Abstract
The hemagglutinin protein (HA) of the influenza virus family is a major antigen for protective immunity. Thus, it is a relevant target for developing vaccines. Here, we describe a human CD4+ T cell epitope in the influenza virus HA that lies in the fusion peptide of the HA. This epitope is well conserved in all 16 subtypes of the HA protein of influenza A virus and the HA protein of influenza B virus. By stimulating peripheral blood mononuclear cells (PBMCs) from a healthy adult donor with peptides covering the entire HA protein based on the sequence of A/Japan/305/1957 (H2N2), we generated a T cell line specific to this epitope. This CD4+ T cell line recognizes target cells infected with influenza A virus seasonal H1N1 and H3N2 strains, a reassortant H2N1 strain, the 2009 pandemic H1N1 strain, and influenza B virus in cytotoxicity assays and intracellular-cytokine-staining assays. It also lysed target cells infected with avian H5N1 virus. We screened healthy adult PBMCs for T cell responses specific to this epitope and found individuals who had ex vivo gamma interferon (IFN-γ) responses to the peptide epitope in enzyme-linked immunospot (ELISPOT) assays. Almost all donors who responded to the epitope had the HLA-DRB1*09 allele, a relatively common HLA allele. Although natural infection or standard vaccination may not induce strong T and B cell responses to this highly conserved epitope in the fusion peptide, it may be possible to develop a vaccination strategy to induce these CD4+ T cells, which are cross-reactive to both influenza A and B viruses.
INTRODUCTION
Influenza remains an important infectious respiratory disease, causing significant morbidity and mortality around the world every year. Influenza A virus is the major type of influenza virus that causes disease in humans, while influenza B virus also causes disease in humans, although to a less severe extent (51). Influenza A virus undergoes frequent antigenic drifts at antibody-combining sites on hemagglutinin (HA) and occasional shifts due to the emergence of currently circulating strains with novel genes reassorted from nonhuman virus strains into human viruses. These antigenic drifts/shifts make it extremely challenging to design vaccines that can protect against emerging antigenically variant influenza viruses. The annual seasonal influenza vaccine requires accurate prediction of influenza virus strains that will circulate in the coming season, and this relies on viral surveillance data (24). When the vaccine strains do not match the actual circulating strains for a given influenza season, the immunity generated by these vaccines is not optimal. To measure vaccine immunogenicity, the hemagglutination inhibition antibody titer in nature or after vaccination is a correlate of protection and a determinant of vaccine efficacy. In addition, both CD8+ and CD4+ T cells to multiple viral proteins, including HA, also contribute to the immune responses to influenza virus (76).
CD4+ T cells are not essential in providing protective immunity in mouse models of influenza infection when both CD8+ T cells and B cells are present (reviewed in reference 9). However, they play important roles in the immune response to influenza virus by maintaining the CD8+ T cell cytotoxic responses and the transition to memory phase (6) and by providing help to antibody-producing B cells (31). Human CD4+ T cell responses to influenza virus are not well understood. A recent study by Wilkinson and colleagues demonstrated that memory influenza virus-specific CD4+ T cells contribute to disease protection in a human influenza virus infection challenge (80). Most of the human CD4+ T cell responses to influenza virus have been mapped to the HA protein (30). In a genome-wide screening of T cell epitopes to the influenza virus proteins that we recently performed, we found that the HA and matrix 1 (M1) proteins contained more CD4+ T cell epitopes than other viral proteins (4). We also found individuals whose T cells responded to the H5 avian HA peptides even though they had not been previously exposed to H5N1 virus. Other groups have also found cross-reactive T cells to H5 HA in individuals who had not been exposed to avian influenza virus (16, 47, 64, 83). These results suggest that cross-reactive CD4+ T cells to the surface glycoprotein HA are generated by infection and/or vaccination. These CD4+ T cells, in turn, have the potential to mediate protection against a different subtype of influenza A virus.
The influenza HA is a major antigenic site of protective immunity. It is also indispensable in the viral life cycle because it is necessary for binding the viral receptor (sialic acid) on target cells and mediating the fusion of viral and cellular membranes (15, 68). HA consists of two subunits, HA1 and HA2, which are products of the enzymatic cleavage of the precursor protein HA0 (51). The majority of the antigenic sites recognized by neutralizing antibodies and subject to antigenic drift are located in the HA1 subunit. The HA2 subunit, on the other hand, is more conserved among the different HA subtypes. Recently, an in silico study was done to evaluate the presence of cross-reactive T cell epitopes in the HA of seasonal influenza H1N1 and H3N2 viruses and the novel pandemic H1N1 virus (20). Duvvuri and colleagues found that CD4+ T cell epitopes within different H1N1 strains were significantly conserved, while there were fewer conserved epitopes between the H3 and H1 subtypes (20). Although their findings suggest that cross-reactive T cells recognizing an epitope across all the different HA subtypes may be elusive, there are regions in the HA2 subunit, in particular the fusion peptide sequence, that are highly conserved in all the different HA subtypes (15).
In this study, we generated a human CD4+ T cell line that recognizes the fusion peptide sequence of the influenza virus HA. This epitope is well conserved in all 16 types of HA of influenza A virus and the HA of the influenza B virus. This CD4+ T cell line was able to lyse target cells that are infected with influenza A H1N1 virus, including the novel pandemic H1N1 and the H2N1 and H3N2 viruses, as well as an influenza B virus. We tested healthy adult peripheral blood mononuclear cells (PBMCs) against the various HA peptides and found that donors who had the HLA-DRB1*09 allele had ex vivo gamma interferon (IFN-γ) responses to the epitope, suggesting that this is a major histocompatibility complex (MHC) restriction allele for the highly conserved sequence.
MATERIALS AND METHODS
Influenza viruses, peptides, and proteins.
Lyophilized 17-mer peptides spanning the HAs of influenza A H1N1, H3N2, and H5N1 and influenza B viruses were obtained from the National Institutes of Health (NIH) Biodefense and Emerging Infections Research Resources Repository (BEI Resources, Manassas, VA) and were previously described (4). Peptides spanning the HA of influenza virus A/Japan/305/1957 (H2N2) were designed to correspond to BEI Resources' H5 HA peptides and were synthesized by AnaSpec, Inc. (San Jose, CA). The peptides were reconstituted by dissolving them in 100% dimethyl sulfoxide at a stock concentration of 10 mg/ml. Peptide pools were made with 9 or 10 nonoverlapping peptides per pool at a concentration of 1.1 or 1.0 mg/ml per peptide. Recombinant HA proteins of A/Singapore/1/1957 (H2N2) and A/Canada/RV444/04 (H7N3) were obtained from BEI Resources. Recombinant HA proteins from A/New Caledonia/20/1999 (H1N1), A/Wisconsin/67/2005 (H3N2), and A/Vietnam 1203/2004 (H5N1) (H1, H3, and H5) were obtained from Protein Biosciences (Meriden, CT). For live-virus infections, the following influenza virus strains were used: A/New Caledonia/20/1999 (H1N1), A/Wisconsin/67/2005 (H3N2), X-27 (a reassortant of A/Rockefeller Institute/5/57 [HA] × A/NWS/34 [NA], subtype H2N1), A/Hong Kong/483/1997 (H5N1), A/California/7/2009 NYMC (H1N1), and B/Malaysia/2506/2004. The H1N1 and H3N2 seasonal virus strains and the influenza B virus were a gift from Michel DeWilde and Robert Ryall of Sanofi Pasteur. The H5N1 strain was provided by Nancy Cox from the World Health Organization Influenza Reference Laboratory at the Centers for Disease Control and Prevention (CDC), and the reassortant H2N1 strain was obtained from BEI Resources. The 2009 pandemic H1N1 strain was provided by Alexander Klimov of the CDC.
All experiments using the live H5N1 strain were performed in a biosafety level 3 (BSL3) laboratory of the University of Massachusetts Medical School according to enhanced BSL3 guidelines (23a).
Study subjects.
Blood samples were obtained from healthy adult donors, and PBMCs were purified by Ficoll-Hypaque density gradient centrifugation as previously described. To determine the HLA class II typing of these donors, DNA from either PBMCs or autologous B lymphoblastoid cell lines (BLCLs) were extracted using the Puregene DNA purification kit (Gentra Systems, Minneapolis, MN). For donors YD10, YD12, and YD13, high-resolution HLA class II typing was performed using the Applied Biosystems (Foster City, CA) high-resolution typing system. HLA class II typing for the rest of the donors described in this paper was done using the Olerup (Stockholm, Sweden) HLA-DR SSP kit.
IFN-γ ELISPOT.
Enzyme-linked immunospot (ELISPOT) assays were performed as previously described (4). Briefly, cryopreserved PBMCs (2 × 105 to 2.5 × 105 cells per well) were seeded onto polyvinylidene difluoride membrane 96-well plates (Millipore, Bedford, MA) precoated with 5 μg/ml anti-IFN-γ monoclonal antibody (clone D1K; Mabtech, Cincinnati, OH) in the presence or absence of peptide or peptide pools. Phytohemagglutinin (Sigma-Aldrich, St. Louis, MO; 1:100) was used as a positive control. After 18 to 24 h of incubation, the cells were removed by washing with phosphate-buffered saline (PBS) plus 0.05% Tween 20. Secondary biotinylated anti-IFN-γ monoclonal antibody (clone 7-B6-1; Mabtech) was added at 2 μg/ml and the plates were incubated for 2 h at room temperature. The plates were washed again, and IFN-γ was detected with avidin-peroxidase (3420-2H; Mabtech) and a substrate kit (NovaRed; Vector Laboratories, Burlingame, CA). The frequency of IFN-γ-producing cells was determined by using the ImmunoSpot S4 Pro Analyzer and the ImmunoSpot Academic V.4 Software (Cellular Technologies Ltd., Shaker Heights, OH). Experiments were performed in triplicate wells.
To determine the phenotype of IFN-γ-producing cells in ELISPOT, we depleted PBMCs of CD4+ or CD8+ cells as previously described (4). Briefly, negative selection was done by using anti-CD4 or anti-CD8 antibody-coated magnetic beads from the MACS cell purification system (Miltenyi Biotec, Bergisch Gladbach, Germany), which resulted in about 80 to 85% purity. The selected cells were then processed according to the manufacturer's protocol and were used in IFN-γ ELISPOT.
Generation of T cell lines.
Influenza A virus-specific T cell lines were established using a limiting-dilution assay, as previously described (4). Briefly, PBMCs that were previously stimulated with the peptide(s) of interest in culture for at least 14 days were plated at a concentration of 1, 3, 10, or 30 cells per well in 96-well round-bottom microtiter plates in 50 μl of AIM-V medium supplemented with 10% fetal bovine serum (FBS) (AIM-V-10), 50 U interleukin 2 (IL-2), and a 1:1,000 dilution of anti-CD3 monoclonal antibody 12F6 (a gift from Johnson Wong) and 1 × 105 irradiated (3,500 rads) allogeneic PBMCs/well. On day 7, 50 μl of fresh AIM-V-10 and 50 U IL-2 were added, and on day 14, fresh AIM-V-10 and IL-2, 1 × 105 irradiated allogeneic PBMCs/well, and a 1:1,000 dilution of the anti-CD3 12F6 were added. The cells were assayed for cytolytic activity using 51Cr assays between days 21 and 28. Cells from wells with influenza A virus peptide-specific cytolytic activity (specific killing of 10% and above at a peptide concentration of 10 μg/ml) were expanded to 48-well plates.
Preparation of APCs for 51Cr release assays and ICS.
Autologous BLCLs were established by taking donor PBMCs and culturing them with Epstein-Barr virus, as previously described (34). The BLCLs were either infected with influenza virus or pulsed with peptide (4). Briefly, virus-infected target BLCLs were incubated for 18 h at 37°C and radiolabeled for 51Cr release assays or were used as antigen-presenting cells (APCs) in intracellular-cytokine staining (ICS). In experiments using inactivated virus, influenza virus was heat inactivated in a water bath at 60°C for 30 min (42) prior to incubation with autologous BLCLs. For peptide-pulsed targets, 10 μg/ml of peptide was added to autologous BLCL suspensions as described previously (4), unless otherwise indicated.
51Cr release assay.
T cell lines or bulk culture effector cells were added to 1.5 × 103 51Cr-labeled target cells at an effector-to-target (E:T) ratio of 10 unless otherwise indicated. After incubation for 4 to 6 h at 37°C, supernatants were harvested (Skatron Instruments, Sterling, VA), and percent specific immune lysis was calculated as follows: [(experimental release − spontaneous release)/(maximum release − spontaneous release)] × 100. Spontaneous lysis—given by the following formula: (spontaneous release/maximum release) × 100—was <30% in all assays. Peptide-pulsed target cells tended to have lower spontaneous lysis than virus-infected target cells. All experiments were performed in triplicate wells. Uninfected and non-peptide-pulsed target cells were used as negative controls.
ICS.
A T cell line derived from a limiting-dilution assay was used as the effector cells in ICS. The cells were washed and resuspended at 5 × 105 cells in RPMI 1640 medium supplemented with 10% FBS (RPMI 10). Autologous BLCLs were used as APCs at an E:T ratio of 10 and were added to the effector cells. They were incubated for 1 h at 37°C in a 5% CO2 incubator, followed by an additional 5 h in the presence of Golgi plug (BD Biosciences, San Jose, CA). The cells were then washed with fluorescence-activated cell sorter (FACS) buffer (2% FBS and 0.1% sodium azide in PBS) and stained using the Live/Dead aqua fixable dead cell stain kit (Invitrogen, Eugene, OR) to identify live and dead cells. The cells were then stained for surface markers, such as CD3–allophycocyanin (APC)-Cy7, CD8–peridinin chlorophyll protein (PerCP)-CY5.5, CD19/CD14-phycoerythrin (PE)-Cy7 (BD Biosciences, San Jose, CA), or CD4-Pacific Blue (eBiosciences, San Diego, CA) for 30 min at 4°C. After washing with FACS buffer, the cells were fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences, San Jose, CA) and stained for the intracellular cytokine IFN-γ (Alexa 700 conjugate; BD Biosciences) or tumor necrosis factor alpha (TNF-α) (APC conjugate; BD Biosciences, San Jose, CA) for 30 min at 4°C. The cells were then washed with Permwash buffer (BD Biosciences, San Jose, CA) and resuspended in 1× BD Stabilizing Fixative (BD Biosciences, San Jose, CA) for flow cytometric analysis. Multiparameter flow cytometric analyses were performed using the Aria flow cytometer. The number of events collected per experiment varied from 100,000 to 300,000. List mode data files were analyzed using FlowJo (version 6.3; TreeStar 6, Inc., Ashland, CA). Graphs were plotted as dot plots of TNF-α- versus IFN-γ-producing T cells in the gated live CD3+ CD4+ cell population.
Peptide binding assay.
A fluorescence polarization assay was used to determine the binding affinities of HA peptides containing the fusion peptide sequence to the HLA-DR1 molecule. The HA306-318 peptide probe (acetyl [Ac]-PRFVKQNTLRLAT) was synthesized (21st Century Biochemicals, Marlboro, MA) and labeled with Alexa 488-tetrafluorophenyl ester (Invitrogen, Eugene, OR). Soluble recombinant HLA-DR1was prepared as previously described (26). Peptide-free HLA-DR1 (100 nM) was mixed together with the Alexa 488-HA peptide probe (25 nM) and various concentrations of unlabeled competitor peptide (from 0.08 to 20 μM). The reactions were done in triplicate wells for each competitor peptide concentration in a 96-well format and incubated for 3 days at 37°C in binding buffer (pH 5.5) containing protease inhibitors and 0.5 mg/ml octylglucoside. Fluorescence was detected using the Alexa 488-fluorescence polarization measurement protocol of the Perkin Elmer 2030 Explorer multilabel plate reader. Fifty percent inhibitory concentrations (IC50s) were obtained by fitting a competition curve of percent inhibition of peptide probe versus the logarithmic value of the concentration of competitor peptide using GraphPad Prism version 5.04.
RESULTS
Donors who were not exposed to the H2N2 virus have T cell responses to H2 HA.
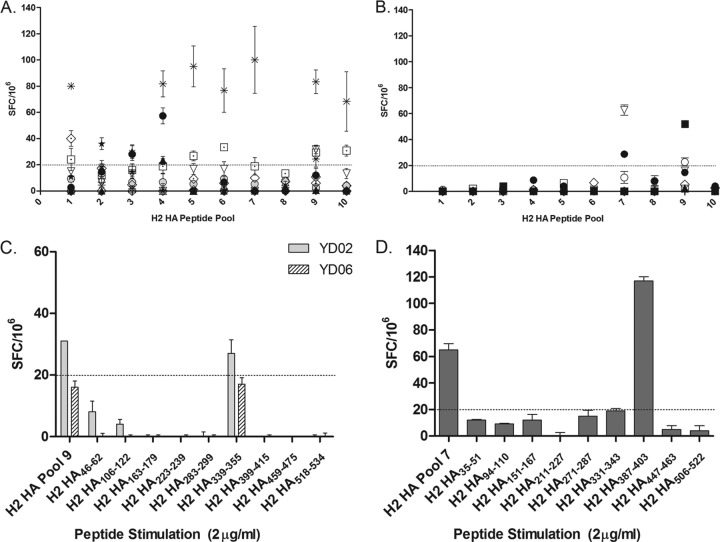
We were originally interested in looking at T cell memory responses to the H2 HA in individuals who were previously exposed to the H2N2 virus, i.e., those who were alive at the time of the 1957 pandemic and during the years H2N2 viruses circulated up to the emergence of H3N2 in 1968. Our group (4) and others (16, 64) previously found individuals who had H5 HA T cell responses: therefore, we hypothesized that prior immunity to H2 HA may contribute to this cross-reactive T cell response because H2 and H5 HA sequences are the closest based on their phylogeny (25). IFN-γ ELISPOT assays were done to quantify the H2 HA memory T cells using H2 HA peptide pools to be able to compare the responses of older (born before 1957) and younger (born after 1968) donors. We expected to detect responses to H2 HA in the PBMCs of the older group, but not in the younger donors. Indeed, 7 of the 11 older individuals' T cells had IFN-γ responses to at least one peptide pool (Fig. 1A), although for most donors, the spot-forming-cell (SFC) values are modestly above our cutoff for a positive IFN-γ response (20 SFCs/106, 3 standard deviations of the negative-control wells [PBMCs without peptides], which we previously set for our ELISPOT assays [4]). To our surprise, younger donors' PBMCs had responses to H2 HA peptide pools, as well (Fig. 1B, pool 7 and pool 9). When we tested the individual peptides in these pools, the PBMCs of donor YD02 had IFN-γ responses to H2 HA339-355 (Fig. 1C). The PBMCs of donor YD06 also had similar responses, although the numbers of spots were below the cutoff for both pool 9 and the H2 HA339-355 peptide. The PBMCs of donor YD04 had responses to H2 HA387-403 (Fig. 1D).
Fig 1.
IFN-γ responses of various donor PBMCs to H2 HA peptide pools and individual peptides. (A and B) Older (A) and younger (B) donor PBMCs were tested in ELISPOT against H2 HA peptide pools to quantify and compare the IFN-γ responses to H2 HA. Each peptide pool contained 9 or 10 nonoverlapping peptides spanning the H2 HA, and each point represents an individual donor. Due to the limited number of PBMCs collected from each donor, the ELISPOT peptide pool screen was done only once, with each pool tested in triplicate wells. Donor PBMCs that had responses to the peptide pools were screened against individual peptides contained in those pools. The peptide pools and individual peptides were tested in three replicate wells with 200,000 to 250,000 cells per well. (C and D) YD02 and YD06 were screened against the individual peptides of H2 HA pool 9, while YD04 was screened against the individual peptides of H2 HA pool 7. Values are given as SFCs/106, which is the mean number of spots from triplicate wells. The data presented are adjusted for background spots given by the wells that had only PBMCs and medium. The error bars indicate the standard errors of the mean of triplicate wells. All donors responded to the positive control, PHA (mean = 2,246.8 SFCs/106). Negative-control wells had mean values of 3.5 SFCs/106 for the older donors and 1.7 SFCs/106 for the younger donors. The dotted lines indicate the cutoff SFC value we established previously. The final concentration of peptide used for all experiments was 2 μg/ml per peptide.
The IFN-γ response to the H2 HA339-355 peptide is well conserved in other HA subtypes and is mediated by CD4+ cells.
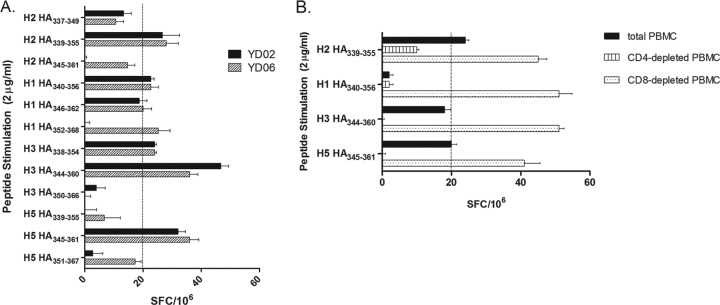
Donors YD02 and YD06 were born in 1977 and 1981, respectively, well after 1968, when H2N2 was last detected in the human population. We thought that the IFN-γ responses in the PBMCs of these donors might be due to cross-reactive memory T cell responses generated by exposure to more recently circulating seasonal influenza virus strains of the H1N1 and H3N2 subtypes. We tested the corresponding peptides located in the H1, H3, and H5 HAs and found that the donors' T cells responded to these peptides (Fig. 2A). Peptides that induced IFN-γ production in PBMCs from donor YD02 all contained the RGLFGAIAGFIEGG (RGIFGAIAGFIENG in H3) amino acid sequence (Table 1), which maps to the fusion peptide of the influenza virus HA (15). Although the H3 HA338-354 peptide, which lacks IENG at its C terminus, induced IFN-γ secretion, other peptides without IEGG at their C termini, such as H2 HA337-349 and H5 HA339-355, did not. CD4+ or CD8+ cells were depleted from YD02 PBMCs to identify the cell population that was responding to the peptides. The depleted PBMCs were tested against the peptides in H1, H2, H3, and H5 HAs that contained the entire RG(L/I)FGAIAGFIE(G/N)G amino acid sequence. IFN-γ was produced in the presence of CD4+ cells in the PBMCs, indicating that the peptide is presented by the MHC class II molecule (Fig. 2B).
Fig 2.
IFN-γ responses to corresponding peptide sequences in other A HAs. (A) YD02 and YD06 PBMCs were screened against the corresponding 17-mer peptides in H1, H3, and H5 HAs that included the partial or the entire RGLFGAIAGFIE(G/N)G amino acid sequence in ELISPOT. The data are representative of two independent ELISPOT experiments. (B) YD02 PBMCs were depleted of either CD4- or CD8-expressing cells by negative selection using magnetic beads and were used in ELISPOT assays to determine the phenotype of IFN-γ-responding cells. SFC values for the positive control, PHA, were greater than 2,000 SFCs/106, while negative-control SFCs were between 2 and 3/106. The error bars indicate the standard errors of the mean of triplicate wells. The dashed lines indicate the cutoff SFC value we established previously.
Table 1.
Summary of influenza A/HA subtypes and B/HA peptides containing the conserved RG(L/I)FGAIAGFIE(G/N)G sequence of the fusion peptide and the corresponding responses of donor YD02 T cells in ELISPOT and 51Cr release assays
| Influenza A virus strain | Peptide name | Peptide sequencea | YD02 PBMCs in ELISPOTb | T cell line from YD02 in CTLsc |
|---|---|---|---|---|
| A/Japan/305/1957 (H2N2) | H2 HA337-349 | IESRGLFGAIAGF | − | − |
| H2 HA339-355 | SRGLFGAIAGFIEGGWQ | + | + | |
| H2 HA345-361 | AIAGFIEGGWQGMVDGW | − | − | |
| A/NewCaledonia/20/1999 (H1N1) | H1 HA340-356 | IQSRGLFGAIAGFIEGG | + | − |
| H1 HA346-362 | FGAIAGFIEGGWTGMVD | − | − | |
| H1HA352-368 | FIEGGWTGMVDGWYGYH | − | − | |
| A/New York/384/2005 (H3N2) | H3 HA338-354 | NVPEKQTRGIFGAIAGF | + | ND |
| H3 HA344-360 | TRGIFGAIAGFIENGWE | + | + | |
| H3 HA350-366 | AIAGFIENGWEGMVDGW | − | − | |
| A/Thailand/4(SP-528)/2004 (H5N1) | H5 HA339-355 | RERRRKKRGLFGAIAGF | − | − |
| H5 HA345-361 | KRGLFGAIAGFIEGGWQ | + | + | |
| H5 HA351-367 | AIAGFIEGGWQGMVDGW | − | − | |
| B/Nanchang/12/1998 (Influenza B) | B/HA354-370 | PAKLLKERGLFGAIAGF | − | − |
| B/HA360-376 | ERGLFGAIAGFIEGGWQ | + | + |
The amino acids that comprise the conserved RG(L/I)FGAIAGFIE(G/N)G sequence are underlined.
Summary of ELISPOT results from Fig. 2A, with the exception of the B/HA peptides, which were determined in a separate experiment. A plus sign is defined as an ELISPOT response greater than or equal to 20 SFC/106.
Summary of CTL results from Fig. 3B. A plus sign is defined as greater than or equal to 15% specific lysis in 51Cr release assays. ND, not done.
In vitro-generated CD4+ T cell lines specific to H2 HA339-355 recognize autologous target cells infected with various influenza A and B viruses.
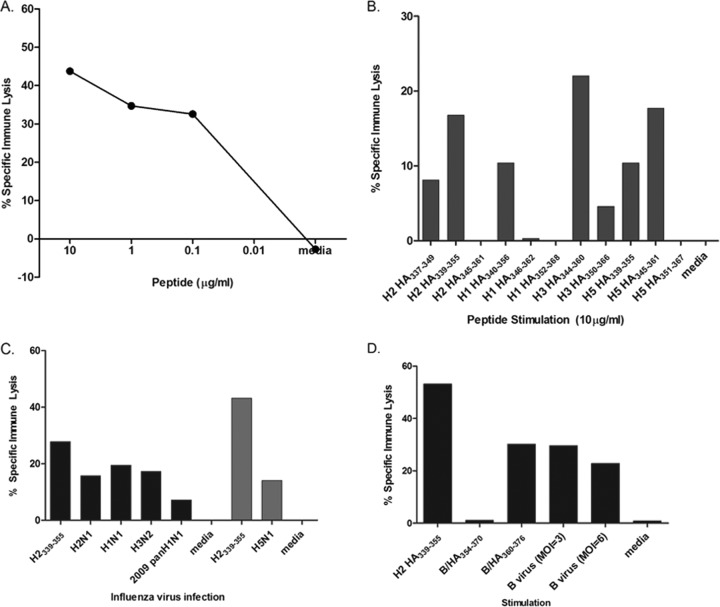
To further characterize the conserved epitope, we generated bulk culture lines by stimulating PBMCs from YD02 with the H2 HA339-355 peptide. A limiting-dilution assay at 1, 3, or 10 cells per well in a 96-well plate was then set up to isolate a T cell line(s) specific to H2 HA339-355. Although cytotoxicity may not be a major function of virus-specific CD4+ T cells, we had previously established both CD8+ and CD4+ T cell lines from a limiting-dilution assay using a standard 51Cr release assay (4, 22, 27, 35, 40, 41, 50). Thus, after establishment, we initially characterized candidate T cell lines by looking at their ability to kill autologous BLCL targets pulsed with the cognate peptide. We found a T cell line that was able to kill target cells pulsed with H2 HA339-355 in a dose-dependent manner (Fig. 3A). When we performed surface staining, more than 97% of the live CD3+cells were CD4+ (see Fig. S1 in the supplemental material). The CD4+ T cell line was also tested against target cells pulsed with corresponding peptides in H1, H3, and H5 HAs (Fig. 3B). As expected, this H2 HA339-355-specific T cell line was able to lyse those peptide-pulsed target cells. The pattern of lysis is also consistent with the IFN-γ responses to the peptides shown in Fig. 2A (summarized in Table 1). The H2 HA339-355 T cell line was also able to lyse target cells pulsed with various recombinant HA proteins (see Fig. S2 in the supplemental material).
Fig 3.
Characterization of the H2 HA339-355-specific T cell line using a standard 51Cr release assay. (A) A dose response curve of the T cell line to H2 HA339-355 peptide was determined using a standard 51Cr release assay. (B and C) The T cell line was also used as effector cells against autologous BLCLs pulsed with the corresponding peptides in other A/HAs (B) or infected with various influenza A virus strains (C). A 51Cr release assay using H5N1-infected targets was done in a separate experiment under enhanced biosafety level 3 conditions (light-gray bars). (D) Responses to the corresponding B HA peptides and B virus infection were also determined in a 51Cr release assay. The percent specific immune lysis was determined by subtracting the percentage of lysis of unpulsed targets from that of the peptide-pulsed targets. H2 HA339-355-pulsed target cells were used as a positive control. All assays were performed in triplicate.
To determine if the T cell line can kill virus-infected targets, we infected autologous BLCLs with seasonal H1N1 and H3N2 strains, including a 2009 pandemic H1N1 strain, an H5N1 strain, and a reassortant H2N1 strain. All of the virus-infected target cells were lysed specifically by the H2 HA339-355 T cell line (Fig. 3C).
The fusion peptide sequence is well conserved in influenza virus HA, including B HA (15). Therefore, we asked if this T cell line is able to recognize this sequence in the influenza B virus HA protein, as well. Indeed, the T cell line killed both peptide-pulsed and B virus-infected targets (Fig. 3D), indicating that this CD4+ T cell epitope is also presented in the context of an influenza B virus infection. These results also confirm the IFN-γ responses we saw in ELISPOT for the B HA peptide containing the fusion peptide sequence (Table 1).
The H2 HA339-355 T cell line produces IFN-γ and TNF-α.
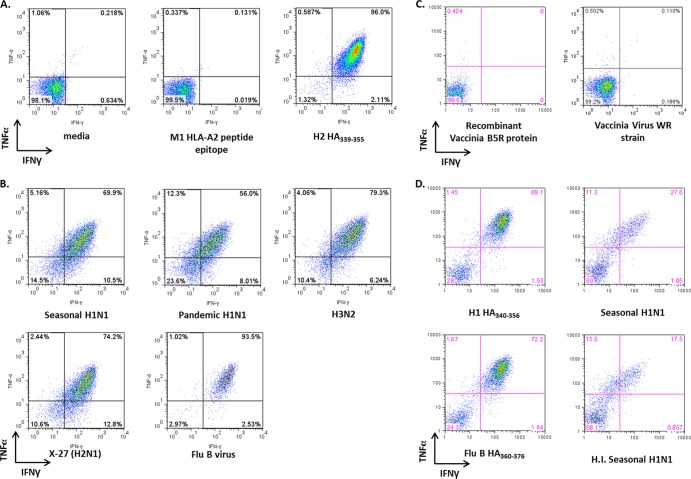
Influenza virus-specific CD4+ T cells have been shown to produce a variety of cytokines, including IFN-γ and TNF-α (49). We performed an ICS assay to identify the cytokines produced by the H2 HA339-355 T cell line upon stimulation with either peptide-pulsed or virus-infected target cells. The T cell line was incubated with either peptide-pulsed or virus-infected autologous BLCLs as APCs in the presence of GolgiPlug for 6 h. The live CD3+ CD4+ T cells (>95%) produced both IFN-γ and TNF-α when they were stimulated with autologous BLCLs that were pulsed with the fusion peptide epitope, but not with the negative-control HLA-A2-restricted M158-66 epitope peptide (Fig. 4A). Autologous BLCLs were also infected with seasonal and pandemic H1N1 and H3N2 strains, a reassortant H2N1, and an influenza B virus strain and were used to stimulate the H2 HA339-355 T cell line. As with peptide stimulation, the CD4+ T cells were double-positive for IFN-γ and TNF-α, with more than 50% of the cells producing both cytokines in response to influenza virus infection (Fig. 4B), but not against target cells infected with vaccinia virus strain WR (at a multiplicity of infection [MOI] of 1) or pulsed with the vaccinia virus B5R protein (Fig. 4C). The H2 HA339-355 T cell line also responded to APCs that were mixed with heat-inactivated influenza virus, suggesting that extracellular influenza virus antigens are taken up through the endocytic pathway and undergo MHC class II presentation (Fig. 4D).
Fig 4.
Cytokine profile of the H2 HA339-355-specific T cell line upon stimulation with peptide-pulsed or virus-infected autologous BLCLs. (A and B) The H2 HA339-355-specific T cell line was incubated with either influenza virus peptide-pulsed (A) or influenza virus-infected (B) BLCLs for 5 to 6 h in the presence of Golgi plug. The T cells were then stained with surface marker and intracellular-cytokine fluorophore-conjugated antibodies to determine the cytokine profile after stimulation. (C) In a separate experiment, the autologous target cells were pulsed with recombinant vaccinia B5R protein or infected with vaccinia virus (strain WR; MOI of 1) to demonstrate the specificity of the H2 HA339-355 T cell line. (D) The T cell line was also incubated with autologous target cells pulsed with influenza virus HA peptides containing the RG(L/I)FGAIAGFIE(G/N)G sequence and with target cells either infected with live virus (seasonal H1N1) or mixed with heat-inactivated influenza virus (H.I. seasonal H1N1). The peptide and recombinant protein concentration used in all experiments was 10 μg/ml. The plots were gated for live, CD3+, CD19−, CD4+/CD8− cells. The cytokine responses to peptide shown in panel A are representative of one out of three experiments. Those in panels B, C, and D were determined in separate single experiments.
The HLA alleles of human donors who have ex vivo responses to the H2 HA339-355 epitope.
The two donors, YD02 and YD06, that responded to the H2 HA339-355 peptide in our initial ELISPOT experiments both expressed the HLA-DRB1*09 allele. We therefore tested additional donor PBMCs that had the HLA-DRB1*09 allele. All five of the additional HLA-DRB1*09-expressing donors' cells had ex vivo IFN-γ responses to the HA peptides containing the RG(L/I)FGAIAGFIE(G/N)G sequence of the fusion peptide (Table 2). To confirm that the T cells responding to the fusion peptide epitope in HLA-DRB1*09 donors were CD4+ T cells, we performed depletion of CD4+ or CD8+ T cells from the PBMCs of donors YD09 and YD12, as we did with the PBMCs of donor YD02 (Fig. 2B). CD4+ T cell depletion, but not CD8+ T cell depletion, resulted in loss of the IFN-γ response in ELISPOT (Table 3).
Table 2.
Ex vivo IFN-γ responses to the fusion peptide epitope in healthy adult donors
| Donor | HLA class II typing | SFC/106a |
||||
|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H5 | B/HA | ||
| YD02 | DRB1*03, DRB1*09:01:02 | 22.7 ± 1.7 | 26.7 ± 2.7 | 46.7 ± 2.7 | 32 ± 5.9 | 14 ± 1.5b |
| YD06 | DRB1*01, DRB1*09:01:02 | 22.7 ± 4.0 | 28 ± 3.2 | 36 ± 2.9 | 36 ± 4.2 | NDc |
| YD08 | DRB1*04, DRB1*13 | 22.2 ± 2.7 | 60 ± 4.0b | 66.7 ± 2 | 22.2 ± 3 | 7.4 ± 2.3 |
| YD09 | DRB1*09, DRB1*12 | 33.3 ± 4 | 46.7 ± 5.5 | 41.7 ± 3.1 | 31.7 ± 3.1 | 48.3 ± 6.6 |
| YD10 | DRB1*09, DRB1*14 | 101.7 ± 4.2 | 96 ± 10.5 | 84.7 ± 11.8 | 71.2 ± 7 | 81.4 ± 9.2 |
| YD11 | DRB1*01, DRB1*09 | 68 ± 7.6 | 40 ± 2.7 | 72 ± 7 | 68 ± 3.6 | 34.7 ± 3.5 |
| YD12 | DRB1*04:06, DRB1*09:01 | 103.3 ± 20.4 | 56.7 ± 2.1 | 53.3 ± 6.4 | 40 ± 7 | 48.3 ± 2.9 |
| YD13 | DRB1*04, DRB1*09 | ND | 66.7 ± 6.1 | ND | 30 ± 3.5 | ND |
The SFC/106 values indicate the IFN-γ response of the donor to the HA peptide containing the conserved RG(L/I)FGAIAGFIE(G/N)G sequence. The cutoff for a positive response is 20 SFC/106. The ELISPOT was performed once for each donor PBMC using triplicate wells to test the HA peptides. The standard error of the mean of triplicate wells was calculated for each HA peptide.
Performed in a separate experiment.
ND, not done.
Table 3.
Ex vivo IFN-γ responses to the fusion peptide epitope in CD4- or CD8-depleted PBMCs
| Donor | H5 HA345-361 peptide (SFC/106)a |
B HA360-376 peptide (SFC/106)a |
||||
|---|---|---|---|---|---|---|
| Whole PBMCs | CD8-expressing cells | CD4-expressing cells | Whole PBMCs | CD8-expressing cells | CD4-expressing cells | |
| YD03b | 2.7 ± 0.6 | 0 | 4 | 0 | 0 | 4 ± 2.1 |
| YD09 | 37.3 ± 12.4 | 0 | 61.3 ± 1.5 | 18.7 ± 2 | 0 ± 0.6 | 60 ± 2.1 |
| YD12 | 14.7 ± 5.8 | 0 | 120 ± 9.5 | 83.3 ± 27.6 | 0 | 165.3 ± 18.2 |
The SFC/106 values indicate the IFN-γ response of the donor to the HA peptide containing the conserved RG(L/I)FGAIAGFIE(G/N)G sequence. The cutoff for a positive response is 20 SFC/106. The ELISPOT was performed once using triplicate wells for each HA peptide, and the standard errors of the mean of triplicate wells were calculated.
Donor YD03 is a nonresponder to the fusion peptide epitope and is used as a negative PBMC control.
An MHC class II binding motif prediction algorithm (http://tools.immuneepitope.org/analyze/html/mhc_II_binding.html) from the Immune Epitope Database (IEDB) (http://www.immuneepitope.org) (53) identified IESRGLFGAIAGFIE as a top 4.52% binder to the HLA-DRB1*09:01 molecule, as well as ESRGLFGAIAGFIEG (top 4.98%) and SRGLFGAIAGFIEGG (top 5.32%), among peptides in the HA protein of the A/Japan/305/1957 (H2N2) strain. An L→I or G→N change in the H3 sequence (RGIFGAIAGFIENG) did not affect the predicted binding scores. Donor YD08 also had ex vivo responses to the H2 HA339-355 epitope but did not have the HLA-DRB1*09 allele (Table 2), suggesting the promiscuity of the epitope. Treatment of autologous BLCLs with blocking antibodies to HLA-DQ and -DP did not inhibit specific immune lysis by the H2 HA339-355 T cell line, while anti-HLA-DR blocked antigen presentation by as much as 70% (data not shown).
We also performed a fluorescence polarization assay to further confirm the affinity of the peptide epitope for the HLA-DR molecule. Since the HLA-DRB1*09 molecule was not available for the assay, we used the HLA-DRB1*01:01 (HLA-DR1) molecule, which has a similar peptide binding motif in position 1 (19, 70). In addition, this H2 HA339-355 peptide was also predicted to be a top 9.36% binder to HLA-DRB1*01:01 using the IEDB MHC class II binding motif prediction algorithm (http://tools.immuneepitope.org/analyze/html/mhc_II_binding.html; 53). We used overlapping H5 HA peptides containing parts of the fusion peptide sequence to determine the optimal 17-mer that can bind to HLA-DR1 (see Fig. S3 in the supplemental material). The H5 HA345-361 (KRGLFGAIAGFIEGGWQ) had an IC50 of ∼893 nM, which indicates that this 17-mer sequence can bind to HLA-DR1, although modestly compared to HA306-318 (see Table S1 in the supplemental material).
DISCUSSION
We established a human CD4+ T cell line recognizing a cross-reactive epitope that is conserved among the different HA types of influenza A virus, and also the HA of influenza B virus. The epitope is located in the fusion peptide sequence of the influenza virus HA. We also found by ELISPOT that adult donors' T cells had ex vivo IFN-γ responses to the different influenza A virus HA and B virus HA peptides that contain the RG(L/I)FGAIAGFIE(G/N)G sequence of the fusion peptide. Analysis of evolutionarily conserved sequences in the different influenza A virus components revealed that the FGAIAGFIE sequence of the fusion peptide is the only region in the HA protein that is 98 to 100% conserved in strains of the different human and avian influenza virus subtypes that circulated between 1997 and 2006 (39), as well as in influenza B viruses (12). This conservation is probably due to the critical role of the domain in triggering fusion and destabilizing target membranes during the fusion process (15). A previous study using a mouse model showed that the stability of the fusion peptide sequence enhanced the immunogenicity of CD4+ T cell epitopes adjacent to the fusion domain (56). Our findings show that the fusion peptide itself contains a CD4+ T cell epitope, as well.
The CD4+ T cell epitope that we have characterized is likely to be restricted by the HLA-DRB1*09 allele, based on our results, although it could possibly bind to other HLA-DR molecules, as well, as was previously shown for the HA306-320 CD4+ T cell epitope (84). We also detected ex vivo T cell responses to the fusion peptide in one donor who is not HLA-DRB1*09, indicating that the fusion peptide epitope is promiscuous. In fact, we have shown that this CD4+ T cell epitope peptide can bind to the HLA-DR1 molecule in a biochemical assay (see Fig. S3 in the supplemental material). Moreover, all seven of the HLA-DRB1*09 donors' PBMCs have ex vivo IFN-γ responses to the fusion peptide epitope in ELISPOT. The ethnic origin of the HLA-DRB1*09 allele is Caucasoid and Oriental (http://www.ebi.ac.uk/cgi-bin/imgt/hla/get_allele.cgi?DRB1*09:01:02), and recent studies have shown that this allele is frequently present in East and Southeast Asian populations (52, 55, 63, 85), in particular the Han Chinese, who comprise more than 90% of the population of mainland China. In a high-resolution genotyping of the HLA-DRB1 locus in individuals from Jiangsu Province, China, HLA-DRB1*09:01 was the most frequent allele (15.69%) (52). Another possibility is that the epitope is also restricted by other HLA-DR haplotypes. The HLA-DR9 serotype has been shown to be in linkage disequilibrium with HLA-DR53 (HLA-DRB4*01:01) (reviewed in references 2, 5, and 37) (we determined only the HLA-DRB1 typing of the donor PBMCs), and it has been reported that one out of six HLA-DRB1*09:01-binding peptides can bind to HLA-DRB4*01:01 (44). Using the IEDB MHC class II binding motif prediction algorithm, the fusion peptide sequence is predicted to be among the top ∼50 to 80% of binders to HLA-DRB4*01:01 compared to HLA-DRB1*09:01, where it is predicted to be among the top 5% of binders in the HA protein. This indicates that the fusion peptide is more likely to bind to the HLA-DRB9*01:01 molecule, since the predicted top 10% highest-scoring peptides are considered to be good binders (79).
There were notable differences in the T cell responses we saw to the various HA peptides. The H2 HA337-349, H5 HA339-355, and B virus HA354-370 peptides, which lack the IEGG sequence at their C termini, were not positive in our ELISPOT assays, while the H3 HA338-354 peptide, which also lacks the IEGG sequence, was positive (Table 1 and Fig. 2A). When the H3 HA338-354 peptide and the H3 HA344-360 peptide were compared (Fig. 2A), the H3 HA344-360 peptide, which has the IENG sequence, stimulated more IFN-γ-producing cells than the H3 HA338-354 peptide without the IENG sequence (as mentioned in Results, an L→I or G→N change in the H3 sequence did not affect the predicted binding scores). These results suggest the presence of at least two overlapping epitopes, which is consistent with the peptide binding prediction that there are three potential overlapping peptides binding to the HLA-DRB1*09:01 molecule.
More CD8+ T cell epitopes to influenza virus have been defined than CD4+ T cell epitopes. The CD4+ T cell epitope PKYVKQNTLKLAT was one of the first human CD4+ T cell epitopes identified (45), and initially, it was thought to be immunodominant (46, 84). It has been used in several studies to characterize and describe the CD4+ T cell responses to influenza virus (11, 17, 29, 48, 54, 66). Subsequent studies have shown that a number of HA-derived CD4+ T cell epitopes can be recognized in infected individuals (30) and in healthy adults (4). Several of these HA-derived CD4+ T cell epitopes are restricted by HLA-DR1 (http://www.immuneepitope.org; 10, 53). In addition, a study looking at CD4 T cell epitopes in the influenza virus HA using an HLA-DR1 transgenic-mouse system show the diversity of the T cell response to HA and that several of these epitopes are located in conserved regions of the HA (57). The same group also looked at the CD4+ T cell memory phase and found that, although the overall memory response to influenza virus remains diverse and directed to several influenza virus proteins, there was a modest but reproducible shift toward HA-derived epitopes (58). This suggests that a majority of CD4+ T cell responses are directed to influenza virus HA, and most of them are in regions that are structurally and functionally conserved. A possible explanation could be that repeated exposure to different virus strains through infection or immunization may selectively stimulate T cells specific to the epitopes located in conserved regions.
The CD4+ T cell epitope that we have described here is also conserved in the HA of influenza B viruses. Influenza A and B viruses are almost the same in structure by electron microscopy, with both having the same number of gene segments that encode the viral proteins (65). At the amino acid level, however, sequence similarities are only 12 to 39% for all proteins, except for basic polymerase 1, which is 60% similar (comparisons were performed between A/New York/348/2003 [H1N1] and B/Florida/4/2006 using CLUSTAL W Multiple Sequence Alignments [http://www.genome.jp/tools/clustalw/] for internal proteins). Influenza B virus has a larger genome, and its membrane channel protein is quite different from that of influenza A virus (65). Although a number of CD4+ and CD8+ T cell epitopes in influenza A viruses have been identified using different strategies (IEDB [http://www.immuneepitope.org]), only a few T cell epitopes have been identified in influenza B viruses. An HLA-A*0201-restricted CD8+ T cell epitope located in the NP (NP85-94) has been studied most extensively (60, 61, 67). There are also two HLA-B8-restricted CD8+ T cell epitopes, also located in the NP (NP30-38 and NP262-271), that have been identified (62) and one HLA-DRB1*0101-restricted CD4+ T cell epitope that is located in the HA (HA308-320) (62). These epitopes were identified by generating peptides from these two viral proteins based on prediction algorithms and using them to stimulate cytotoxic T lymphocyte responses in PBMCs from a limited number of donors. In addition, the HA protein sequence of influenza B virus has only 26 to 27% sequence similarity with the H1, H3, and H5 HAs of influenza A virus (BLAST [http://blast.ncbi.nlm.nih.gov/Blast.cgi] results comparing HA protein sequences of B/Florida/4/2006 and A/New Caledonia/20/1999 [H1N1], A/New York/384/2005 [H3N2], and A/Thailand/SP [528]/2004 [H5N1]).
The ex vivo responses to the fusion peptide epitope in our donor PBMCs suggest that this CD4+ T cell response is present in the memory compartment and may be attributed to prior exposure to influenza virus antigens via infection or vaccination. This type of cross-reactive immunity is called influenza virus heterosubtypic immunity (HSI) (reviewed in reference 33). HSI has been demonstrated in animal experiments but is challenging to demonstrate in humans. The existing literature on the topic includes a retrospective analysis (23) and an epidemiological study done at the brink of the 1957 H2N2 pandemic that compare immune responses in individuals who had prior H1N1 infection and their propensity to become ill after exposure to the novel H2N2 virus (69). Both studies suggest the impact of accumulated HSI in the event of a pandemic, where partial protection against H2N2 was seen in individuals who were previously infected with H1N1. Epstein suggests the contribution of CD4+ and CD8+ T cells in mediating HSI, since these individuals did not have antibodies to H2 HA that could have protected them from H2N2 infection. It is interesting to speculate that a subset of the population could have maintained a memory response to this particular HA epitope by repeated exposure to influenza virus antigens via natural infection and/or vaccination. In light of the recent influenza pandemic, HSI may be a contributing factor to its mildness and lower mortality rates compared to previous pandemics. Aside from the detection of preexisting antibodies to HA in older individuals (38), there was also preexisting T cell immunity to the pandemic strain, as exemplified by the presence of both CD8+ and CD4+ T cell epitopes generated from previous encounters with seasonal influenza virus strains (1, 18, 28, 59, 82). Thus, HSI significantly impacted the course of the recent influenza pandemic.
Although CD4+ T cells may be dispensable in viral clearance and protection, as shown in mouse studies, optimal humoral and cellular immunities to influenza require the activation of CD4+ T helper cells (reviewed in reference 9). Indeed, a recent study by Wilkinson and colleagues demonstrated that preexisting influenza virus-specific CD4+ T cells were correlated with protection against influenza virus challenge in humans (80). McKinstry and colleagues suggest several mechanisms by which CD4+ T cells can contribute to heterosubtypic immunity (reviewed in references 49 and 75). Influenza virus-specific CD4+ T cells can provide help to B cells and cross-reactive CD8+ T cells during infection, and they may also regulate early innate immune responses by indirectly upregulating inflammatory cytokines and chemokines during the early stage of infection (73). They can also act as antiviral effectors themselves during a heterosubtypic infection (49). CD4+ T cells can also have a cytotoxic effector function via a perforin-mediated mechanism in a mouse model of influenza virus infection (8). Teijaro et al. have shown that memory CD4+ T cells specific to the H1N1 influenza virus provide a protective immune response in the lungs after a lethal challenge in mice, suggesting that CD4+ T cells can be effectors as well, independent of T helper mechanisms (77).
Cross-reactive CD4+ T cells may also play important roles in generating robust antibody responses to influenza virus, although the mechanisms are still not clear. With the recent identification and characterization of several cross-reactive antibodies to the more conserved HA2 domain or the stalk region of HA (14, 21, 74, 81) and the novel approach of generating influenza virus vaccines based on the stalk region (including fusion peptide) (72), the presence of the cross-reactive CD4+ T cell epitope in the fusion peptide may be helpful to induce higher levels of cross-reactive antibody responses, at least for individuals with the HLA-DRB1*09 allele (and other alleles with similar peptide binding motifs). Several groups have also shown that these cross-reactive antibodies can be detected in human sera (13, 43, 78). Specific activation of these helper T cells should be considered in designing or improving vaccination strategies against influenza virus. CD4+ T cell epitopes tend to be more promiscuous than CD8+ T cell epitopes due to the nature of MHC class II molecules (36). This may be advantageous when considering epitopes to be included in an improved vaccine for influenza virus, since more than one HLA molecule can present the peptide. In addition, a peptide conjugate vaccine based on the fusion peptide of the precursor HA of the B virus was shown to elicit protection in mice against lethal challenge of various strains of influenza B virus and can potentially be extended to influenza A virus strains (7). It is also interesting to note that a B cell epitope (3) and an HLA-A2-restricted CD8+ T cell epitope (32) have been previously described. A more recent study examined the presence of HA2-specific antibodies in acute- and convalescent-phase sera from adults with confirmed H3N2 infection (71). They found that a third (15/45) of the subjects had antibodies specific to the N-terminal residues 1 to 38 of HA2, which includes the fusion peptide sequence. These data indicate that the fusion peptide of the influenza virus HA is a relevant target of the immune response to influenza virus.
In conclusion, we have identified and described a cross-reactive human CD4+ T cell epitope in the influenza virus HA that is highly conserved in all the influenza A and B virus HA proteins. This epitope is recognized by individuals who have the HLA-DRB1*09 allele. Our in vitro experiments show that the CD4+ T cell response to this epitope is characterized by the production of IFN-γ and TNF-α. To our knowledge, this is the first influenza virus CD4+ T cell epitope in the HA protein that is cross-reactive to influenza A and B viruses. Although natural infection or standard vaccination may not induce strong T and B cell responses to this very conserved epitope in the fusion peptide, it may be possible to develop a vaccination strategy to induce these CD4+ T cells, which are cross-reactive to both influenza A and B viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mary Dawn T. Co, Jeffrey S. Kennedy, Karen Longtine, Melissa O'Neill, and Jaclyn Longtine for their help in obtaining the human PBMC samples that were used in this study. We thank Christine Turcotte, Lynne Burns, and Pamela Pazoles for assistance with HLA typing and Michel DeWilde and Robert Ryall of Sanofi Pasteur and Nancy Cox and Alexander Klimov of the CDC for the influenza virus strains used in this study. We thank Lawrence Stern for reagents and discussion. We also thank anonymous reviewers for helpful suggestions. The following reagents were obtained through BEI Resources: peptide arrays previously described (4), control peptides for major histocompatibility complex class I and II epitopes of influenza virus A and B proteins, and the peptides listed in Table 1 except for the H2 HA peptides.
This work was supported by NIH/National Institute of Allergy and Infectious Diseases (NIAID) grant U19 AI-057319.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH/NIAID.
Footnotes
Published ahead of print 20 June 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alam S, Sant AJ. 2011. Infection with seasonal influenza virus elicits CD4 T cells specific for genetically conserved epitopes that can be rapidly mobilized for protective immunity to pandemic H1N1 influenza virus. J. Virol. 85:13310–13321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersson G. 1998. Evolution of the human HLA-DR region. Front. Biosci. 3:d739–d745 [DOI] [PubMed] [Google Scholar]
- 3. Atassi MZ, Webster RG. 1983. Localization, synthesis, and activity of an antigenic site on influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 80:840–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babon JAB, et al. 2009. Genome-wide screening of human T-cell epitopes in influenza A virus reveals a broad spectrum of CD4+ T-cell responses to internal proteins, hemagglutinins, and neuraminidases. Hum. Immunol. 70:711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bell JI, et al. 1987. Allelic variation in the DR subregion of the human major histocompatibility complex. Proc. Natl. Acad. Sci. U. S. A. 84:6234–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. 2002. Compromised influenza virus-specific CD8+-T-cell memory in CD4+-T-cell-deficient mice. J. Virol. 76:12388–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bianchi E, et al. 2005. Universal influenza B vaccine based on the maturational cleavage site of the hemagglutinin precursor. J. Virol. 79:7380–7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown DM, Dilzer AM, Meents DL, Swain SL. 2006. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 177:2888–2898 [DOI] [PubMed] [Google Scholar]
- 9. Brown DM, Roman E, Swain SL. 2004. CD4 T cell responses to influenza infection. Semin. Immunol. 16:171–177 [DOI] [PubMed] [Google Scholar]
- 10. Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. 2007. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc. Natl. Acad. Sci. U. S. A. 104:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cameron TO, Cochran JR, Yassine-Diab B, Sékaly R-P, Stern LJ. 2001. Cutting edge: detection of antigen-specific CD4+ T cells by HLA-DR1 oligomers is dependent on the T cell activation state. J. Immunol. 166:741–745 [DOI] [PubMed] [Google Scholar]
- 12. Chun S, et al. 2008. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine 26:6068–6076 [DOI] [PubMed] [Google Scholar]
- 13. Corti D, et al. 2010. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 120:1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corti D, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856 [DOI] [PubMed] [Google Scholar]
- 15. Cross KJ, Langley WA, Russell RJ, Skehel JJ, Steinhauer DA. 2009. Composition and functions of the influenza fusion peptide. Protein Pept. Lett. 16:766–778 [DOI] [PubMed] [Google Scholar]
- 16. Cusick MF, Wang S, Eckels DD. 2009. In vitro responses to avian influenza H5 by human CD4 T cells. J. Immunol. 183:6432–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Danke NA, Kwok WW. 2003. HLA class II-restricted CD4+ T cell responses directed against influenza viral antigens postinfluenza vaccination. J. Immunol. 171:3163–3169 [DOI] [PubMed] [Google Scholar]
- 18. De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. 2009. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008–2009 conventional influenza vaccine. Vaccine 27:5740–5747 [DOI] [PubMed] [Google Scholar]
- 19. Doytchinova IA, Flower DR. 2005. In silico identification of supertypes for class II MHCs. J. Immunol. 174:7085–7095 [DOI] [PubMed] [Google Scholar]
- 20. Duvvuri VR, et al. 2010. Highly conserved cross-reactive CD4+ T-cell HA-epitopes of seasonal and the 2009 pandemic influenza viruses. Influenza Other Respi. Viruses 4:249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ekiert DC, et al. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ennis FA, et al. 1997. Hantavirus pulmonary syndrome: CD8+ and CD4+ cytotoxic T lymphocytes to epitopes on sin nombre virus nucleocapsid protein isolated during acute illness. Virology 238:380–390 [DOI] [PubMed] [Google Scholar]
- 23. Epstein S. 2006. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J. Infect. Dis. 193:49–53 [DOI] [PubMed] [Google Scholar]
- 23a. Federal Register 2009. Notice of changes to the NIH guidelines. Fed. Regist. 74:48275–48280 http://oba.od.nih.gov/oba/rac/fractions/74_FR_48275.pdf [Google Scholar]
- 24. Fiore AE, et al. 2009. Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Recommend Rep. 58:1–52 [PubMed] [Google Scholar]
- 25. Fouchier RAM, et al. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frayser M, Sato AK, Xu L, Stern LJ. 1999. Empty and peptide-loaded class II major histocompatibility complex proteins produced by expression in Escherichia coli and folding in vitro. Protein Expr. Purif. 15:105–114 [DOI] [PubMed] [Google Scholar]
- 27. Gagnon SJ, Ennis FA, Rothman AL. 1999. Bystander target cell lysis and cytokine production by dengue virus-specific human CD4+ cytotoxic T-lymphocyte clones. J. Virol. 73:3623–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ge X, et al. 2010. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J. Virol. 84:3312–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gelder C, et al. 1998. Six unrelated HLA-DR-matched adults recognize identical CD4+ T cell epitopes from influenza A haemagglutinin that are not simply peptides with high HLA-DR binding affinities. Int. Immunol. 10:211–222 [DOI] [PubMed] [Google Scholar]
- 30. Gelder CM, Welsh KI, Faith A, Lamb JR, Askonas BA. 1995. Human CD4+ T-cell repertoire of responses to influenza A virus hemagglutinin after recent natural infection. J. Virol. 69:7497–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerhard W. 2001. The role of the antibody response in influenza virus infection. Curr. Top. Microbiol. Immunol. 260:171–190 [DOI] [PubMed] [Google Scholar]
- 32. Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A. 2000. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum. Immunol. 61:438–452 [DOI] [PubMed] [Google Scholar]
- 33. Grebe KM, Yewdell JW, Bennink JR. 2008. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect. 10:1024–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Green S, et al. 1993. Dengue virus-specific human CD4+ T-lymphocyte responses in a recipient of an experimental live-attenuated dengue virus type 1 vaccine: bulk culture proliferation, clonal analysis, and precursor frequency determination. J. Virol. 67:5962–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Green S, Kurane I, Pincus S, Paoletti E, Ennis FA. 1997. Recognition of dengue virus NS1-NS2a proteins by human CD4+ cytotoxic T lymphocyte clones. Virology 234:383–386 [DOI] [PubMed] [Google Scholar]
- 36. Greenbaum J, et al. 2011. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 63:325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gregersen PK, et al. 1986. Polymorphism of HLA-DR beta chains in DR4, -7, and -9 haplotypes: implications for the mechanisms of allelic variation. Proc. Natl. Acad. Sci. U. S. A. 83:9149–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hancock K, et al. 2009. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361:1945–1952 [DOI] [PubMed] [Google Scholar]
- 39. Heiny AT, et al. 2007. Evolutionarily conserved protein sequences of influenza A viruses, avian and human, as vaccine targets. PLoS One 2:e1190 doi:10.1371/journal.pone.0001190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jameson J, Cruz J, Ennis FA. 1998. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 72:8682–8689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jameson J, Cruz J, Terajima M, Ennis FA. 1999. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J. Immunol. 162:7578–7583 [PubMed] [Google Scholar]
- 42. Jonges M, et al. 2010. Influenza virus inactivation for studies of antigenicity and phenotypic neuraminidase inhibitor resistance profiling. J. Clin. Microbiol. 48:928–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kashyap AK, et al. 2008. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. U. S. A. 105:5986–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kobayashi H, et al. 1996. Analysis of anchor residues in a naturally processed HLA-DR53 ligand. Immunogenetics 44:366–371 [DOI] [PubMed] [Google Scholar]
- 45. Lamb JR, Eckels DD, Lake P, Woody JN, Green N. 1982. Human T-cell clones recognize chemically synthesized peptides of influenza haemagglutinin. Nature 300:66–69 [DOI] [PubMed] [Google Scholar]
- 46. Lamb JR, Green N. 1983. Analysis of the antigen specificity of influenza haemagglutinin-immune human T lymphocyte clones: identification of an immunodominant region for T cells. Immunology 50:659–666 [PMC free article] [PubMed] [Google Scholar]
- 47. Lee LY, et al. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118:3478–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lucas M, et al. 2004. Ex Vivo phenotype and frequency of influenza virus-specific CD4 memory T cells. J. Virol. 78:7284–7287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McKinstry KK, Strutt TM, Swain SL. 2011. Hallmarks of CD4 T cell immunity against influenza. J. Intern. Med. 269:507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mitra-Kaushik S, Cruz J, Stern LJ, Ennis FA, Terajima M. 2007. Human cytotoxic CD4+ T cells recognize HLA-DR1-restricted epitopes on vaccinia virus proteins A24R and D1R conserved among poxviruses. J. Immunol. 179:1303–1312 [DOI] [PubMed] [Google Scholar]
- 51. Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1648–1689 In Knipe DM, et al. (ed), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 52. Pan QQ, et al. 2011. The distribution of human leukocyte antigen-A, -B, and -DRB1 alleles and haplotypes based on high-resolution genotyping of 167 families from Jiangsu Province, China. Hum. Immunol. 72:672–676 [DOI] [PubMed] [Google Scholar]
- 53. Peters B, et al. 2005. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 3:e91 doi:10.1371/journal.pbio.0030091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prevost-Blondel A, et al. 1995. Preferential usage of the T-cell receptor by influenza virus hemagglutinin-specific human CD4+ T lymphocytes: in vitro life span of clonotypic T cells. J. Virol. 69:8046–8050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qin Qin P, et al. 2011. Distribution of human leucocyte antigen-A, -B and -DR alleles and haplotypes at high resolution in the population from Jiangsu province of China. Int. J. Immunogenet. 38:475–481 [DOI] [PubMed] [Google Scholar]
- 56. Rajnavolgyi E, et al. 1997. Characterizing immunodominant and protective influenza hemagglutinin epitopes by functional activity and relative binding to major histocompatibility complex class II sites. Eur. J. Immunol. 27:3105–3114 [DOI] [PubMed] [Google Scholar]
- 57. Richards KA, et al. 2007. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J. Virol. 81:7608–7619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Richards KA, Chaves FA, Sant AJ. 2011. The memory phase of the CD4 T-cell response to influenza virus infection maintains its diverse antigen specificity. Immunology 133:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Richards KA, Topham D, Chaves FA, Sant AJ. 2010. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can. directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J. Immunol. 185:4998–5002 [DOI] [PubMed] [Google Scholar]
- 60. Robbins PA, Garboczi DN, Strominger JL. 1995. HLA-A*0201 complexes with two 10-mer peptides differing at the P2 anchor residue have distinct refolding kinetics. J. Immunol. 154:703–709 [PubMed] [Google Scholar]
- 61. Robbins PA, et al. 1989. Comparison between two peptide epitopes presented to cytotoxic T lymphocytes by HLA-A2. Evidence for discrete locations within HLA-A2. J. Immunol. 143:4098–4103 [PubMed] [Google Scholar]
- 62. Robbins PA, Rota PA, Shapiro SZ. 1997. A broad cytotoxic T lymphocyte response to influenza type B virus presented by multiple HLA molecules. Int. Immunol. 9:815–823 [DOI] [PubMed] [Google Scholar]
- 63. Romphruk AV, et al. 2010. HLA class I and II alleles and haplotypes in ethnic Northeast Thais. Tissue Antigens 75:701–711 [DOI] [PubMed] [Google Scholar]
- 64. Roti M, et al. 2008. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J. Immunol. 180:1758–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ruigrok RW. 1998. Structure of influenza A, B and C viruses, p 29–42 In Nicholson K, Webster RG, Hay AJ. (ed), Textbook of influenza. Blackwell Science Ltd, Oxford, United Kingdom [Google Scholar]
- 66. Scriba TJ, et al. 2005. Ultrasensitive detection and phenotyping of CD4+ T cells with optimized HLA class II tetramer staining. J. Immunol. 175:6334–6343 [DOI] [PubMed] [Google Scholar]
- 67. Silver ML, Parker KC, Wiley DC. 1991. Reconstitution by MHC-restricted peptides of HLA-A2 heavy chain with [beta]2-microglobulin, in vitro. Nature 350:619–622 [DOI] [PubMed] [Google Scholar]
- 68. Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 69. Slepushkin AN. 1959. The effect of a previous attack of A1 influenza on susceptibility to A2 virus during the 1957 outbreak. Bull. World Health Organ. 20:297–301 [PMC free article] [PubMed] [Google Scholar]
- 70. Southwood S, et al. 1998. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 160:3363–3373 [PubMed] [Google Scholar]
- 71. Stanekova Z, et al. 2012. Epitope specificity of anti-HA2 antibodies induced in humans during influenza infection. Influenza Other Respi. Viruses doi:10.1111/j.1750-2659.2011.00328.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Steel J, et al. 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1:e00018–00010 doi:10.1128/mBio.00018-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Strutt TM, et al. 2010. Memory CD4+ T cells induce innate responses independently of pathogen. Nat. Med. 16:558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sui J, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Swain SL, et al. 2006. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunol. Rev. 211:8–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Swain SL, Dutton RW, Woodland DL. 2004. T cell responses to influenza virus infection: effector and memory cells. Viral Immunol. 17:197–209 [DOI] [PubMed] [Google Scholar]
- 77. Teijaro JR, Verhoeven D, Page CA, Turner D, Farber DL. 2010. Memory CD4 T cells direct protective responses to influenza virus in the lungs through helper-independent mechanisms. J. Virol. 84:9217–9226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Throsby M, et al. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3:e3942 doi:10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang P, et al. 2008. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 4:e1000048 doi:10.1371/journal.pcbi.1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wilkinson TM, et al. 2012. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 18:274–280 [DOI] [PubMed] [Google Scholar]
- 81. Wrammert J, et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208:181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xing Z, Cardona CJ. 2009. Preexisting immunity to pandemic (H1N1). Emerg. Infect. Dis. 15:1847–1849 (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yang J, et al. 2009. H5N1 strain-specific Hemagglutinin CD4+ T cell epitopes restricted by HLA DR4. Vaccine 27:3862–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zeliszewski D, et al. 1994. Molecular basis for degenerate T-cell recognition of one peptide in the context of several DR molecules. Hum. Immunol. 41:28–33 [DOI] [PubMed] [Google Scholar]
- 85. Zhu F, et al. 2011. Analysis of the complete cDNA sequences of HLA-DRB1 alleles with group-specific amplification primers in the Chinese Han population. Tissue Antigens 77:329–332 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.