Abstract
Clearance of hepatitis D virus (HDV) viremia leads to disease remission. Large hepatitis delta antigen (L-HDAg) has been reported to activate transforming growth factor β, which may induce epithelial-mesenchymal transition (EMT) and fibrogenesis. This study analyzed serum HDV RNA “quasispecies” in HDV-infected patients at two stages of infection: before and after alanine aminotransferase (ALT) elevations. Included in the study were four patients who went into remission after ALT elevation and three patients who did not go into remission and progressed to cirrhosis or hepatocellular carcinoma. Full-length HDV cDNA clones were obtained from the most abundant HDV RNA species at the pre- and post-ALT elevation stages. Using an in vitro model consisting of Huh-7 cells transfected with cloned HDV cDNAs, the pre- or post-ALT elevation dominant HDV RNA species were characterized for (i) their replication capacity by measuring HDV RNA and HDAg levels in transfected cells and (ii) their capacity to induce EMT by measuring the levels of the mesenchymal-cell-specific protein vimentin, the EMT regulators twist and snail, and the epithelial-cell-specific protein E-cadherin. Results show that in patients in remission, the post-ALT elevation dominant HDV RNA species had a lower replication capacity in vitro and lower EMT activity than their pre-ALT elevation counterparts. This was not true of patients who did not go into remission. The expression of L-HDAg, but not small HDAg, increased the expression of the EMT-related proteins. It is concluded that in chronically infected patients, HDV quasispecies with a low replication capacity and low EMT activity are associated with disease remission.
INTRODUCTION
Hepatitis D virus (HDV) has an outer envelope of hepatitis B surface antigen (HBsAg), which is essential for virus assembly, secretion, and infection (16, 23, 29). The inner component of the HDV virion is the ribonucleoprotein that includes HDV RNA and HDV proteins (29). HDV encodes the small and large hepatitis delta antigens (S-HDAg and L-HDAg, respectively) (2, 4, 29). S-HDAg is essential for HDV RNA replication, while L-HDAg is indispensable for HDV virion assembly (4, 29). HDV superinfection in chronic hepatitis B patients results in various outcomes, including remission, chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) (9, 10, 12, 24, 32). However, the mechanism that leads to these diverse outcomes is still obscure.
Active replication and the high evolutionary rate of viral genomes are the two important characteristics of the RNA virus life cycle by which they may evade attacks by the host immune system. Similar to other RNA viruses, HDV has a high evolutionary rate (8, 17). Consequently, HDV RNA genomes in a chronic hepatitis D (CHD) patient are composed of a population of RNA molecules with closely related but slightly different nucleotide sequences, called quasispecies (8, 17). Changes in HDV quasispecies and replacement by certain dominant species are observed during clinical courses of CHD and may play an important role in viral escape from host immune attack and in clinical relapse (31, 36). The consequences of such selection for the disease course are still unclear.
Several lines of evidence indicate that HDV may not be directly cytopathic because a cell line with stable HDV expression and transgenic mice with HDV do not show obvious cytopathic injury; however, some studies have suggested the possibility of cytotoxicity or deleterious effects of HDV on cell proliferation (5, 11, 20, 22, 30). HDV may indirectly induce immune-mediated liver injury (15, 21). In previous studies, selection of HDV RNA quasispecies has been observed after ALT elevation (15, 31, 36). Furthermore, there are alterations in amino acid sequences in the immunogenic epitopes of emergent dominant HDV quasispecies. The selection of HDV RNA quasispecies may be due to immune selection or a growth advantage.
Aside from the viral genotype and viral load, other confounding factors, like transforming growth factor β (TGF-β), may also influence disease outcomes. TGF-β plays an important role in liver fibrosis and cirrhosis (3). Choi et al. reported that L-HDAg may induce liver fibrosis through TGF-β-induced signal transduction (6). Activation of specific receptors by TGF-β has also been shown to provoke epithelial-to-mesenchymal transitions (EMT) in many types of epithelial cells in culture (18). Several lines of evidence imply increased TGF-β signaling as a key effector of EMT in cancer progression and metastasis (7). The EMT and mesenchymal-to-epithelial transitions are known to occur when tissues are constructed during embryogenesis/development. They are also involved in organ fibrosis (38).
When injury and inflammation persist, EMT activates fibroblastic cells that accumulate and cause progressive fibrosis (38). During EMT, levels of epithelial-cell-specific proteins, such as E-cadherin, are decreased while those of mesenchymal-cell-specific proteins, including α-smooth-muscle actin and vimentin, are increased (38). Recent studies suggest that expression of twist and snail in HCC is associated with EMT and recurrence of HCC after tumor resection (37). In this study, we hypothesized that in chronically HDV-infected patients, the emergence of HDV RNA quasispecies presenting low replication capacity and low levels of EMT activity may be predictive of disease remission.
MATERIALS AND METHODS
Patients.
Four patients with disease remission and normal ALT levels and three with persistently elevated ALT levels, cirrhosis, or HCC were randomly selected from HDV-infected patients with serial serum samples available for HBV DNA, HBsAg, and HDV RNA analyses (Table 1). The patients included were all positive for serum HBsAg (Ausria II-125; Abbott Laboratories, North Chicago, IL) and antibody to HDV antigen (anti-HDV) (anti-Delta; Abbott Laboratories). In addition, they were all negative for immunoglobulin M antibody to hepatitis B core antigen (CORAB-M; Abbott Laboratories), hepatitis C virus (Abbott Laboratories), and hepatitis A virus (HAVABM; Abbott Laboratories). Serum ALT levels were measured with a systemic multiautoanalyzer (Technicon SMAC; Technicon Instruments Corp., Tarrytown, NY). The clinical profiles and serological data of patients are shown in Table 1. Of the seven patients, patients I, II, and III had been treated with short-acting alpha interferon 2b (Intron A; Schering Corporation) at 5 million IU thrice weekly for 1 year. This study was approved by the Institutional Review Board.
TABLE 1.
Correlation of serum HBV DNA, HDV RNA, and HBsAg levels and age with disease courses
| Patient group, no. (sex), and age (yr) | HBV type | HDV type | Outcome | ALT (U/liter) | ASTa (U/liter) | HBV DNA (copies/ml) | HBsAg (IU/ml) | HDV RNA (copies/ml) |
|---|---|---|---|---|---|---|---|---|
| Disease remission | ||||||||
| I (M) | ||||||||
| 50b | B | 1 | CHD | 360 | 176 | 19,200 | 184.2 | 36,320 |
| 51c | CHD | 128 | 48 | 38,700 | 118.6 | 552,253 | ||
| 53d | CHD | 295 | 129 | 346,000 | 631.3 | 10,882,490 | ||
| 56 | Remission | 41 | 28 | <300 | 0.1 | <400 | ||
| 66 | Remission | 20 | 16 | <300 | 0 | <400 | ||
| II (M) | ||||||||
| 28c | B | 2 | CHD | 107 | 80 | <300 | 2670 | 82,200,000 |
| 40d | CHD | 129 | 76 | <300 | 1831.6 | 19,800 | ||
| 42 | Remission | 14 | 21 | <300 | 0.1 | <400 | ||
| 43 | Remission | 33 | 21 | <300 | 0 | <400 | ||
| III (M) | ||||||||
| 33c | NDe | 4 | AHDf | 1,235 | 645 | <300 | 6,458 | 32,200,000 |
| 44d | CHD | 61 | 55 | <300 | 3,643 | 2,05,200 | ||
| 50 | Remission | 16 | 23 | <300 | 0.1 | <400 | ||
| 61 | Remission | 21 | 23 | <300 | 0.1 | <400 | ||
| IV (M) | ||||||||
| 27c | B | 4 | AHD | 926 | 502 | 987 | 784 | 2,755,440 |
| 29d | CHD | 117 | 83 | <300 | 138.4 | 185,884 | ||
| 44 | Remission | 14 | 19 | <300 | 0.2 | <400 | ||
| 45 | Remission | 22 | 23 | <300 | 0.2 | <400 | ||
| Disease progressiong | ||||||||
| V (F) | ||||||||
| 24c | C | 4 | CHD | 179 | 153 | 14,900 | 11,456 | 303,200,000 |
| 29d | CHD | 79 | 62 | 189,800 | 7,825 | 89,600,000 | ||
| 33 | CHD | 56 | 48 | 66,348 | 9,875.2 | 29,300,000 | ||
| VI (M) | ||||||||
| 48c | B | 1 | Cirrhosis | 32 | 26 | <300 | 945.4 | 4,076,000 |
| 53d | Cirrhosis | 44 | 36 | <300 | 829.2 | 1,064,000 | ||
| 56 | Cirrhosis | 17 | 16 | <300 | 542.1 | 3,236 | ||
| 63 | Cirrhosis | 25 | 19 | 1,164 | 77.8 | <400 | ||
| 67 | Cirrhosis | 22 | 20 | <300 | 73.8 | <400 | ||
| VII (F) | ||||||||
| 52c | B | 4 | HCC | 41 | 23 | <300 | 309.6 | 840,000 |
| 56d | HCC | 45 | 41 | 2,060 | 841.5 | 1,968,560 | ||
| 58 | HCC | 55 | 50 | 3,270 | 1,372 | 1,138,134 |
AST, aspartate transaminase.
First time point.
Early time point.
Late time point.
ND, nondetectable.
AHD, acute hepatitis D.
With persistently elevated ALT or adverse outcomes (cirrhosis or HCC).
Detection of original and novel dominant HDV strains and determination of variant-specific restriction sites.
The original and novel dominant HDV quasispecies before (early time point) and after (late time point) serum ALT elevation were cloned from the sera of CHD patients. Detection of dominant viral strain RNA by reverse transcription (RT)-PCR and PCR cloning were performed as previously reported (32). More than 10 colonies from each time point of each case were randomly selected, and their sequences were analyzed with an ABI 373A sequencer (Perkin-Elmer Cetus Corp., Norwalk, CT). The sequences of the original and novel dominant HDV quasispecies were compared, and restriction enzymes that were able to differentiate these quasispecies were determined by large-scale screening as described in the following section.
Identification and functional analysis of HDV RNA quasispecies.
In order to have equal chances to detect all of the HDV quasispecies, the primers for RT-PCR were synthesized on the basis of the most conserved parts of the HDAg sequences of the original and novel dominant quasispecies. Primer 120 (5′-ATG CCA TGC CGA CCC GAA GAG GAA-3′) and primer 214 (5′-CTC AGG GGA GGG TTC TCC GAC A-3′) were synthesized according to the conserved region of the hepatitis delta antigen coding region. The amplified PCR products derived from HDV-infected serum obtained at different time points were ligated into plasmid vector pCR2 (Original TA Cloning Kit; Invitrogen Corporation, Carlsbad, CA) and then transformed into competent Escherichia coli strain DH5α (Gibco BRL, Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions (32). More than 100 HDV colonies from each case at each time point were randomly selected and subjected to restriction fragment length polymorphism (RFLP) differentiation based on quasispecies-specific restriction enzyme digestion. The determination of quasispecies-specific restriction enzymes was based on comparisons of the nucleotide sequences of dominant HDV species obtained at the pre- and post-ALT elevation stages. For patient I (infected with HDV genotype 1), the 446-bp PCR products amplified with primers 214 and 120 were cut with restriction enzymes BlpI and HpyCH4V (TGCA), respectively. The BlpI-cut PCR products of the original dominant species generated 200- and 246-bp fragments, while the PCR products of the novel dominant species could not be cut by restriction enzyme BlpI. Using the same method, the PCR products cut by HpyCH4V showed inverse results. For patient II (infected with HDV genotype 2), the PCR products cut by restriction enzyme FokI also gave two different RFLP patterns. The PCR products amplified from the original dominant species cut by FokI generated a 312-bp fragment, while a 451-bp fragment was generated from the cleaved PCR products of the novel dominant species isolated after exacerbation. For patient III (infected with HDV genotype 4), the PCR products cut with AluI generated 230-, 140-, and 75-bp fragments, indicating the original dominant species before ALT elevation, while the cut PCR products of 370- and 75-bp fragments indicated the novel dominant species.
Competition analysis via large-scale screening of secreted HDV virions using variant-specific RFLP.
For competition analysis of the respective amounts of the original and novel dominant HDV species, Huh-7 cells were cotransfected with paired HDV-producing plasmids generated at the pre- or post-ALT elevation stage (from the same patient) and an HBV-producing plasmid. Viral particles from 9 ml of day 9 culture supernatant were pelleted, and HDV RNA was extracted as described previously (26). To remove cell debris, the culture media were centrifuged at 3,000 rpm at 4°C for 15 min. The viral particles were pelleted by high-speed centrifugation at 40,000 rpm at 4°C for 5 h in an SW41 rotor (Beckman) through a 20% sucrose cushion. The HDV cDNA was generated in the presence of reverse transcriptase (SUPERSCRIPT; GIBCO BRL, Life Technologies, Rockville, MD) and ligated into pCRR2.1-TOPO (Invitrogen Corporation, Carlsbad, CA). More than 100 colonies from each case were randomly selected and subjected to variant-specific RFLP differentiation.
Primer 120 and primer 214 were synthesized accordingly. For patient I (infected with HDV genotype 1), patient II (infected with HDV genotype 2), and patient III (infected with HDV genotype 4), the same restriction enzymes were used as described in the previous section. For patient V (infected with HDV genotype 4), with persistent hepatic inflammation, the 446-bp PCR products amplified with primers 214 and 120 were cut with restriction enzyme BspQI. The PCR products of the original dominant species cut by BspQI generated 89- and 357-bp fragments, while the PCR products of the novel dominant HDV species could not be cut by this enzyme.
Plasmids for HDV, L-HDAg, and S-HDAg expression.
The dominant HDV species expression plasmids were isolated and constructed from the pre- and post-ALT elevation stages of the disease course, respectively, of each patient. To generate the replicative form of HDV cDNA, plasmids expressing HDV genotype 1 (TWD2577-66, TW3678-25, TW1435-47, TW5132-24), genotype 2 (1079, m7049), and genotype 4 (TWD62-16, TW2621-56, TW1025-14, TW3038-25, 8800, m9932) were constructed as previously described (14). Briefly, for cloning of the entire HDV genome, the overlapping subgenomic clones were stitched by further PCR. Using a strategy of two-copy insertion, the 1.7-kb XbaI fragment was isolated and inserted as a tandem repeat into the XbaI site of pCDNA3.1(−) (Invitrogen Corp., Carlsbad, CA) to generate pHDV-D2G. Plasmids containing the L-HDAg or S-HDAg coding fragment were cloned into pCMV-EBNA (Clontech Laboratories, Palo Alto, CA) and constructed as previously described (14).
Plasmids for HBV expression.
HBV-producing plasmids were isolated and constructed as previously described (26). A partial HBV genome sequence (approximately 3,056 bp) which contains the complete HBV envelope open reading frame was amplified using primers B2600 NheI (5′-CCG CTA GCC TTA CAG TAA ATG AAA A-3′) and B25R (5′-TCC CAC CTT ATG TGT CCA-3′) via PCR and then cloned into the NheI/HindIII sites of vector plasmid pcDNA3.1(−) (Invitrogen, San Diego, CA). Another partial HBV genome sequence (approximately 1,859 bp) was amplified with primers B980 NheI (5′-GGA AAG TAT GCT AGC GAA TTG TGG-3′) and B2839 BstEII (5′-CCA AGA ATA TGG TGA CCC-3′) by PCR and then cloned into the NheI/BstEII sites of the plasmid containing the previously described 3-kb partial HBV sequence to become a replicative-form HBV construct. In all HBV recombinant plasmids, a cytomegalovirus promoter drives the transcription of the pregenomic RNA.
Culture and transfection of Huh-7 cells.
The well-differentiated HCC cell line Huh-7 was used (33). The cells (106) were transfected with FuGENE HD transfection reagent (Roche, Inc.) according to the supplier's instructions. For analysis of snail, twist, vimentin, and E-cadherin expression, 15 to 30 μg of L-HDAg- or S-HDAg-expressing plasmid DNA was transfected into Huh-7 cells and the medium was changed at 6 h posttransfection. Subsequently, the medium was replaced and collected on days 3, 6, and 9 as previously described (26).
Western blot analysis.
The isolated proteins were separated by SDS-PAGE, blotted onto nitrocellulose membranes, and stained for HDAgs with anti-HDV-positive human serum (1:5,000), for HBsAgs with monoclonal antibody A10F1 (1:2,000), or for the reference protein heat shock cognate 70 (Hsc70) with monoclonal antibody HSC70 (B-6; 1:5,000; Santa Cruz Biotechnology, Santa Cruz, CA) (26). For analysis of the expression of snail, twist, vimentin, and E-cadherin, cell lysates were harvested on days 6 and 9 posttransfection. The membranes were probed with a primary antibody incubated at 4°C overnight, followed by the addition of a secondary antibody. Snail was detected by using rabbit anti-snail antibody (C15D3; 1:1,000; Cell Signaling Technology, Inc.), twist was detected by using rabbit anti-twist antibody (ab50581; 1:1,000; Abcam, Cambridge, MA), E-cadherin was detected by using rabbit anti-E-cadherin antibody (4065; 1:1,000; Cell Signaling Technology, Inc.), and vimentin was detected by using mouse anti-vimentin antibody (V6630; 1:1,000; Sigma-Aldrich). The relative quantitative values of EMT markers were estimated by using the AlphaImager 2000 Documentation Analysis System and the AlphaImager 2000 software package.
Northern blot analysis.
A total of 20 μg of RNA was analyzed by Northern blotting as previously described (26). After fixation by UV illumination, RNA was hybridized with digoxigenin (DIG)-labeled cDNA probes derived from different genotypes of HDV templates as previously described (26). Hybridization was performed by incubation with Dig-Hyb solution (DIG labeling and detection kit; Roche Diagnostics System, Basel, Switzerland) at 55°C overnight. For controls, hybridization with the DIG-labeled glyceraldehyde-3-phosphate dehydrogenase (G3PDH) gene was performed simultaneously.
Indirect double immunofluorescence staining.
Immunofluorescence staining was modified and performed as previously described (26). Before DNA transfection, Huh-7 cells were cultured overnight on coverslips. After transfection with an L-HDAg- or S-HDAg-expressing plasmid, the cells were fixed and permeated with acetone at 4°C for 20 min and then blocked with phosphate-buffered saline (PBS) containing 1% bovine serum albumin at room temperature for 30 min. The cells were incubated with polyclonal human anti-delta serum diluted 1:200 and mouse monoclonal antibody to vimentin (V6630; Sigma-Aldrich) or rabbit monoclonal antibody to E-cadherin (4065; Cell Signaling Technology, Inc.), snail (C15D3; Cell Signaling Technology, Inc.), twist (ab50581; Abcam, Cambridge, MA, or Santa Cruz Biotechnology, Inc.), and β-catenin (ab6302; Abcam, Cambridge, MA, or Santa Cruz Biotechnology, Inc.) primary antibodies diluted 1:200 in the same buffer at room temperature for 30 min. After the cells were washed thrice with PBS, they were incubated with rhodamine-conjugated rabbit anti-human IgG (ab6756; Abcam Cambridge Science Park, Cambridge, United Kingdom) and fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG (ab6724; Abcam Cambridge Science Park, Cambridge, United Kingdom) secondary antibodies at room temperature for 30 min. Both conjugates were diluted 1:200 in the same buffer. Finally, the coverslips were washed thoroughly with PBS and then mounted with 50% glycerol. Photographs were taken using a confocal fluorescence microscope (Axiovert 200M; Zeiss) with oil objective (63×, numerical aperture 1.4; Zeiss). A 488-nm argon laser (45 mW; LASOS) and a 543-nm He-Ne laser (2.5 mW; LASOS) were used to excite HDAg-rhodamine and EMT marker-FITC, respectively.
Quantification of TGF-β1.
Total TGF-β was measured with a commercially available kit (DB100B; R&D Systems, Minneapolis, MN) as the manufacturer recommends. Briefly, Huh-7 cells transfected with L-HDAg- or S-HDAg-expressing plasmid DNA were incubated in Dulbecco modified Eagle medium (Life Technologies, Grand Island, NY) supplemented with 2% fetal calf serum (FBS), and the supernatants were collected after 3 days of incubation. Because ordinary 10% FBS contains TGF-β, 2% FBS was used in this experiment to minimize the interfering effects of preexisting TGF-β in 10% FBS. For total TGF-β measurements, the supernatants were preactivated by the addition of acid. Aliquots of samples or standards were added to a 96-well microplate, covered with an adhesive strip, and incubated for 2 h at room temperature. After three washes, the detection antibody was added and the mixture was incubated at room temperature for 2 h. After three washes, the substrate solution was added and the mixture was incubated in the dark for 30 min. After the addition of stop solution, the optical density of each well was determined immediately with a microplate reader set to 450 nm with wavelength correction set to 540 nm.
Quantitative analysis of HBsAgs, HBV DNA, and HDV RNA.
HBsAg in spent medium or human serum was quantified by the ARCHITECT HBsAg system kit (Abbott Laboratories), while HBV DNA was measured with COBAS Amplicor HBV monitor (Roche Diagnostic Systems, Basel, Switzerland). The detection limit of this assay is 300 copies/ml.
For quantitative HDV RNA analysis, total RNA were isolated from patient serum as previously described (26). Twenty microliters of a final cDNA solution was obtained from 50 μl of serum. For synthesis of standard HDV cDNA, a subgenomic fragment was amplified by PCR with primer 214 and primer 120. The PCR products were inserted into the pCRII vector by TA cloning. The resulting plasmids were used as the standards for HDV cDNA quantification (plasmid 1078-5 is for HDV type 1, plasmid 110-A2 is for type 2, and plasmid D62-28 is for type 4). The purified plasmids were quantified by measuring optical density at a wavelength of 260 nm. Real-time PCR was performed by using the BD QTaq DNA Polymerase Mix (Clontech Laboratories). Purified cDNA (5 μl) was added to a 20-μl PCR mixture containing 12.5 μl of BD QTaq DNA Polymerase Mix, 900 nM each primer, and 250 nM fluorescent TaqMan MGB probe. The standard curve for this assay was calculated by using a series of 10-fold dilutions of (5 to 5 × 106 copies/reaction) of previously titrated plasmids (p1078-5 for HDV type 1, p110-A2 for type 2, and pD62-28 for type 4). The reaction protocol consisted of one initiating step of 3 min at 95°C, followed by 40 amplification cycles consisting of 15 s at 95°C and 1 min at 60°C. The reactions, data acquisition, and analyses were performed with the ABI PRISM 7700 sequence detection system (Applied Biosystems, Courtaboeuf, France).
The sensitivity and linearity of the real-time PCR method for the quantitative assay of HDV RNA were examined. The plasmids containing HDAg coding sequences were used as standards to quantify HDV RNA in sera. Tenfold serial dilutions ranging from 5 to 5 × 106 copies were tested in triplicate, and the mean cycle threshold values were plotted against the copy number to establish a standard curve. The correlation coefficients were repeatedly >0.995, and the slopes ranged from −3.1 to −3.4. Dilutions corresponding to inputs of 5 copies per reaction were repeatedly detected. Thus, based on the dilution factors used during the RNA extraction and RT procedures, the sensitivity of the assay to detect HDV RNA in clinical samples was 400 copies/ml of serum and the linearity of quantification ranged from 2 × 103 to 2 × 109 copies/ml.
Statistical analysis.
All statistical analyses were performed with SPSS 17.0 for Windows (SPSS, Inc., Chicago, IL). Student's t test was performed to compare continuous variables. A two-tailed P value of <0.05 was considered statistically significant.
RESULTS
Replacement of dominant species of HDV in CHD.
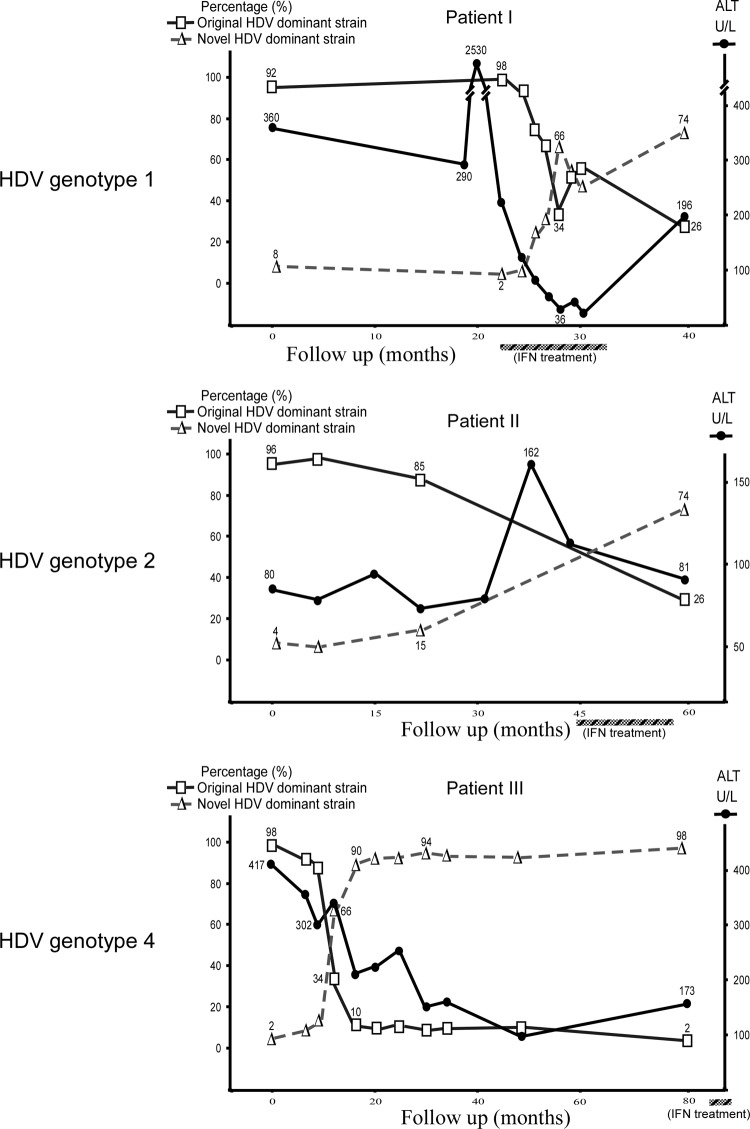
To determine the selection of quasispecies in patients, large-scale screening of at least 100 colonies was carried out by using variant-specific RFLP analysis. Percentages of the original HDV dominant species of genotypes 1, 2, or 4 in the early stage of the disease course were very high (about 90 to 95%) (Fig. 1). Following marked elevations of ALT, the percentages of the original HDV dominant species decreased and they finally became minor ones. In contrast, the percentages of the novel HDV dominant species increased from preexisting minor variants in the original quasispecies after hepatitis flares.
Fig 1.
Replacement of the original dominant species by novel dominant species during the disease courses of three patients chronically infected with different genotypes of HDV. At least 100 HDV colonies were randomly selected by large-scale screening carried out using quasispecies-specific RFLP analysis at each time point for each case. The original dominant HDV strain and the novel dominant strain are indicated by the symbols □ and △, respectively. The values on each curve are relative percentages of the original and novel dominant HDV species. The percentages of the novel dominant species gradually increased as the percentages of the original dominant species decreased after ALT elevations (U/L, units per liter). The periods of IFN-α treatment are marked by the hatched bars below the graphs.
Comparison of viral replication and HDAg expression levels of the original and novel dominant HDV species.
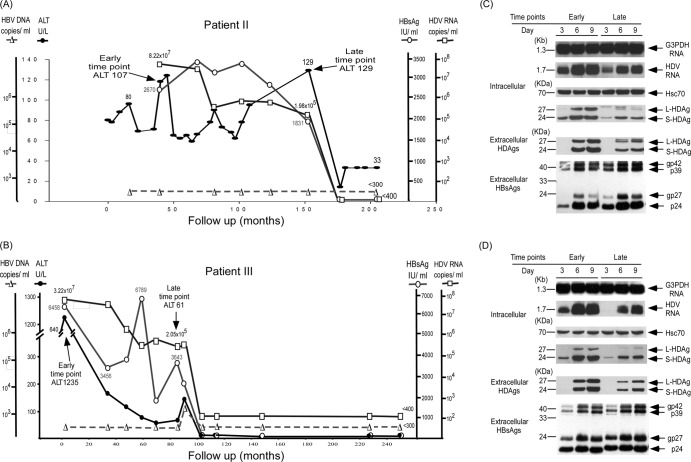
To clarify if the novel dominant species emerged because of a growth advantage, seven pairs of original and novel dominant quasispecies separated by hepatitis flares were cloned from seven patients (labeled I to VII) and cotransfected with an HBV-producing plasmid (a genotype B or C HBV-producing plasmid according to the patient's clinical data) into the Huh-7 human hepatoma cell line to evaluate the replication activity of the dominant HDV strains at early and late time points of the clinical courses of different patients. HBV serves as the helper for HDV virion production. Representative patients II and III, who went into biochemical remission in the absence of cirrhosis or HCC, are first illustrated in Fig. 2A and B. These two patients had been treated with short-acting alpha interferon 2b at 5 million IU thrice weekly for 1 year. Their serum ALT levels remained elevated, and they fluctuated after interferon treatment in patient II. Remission did not occur until more than 110 months after treatment was stopped and was unlikely to have been the result of direct effects of treatment (Fig. 2A). In patient III, remission occurred immediately after interferon treatment (Fig. 2B). In patient I, remission occurred 26 months after interferon treatment (Fig. 3A). The intracellular HDV replication, HDAg expression, and secreted HDV virions of the novel dominant HDV strains late in the clinical course were lower than those of the original dominant strains (Fig. 2C and D). The novel dominant HDV species with a lower relocation capacity was detected about 90 months after the cessation of interferon treatment and 24 months before remission in patient II and about 8 months before the start of interferon treatment, about 15 months before remission, in patient III (Fig. 2A and B).
Fig 2.
Correlation of clinical courses with HDV replication and HDAg expression of novel dominant HDV variants after ALT elevation in patients in remission. The clinical courses of patient II (A) and patient III (B) are shown, and arrows mark the time points of blood sampling to detect the original and novel dominant HDV quasispecies. Comparison of intracellular HDV RNA replication, HDAg expression, and secreted HDV virions in culture medium of original and novel dominant HDV variants isolated from patient II (C) and patient III (D) at early and late time points on days 3, 6, and 9 posttransfection with an HBV expression plasmid. The equal loading of RNA and protein samples was assessed by hybridization with a DIG-labeled DNA probe for G3PDH and a monoclonal antibody specific for heat shock protein Hsc70, respectively.
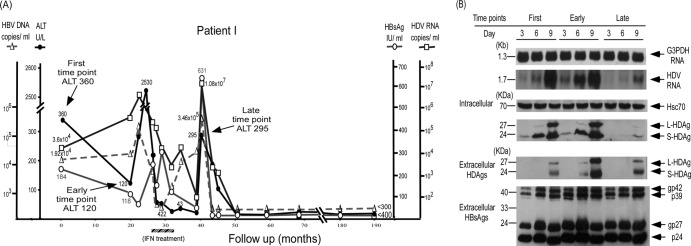
Fig 3.
Correlation of clinical courses with HDV replication and HDAg expression of novel dominant HDV species after elevation of ALT levels in patient I, who went into disease remission. (A) The clinical course of patient I is shown. Arrows mark the time points of blood sampling to detect the dominant HDV species at the first, early, and late time points of the disease course. The period of IFN-α treatment is marked by the hatched bar below the graph. (B) Comparison of intracellular HDV RNA replication, HDAg expression, and secreted HDV virions in culture medium among the dominant HDV species isolated from patient I at different time points on days 3, 6, and 9 posttransfection with an HBV expression plasmid. The equal loading of RNA and protein samples was assessed by hybridization with a DIG-labeled DNA probe for G3PDH and a monoclonal antibody specific for heat shock protein Hsc70, respectively.
Three serum samples obtained from patient I at different time points were available for analysis of dominant HDV species. The dominant HDV species at the first and early time points showed similar replication capacities, and ALT remained elevated and fluctuated after the first time point (Fig. 3A). The HDV species at the late time point showed a much lower replication capacity, as evidenced by intracellular HDV RNA, HDAg, and secreted HDV virion levels lower than those of HDV species at the first and the second time points (Fig. 3B). The novel dominant HDV species with a lower replication capacity was detected about 8 months after interferon treatment and 19 months before remission. Patient IV had not received interferon or any other antiviral treatment. Similar to the findings on patients I, II, and III, a novel dominant HDV species with a lower replication capacity was detected long before disease remission (Table 1).
In order to investigate if there was mutual interference between the original and novel dominant strains, Huh-7 cells were cotransfected with the same amounts of paired HDV-producing plasmids (the original and novel strains from the same patient) and a genotype B or C HBV-producing plasmid, according to the patient's clinical data. The HDV RNA quasispecies were analyzed from HDV particles harvested at day 9 posttransfection. Of the 129 colonies analyzed by quasispecies-specific RFLP patterns, 125 were of the original dominant quasispecies and only 4 were of the novel dominant quasispecies of patient III, indicating that the latter did not have a growth advantage and that the latter did not emerge because of its interference with the former. Similar competition analysis results were observed in patient II.
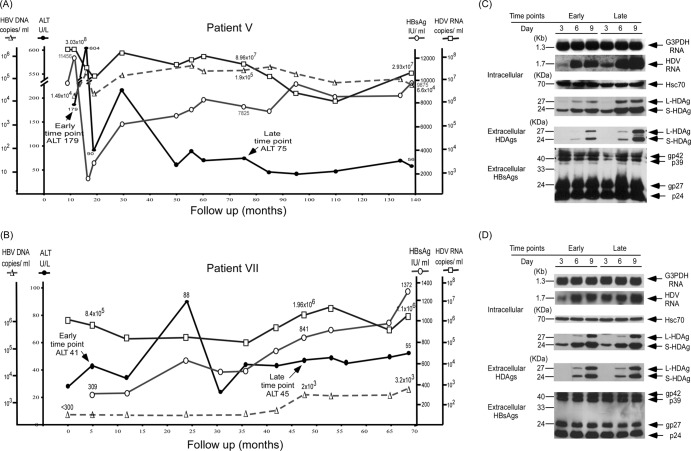
Patients V and VII had representative cases with persistently elevated ALT levels during long-term follow-up (Fig. 4A and B). The intracellular HDV replication, HDAg expression, and secreted HDV virions of the novel dominant HDV species at the post-ALT elevation stage were not decreased compared to those of the original dominant HDV species at the pre-ALT elevation stage (Fig. 4C and D). Competition analysis by cotransfection of the expression plasmids of the original and novel HDV dominant species of patient V showed 37 colonies of the original species and 69 colonies of the novel dominant species in the day 9 culture medium based on quasispecies-specific RFLP patterns. Patients VII, who failed to clear HDV or select HDV species with less active replication at ages older than 58 years, developed cirrhosis and HCC (Fig. 4B; Table 1). In patient VI (Table 1), although the replicative capacity of the novel dominant HDV species at the second time point analyzed was lower than that of the original dominant HDV species, it was still very active, as shown by a high serum viral load of 1,064,000 copies/ml. A high serum viral load of 234,000 copies/ml was still detected 45 months after that. HDV viremia was finally cleared at the age of 63 years by patent VI, who had already developed cirrhosis. Ineffective clearance of HDV at a younger age might account for the development of cirrhosis. However, he finally cleared both HBV and HDV and went into biochemical remission and had clinically inactive cirrhosis.
Fig 4.
Correlation of clinical courses with HDV replication and HDAg expression of novel dominant HDV variants after ALT elevation in patients with persistently elevated ALT levels and adverse outcomes. Shown are the clinical courses of patient V (A) and patient VII (B) and a comparison of intracellular HDV RNA replication, HDAg expression, and secreted HDV virions in culture medium of the original and novel dominant HDV quasispecies isolated from patient V (C) and patient VII (D) at the early and late time points on days 3, 6, and 9 posttransfection.
There was a marked reduction or clearance of serum HBsAg in patients with disease remission, but the clearance of HBsAg levels usually occurred much later than the emergence of novel dominant HDV quasispecies with less active replication. Serum HBsAg remained at high levels in patients with active liver disease and adverse outcomes (cirrhosis or HCC) (Table 1).
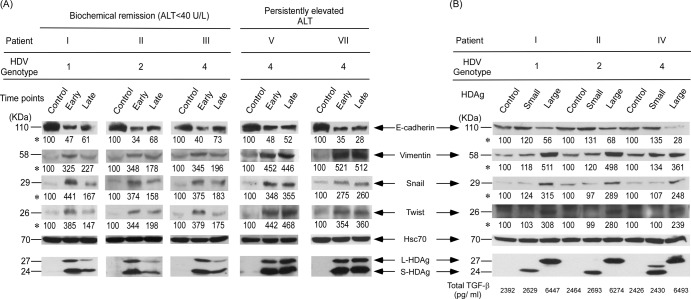
Correlation of HDV replication, assembly, and expression of mesenchymal-cell-specific proteins with disease outcomes.
To determine whether selection of a novel dominant HDV strain may result in different EMT activity of infected hepatocytes in CHD patients, the expression plasmids of the original and novel dominant HDV strains were transfected into the Huh-7 human hepatoma cell line. In the three patients (I, II, and III) infected with different genotypes of HDV who went into disease remission during follow-up, stronger expression (1.5- to 2.7-fold) of the mesenchymal-cell-specific protein vimentin and the EMT transcription factors twist and snail was found in cells transfected with the original dominant HDV strain expression plasmids than in cells transfected with the novel dominant HDV quasispecies (Fig. 5A, left side). In contrast, the epithelial-cell-specific protein E-cadherin was increased (1.2 to 2 times) in cells transfected with the novel dominant HDV expression plasmids (Fig. 5A, left side) with lower replication and assembly efficiency, as shown above.
Fig 5.
Expression profiles of EMT markers determined by Western blotting. (A) EMT factor protein expression in Huh-7 cells transfected with the empty vector (pcDNA3.1) or the original or novel dominant HDV expression plasmid. (B) S-HDAg- or L-HDAg-expressing plasmids. Quantification of TGF-β1 was done with supernatants incubated under 2% FBS collected at the end of day 3. The basal level of TGF-β1 in culture medium without incubation with Huh-7 cells was 210 pg/ml. Intensities of bands shown for each EMT marker panel are indicated by asterisks. Statistical significance, P < 0.05 compared with the control, was determined by the two-tailed Student t test.
However, the amounts of vimentin, twist, and snail remained high and were not significantly different between the cells transfected with the original and the novel dominant HDV expression plasmids in patients V and VII with persistently elevated ALT levels during long-term follow-up (Fig. 5A, right side). These results suggest that HDV with high replication and assembly efficiency tended to induce higher EMT activity and might contribute to the development of fibrosis or cirrhosis in CHD patients during long-term follow-up.
L-HDAg, but not S-HDAg, played a major role in inducing EMT.
To differentiate the roles of L-HDAg and S-HDAg in EMT, Huh-7 cells were transfected with three different genotypes of L-HDAg and S-HDAg expression plasmids. The intracellular vimentin, twist, and snail expression levels were markedly increased at 2.4- to 5.1-fold over the control levels when the L-HDAg expression plasmids of genotypes 1, 2, and 4 were transfected into Huh-7 cells (Fig. 5B). In addition, the corresponding E-cadherin expression level in cell lysates was decreased to 68 and 28% of the control level, respectively. However, the intracellular vimentin, twist, snail, and E-cadherin levels were not significantly altered compared to those of the controls when the S-HDAg expression plasmid was transfected into Huh-7 cells. The amounts of TGF-β in the culture medium of Huh-7 cells transfected with an L-HDAg expression plasmid was nearly 3-fold higher than those in the culture medium of cells transfected with an S-HDAg expression plasmid or vectors. The experiments were done in triplicate, and the amount of TGF-β in the culture medium of transfected cells expressing L-HDAg was significantly higher than that in the medium of transfected cells expressing S-HDAg or a negative control (P < 0.05).
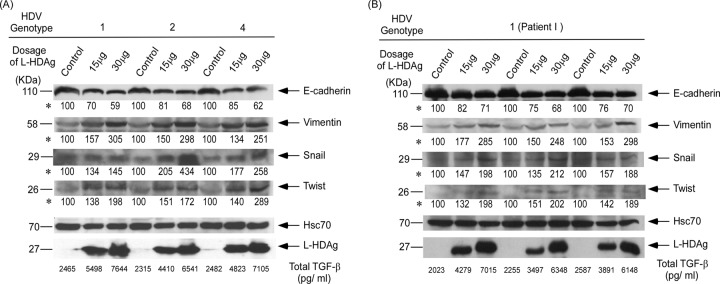
To determine if a dose-dependent response of EMT activity was induced by L-HDAg, different doses of plasmids encoding L-HDAg of genotype 1, 2, or 4 were transfected into Huh-7 cells. When the amount of L-HDAg expression plasmid was increased (from 15 to 30 μg), the expression of vimentin, twist, and snail was also increased (Fig. 6A). However, the expression of E-cadherin was downregulated when the amount of L-HDAg expression plasmid was increased. The experiments were done in triplicate, and the results showed that there was a significant dose-dependent response of TGF-β secretion by transfected cells expressing L-HDAg (P < 0.05). The changes in EMT markers also showed consistent results (P < 0.05) (Fig. 6B).
Fig 6.
Expression levels of induced EMT markers at different doses of L-HDAg-expressing plasmids. Snail, twist, E-cadherin, and vimentin levels in Huh-7 cells transfected with the empty vector (pcDNA3.1) and different doses of L-HDAg-expressing plasmids from three different HDV genotypes (A) and triplicates of the same genotype (B) were analyzed by Western blotting. Heat shock protein Hsc70 was used as a loading control in each lane for the quantification of TGF-β1 in the supernatants collected after 3 days of incubation under 2% FBS. The basal level of TGF-β1 in culture medium without incubation with Huh-7 cells was 210 pg/ml. Intensities of bands shown for each EMT marker panel are indicated by asterisks. Statistical significance, P < 0.05 compared with control, was determined by the two-tailed Student t test.
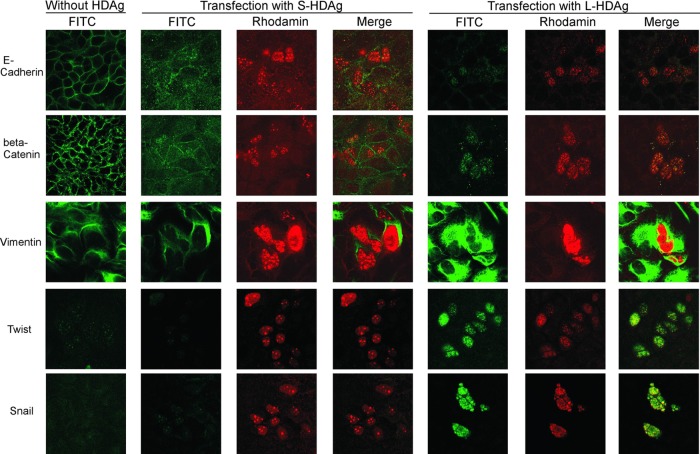
The indirect double immunofluorescence staining of HDAgs, epithelial markers, and mesenchymal markers was performed at day 3 posttransfection. L-HDAg and S-HDAg are shown in nuclei with rhodamine staining in red, while epithelial and mesenchymal markers are shown in the cell membrane, cytoplasm, or nuclei with FITC staining in green (Fig. 7). The right panel of Fig. 7 shows a significant increase in the intensity of the green fluorescence of vimentin, twist, and snail in cells transfected with the L-HDAg expression plasmid, compared to that in cells transfected with the S-HDAg expression plasmid (middle panel) or controls (left panel). In contrast, the immunostaining of membranous E-cadherin was largely absent in the well containing cells transfected with the L-HDAg expression plasmid or in the cells without coexpression of L-HDAg. Huh-7 cells transfected with L-HDAg appeared to secrete larger amounts of TGF-β than cells transfected with S-HDAg or controls (Fig. 5 and 6). TGF-β may induce EMT activity in neighboring cells without the expression of L-HDAg via paracrine effects.
Fig 7.
Immunofluorescence staining of expressed EMT markers in Huh-7 cells transfected with large or small HDAg expression plasmids. S-HDAg and L-HDAg expression plasmids were transfected to Huh-7 cells (middle and right panels, respectively). The cells were doubly stained with human anti-HDV antibody and mouse or rabbit anti-EMT markers (E-cadherin, β-catenin, vimentin, twist, snail) after 3 days of transfection. Rhodamine-conjugated rabbit anti-human IgG, FITC-conjugated sheep anti-mouse IgG, or FITC-conjugated goat anti-rabbit IgG secondary antibodies were used to show the HDAg and EMT markers in red and green immunofluorescence stainings. Photographs (original magnification, ×630) were taken with a confocal fluorescence microscope (Axiovert 200M; Zeiss).
β-Catenin, a central component of the cadherin cell adhesion complex and the Wnt/β-catenin signaling pathway, played important roles during HCC genesis and development. Interestingly, the β-catenin immunoreactivity signal was localized in the nuclei of cells transfected with the L-HDAg expression plasmid. There were no significant differences in epithelial or mesenchymal markers between cells transfected with the S-HDAg expression plasmid or controls (left and middle panels). Together, these results suggested that L-HDAg, but not S-HDAg, played a major role in inducing EMT to contribute to liver fibrosis.
DISCUSSION
This study produced four novel findings. First, quasispecies that are detected in minor proportions prior to a hepatitis flare become dominant after a flare. Second, the replacement by the dominant HDV quasispecies was not due to a growth advantages of the latter or viral interference. Third, the emergence of novel dominant HDV strains with less efficient replication and assembly may be associated with disease remission and vice versa. Fourth, L-HDAg, but not S-HDAg, plays a key role in inducing EMT in the liver and may contribute to the development of fibrosis.
The findings of this study on patients who went into disease remission indicate that the emergence of novel dominant clones with lower replication efficiency is apparently not due to a growth advantage. Because the emergence of novel dominant HDV strains after a marked elevation of ALT levels may represent host immune attacks on virus-containing hepatocytes, it is reasonable to assume that they are positively selected after immune clearance of the original dominant strains, as reported previously (15, 31). Either interferon-induced or spontaneous remission is associated with the emergence of novel dominant HDV species with a lower replication capacity and EMT-inducing activity. Therefore, a novel hypothesis is derived that positive selection of less efficiently replicating HDV by the host may predict disease remission several years earlier than final clearance of HDV. The final eradication of HDV may be a continuous process of selection of species with a lower replication capacity from quasispecies. In contrast, if the host fails to negatively select HDV quasispecies with active replicating and EMT-inducing activity, it may result in persistent hepatic inflammation and unfavorable outcomes, such as liver cirrhosis, liver failure, or HCC. Eight HDV genotypes have been described, and only HDV genotypes 1, 2 and 4 are found in Taiwan (19, 35). In this study, the above-mentioned hypothesis was verified in patients infected with genotype 1, 2, or 4 HDV. Therefore, the correlation of a less efficiently replicating HDV and less EMT-inducing HDV with disease amelioration appears to be independent of the genotype. This would have to be confirmed for genotypes 3, 5, 6, 7, and 8.
However, the timing of positive selection of a less efficiently replicating HDV quasispecies is critical. If the selection occurs in old age, patients may have already developed cirrhosis or HCC before the final clearance of HDV, which is consistent with a previous report that age is a significant factor associated with outcome (28). In addition to the selection of HDV quasispecies contributing to outcome (28, 32), host factors related to immune selection and viral clearance, HBV replication status, and HBsAg expression may also play important roles in determining outcomes of CHD.
According to previous studies, the genetic variations in HDV at different time points before and after serum ALT elevation are shaped and selected by host immunity (31). Comparison of the HDAg domains of original and novel dominant strains shows that there are changes in amino acids of B- or T-cell epitopes that overlap functional domains critical for HDV replication (data not shown). The alteration of functional domains may thus negatively affect HDV replication. Lower replication and assembly efficiencies of novel dominant strains are expected to provoke a less intense immune response from the host, which may subsequently lead to disease amelioration. Further substitution and mutagenesis experiments with original and novel dominant strains will be carried out in the future to determine which functional domains of HDV replication are also the hot spot targets of immune attacks. The information obtained may be critical for the development of therapeutic vaccines.
Recently, it has been reported that an important source of myofibroblasts is hepatocytes that differentiate into myofibroblasts by EMT and subsequently contribute to the development of liver fibrosis (38). Interestingly, the results of this study are the first to indicate that novel dominant HDV strains with higher replication activity correlate with higher EMT expression and vice versa. Several studies have demonstrated that TGF-β is a master regulator of EMT through Smad2/3 complex activation and stimulates hepatic stellate cell activation and collagen production (13, 38). A recent study indicates that L-HDAg, but not S-HDAg, potentiates TGF-β- and c-Jun-induced signal activation and that the isoprenylation of L-HDAg plays a major role in signaling (6). However, the correlation between L-HDAg expression and disease outcomes has not been further investigated. This study demonstrates for the first time that L-HDAg, but not S-HDAg, plays a major role in activating TGF-β and EMT of hepatocytes and may be associated with the progression to cirrhosis and HCC. HSCs are the main source of fibrogenic cells in the liver by TGF-β1 activation and stimulation of the extracellular matrix synthesis (1). The findings in this study suggest the possibility of stimulation of stellate cells by TGF-β induced by hepatocytes expressing L-HDAg. Furthermore, the dosage experiments strengthen the correlation of L-HDAg expression and EMT activity. In a previous study, the amounts of HBsAg expression correlated with the amounts of secreted HDV virions (26). Of note, this study further indicates that the amount of HBsAg not only correlates with secreted HDV virions but also links to disease outcomes.
In summary, CHD may be ameliorated if HDV RNA quasispecies with low replication efficiency are positively selected and become novel dominant quasispecies provoking a less intense immune response and less potent EMT activity. Clearance or reduction of HBsAg may further contribute to disease amelioration by reducing HBV and HDV packaging. On the other hand, if the original dominant strains cannot be negatively selected by immune attacks, or if the positively selected novel HDV dominant variants retain a high level of replication, it may provoke intense hepatic inflammation and strong EMT activity that finally leads to cirrhosis and HCC. The adverse outcomes may ensue with or without the presence of active HBV replication (25, 26, 27, 28, 34). The findings of this study suggest that the suppression of L-HDAg expression, e.g., by a prenylation inhibitor, or the inhibition of EMT activity in addition to inhibition of HBV and HDV replication may be important to control the progression of CHD.
ACKNOWLEDGMENTS
We are grateful to Sheng-Chung Lee for kindly providing the monoclonal antibody (A10F1) against HBsAg and to Michael M. Lai for his instructive comments and English editing of the manuscript. We also acknowledge the technical support of the Sequencing Core, National Yang-Ming University Genome Research Center (YMGC).
The Sequencing Core Facility is supported by the National Research Program for Genome Medicine (NRPGM), National Science Council of Taiwan. This study was supported by grants from the National Science Council (NSC 97-2628-B-010-008-MY3, NSC 99-3112-B-010-007, and NSC 100-2314-B-056-MY3), in part by the Center of Excellence for Cancer Research at the Taipei Veterans General Hospital (DOH100-TD-C-111-007), and by a grant from Yang-Ming University (99A-C-T501, Ministry of Education, Aim for the Top University Plan).
Footnotes
Published ahead of print 6 June 2012
REFERENCES
- 1. Bataller R, Brenner DA. 2005. Liver fibrosis. J. Clin. Invest. 115:209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casey JL, Gerin JL. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 69:7593–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castilla A, Prieto J, Fausto N. 1991. Transforming growth factors beta 1 and alpha in chronic liver disease. Effects of interferon alpha therapy. N. Engl. J. Med. 324:933–940 [DOI] [PubMed] [Google Scholar]
- 4. Chao M, Hsieh SY, Taylor J. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen PJ, et al. 1990. Continuous expression and replication of the hepatitis δ virus genome in Hep G2 hepatoblastoma cells transfected with cloned viral DNA. Proc. Natl. Acad. Sci. U. S. A. 87:5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi SH, Jeong SH, Hwang SB. 2007. Large hepatitis delta antigen modulates transforming growth factor-beta signaling cascades: implication of hepatitis delta virus-induced liver fibrosis. Gastroenterology 132:343–357 [DOI] [PubMed] [Google Scholar]
- 7. Choi SS, Diehl AM. 2009. Epithelial-to-mesenchymal transitions in the liver. Hepatology 50:2007–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Domingo E, et al. 1985. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene 40:1–8 [DOI] [PubMed] [Google Scholar]
- 9. Govindarajan S, Chin KP, Redeker AG, Peters RL. 1984. Fulminant B viral hepatitis: role of delta agent. Gastroenterology 86:1417–1420 [PubMed] [Google Scholar]
- 10. Govindarajan S, De Cock KM, Redeker AG. 1986. Natural course of delta superinfection in chronic hepatitis B virus-infected patients: histopathologic study with multiple liver biopsies. Hepatology 6:640–644 [DOI] [PubMed] [Google Scholar]
- 11. Guilhot S, et al. 1994. Expression of hepatitis delta virus large and small antigens in transgenic mice. J. Virol. 68:1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hadziyannis SJ, Hatzakis A, Paipaioannou C, Anastassakos C, Vassiliadis E. 1987. Endemic hepatitis delta virus infection in a Greek community. Prog. Clin. Biol. Res. 234:181–202 [PubMed] [Google Scholar]
- 13. Hellerbrand C, Stefanovic B, Giordano F, Burchardt ER, Brenner DA. 1999. The role of TGF beta1 in initiating hepatic stellate cell activation in vivo. J. Hepatol. 30:77–87 [DOI] [PubMed] [Google Scholar]
- 14. Hsu SC, et al. 2002. Varied assembly and RNA editing efficiencies between genotypes I and II hepatitis D virus and their implications. Hepatology 35:665–672 [DOI] [PubMed] [Google Scholar]
- 15. Huang YH, Tao MH, Hu CP, Syu WJ, Wu JC. 2004. Identification of novel HLA-A*0201-restricted CD8+ T-cell epitopes on hepatitis delta virus. J. Gen. Virol. 85:3089–3098 [DOI] [PubMed] [Google Scholar]
- 16. Hughes SA, Wedemeyer H, Harrison PM. 2011. Hepatitis delta virus. Lancet 378:73–85 [DOI] [PubMed] [Google Scholar]
- 17. Imazeki F, Omata M, Ohto M. 1990. Heterogeneity and evolution rates of delta virus RNA sequences. J. Virol. 64:5594–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lamouille S, Derynck R. 2007. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 178:437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Gal F, et al. 2006. Eighth major clades for hepatitis delta virus. Emerg. Infect. Dis. 12:1447–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macnaughton TB, Gowans EJ, Jilbert AR, Burrell CJ. 1990. Hepatitis delta virus RNA, protein synthesis and associated cytotoxicity in a stably transfected cell line. Virology 177:692–698 [DOI] [PubMed] [Google Scholar]
- 21. Nisini R, et al. 1997. Human CD4+ T-cell response to hepatitis delta virus: identification of multiple epitopes and characterization of T-helper cytokine profiles. J. Virol. 71:2241–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ottobrelli A, et al. 1991. Patterns of hepatitis delta virus re-infection and disease in liver transplantation. Gastroenterology 101:1649–1655 [DOI] [PubMed] [Google Scholar]
- 23. Rizzetto M, et al. 1980. Transmission of the hepatitis B virus-associated delta antigen to chimpanzees. J. Infect. Dis. 141:590–602 [DOI] [PubMed] [Google Scholar]
- 24. Rizzetto M, et al. 1983. Chronic hepatitis in carriers of hepatitis B surface antigen, with intrahepatic expression of the delta antigen. An active and progressive disease unresponsive to immunosuppressive treatment. Ann. Intern. Med. 98:437–441 [DOI] [PubMed] [Google Scholar]
- 25. Schaper M, et al. 2010. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. J. Hepatol. 52:658–664 [DOI] [PubMed] [Google Scholar]
- 26. Shih HH, et al. 2008. Hepatitis B surface antigen levels and sequences of natural hepatitis B virus variants influence the assembly and secretion of hepatitis D virus. J. Virol. 82:2250–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smedile A, et al. 1991. Hepatitis B virus replication modulates pathogenesis of hepatitis D virus in chronic hepatitis D. Hepatology 13:413–416 [PubMed] [Google Scholar]
- 28. Su CW, et al. 2006. Genotypes and viremia of hepatitis B and D viruses are associated with outcomes of chronic hepatitis D patients. Gastroenterology 130:1625–1635 [DOI] [PubMed] [Google Scholar]
- 29. Taylor JM. 2003. Replication of human hepatitis delta virus: recent developments. Trends Microbiol. 11:185–190 [DOI] [PubMed] [Google Scholar]
- 30. Wang D, Pearlberg J, Liu YT, Ganem D. 2001. Deleterious effects of hepatitis delta virus replication on host cell proliferation. J. Virol. 75:3600–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang SY, et al. 2007. Positive selection of hepatitis delta antigen in chronic hepatitis D patients. J. Virol. 81:4438–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu JC, et al. 1995. Genotyping of hepatitis D virus by restriction-fragment length polymorphism and relation to outcome of hepatitis D. Lancet 346:939–941 [DOI] [PubMed] [Google Scholar]
- 33. Wu JC, et al. 1991. Production of hepatitis D virus and suppression of helper hepatitis B virus in a human hepatoma cell line. J. Virol. 65:1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu JC, et al. 1995. Natural history of hepatitis D viral superinfection: significance of viremia detected by polymerase chain reaction. Gastroenterology 108:796–802 [DOI] [PubMed] [Google Scholar]
- 35. Wu JC, Chiang TY, Sheen IJ. 1998. Characterization and phylogenetic analysis of a novel hepatitis D virus strain discovered by restriction fragment length polymorphism analysis. J. Gen. Virol. 79:1105–1113 [DOI] [PubMed] [Google Scholar]
- 36. Wu JC, et al. 1999. Recombination of hepatitis D virus RNA sequences and its implications. Mol. Biol. Evol. 16:1622–1632 [DOI] [PubMed] [Google Scholar]
- 37. Yang MH, et al. 2009. A comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology 50:1464–1474 [DOI] [PubMed] [Google Scholar]
- 38. Zeisberg M, et al. 2007. Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J. Biol. Chem. 282:23337–23347 [DOI] [PubMed] [Google Scholar]