resistance to nourseothricin and allowed to pop out the SAT1 marker as described above. Reintegration of HSP104 in its original locus was verified via PCR using primer pair HSP104-upstream and HSP104-downstream2, which revealed the presence of the 1.2-kb band relative to disrupted hsp104 together with an ∼4.0-kb band corresponding to reinserted HSP104. Reinsertion was confirmed by sequencing.
Strains containing the tetracycline-inducible HSP104 allele (Ptet-HSP104) were constructed by transformation of C. albicans with an ApaI-SacII DNA fragment extracted from plasmid pAFC80a and selection on YPD containing 200 mg/liter nourseothricin. After restreak purification, transformants were checked for integration of the Ptet-HSP104 cassette into the ADH1 locus via PCR, using primer pair tetO7_check and CaADH1_check. Transformation of AFA48b with the Ptet-Hsp104 cassette generated strain AFA58a.
Western blotting.
Liquid yeast cultures grown overnight in minimal medium were diluted to an optical density at 600 nm (OD600) of 0.2 in prewarmed medium and outgrown for 5 h at 30°C with shaking. After this period, cultures were split in two: one half was immediately processed for total protein extraction (see below), whereas the other was incubated for 30 min at 41°C, with shaking, and subsequently extracted. For experiments with stationary-phase cells, a similar procedure, without the outgrowth period, was applied to cultures grown to an OD600 of ∼23. Total proteins were extracted by vortexing C. albicans cells with 0.5-mm glass beads in cold HEPES/K-acetate buffer (20 mM HEPES-KOH [pH 6.8], 50 mM potassium acetate, 200 mM sorbitol, 1 mM EDTA) (28), in the presence of Complete EDTA-free protease inhibitors (Roche). Protein concentrations were determined by Bradford analysis, and 20 μg of proteins per sample was separated on NuPAGE 4 to 12% bis-Tris/morpholinepropanesulfonic acid (MOPS) gels (Invitrogen). Proteins were subsequently blotted onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore). Blots were blocked with 5% nonfat milk in Tris-buffered saline (TBS) with 0.1% Tween and probed with an anti-Hsp104 antibody (a kind gift of M. Tuite) diluted 1:2,000, followed by incubation with a secondary horseradish peroxidase (HRP)-conjugated anti-rabbit antibody. Blots were also probed with a commercial anti-histone H3 antibody (Abcam) diluted 1:3,000 for loading controls. Blots were quantitated with the ImageJ 1.43u software (http://rsbweb.nih.gov/ij/).
Thermotolerance experiments.
For thermotolerance experiments with exponential-phase cells, cells were grown to mid-log phase at 30°C in SD medium, diluted in prewarmed SD medium at an OD600 of 0.2, and outgrown until cultures reached an OD600 of 0.7 (∼4 to 6 h). Aliquots of cultures were withdrawn, adjusted to an OD600 of 0.7 in sterile 2-ml screw-cap tubes, and shifted to 50°C, with shaking, for the times indicated in the figures. Aliquots of cells withdrawn at the indicated times, and at the beginning of the heat treatment as a control, were serially diluted 1:10 in sterile 96-well plates and spot tested on YPD plates using a spotter. For preadaptation experiments, cells were pretreated at 41°C, with shaking, for 30 or 150 min prior to the heat treatment.
For experiments with stationary-phase cells, cells were grown at 30°C in SD medium to an OD600 of ∼23, adjusted to the OD600 of the culture with the lowest OD value, and subjected to heat treatment for the indicated times. Cell dilutions and spot tests were as described for exponential-phase cells.
For thermotolerance experiments with strains carrying Ptet-HSP104, experiments were conducted as for those with exponential-phase cells except for the fact that 50 mg/liter doxycycline was added to all cultures when cells were diluted at an OD600 of 0.2 and outgrown.
In vitro biofilm assays.
Adhesions and biofilms were allowed to form on flat-bottom 96-well polystyrene plates as described by Ramage et al. (25), and on polyurethane catheters as described previously (27). Briefly, C. albicans strains were grown overnight on YPD plates at 37°C, washed, and resuspended in phosphate-buffered saline (PBS). Polyurethane, 1-cm-long triple-lumen intravenous catheters (2.4-mm diameter) (Arrow International, Inc.) were incubated overnight in fetal bovine serum (Sigma) at 37°C. The final concentration of the cells used for biofilm formation in 96-well plate was 1 × 106 cells/ml, whereas the initial inoculum applied for biofilm production on polyurethane catheters was 5 × 104 cells/ml. Both cell suspensions were prepared by counting cells suspended in RPMI 1640 medium (with glutamine and phenol red) (Sigma) buffered with 3-(N-morpholino)propanesulfonic acid. The pH of the medium was adjusted to 7.0 with 1 M NaOH. Cells were allowed to adhere during a 90-min period of adhesion at 37°C. Afterwards, nonattached Candida cells were removed by two washing steps with PBS. Adhesion was quantified immediately for 90 min, whereas mature biofilms were left to develop for 48 h at 37°C. Prior to the biofilm quantification, each well or catheter was washed twice with PBS.
Adhered cells and biofilms preformed on 96-well plate were evaluated for their metabolic activity by the reduction of XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] (25). Adhesion and biofilm formation on polyurethane catheters were quantified by the amount of CFU, as described previously (27).
During biofilm assays performed on 96-well polystyrene plates, each strain was tested in quadruplicate, and the experiment was reproduced four times. Candida biofilm formation on polyurethane catheters was repeated three times independently; 8 catheters per strain were analyzed in total. Statistical analysis was carried out using Student's t test. Values were considered to be statistically significant when the P value was <0.05.
SEM.
Mature (48-h) C. albicans biofilms used for scanning electron microscopy (SEM) were developed on highly adhesive round tissue culture coverslips (diameter, 13 mm; Sarstedt, Germany). One milliliter of RPMI 1640 culture (106 cells) was added to wells containing coverslips (one per well) and incubated at 37°C for 90 min. The coverslips were then gently washed twice with PBS, placed in a clean 24-well tissue culture plate, and covered with fresh medium for additional 48 h. Mature biofilms were further washed twice with PBS before visualization or quantification by using XTT. Air-dried mounted samples were sputter coated with gold and viewed in an XL30 ESEM FEG scanning electron microscope (Philips). Experiments were repeated three times independently, each time using two coverslips per strain.
Worm survival assays.
In vivo experiments using the Caenorhabditis elegans/C. albicans model system were based on the procedure established by Breger et al. (2) and were performed as previously described (35), with minor modifications. Briefly, larvae of glp-4Δ/sek-1Δ mutants of C. elegans were grown to the L4 stage on NGM agar plates containing a surface lawn of overnight-grown OP50 Escherichia coli. Worms were collected, washed with M9 buffer, and incubated for 2 h on YPD agar plates containing freshly grown surface lawns of strains SC5314, AFA46b, AFA48b, and AFA54a of C. albicans. Forty to 50 worms were then suspended in 0.25 ml M9 buffer in separate wells of 24-well plates, and their survival was monitored daily. Worm survival was expressed as a percentage of their viability at day zero. The data shown represent the mean and standard error of at least quintuple measurements. Results were analyzed for statistical significance by Student's t test. Values were considered to be statistically significant when the P value was <0.05.
RESULTS
Hsp104 is induced in response to elevated temperature and stationary phase.
Zenthon et al. reported previously on induction of Hsp104 in S. cerevisiae and C. albicans in response to heat treatment at 37°C (40). As a human commensal, C. albicans grows at the optimal temperature of 37°C. Therefore, we sought to verify whether Hsp104 of C. albicans was also induced in response to exposure to a detrimental, yet nonlethal, high temperature. Hsp104 was expressed at very low levels in exponential-phase cells of the wild-type strain SC5314 grown at 30°C but increased considerably upon treatment of cells at 41°C for 30 min (Fig. 1A), confirming previous observations on expression of the corresponding gene (11, 18). Contrary to the situation in exponential-phase cells, Hsp104 was expressed in stationary-phase cells at levels comparable to those in heat-treated exponential-phase cells, and subsequent heat treatment did not lead to a further increase (Fig. 1B), suggesting that increased expression of Hsp104 in response to elevated temperatures and in stationary phase may be regulated by the same mechanism(s).
Fig 1.
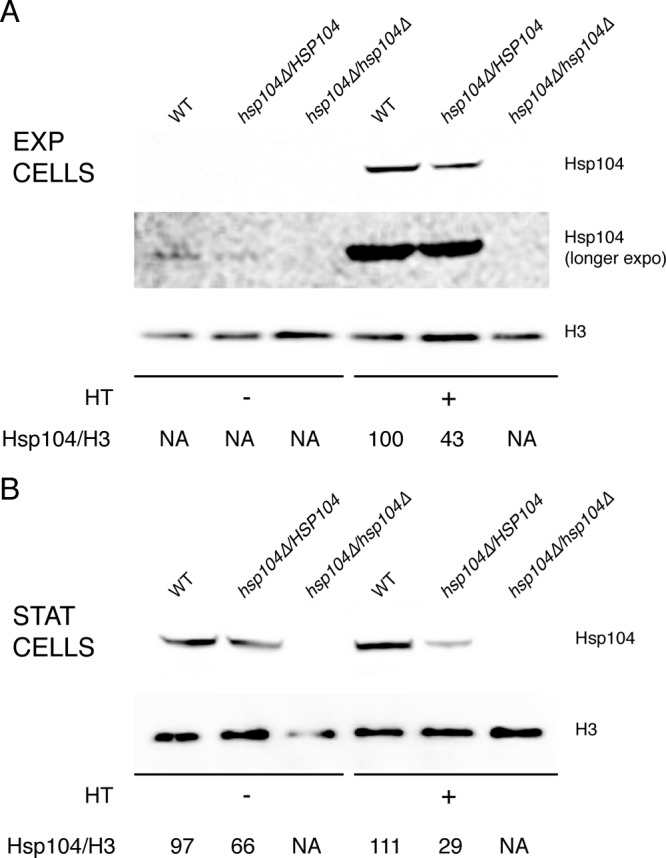
Hsp104 is induced in response to elevated temperature and stationary phase. Levels of Hsp104 were analyzed by Western blotting in wild-type (WT) SC5314 and in hetero- and homozygous hsp104Δ cells prior to and after heat treatment (HT) at 41°C for 30 min. Blots were also probed with an antibody against histone H3 for loading controls. (A) Exponential-phase cells. (B) Stationary-phase cells. See Materials and Methods for experimental details.
Hsp104 is necessary for thermotolerance.
The two HSP104 alleles of Candida albicans were disrupted in the reference strain SC5314 using the SAT1 flipper method (26), generating prototrophic strains with hetero- and homozygous disruptions. Disruption of HSP104 was verified by PCR and confirmed by Western blotting using a polyclonal antibody against Hsp104 of C. albicans. As expected, the band related to Hsp104 was not present in Western blots from the homozygous mutant (Fig. 1); in addition, the intensity of the Hsp104 signal in the heterozygous mutant was reduced with respect to that of the wild type, suggesting the absence of compensating regulatory mechanisms. Cells devoid of Hsp104 showed no other major alterations: the growth rates of the mutants at 37 or 30°C in minimal and rich media were comparable to those of the wild-type strain, and germ tube formation/development and colony morphology in Spider or 10% serum medium were found to be unaltered, as was growth under microaerophilic/embedding conditions in Spider and sucrose media (our unpublished observations).
Given the important role of Hsp104 in thermotolerance in Saccharomyces cerevisiae and the fact that the C. albicans ortholog is induced by elevated temperature, the possible role of Hsp104 in thermotolerance of C. albicans was investigated. Exponential-phase cells of the wild-type and mutant strains were subjected to the lethal temperature of 50°C for increasing amounts of time in minimal glucose medium. After treatment, cell viability was monitored by spot tests on YPD plates. As illustrated in Fig. 2A, exponentially growing cells of the C. albicans hsp104Δ/hsp104Δ homozygous mutant were very sensitive to heat, since the viability of the culture was greatly reduced after a brief (10-min) heat treatment that did not affect wild-type cells. Interestingly, cells of the heterozygous hsp104Δ/HSP104 strain showed an intermediate phenotype (Fig. 2A), a result correlating with the intermediate Hsp104 expression levels of this strain reported in Fig. 1. Experiments performed with cells grown in RPMI or minimal acetate medium produced similar results (not shown).
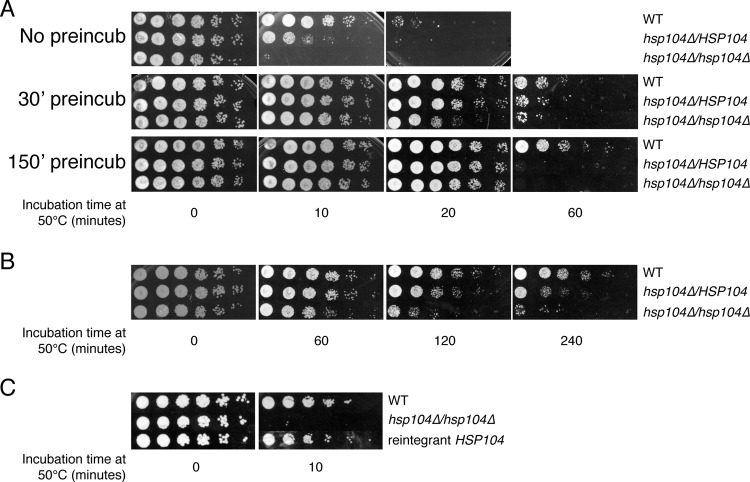
Fig 2.
Hsp104 is necessary for thermotolerance. Ten-fold serial dilutions of wild-type and hetero- and homozygous hsp104Δ cells were spotted on YPD plates after exposure at the lethal temperature of 50°C for the indicated time periods. (A) Exponential-phase cells. Results obtained upon preconditioning cells at 41°C for 30 or 150 min prior to the heat shock are also shown. (A) Similar experiments conducted with stationary-phase cells. (C) Reintroduction of HSP104 restores normal susceptibility to heat stress.
Preadaptation of exponentially grown cells to the elevated, nonlethal temperature of 41°C for short (30-min) or long (150-min) periods increased the general resistance of cells to a subsequent lethal heat stress (Fig. 2A), indicating that cells had become more thermotolerant due to the pretreatment. Notably, this increased thermotolerance correlated with induction of Hsp104 by elevated temperature as shown in Fig. 1. Nevertheless, hetero- and homozygous hsp104Δ mutants were still hypersusceptible to the lethal temperature, clearly demonstrating that Hsp104 is required for thermotolerance of C. albicans (Fig. 2A). Stationary-phase cells were found to be considerably more resistant to the thermal insult (Fig. 2B), and this resistance was diminished in the absence of Hsp104. Together with elevated expression of Hsp104 in stationary-phase cells compared with exponential-phase cells (Fig. 1B), these results indicate that Hsp104 plays a major role in regulating resistance to thermal stress in stationary-phase cells (see also below). Reintegration of the HSP104 gene, in its original locus, in the homozygous mutant restored resistance of these cells to thermal stress (Fig. 2C), confirming that the observed phenotype of hsp104Δ mutants can be attributed solely to the absence of Hsp104, ruling out the possibility of secondary cryptic genetic alterations. In conclusion, HSP104 appears to be a haploinsufficient gene, whose protein product is required for thermotolerance in C. albicans, similarly to its role in baker's yeast.
Hsp104 is sufficient for thermotolerance.
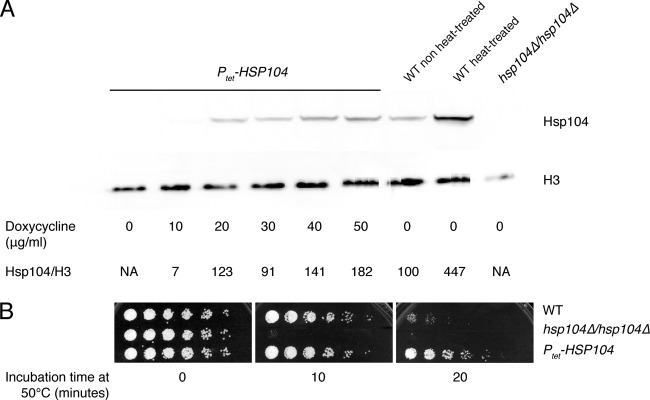
As described above, short pretreatment of cells at elevated temperature increased Hsp104 expression levels and provided a protective effect, suggesting a role in thermotolerance for this protein. To clarify whether protection can be provided solely by increased expression of Hsp104 under these conditions or whether it is the result of the coordinated action of a multiple number of independent factors, experiments were performed using a strain whose only HSP104 gene is under the control of a tetracycline-inducible promoter (Ptet-HSP104). Ptet-HSP104 was integrated at the ADH1 locus of the hsp104Δ/hsp104Δ homozygous mutant, resulting in a strain in which expression of HSP104 is repressed under normal conditions and can be induced by the addition of doxycycline to the medium. As expected, Hsp104 was undetectable in extracts from these cells grown in the absence of doxycycline (Fig. 3A). Addition of 10 μg/ml doxycycline to the medium led to very low, almost undetectable, levels of Hsp104, whereas higher levels of the protein were detected using larger amounts of doxycycline (Fig. 3A); in the presence of concentrations of doxycycline as high as 50 μg/ml, the amount of Hsp104 in cell extracts was approximately double that expressed by wild-type cells under normal conditions of growth at 30°C, although still less than that in heat-treated cells.
Fig 3.
Hsp104 is sufficient for thermotolerance. (A) Doxycycline-regulated expression of HSP104 in strain AFA58a, carrying ectopic Ptet-HSP104, as analyzed by Western blotting. Results from the wild-type strain SC5314, before and after heat treatment, and the null mutant are shown as controls. (B) Expression of HSP104 was induced with 50 μg/ml doxycycline prior to administration of the heat shock. Ten-fold serial dilutions of wild-type, homozygous hsp104Δ, and Ptet-HSP104 cells were spotted on YPD plates after exposure at the lethal temperature of 50°C for the indicated time periods. Cells of the wild-type and homozygous hsp104Δ strains were also treated with doxycycline as a control.
Turning on expression of HSP104 prior to administration of the heat shock resulted in cells acquiring a greater thermotolerance with respect to cells of the wild-type strain (Fig. 3B). These results demonstrate that the induction of moderately high levels of Hsp104 prior to the heat shock is sufficient to provide protection to C. albicans, suggesting that Hsp104 alone is capable of orchestrating the cells' response to elevated temperatures.
Hsp104 is required for efficient biofilm formation.
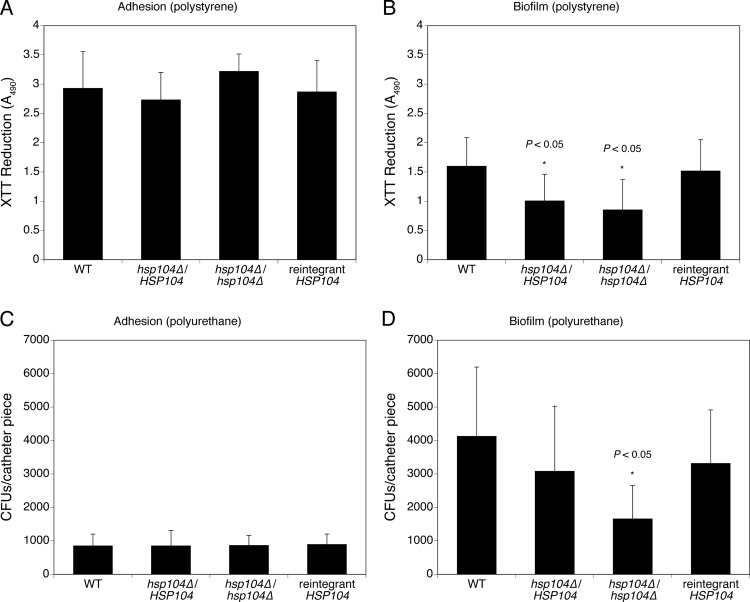
The ability of C. albicans to form biofilms is an important medical aspect of this fungus. In this study, we investigated the capability of the hsp104Δ hetero- and homozygous mutants to adhere and form mature biofilms, using two established protocols (25, 27). In the 96-well polystyrene plate method, adhesion and biofilms were assessed by the XTT reduction assay measured at 490 nm, whereas in the polyurethane catheter model, the total count of cells recovered from catheters was used. Figure 4A and B show the average XTT-measured metabolic activities of adhering and biofilm-forming cells, respectively. As can be seen, differences in adhesion on polystyrene between the wild type, the hsp104Δ mutants, and the reintegrant strain are only minimal and statistically not significant (P > 0.05) (Fig. 4A). In contrast, measurement of the metabolic activity of mature biofilms allowed to form on the same material revealed a statistically significant (P < 0.05) decrease in both hsp104Δ hetero- and homozygous mutants compared to the wild type (Fig. 4B), indicating that Hsp104 may influence the formation of biofilm structures.
Fig 4.
Genetic deprivation of Hsp104 results in reduced in vitro biofilm formation. C. albicans adhesion and mature biofilm formation were assessed in 96-well polystyrene plates (A and B) and polyurethane catheter fragments (C and D) in RPMI 1640 at 37°C. Standard deviations were calculated from four independent experiments performed in polystyrene and three experiments performed on polyurethane.
Similar results were obtained using the polyurethane catheter model (Fig. 4C and D). Again, no difference was recorded among strains in regard to their ability to adhere on these substrates (Fig. 4C), whereas a marked decrease in viable cell counts was observed for the homozygous hsp104Δ mutant relative to the wild-type strain (P < 0.05) (Fig. 4D). These results are consistent with the ones described above and confirmed a role for Hsp104 in the process of biofilm maturation.
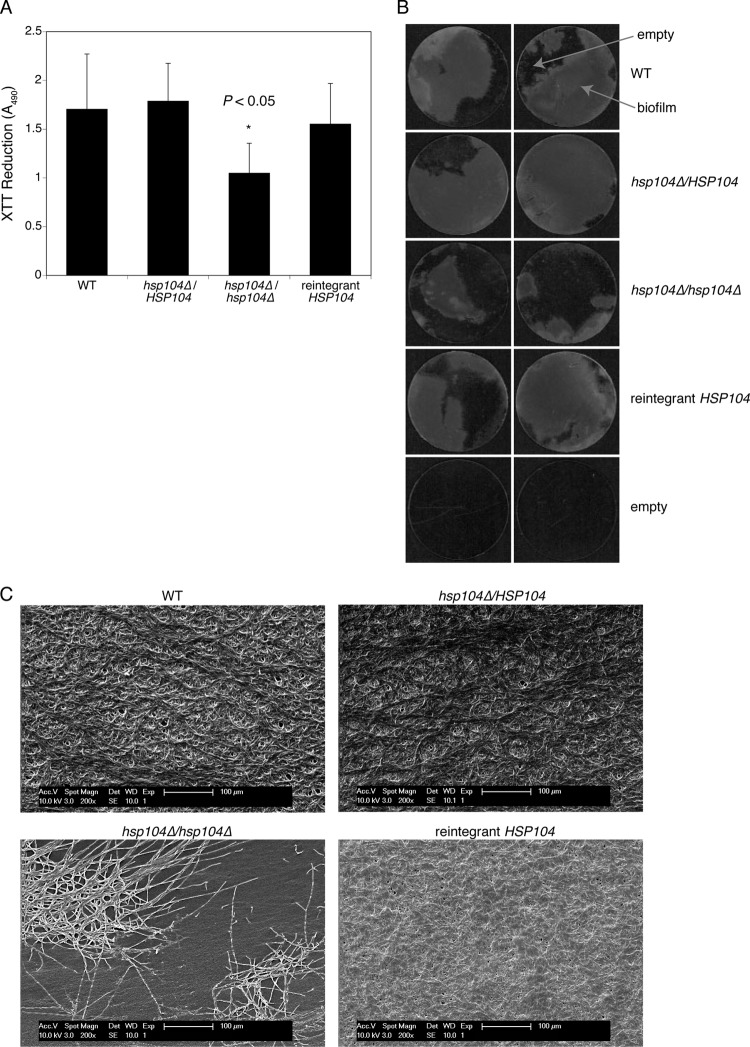
We next sought to evaluate the effect of the absence of Hsp104 on biofilm architecture. Mature biofilms were allowed to form on highly adhesive tissue culture coverslips in order to mimic the surface of polystyrene plates. Measurements of biofilm formation by way of the XTT reduction method confirmed a defect in biofilm formation from the homozygous hsp104Δ mutant (Fig. 5A). The failure to observe defective biofilm formation for the heterozygous hsp104Δ mutant in these experiments may be due to the slightly different experimental procedures used. It is important to point out here that evaluation of the biofilm formed on tissue culture coverslips had the only purpose of providing a reliable experimental background for the microscopic observations reported next. Coverslips from separate experiments were used for imaging of the biofilm presence and architecture. Figure 5B shows two representative coverslips per strain; as can be seen, biofilms formed by the wild-type, heterozygous hsp104Δ mutant and the reintegrant strain covered the vast majority of the coverslip surface, whereas most of the coverslip areas where biofilms of the hsp104Δ homozygous mutant had been allowed to develop appeared empty. SEM micrographs were taken to obtain a closer look at the architecture of biofilms formed by strains with a different content of Hsp104. These pictures provided a more detailed structural characterization of the upper layers of biofilms (Fig. 5C). Mature biofilms formed by the heterozygous mutant and the reintegrant strains were indistinguishable from those formed by wild-type cells, with dense and compact layers of hyphae embedded in material that can be considered extracellular matrix. In contrast, whereas the majority of the surface of coverslips incubated with the homozygous hsp104Δ mutant remained uncovered, SEM micrographs of the covered areas revealed the presence of rudimentary biofilm only, with dense clumps of hyphal cells rather than a compact biofilm layer (Fig. 5C). Taken together, these results provide structural support to the data obtained using the polystyrene and polyurethane biofilm systems illustrated in Fig. 4.
Fig 5.
Impact of genetic deprivation of Hsp104 on biofilm architecture. (A) C. albicans biofilms were developed on highly adhesive round tissue culture coverslips, as a validation of subsequent experiments. Quantitative analyses of mature biofilms were documented by XTT reduction assay measured at 490 nm. Standard deviations were calculated from three independent experiments, each conducted with two coverslips per strain. (B) Pictures of mature biofilms formed on the surface of round tissue culture coverslips. (C) Scanning electron microscopy pictures of mature biofilms formed on the surface of round tissue culture coverslips. See text for details. Magnification, ×200.
Virulence in the Caenorhabditis elegans infection model is attenuated in the absence of Hsp104.
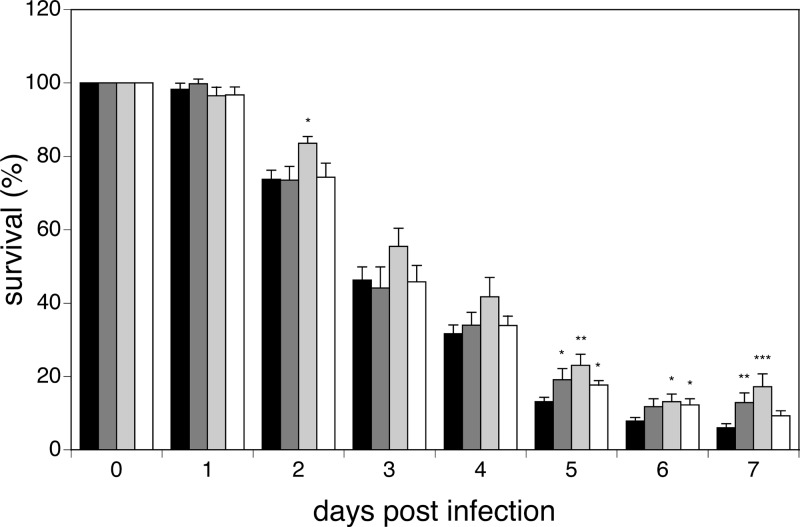
In order to obtain information on a possible contribution of Hsp104 to the virulence potential of C. albicans, independently of its role in thermotolerance, we tested the hsp104Δ mutants in the worm infection model. Infection experiments using the nematode C. elegans as a host are well suited for this purpose, since they are conducted at 25°C using a temperature-sensitive mutant (2). Hence, we followed the survival of worms after infection with the hsp104Δ hetero- and homozygous mutants, as well as with the reconstituted strain. Worms infected with the homozygous mutant showed a significant (P < 0.05) increase in survival relative to animals infected with the wild-type reference strain or with the heterozygous and reconstituted strains (Fig. 6). At day 2 postinfection, the survival of animals infected with the wild-type, heterozygous, and reconstituted strains was 73.8%, 73.5%, and 74.3%, respectively, whereas that of animals infected with the Hsp104 homozygous strain was 83.5%. Such difference was maintained throughout the experiment, and at its end at day 7, viable worms were 6%, 12.9%, and 9.3% for the wild-type, heterozygous, and reconstituted strains, respectively, and 17.2% for the homozygous strain (Fig. 6). These results demonstrate that the absence of Hsp104 has a detectable impact on the virulence potential of C. albicans in the worm infection model.
Fig 6.
Genetic deprivation of Hsp104 attenuates virulence of C. albicans in the worm infection model. Survival of C. elegans infected with the wild type (black bars), heterozygous (dark gray bars) and homozygous (light gray bars) hsp104Δ mutants, and reconstituted HSP104 strain (white bars) is shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
This study reports on the consequences of the absence of the molecular disaggregase Hsp104 in Candida albicans. Expression of this protein is minimal in exponentially growing cells and is induced upon thermal stress and in stationary phase. Disruption of one HSP104 allele was observed to be sufficient to render cells of the wild-type strain SC5314 more sensitive to short exposures to the lethal temperature of 50°C. This observation correlates with decreased total expression of Hsp104 in the heterozygous deletion mutant, demonstrating that HSP104 is a haploinsufficient gene. Disruption of the second HSP104 allele further aggravated the tolerance of C. albicans to thermal stress, rendering the corresponding cells extremely sensitive. Wild-type cells in stationary phase were found to be more resistant than exponential-phase cells against exposure to lethal high temperature. However, exponential-phase cells could be rendered more resistant following a pretreatment of 30 min at the elevated, harmless temperature of 41°C, which induces Hsp104 expression. Therefore, the establishment of acquired thermotolerance appears to be dependent on the presence of Hsp104 in stationary-phase cells as well as in heat-stressed exponential-phase cells. Ectopic expression of Hsp104 from a regulatable promoter, in the absence of both endogenous wild-type alleles, clearly established that inducing Hsp104 prior to exposure to the lethal temperature is sufficient to confer cellular protection against the thermal insult, indicating that Hsp104 plays a central role in governing the cellular defense against thermal stress. Interestingly, Hsp104 was recently identified in a proteomics screen for proteins sumoylated in response to stress in C. albicans (17). The authors of that study highlighted the importance of this posttranslational modification, showing that an alteration of the Hsp104 consensus target for sumoylation leads to higher susceptibility to elevated temperatures.
A general consideration in the fungal stress response envisages the possibility that fungi may have evolved response mechanisms that are more suited to counteract stress they may encounter in their environment, rather than having a generally conserved response (20). In line with this, the heat shock regulon of an organism obligately associated with warm-blooded animals, such as C. albicans, has been proposed to modulate growth of the fungus in response to minor temperature perturbations occurring in its host (3). Fine-tuning the expression of several molecular chaperones, among which is Hsp104, appears to be accomplished by the transcription factor Hsf1 in C. albicans (18).
Although the expression of HSP104 seems to be regulated similarly in C. albicans and in the nonpathogenic yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe (12), some intriguing observations suggest that Hsp104 may play a previously unrecognized role in processes of medical interest such as biofilm formation and filamentation/virulence, independently from its involvement in cellular defense against thermal stress. First, upregulation of HSP12, HSP104, and SSA4 has been shown to occur in the livers of animal experimental models infected with C. albicans (36). Second, elevated expression of HSP70, HSP82, HSP90, and HSP104 was reported to be associated with the yeast-pathogenic phase of the human fungal pathogen Paracoccidioides brasiliensis (16, 19). Third, Cao et al. (5) reported downregulation of HSP70, HSP90, HSP104, MSI3, and SSA2 upon exposure to farnesol, a quorum-sensing molecule that inhibits biofilm formation. Finally, coregulation of HSP104 with genes associated with hypha formation and virulence in several mutants was recently observed (14).
In agreement with the hypothesis suggested by these observations, we found, using two different models, that mutants devoid of Hsp104 are defective in biofilm formation. Biofilms formed by the hsp104Δ/hsp104Δ strain showed a rudimentary structure (Fig. 5C) and were more easily removed from the substrate during the washing procedure (our unpublished observations), indicating that biofilms developed in the absence of Hsp104 may be less rigid and characterized by a looser structure. Considering that adhesion to the substrate was not compromised in the absence of Hsp104 in any of the experimental models used (Fig. 4A and C), it is tempting to speculate that Hsp104 may have a direct or indirect role in cell-cell adhesion. Although targeted experiments will be needed to validate this hypothesis, it is interesting to note in this respect that Hsp104 has been described to be present on the surface of C. albicans cells (23).
Increasing evidence that biofilms are critical to the development of clinical infections is accumulating, suggesting that they might represent an important pathogenicity factor (reviewed in references 13 and 24). Infection experiments using the C. elegans model evidenced a role of Hsp104 in virulence that can be separated from its role in governing thermotolerance, since in contrast to the case for other animal models, these worm infection experiments were conducted at 25°C. The worm system appeared to be particularly well suited for detection of the small but significant reduction in infectivity of C. albicans cells devoid of Hsp104, given its higher throughput compared to the mouse systemic infection model.
In conclusion, even if Hsp104 does not appear to play a role in traditional determinants such as cell fitness and hypha formation, at least under the in vitro conditions tested, the attenuation of virulence in the C. elegans system and the reduction of biofilm formation in the polyurethane and polystyrene models point to a role of this protein in counteracting cellular stress that goes beyond the response to high temperatures. Taken together, the data presented in this paper contribute to the general knowledge about C. albicans pathogenicity, indicating a role for Hsp104 in infection and biofilm formation. This role does not appear to be of sufficient magnitude to justify the use of Hsp104 as a target for antifungals, an idea made attractive by the unique distribution of Hsp104 in the fungal kingdom. However, the general idea of targeting the pathogen's stress response to decrease its virulence potential is supported and reinforced by our observations. Considering the known role of Hsp104 in resolving protein aggregates and modulating protein turnover in other systems (1), it would be interesting to test, in future studies, the possibility that the environmental stress conditions encountered by C. albicans during biofilm formation and infection of host cells impose a cost on the equilibrium between protein synthesis and folding/misfolding.
ACKNOWLEDGMENTS
We are grateful to M. Tuite for his generous gift of antibody against C. albicans Hsp104. We thank Ilse Palmans for excellent technical assistance and Nico Vangoethem for help with the preparation of figures.
This work was funded by was grants from the Flemish Science Foundation (FWO) (G.0804.11 and WO.026.11N to P.V.D. and G.0896.10 to K.T. and B.P.A.C.), the research fund of KU Leuven (GOA/2007/08 to P.V.D.), and the Industrial Research Fund of KU Leuven (IOF-M to K.T.). S.K. is supported by a KU Leuven postdoctoral grant (PMDK/11/089), and G.G. was the recipient of a predoctoral fellowship from IWT-Vlaanderen.
Footnotes
Published ahead of print 25 May 2012
REFERENCES
- 1. Bosl B, Grimminger V, Walter S. 2006. The molecular chaperone Hsp104—a molecular machine for protein disaggregation. J. Struct. Biol. 156: 139–148 [DOI] [PubMed] [Google Scholar]
- 2. Breger J, et al. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3: e18 doi:10.1371/journal.ppat.0030018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown AJ, Leach MD, Nicholls S. 2010. The relevance of heat shock regulation in fungal pathogens of humans. Virulence 1: 330–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown AJ, Odds FC, Gow NA. 2007. Infection-related gene expression in Candida albicans. Curr. Opin. Microbiol. 10: 307–313 [DOI] [PubMed] [Google Scholar]
- 5. Cao YY, et al. 2005. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob. Agents Chemother. 49: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884 [DOI] [PubMed] [Google Scholar]
- 7. Cox BS, Byrne LJ, Tuite MF. 2007. Prion stability. Prion 1: 170–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eggimann P, Garbino J, Pittet D. 2003. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 3: 685–702 [DOI] [PubMed] [Google Scholar]
- 10. Engel A, Shewmaker F, Edskes HK, Dyda F, Wickner RB. 2011. Amyloid of the Candida albicans Ure2p prion domain is infectious and has an in-register parallel beta-sheet structure. Biochemistry 50: 5971–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enjalbert B, Nantel A, Whiteway M. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14: 1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Enjalbert B, et al. 2006. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17: 1018–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fanning S, Mitchell AP. 2012. Fungal biofilms. PLoS Pathog. 8: e1002585 doi:10.1371/journal.ppat.1002585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finkel JS, et al. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8: e1002525 doi:10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a. Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198: 179–182 [DOI] [PubMed] [Google Scholar]
- 15. Glover JR, Lindquist S. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- 16. Goldman GH, dos Reis Marques E, et al. 2003. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot. Cell 2: 34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leach MD, Stead DA, Argo E, Brown AJ. 2011. Identification of sumoylation targets, combined with inactivation of SMT3, reveals the impact of sumoylation upon growth, morphology and stress resistance in the pathogen Candida albicans. Mol. Biol. Cell 22: 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicholls S, Leach MD, Priest CL, Brown AJ. 2009. Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol. Microbiol. 74: 844–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicola AM, et al. 2008. The stress responsive and morphologically regulated hsp90 gene from Paracoccidioides brasiliensis is essential to cell viability. BMC Microbiol. 8: 158 doi:10.1186/1471-2180-8-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nikolaou E, et al. 2009. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol. Biol. 9: 44 doi:10.1186/1471-2148-9-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Odds FC. 1994. Pathogenesis of Candida infections. J. Am. Acad. Dermatol. 31: S2–S5 [DOI] [PubMed] [Google Scholar]
- 22. Park YN, Morschhauser J. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4: 1328–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pitarch A, Sanchez M, Nombela C, Gil C. 2002. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteomics 1: 967–982 [DOI] [PubMed] [Google Scholar]
- 24. Ramage G, Rajendran R, Sherry L, Williams C. 2012. Fungal biofilm resistance. Int. J. Microbiol. 2012: 528521 doi:10.1155/2012/528521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramage G, Vandewalle K, Wickes BL, Lopez-Ribot JL. 2001. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 18: 163–170 [PubMed] [Google Scholar]
- 26. Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341: 119–127 [DOI] [PubMed] [Google Scholar]
- 27. Ricicova M, et al. 2010. Candida albicans biofilm formation in a new in vivo rat model. Microbiology 156: 909–919 [DOI] [PubMed] [Google Scholar]
- 28. Rieder SE, Emr SD. 2001. Isolation of subcellular fractions from the yeast Saccharomyces cerevisiae. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.8 [DOI] [PubMed] [Google Scholar]
- 29. Sanchez Y, Lindquist SL. 1990. HSP104 required for induced thermotolerance. Science 248: 1112–1115 [DOI] [PubMed] [Google Scholar]
- 30. Sanchez Y, Taulien J, Borkovich KA, Lindquist S. 1992. Hsp104 is required for tolerance to many forms of stress. EMBO J. 11: 2357–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schirmer EC, Lindquist S, Vierling E. 1994. An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell 6: 1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmitt M, Neupert W, Langer T. 1996. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J. Cell Biol. 134: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Senechal P, Arseneault G, Leroux A, Lindquist S, Rokeach LA. 2009. The Schizosaccharomyces pombe Hsp104 disaggregase is unable to propagate the [PSI+] prion. PLoS One 4: e6939 doi:10.1371/journal.pone.0006939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singer MA, Lindquist S. 1998. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1: 639–648 [DOI] [PubMed] [Google Scholar]
- 35. Thevissen K, et al. 2011. Novel fungicidal benzylsulfanyl-phenylguanidines. Bioorg. Med. Chem. Lett. 21: 3686–3692 [DOI] [PubMed] [Google Scholar]
- 36. Thewes S, et al. 2007. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol. Microbiol. 63: 1606–1628 [DOI] [PubMed] [Google Scholar]
- 37. Tuite M, Stojanovski K, Ness F, Merritt G, Koloteva-Levine N. 2008. Cellular factors important for the de novo formation of yeast prions. Biochem. Soc. Trans. 36: 1083–1087 [DOI] [PubMed] [Google Scholar]
- 38. Vacher C, Garcia-Oroz L, Rubinsztein DC. 2005. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum. Mol. Genet. 14: 3425–3433 [DOI] [PubMed] [Google Scholar]
- 39. Wilson D, Fiori A, De Brucker K, Van Dijck P, Stateva L. 2010. Candida albicans Pde1p and Gpa2p comprise a regulatory module mediating agonist-induced cAMP signalling and environmental adaptation. Fungal Genet. Biol. 47: 742–752 [DOI] [PubMed] [Google Scholar]
- 40. Zenthon JF, Ness F, Cox B, Tuite MF. 2006. The [PSI+] prion of Saccharomyces cerevisiae can be propagated by an Hsp104 ortholog from Candida albicans. Eukaryot. Cell 5: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]