Abstract
The high-risk human papillomavirus type 16 (HPV-16) E5 protein (16E5) induces tumors in a transgenic mouse model and may contribute to early stages of cervical carcinogenesis. Although high-risk E5 expression is generally thought to be lost during the progression to cervical carcinoma following integration of HPV DNA into the host genome, episomal viral DNA has been documented in a subset of HPV-16-positive malignant lesions. Numerous studies have shown that transcripts that could potentially encode 16E5 are present in cervical biopsy specimens and cervical cancer cell lines, but the presence of E5 protein has been demonstrated in only two reports that have not been corroborated. In the present study, we show that trypsin cleavage of 16E5 generates a unique four-amino-acid C-terminal peptide (FLIT) that serves as a marker for E5 expression in transfected cells and epithelial cell lines containing integrated and episomal HPV-16 DNA. Following trypsin cleavage, reversed-phase chromatography and mass spectrometry (MS) were used to detect FLIT. Immunoprecipitation assays using a newly generated anti-16E5 antibody confirmed that 16E5 was solely responsible for the FLIT signal, and deuterated FLIT peptide provided an internal standard that enabled us to quantify the number of 16E5 molecules per cell. We show that 16E5 is expressed in the Caski but not in the SiHa cervical cancer cell line, suggesting that 16E5 may contribute to the malignant phenotype of some cervical cancers, even in cells exclusively containing an integrated HPV genome.
INTRODUCTION
High-risk human papillomavirus type 16 (HPV-16) is the causative agent of a majority of cervical cancers worldwide (12). HPV-16 encodes three transforming proteins: E5, E6, and E7. The E6 and E7 oncoproteins are required for the immortalization of human genital keratinocytes and are known to inactivate the p53 and pRb tumor suppressors and to induce telomerase (8, 36, 40, 49, 61, 62). The HPV-16 E5 oncoprotein (16E5) may contribute to early steps of tumor initiation (7, 55) and promotes neoplasia in a transgenic mouse model (2, 20, 46).
To date, many phenotypes have been attributed to 16E5, including potentiation of the epidermal growth factor receptor signaling pathway (15, 20, 43, 54), enhancement of cell immortalization and transformation by E6/E7 (53, 60), induction of koilocytosis (29), inhibition of TRAIL- and FasL-mediated apoptosis (26), interactions with BAP31 (45) and karyopherin β3 (28), and interference with the intracellular trafficking of endocytic vesicles (57, 58) and HLA-I (3, 21).
Integration of HPV DNA into the host genome is thought to be a key event in neoplastic progression. Integration can result in the rearrangement or regional loss of both host and viral genes. Although there are no defined sites for HPV integration, certain “hot spots” (termed common fragile sites [CFS]) exist at which integration is more likely to occur (59). Upon integration, the HPV genome is almost always rearranged in the E2-coding region, resulting in deletion of the E2 open reading frame (ORF) or its separation from the early viral promoter (5, 13, 14). The E2 protein, when present, regulates E6 and E7 expression, and its loss elevates E6 and E7 levels (13, 48). Cells that highly express E6 and E7 from integrated genes have a growth advantage over cells that maintain HPV DNA episomally (25). The same genetic changes that alter E2 expression during viral integration are also believed to disrupt E5 expression (52). It is for this reason that E5 is thought to function mainly during the productive life cycle of the virus (when the viral genome is maintained episomally).
Unlike high-risk HPV-18, which is almost exclusively integrated in cervical cancer (18, 64), the HPV-16 genome has been found to persist episomally in 26 to 76% of malignant lesions (9, 11, 16, 22, 37, 44). Other evidence suggests that 16E5 also may be expressed from integrated viral DNA. Two HPV-16-positive cervical cancer cell lines, SiHa and Caski, harbor 1 or 2 and approximately 500 integrated copies of the HPV-16 genome, respectively (38, 41, 63). Caski cells retain an intact HPV-16 genome, while SiHa cells exhibit disruption of the E1 and E2 ORFs (41). In Caski cells, a 16E5 ORF (5) and spliced mRNA transcripts potentially encoding 16E5 (50) have been identified. In addition, one study used in situ gene-specific hybridization to show the presence of a 16E5 ORF in five carcinomas and one invasive carcinoma, which all contained integrated viral genomes (51). Unfortunately, these reports demonstrate the presence of an E5 ORF or transcript, but not 16E5 protein. Two studies have used antibodies to detect 16E5 protein in cervical tissue (9, 27); however, these results have not been verified, and attempts to recreate the antibodies have been unsuccessful due to the highly hydrophobic, nonimmunogenic nature of 16E5.
In the present study, we showed that trypsin cleavage of 16E5 generates a unique four-amino-acid C-terminal peptide (FLIT) that serves as a marker for E5 expression in transfected cells and epithelial cell lines. Following trypsin cleavage, reversed-phase chromatography and mass spectrometry (MS) were used to detect FLIT. Immunoprecipitation assays using 16E5 antiserum that we developed confirmed the specificity of FLIT detection. The addition of a defined amount of deuterated FLIT to samples prior to chromatography and MS provided an internal standard that enabled us to quantitatively determine the number of 16E5 molecules per cell.
MATERIALS AND METHODS
Cells and viruses.
Retroviruses encoding HPV-16 E6 and E7 in the vector pBabePuro (39) or encoding wild-type (wt) 16E5 in the vector pLXSN were generated using the Phoenix cell system (42). wt and codon-optimized 16E5 also were cloned into the pJS55 expression vector as described previously (17). For some experiments, E5 proteins were N-terminally tagged with the AU1 epitope DTYRYI (34).
Primary human ectocervical cells (HECs) were derived from cervical tissue after hysterectomy for benign uterine disease (4) and were immortalized by infection with HPV-16 E6/E7-encoding retrovirus and selection in the presence of puromycin (0.5 μg/ml). 16E5-expressing cell lines were generated from immortalized HECs by infection with retroviruses encoding E5 and selection in the presence of Geneticin G418 (100 μg/ml). HEC lines were grown at 37°C with 5% CO2 in keratinocyte growth medium (KGM) (Invitrogen, Carlsbad, CA), supplemented with gentamicin sulfate (10 μg/ml). To maintain constant levels of E5 expression, G418 was administered to cell cultures at alternating passages.
COS-1, C33A, SiHa, and Caski cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin G, and 100 μg/ml streptomycin sulfate (Invitrogen).
Normal immortalized human keratinocyte (NIKS) cells, which are spontaneously immortalized but not transformed, were cultured on a layer of irradiated murine fibroblasts as previously described (1). NIKS-4H and NIKS-2L clonal cell lines harboring episomal HPV-16 genomes were generated by Effectine-mediated cotransfection (Qiagen, Valencia, CA) of NIKS cells with circularized HPV-16 DNA (W12) and a blastocidin resistance plasmid, followed by the selection and expansion of individual colonies (24).
Transfection of COS cells.
COS cells were grown to 90% confluence in antibiotic-free Opti-MEM (Invitrogen) containing 4% FBS. For each 10-cm tissue culture dish, 72 μg DNA (in pJS55) was mixed with 9 ml serum-free Opti-MEM and 90 μl Lipofectamine 2000 transfection reagent (Invitrogen), added to the cells, and left for 4 h. Lysates were prepared 24 h after transfection by washing the dishes with phosphate-buffered saline (PBS) and scraping the cells into 0.8 ml radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (56) at 4°C. For Western blotting, RIPA lysates were mixed with an equal volume of 2× SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (30) and heated at 110°C for 10 to 15 min.
16E5 antiserum, immunoprecipitation, and Western blotting.
A peptide consisting of amino acids 75 to 83 of 16E5 repeated 3 times (separated by short stretches of polyethylene glycol) was used to generate rabbit polyclonal antiserum (New England Peptide, Gardner, MA) to immunoprecipitate 16E5. AU1 epitope-tagged 16E5 was detected on anti-AU1 immunoblots as described previously (56) using an anti-AU1 mouse monoclonal antibody (Covance, Emeryville, CA). When 16E5 immunoprecipitates were analyzed by mass spectrometry, proteins were eluted from immobilized protein A beads (Thermo Scientific, Rockford, IL) by boiling in distilled water, rather than in 2× SDS-PAGE sample buffer, to avoid high levels of salt and detergent.
Protein mass spectrometry.
Cells were grown to 80% confluence on 10-cm tissue culture dishes, washed with PBS, and scraped into 0.5 to 1.0 ml RIPA buffer with protease inhibitors at 4°C. Lysates were frozen immediately on dry ice and stored at −80°C until use. Samples were thawed in a 37°C water bath and centrifuged at 14,000 × g for 15 min. Protein concentrations were determined using the Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, CA), according to the manufacturer's instructions, with bovine IgG as the standard. Thirty to 40 μg of protein from each sample was loaded on a 4 to 12% SDS polyacrylamide gel and separated in MES (morpholineethanesulfonic acid) buffer. After separation, no staining was performed (E5 was below the detection limit of Coomassie blue staining), and sections of the gel corresponding to 8 to 14 kDa were excised from each lane. These were dehydrated by incubating with 200 μl of acetonitrile (ACN) for 10 min. The ACN was removed, followed by rehydration of the gel sections in reducing buffer (10 mM dithiothreitol in 50 mM NH4HCO3) for 1 h at 56°C. The samples were then washed twice with 50 mM NH4HCO3, followed by incubation for 30 min in alkylating buffer (55 mM iodoacetamide in 50 mM NH4HCO3). Gel sections were washed one last time with 50 mM NH4HCO3 and then dehydrated with ACN as described above. After removal of the ACN, the samples were vacuum centrifuged until completely dehydrated. These were rehydrated in 200 μl of 50 mM NH4HCO3 containing 2.5 μg TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-modified trypsin, incubated overnight at 37°C, and subjected to ultrasonic disruption for 30 min to ensure quantitative recovery of the tryptic peptides. Ten microliters of each digest was transferred to a new tube and mixed with 89 μl of solvent A (0.1% formic acid in ultrapure water) and 1 μl of a 10-pg/μl solution of deuterated peptide standard, D5-Phe-Leu-Ile-Thr (D5-FLIT) (IsoSciences, King of Prussia, PA). Five microliters of this mixture was analyzed by MS.
Tryptic digests were loaded via the injection loop onto a 180-μm by 2-cm nanoACQUITY UltraPerformance liquid chromatography (UPLC) symmetry trap column packed with 5 μm C18 resin (Waters, Milford, MA) at a trapping flow rate of 5 μl/min. The digests were then separated on a 75-μm-diameter by 15- or 25-cm-length nanoACQUITY 1.7-μm BEH 300 C18 resin reversed-phase chromatography column maintained at 50°C (to lower back pressure). The retention times for the FLIT peptide were 25 min on the 15-cm column and 29 min on the 25-cm column. The separation was performed with solvent A and solvent B (0.1% formic acid in ACN), using the following gradient: 1% B for 1 min, 1 to 10% B over 5 min, 10 to 60% B over 30 min, 60 to 99% B over 1 min, 99% B for 18 min (to avoid carryover), 99 to 1% B over 3 min, and 1% B for 8 min.
Multiple-reaction monitoring (MRM) data were collected on a 4000 QTRAP hybrid triple-quadrupole/linear ion trap mass spectrometer (AB Sciex, Foster City, CA) interfaced with the nanoACQUITY UPLC system. MRM data acquisition was performed with a spray voltage of 2,300 V, an interface heater temperature of 150°C, a curtain gas pressure of 13 lb/in2, an ion source gas pressure of 25 lb/in2, a declustering potential of 60 V, an entrance potential of 10 V, collision cell exit potential of 10 V, and a pause time of 5 ms. The collision gas was set at “high.”
Synthetic FLIT and D5-FLIT were analyzed using a nanoUPLC-Q-TOF mass spectrometer (QStar Elite, AB Sciex) to identify the transitions associated with breakdown of the peptides. Six MRM transitions were detected using unit resolution in both the Q1 and Q3 quadrupoles and a 75-ms dwell time; these included 3 FLIT “analyte” transitions (493.3/233.1, 493.3/261.1, and 493.3/374.2) and 3 D5-FLIT “internal standard” transitions (498.3/233.1, 498.3/266.1, and 498.3/379.2). Data analysis was performed using MultiQuant version 1.1 software (AB Sciex). The most abundant transition (with a good signal-to-noise ratio) was used for quantitation of endogenous FLIT and the D5-FLIT internal standard in tryptic digests, which yielded ng of 16E5 per ml of lysate. To calculate molecules of 16E5 per cell, identical cell cultures were harvested by treatment with trypsin-EDTA (Invitrogen) and analyzed using a Beckman/Coulter Z1 particle counter (Beckman/Coulter, Miami, FL) to determine the number of cells per ml of lysate.
RNA extraction and quantitative real-time PCR.
RNA was isolated from 10-cm tissue culture dishes at 80% confluence and treated with DNase using the RNAaqueous-4PCR kit according to the manufacturer's protocol (Ambion, Austin, TX). A RETROScript kit (Ambion) was used for reverse transcriptase PCR (RT-PCR). RNA was denatured for 3 min at 80°C in the presence of oligo(dT) and random hexamers. This was followed by the reverse transcriptase step, consisting of 60 min at 45°C and 10 min at 92°C. cDNA samples were diluted to 75 ng/μl and stored at −20°C. Quantitative real-time PCR mixtures contained (in a total volume of 20 μl) 0.8 μl cDNA (75 ng/μl), 10 μl 2× Bio-Rad IQ SYBR green Supermix, 0.125 μl forward primer (20 μM), 0.125 μl reverse primer (20 μM), and 8.95 μl distilled water. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the control. Three replicates of each sample were run in triplicate on a 96-well plate. Reaction products were annealed and analyzed using a Bio-Rad iCycler and accompanying software. The following primers were used: E5 forward, 5′-CTTTGCTTTTGTGTGCTTTTGTGTG-3′; E5 reverse, 5′-AAAGCGTGCATGTGTATGTATTAAA-3′; E1^E4 spliced transcript forward, 5′-TGGCTGATCCTGCAGCAGC-3′; E1^E4 spliced transcript reverse, 5′-AGGCGACGGCTTTGGTATG-3′; GAPDH forward, 5′-TCTCCTCTGACTTCAACAGC-3′; and GAPDH reverse, 5′-GAAATGAGCTTGACAAAGTG-3′.
RESULTS
FLIT peptide as a marker for 16E5.
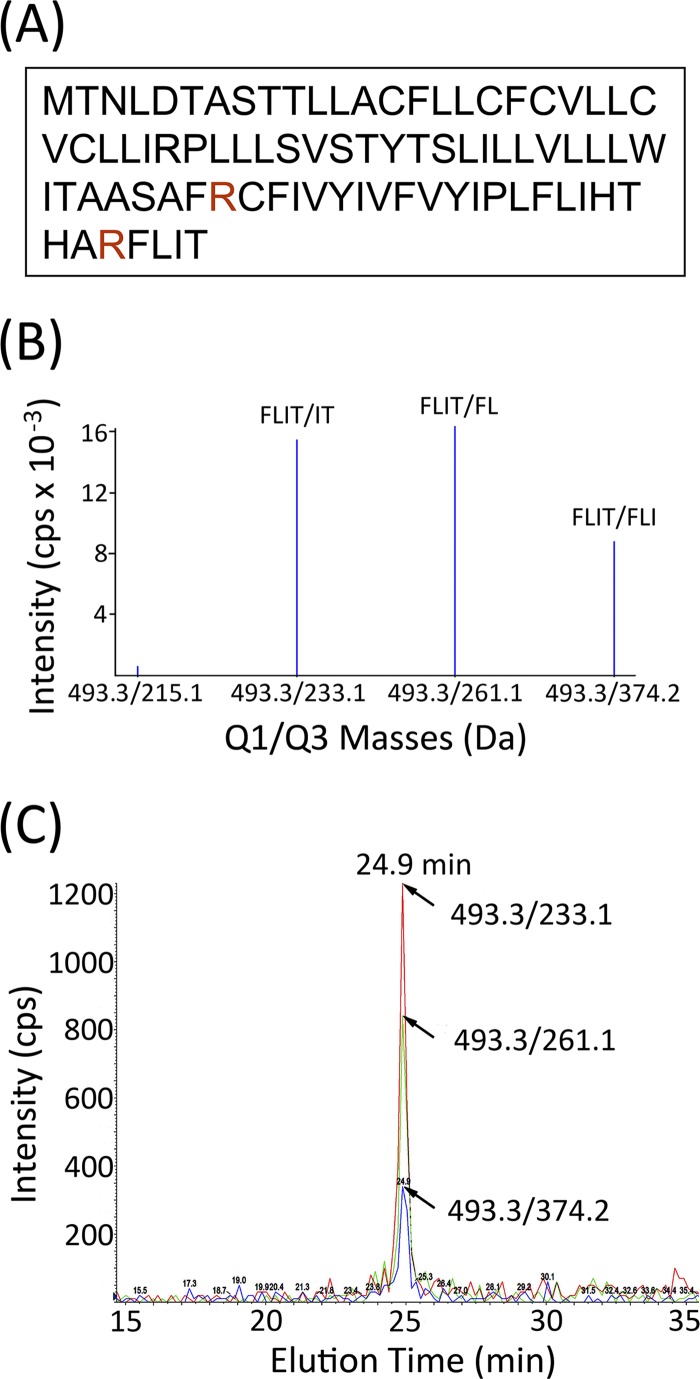
16E5 is a small (83-amino-acid), highly hydrophobic protein that contains three arginine residues and no lysine residues. Since one arginine is followed by proline, it is not a reliable cleavage site for trypsin (47); thus, a tryptic digest of 16E5 yields three peptides (58, 21, and 4 amino acids) (Fig. 1A). The four-amino-acid peptide FLIT has a favorable charge-to-mass ratio for MS and was investigated as a potential marker for 16E5. Since trypsin cleavage could be preceded by SDS-PAGE to exclude proteins smaller than 8 kDa and larger than14 kDa, a BLAST search for all permutations of C-terminal KFLIT and RFLIT (a total of 48 possibilities) in 8- to 14-kDa proteins was conducted and revealed no cellular proteins containing this sequence. Trypsin cleavage of any non-C-terminal KFLIT or RFLIT was not considered, as it would generate FLITK or FLITR, rather than FLIT. Therefore, a quantitative MS-based assay was developed to measure levels of this unique 16E5 marker in cell lysates.
Fig 1.
Characterization of the FLIT peptide as a marker for 16E5. (A) Amino acid sequence of 16E5. The protein is cleaved by trypsin after the 2 arginine residues indicated in red, which generates 3 peptides, including C-terminal FLIT. (B) MRM shows breakdown of the parental FLIT ion (493.3 Da) to three major secondary ions during MS: IT (233.1 Da), FL (261.1 Da), and FLI (374.2 Da). (C) FLIT chromatogram showing all 3 transitions eluting at 24.9 min. MRM was performed on synthetic FLIT peptide that was eluted from a C18 UPLC nanocolumn (15-cm length) with an increasing acetonitrile gradient.
Purified synthetic FLIT peptide was analyzed by MS to identify secondary ions that result from breakdown of the parental ion. MRM revealed three major transitions (Fig. 1B), corresponding to breakdown of the parental ion FLIT (493.3 Da) to IT (233.1 Da), FL (261.1 Da), and FLI (374.2 Da). When purified FLIT peptide was loaded onto a 15-cm C18 reversed-phase UPLC nanocolumn in 0.1% formic acid and eluted with an acetonitrile gradient, all three FLIT transitions exhibited a retention time of 24.9 min (Fig. 1C), thereby indicating where FLIT would chromatograph in tryptic digests of cellular samples.
16E5 expression in transfected COS cells.
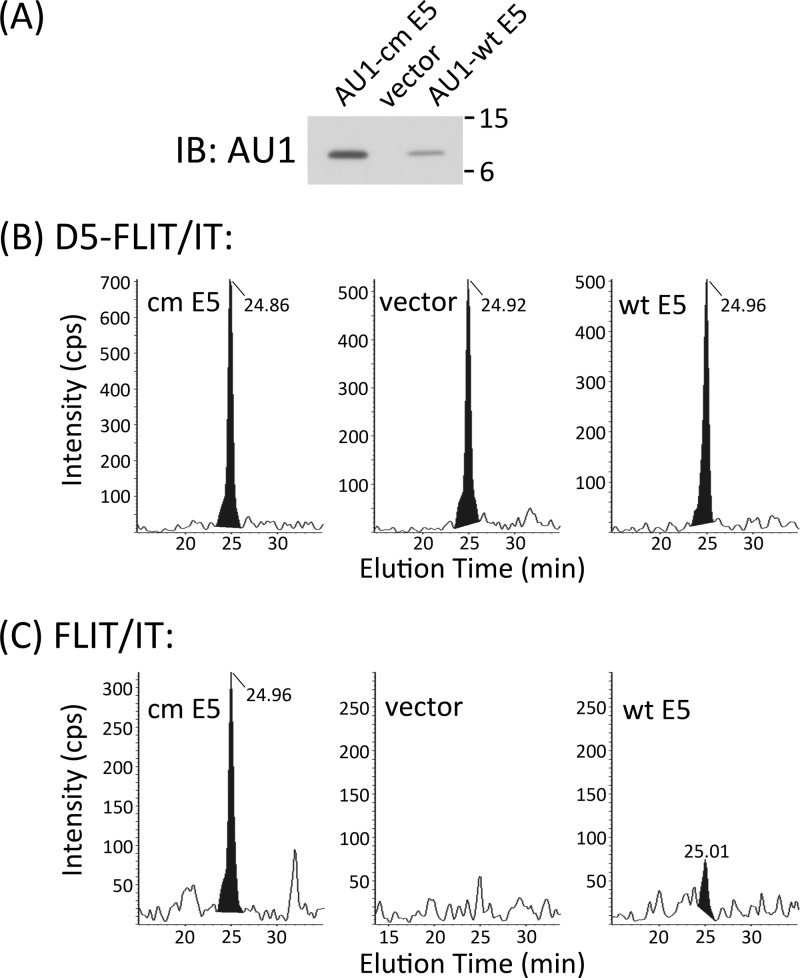
Having characterized the chromatographic profile and MS breakdown of synthetic FLIT, we attempted to detect FLIT in tryptic digests of transfected COS cells, where 16E5 is expressed at a high level. We transiently transfected COS cells with the pJS55 empty vector and with pJS55 encoding AU1 epitope-tagged wt 16E5 or epitope-tagged 16E5 in which infrequently used codons are converted to those more common in mammalian genes in order to increase the level of expression (17). An anti-AU1 Western blot of lysates prepared 24 h after transfection showed that codon-modified (cm) 16E5 was expressed at a 3.3-fold-higher level than the wt protein and that E5 was not present in cells transfected with the empty expression vector (Fig. 2A).
Fig 2.
Quantification of the 16E5 protein and 16E5 marker FLIT in transfected COS cells. (A) Anti-AU1 Western blot (IB) of COS cells 24 h after transfection with an empty expression vector or the same vector encoding AU1 epitope-tagged wt 16E5 or 16E5 which has been codon modified (cm) to optimize expression. (B) Chromatograms showing the D5-FLIT/IT transition for 0.5 pg of deuterated FLIT standard which was added to each of the tryptic digests described below. (C) Chromatograms showing the FLIT/IT transition for endogenous FLIT peptide in tryptic digests of COS cell lysates prepared 24 h after transfection as for panel A. Note the absence of a FLIT peak in cells transfected with the empty expression vector. Levels of 16E5 expression were calculated from the area under the peak of FLIT/IT transitions compared to the area of corresponding D5-FLIT/FLIT transitions. All retention times of the deuterated internal standards and endogenous FLIT were 24.95 ± 0.1 min (15-cm column).
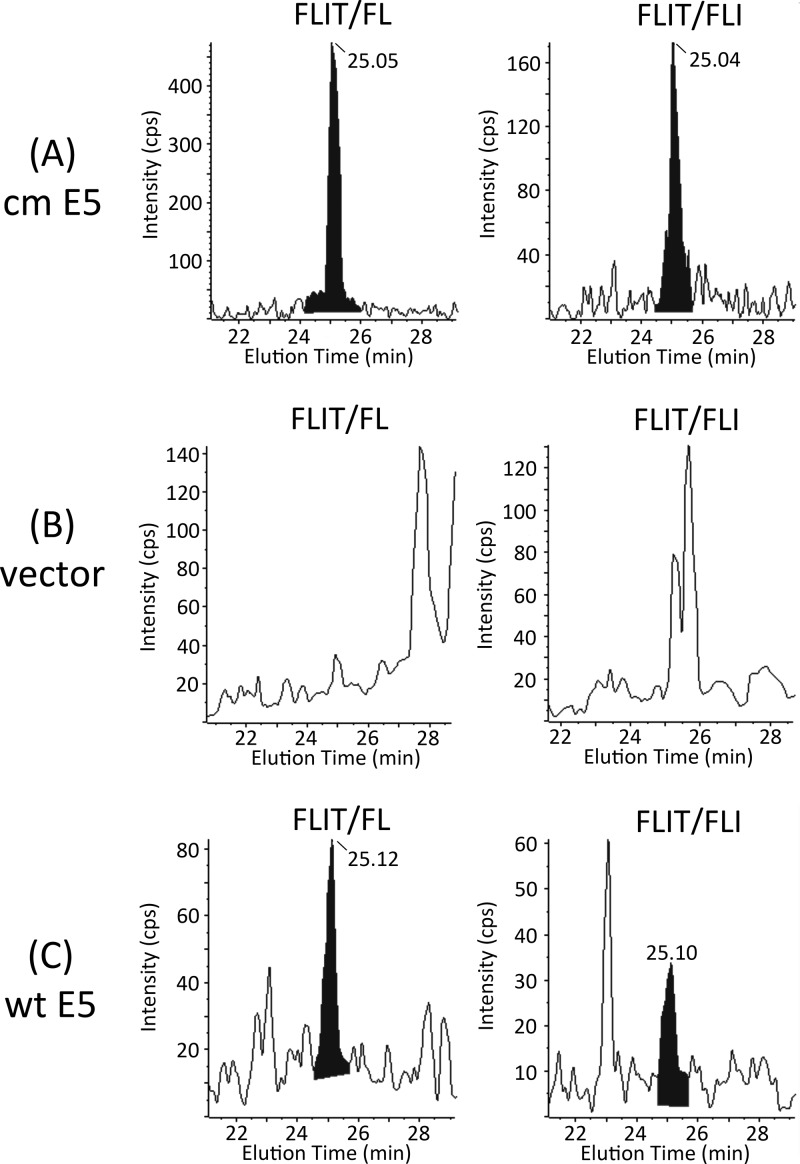
COS cell lysates were enriched for 16E5 (molecular mass, 9.4 kDa) by fractionation on a 4 to 12% SDS polyacrylamide gel. (Since AU1 is appended to the N terminus of 16E5, trypsin treatment still generates a C-terminal FLIT peptide.) Sections of unstained gel corresponding to 8 to 14 kDa were excised from each lane and digested with trypsin overnight. After ultrasonic disruption, a portion of each sample was combined with 0.5 pg of deuterated FLIT peptide (D5-FLIT), in which 5 hydrogen atoms on the N-terminal phenylalanine are replaced with deuterium. MRM can distinguish breakdown of this peptide from that of endogenous FLIT, since it undergoes distinct transitions to secondary ions: D5-FLIT/IT (498.3 kDa/233.1 kDa), D5-FLIT/D5-FL (498.3 kDa/266.1 kDa), and D5-FLIT/D5-FLI (498.3 kDa/379.2 kDa). Thus, absolute levels of endogenous FLIT (and 16E5) can be quantified relative to the known amount of internal deuterated standard. As shown in Fig. 2B and C, D5-FLIT and endogenous FLIT coeluted from the C18 UPLC nanocolumn. However, while D5-FLIT was detected in all three samples (Fig. 2B), FLIT was not present in tryptic digests of COS cells transfected with the empty expression vector (Fig. 2C). A comparison of the areas under these peaks indicated that cells transfected with cm 16E5 DNA contained 26.5 ng of E5 protein per ml of lysate, while cells transfected with wt 16E5 DNA contained 7.18 ng/ml. Considering the number of cells per ml of each lysate (determined by counting cells from identical cultures that were plated and transfected at the same time as those which were lysed) and a transfection efficiency of 50%, cm 16E5 was present at a level of 554,000 molecules/cell and wt 16E5 at 133,000 molecules/cell. This 4.2-fold difference in expression is similar to the 3.3-fold difference obtained from the anti-AU1 Western blot and demonstrates that 16E5 can be quantified using our MS-based assay. The same results were obtained by analysis of FLIT/FL and FLIT/FLI transitions for the endogenous FLIT peptide (Fig. 3).
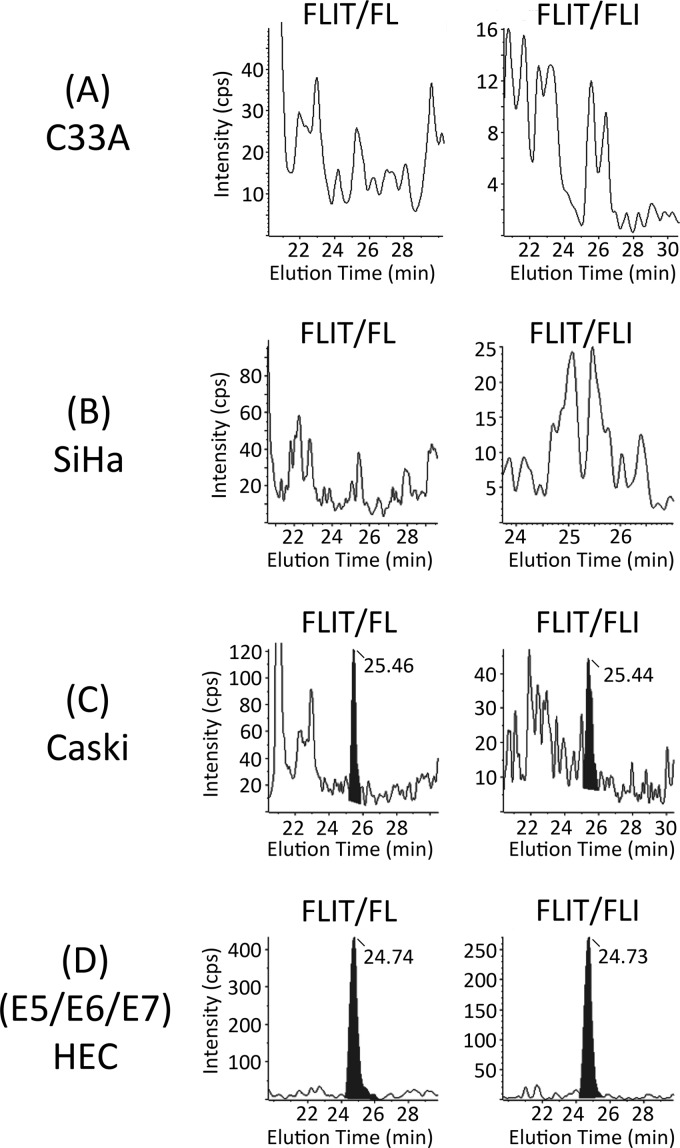
Fig 3.
Chromatograms of FLIT/FL and FLIT/FLI transitions for endogenous FLIT peptide in tryptic digests of COS cell lysates prepared 24 h after transfection with DNA encoding 16E5 which has been codon modified (cm) to optimize expression (A), the empty expression vector (B), or wt 16E5 (C). FLIT retention times were 25.08 ± 0.04 min (15-cm column).
16E5 protein levels in immortalized and transformed cervical cell lines.
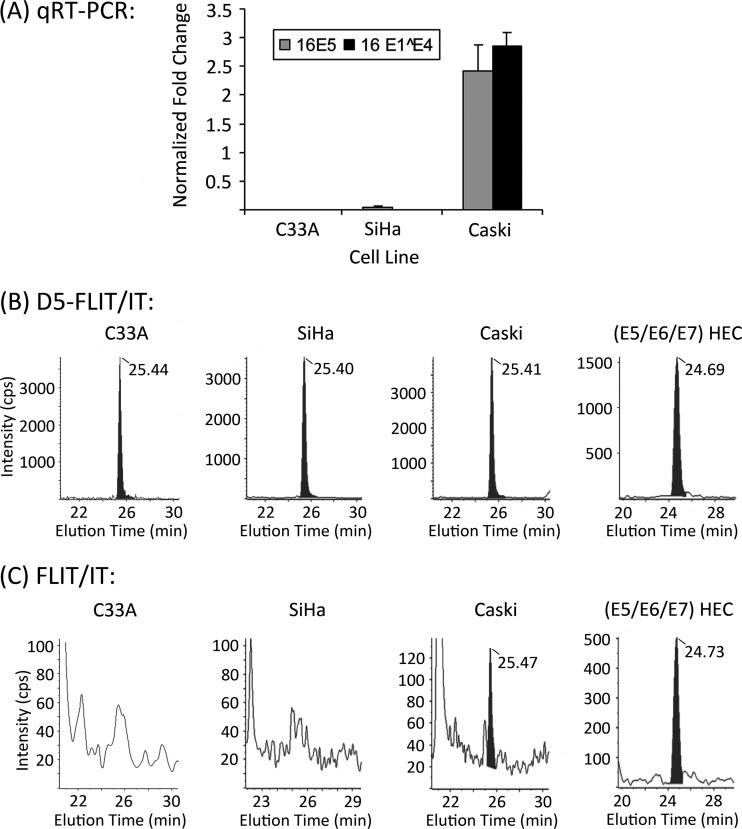
Previous reports suggest that two HPV-16-positive cervical cancer cell lines, SiHa and Caski, have either detectable E5 ORFs or potentially E5-coding mRNA transcripts (5, 6, 50). We performed quantitative real-time PCR on these cell lines using primers for 16E5, as well as primers for the HPV-16 E1^E4 spliced transcript of the E1 and E4 ORFs, which is upstream of E5. The HPV-negative cervical cancer cell line C33A was included as a negative control. The presence of 16E5 mRNA was confirmed in Caski cells, as was the E1^E4 transcript. In contrast, neither mRNA was detected in SiHa or C33A cells (Fig. 4A).
Fig 4.
16E5 levels in immortalized and transformed cervical cell lines. (A) Real-time PCR showing levels of E5 and E1^E4 transcripts relative to GAPDH in HPV-16-negative (C33A) and HPV-16-positive (SiHa and Caski) cervical cancer cell lines. Error bars represent means ± standard errors of the means (SEM) (n = 3). (B) Chromatograms of D5-FLIT/FLIT transitions for the deuterated internal standard added to tryptic digests of the cervical cancer cell lines in panel A and for HPV-16 E6/E7-immortalized HECs that stably express wt 16E5. (C) Chromatograms of FLIT/IT transitions for endogenous FLIT peptide in the tryptic digests. No endogenous FLIT was detected in C33A and SiHa cells. All retention times of the deuterated internal standards and endogenous FLIT were 25.08 ± 0.4 min (15-cm column).
MS was then used to quantify the FLIT 16E5 peptide in tryptic digests of these cell lines. In addition, HPV-16 E6/E7-immortalized HECs that stably express wt 16E5 were included as a positive control. Figure 4B shows D5-FLIT/IT transition data for the D5-FLIT internal standard added to all samples. Endogenous FLIT was present in the (16E5/E6/E7) HECs and in Caski cells (Fig. 4C), which also contained E5 mRNA. E5 was not detected in the negative control (C33A cells) or in SiHa cells (Fig. 4C). A comparison of the areas under the peaks of endogenous FLIT and D5-FLIT showed 105,000 copies of 16E5 protein per cell in (16E5/E6/E7) HECs and 12,700 copies per Caski cell. The FLIT/FL and FLIT/FLI transitions of endogenous FLIT peptide agree with these results (Fig. 5). Since the HEC line was generated by infection with an E5-encoding retrovirus and G418 selection, the observed level of E5 expression likely reflects efficient transcription from the long terminal repeat (LTR) retroviral promoter at multiple integration sites. In contrast, the presence of HPV-16 E1^E4 spliced transcript in Caski cells suggests that E5 expression may be driven by the HPV-16 promoter. Importantly, the detection of 16E5 protein in Caski cells shows that 16E5 can be expressed in cells that exclusively contain an integrated HPV genome.
Fig 5.
Chromatograms of FLIT/FL and FLIT/FLI transitions for endogenous FLIT peptide in tryptic digests of C33A (A), SiHa (B), Caski (C), and HPV-16 E6/E7-immortalized HECs that stably express wt 16E5 (D). FLIT retention times were 25.10 ± 0.4 min (15-cm column).
Confirmation of 16E5 protein in Caski cells using E5 immunoprecipitation.
Concurrent with our MS studies, we generated rabbit antiserum directed against the 9 C-terminal amino acids of 16E5. Although the E5 antiserum was not effective for Western blotting or immunofluorescence microscopy (data not shown), it was useful for immunoprecipitation. This was demonstrated by immunoprecipitating 16E5 from COS cells that were transfected with the empty pJS55 expression vector or with pJS55 encoding AU1 epitope-tagged or untagged 16E5. As shown in Fig. 6A, an anti-AU1 Western blot of the immunoprecipitates detected AU1-16E5 in cells transfected with this construct when the E5 antiserum was used for immunoprecipitation. AU1 was not detected when preimmune serum was used for immunoprecipitation or in anti-E5 immunoprecipitates from cells transfected with untagged 16E5 or the empty expression vector (Fig. 6A).
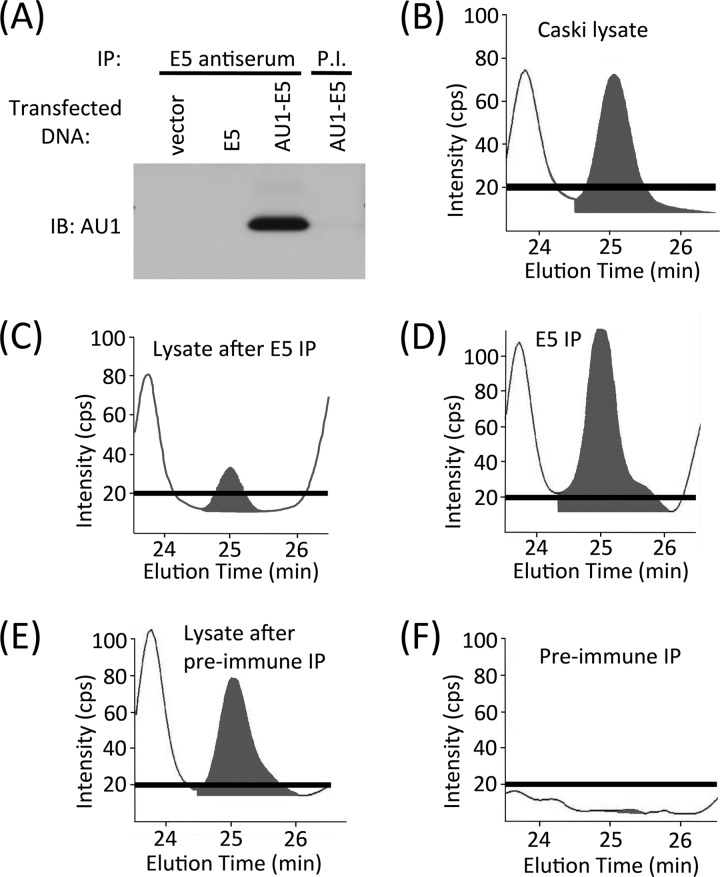
Fig 6.
Immunoprecipitation confirms that the MS-detected FLIT peptide in Caski cells originates from 16E5. (A) 16E5 antiserum immunoprecipitates the 16E5 protein. COS cells were transfected with an empty expression vector or with the same vector encoding 16E5 with or without an AU1 epitope tag. Twenty-four hours later, the cells were lysed and immunoprecipitations (IP) were performed (on equal amounts of cell protein) using 16E5 antiserum or preimmune serum from the same rabbit (P.I.). Immunoprecipitated proteins were analyzed on Western blots labeled with an anti-AU1 monoclonal antibody (IB: AU1). (B to F) 16E5 antiserum “clears” Caski cell lysates of FLIT peptide. A Caski cell lysate was digested with trypsin and analyzed by UPLC and MS without immunoprecipitation (B) or following immunoprecipitation with 16E5 antiserum (C) or preimmune serum (E). Proteins immunoprecipitated by the 16E5 antiserum (D) or preimmune serum (F) were eluted, digested with trypsin, and analyzed by UPLC and MS. The black bar indicates the signal intensity of the blank (no sample).
To confirm that the endogenous FLIT peptide detected by MS in tryptic digests of Caski cells was indicative of 16E5 expression, E5 antiserum was used to deplete Caski cell lysates of FLIT. A RIPA lysate of Caski cells was divided into three aliquots; two were immunoprecipitated with either E5 antiserum or preimmune serum, and the third was not subjected to immunoprecipitation. All lysates (and immunoprecipitates) were then digested with trypsin and analyzed by MS. The results showed that immunoprecipitation with E5 antiserum largely depleted FLIT from the lysate (Fig. 6B and C) and that FLIT was present in the immunoprecipitate (Fig. 6D). In contrast, immunoprecipitation with preimmune serum did not reduce FLIT in the lysate (Fig. 6B and E), and FLIT was not detected in the immunoprecipitate (Fig. 6F). These E5 immunoprecipitation experiments clearly demonstrate that 16E5 is solely responsible for the FLIT peptide detected by MS.
E5 expression from HPV-16 episomes.
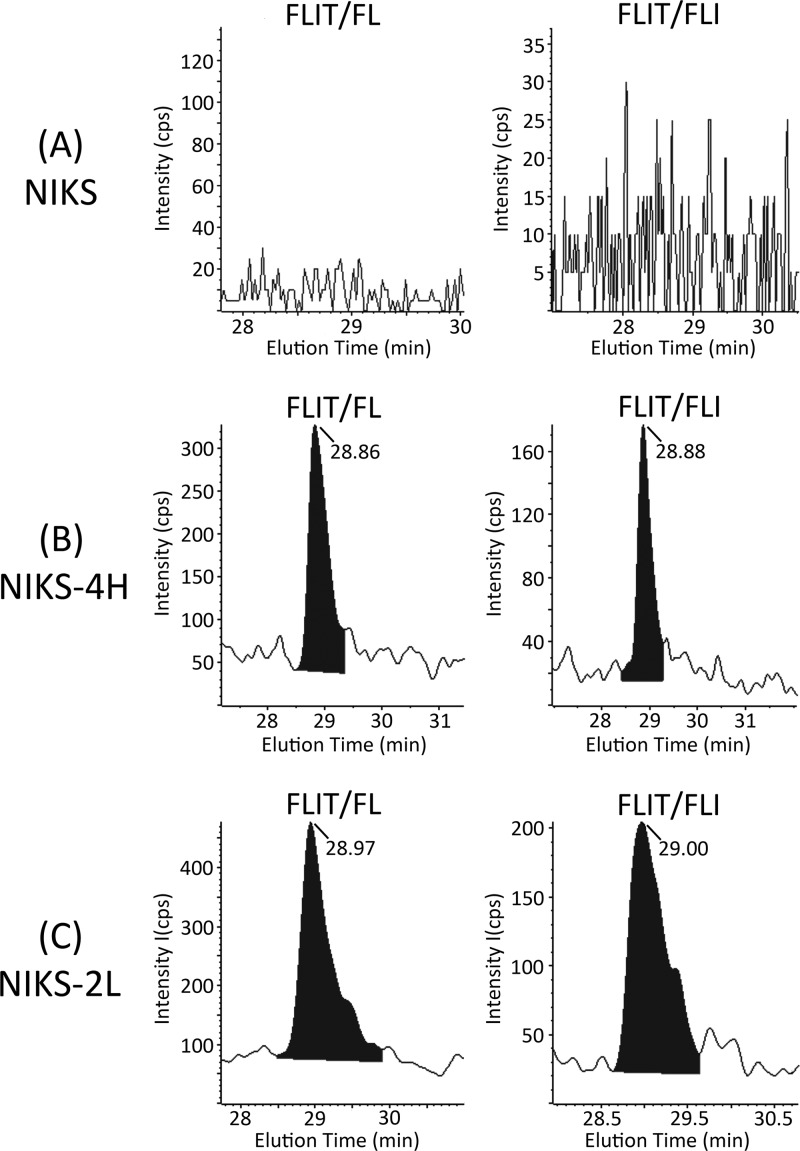
During early stages of infection, the HPV genome is maintained as multiple episomal copies. The viral early promoter regulates transcription of a polycistronic mRNA which encodes the E1, E2, E4, E5, E6, and E7 early proteins (18). To measure levels of 16E5 protein when expressed from episomes under the control of the early viral promoter, two clonal cell lines harboring episomal copies of the HPV-16 genome, NIKS-2L and NIKS-4H, were used for MS analysis. These cell lines replicate both the pathology and pattern of viral gene expression of low-grade (NIKS-2L) or high-grade (NIKS-4H) squamous intraepithelial lesions (LSIL and HSIL) in organotypic raft cultures (24). Parental NIKS cells also were analyzed as a negative control. Comparison of the areas under the peaks of the D5-FLIT/IT internal standard (Fig. 7A) and FLIT/IT analyte (Fig. 7B) indicated that NIKS-4H contained 21,400 molecules of 16E5 protein per cell and that NIKS-2L contained 48,300 molecules per cell. 16E5 was not detected in the parental NIKS line. The FLIT/FL and FLIT/FLI transitions of endogenous FLIT peptide agree with these results (Fig. 8). Therefore, wt 16E5 is expressed at similar levels in our E6/E7-immortalized HECs and the NIKS-2L cells that harbor episomal HPV-16 DNA (2.2-fold difference).
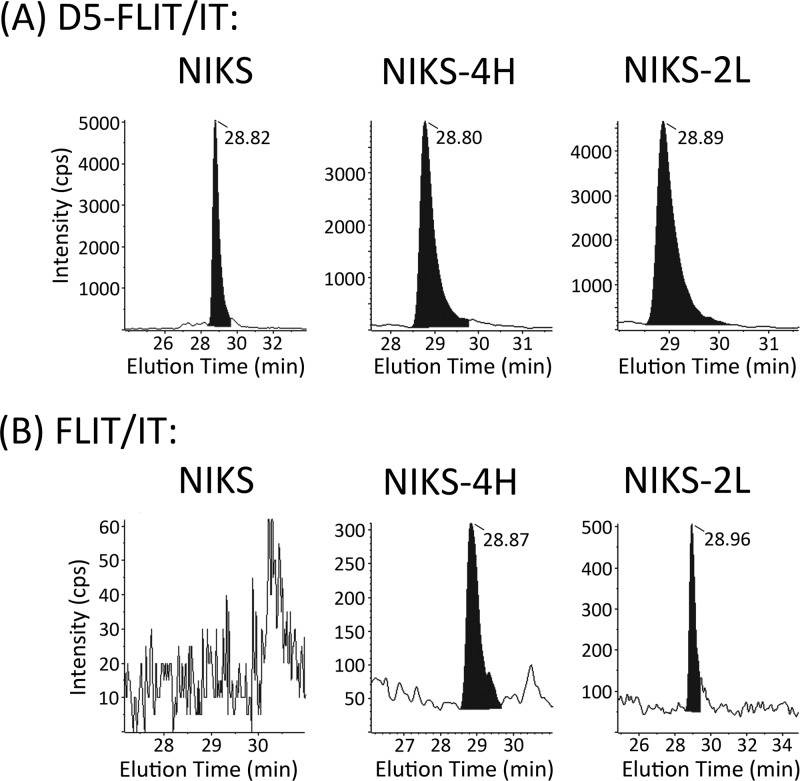
Fig 7.
Levels of 16E5 expressed from epithelial cell lines harboring episomal HPV-16 genomes. (A) Chromatograms of D5-FLIT/FLIT transitions for deuterated internal standard added to tryptic digests of parental NIKS cells or NIKS-4H and NIKS-2L clonal cell lines harboring episomal copies of the HPV-16 genome. (B) Chromatograms of FLIT/IT transitions for endogenous FLIT peptide in the tryptic digests. No endogenous FLIT was detected in the parental NIKS cells. All retention times of the deuterated internal standards and endogenous FLIT were 28.88 ± 0.1 min (25-cm column).
Fig 8.
Chromatograms of FLIT/FL and FLIT/FLI transitions for endogenous FLIT peptide in tryptic digests of parental NIKS cells (A) or NIKS-4H (B) and NIKS-2L (C) clonal cell lines harboring episomal copies of the HPV-16 genome. FLIT retention times were 28.93 ± 0.1 min (25-cm column).
DISCUSSION
Although E5 expression is generally thought to be lost upon integration of HPV DNA into the host genome, some studies have demonstrated the presence of an E5 ORF and transcripts that may encode E5 in cervical cancer cell lines with integrated HPV DNA (5, 6, 41, 50, 51). However, since all HPV early genes are encoded by spliced polycistronic mRNAs that contain a 3′-terminal E5 ORF (18), it cannot be assumed that any of these transcripts are actually translated into E5 protein. To date, there is no direct evidence that the E5 protein is present in cervical cancer cell lines. Due to the highly hydrophobic, nonimmunogenic properties of 16E5, only two studies have used antibodies to detect 16E5 protein in cervical tissue (9, 27). Attempts to recreate these antibodies have not been successful, and the results have not been confirmed.
Our approach has been to develop a direct, nonimmunologic method for quantitatively detecting the 16E5 protein in cells. We used MS to establish for the first time that E5 is expressed in Caski cervical cancer cells that contain an integrated HPV-16 genome. In addition, we determined levels of the E5 protein in E6/E7-immortalized cervical cells stably transduced with wt 16E5 and in keratinocytes that contain episomal HPV-16 DNA. While MS has been used previously to identify interactions between viral and cellular proteins (23, 19) and to identify proteins and their modifications (32, 33), we are unaware of its use to identify specific HPV proteins in clinical specimens and derived cell lines. It is noteworthy that episomal E5 expression is 2.3-fold higher in the LSIL-like NIKS-2L cell line than in the HSIL-like NIKS-4H. NIKS-2L is less proliferative than NIKS-4H at confluence (24), and the higher level of E5 observed in NIKS-2L at near confluence in the present study may be related to the initial signs of differentiation and episomal copy number rise as the cells move toward genome amplification.
In vivo studies have shown that transgenic mice expressing HPV-16 E5, E6, and E7 (2) develop larger tumors than mice expressing E6 and E7 alone (48). Since we unequivocally show that 16E5 can be present with E6/E7 in the late stages of human cervical cancer, even following integration of the viral genome, it will now be interesting to determine if cervical cancers (and other HPV-induced cancers) differ in their aggressiveness and/or response to therapy depending upon whether E5 is expressed. This expression will depend on the precise integration site of the HPV genome, since the E2, E4, and E5 ORFs frequently are disrupted upon integration (5, 13, 14, 52). It also is possible that HPV-16 E5 may be expressed from an episomal viral genome, which has been reported to persist in 26 to 76% of malignant cervical lesions (9, 11, 16, 22, 37, 44).
In this regard, the high sensitivity of our MS-based assay should allow quantitative detection of 16E5 in archived paraffin-embedded biopsy specimens in order to correlate its expression with clinical outcomes. After deparaffinization and rehydration, the proteins present in sections can be digested with trypsin in situ and the resulting peptides recovered for MS analysis using hydrophobic magnetic beads (31). The MS assay also will make possible prospective in vitro studies, in which the effect of 16E5 expression on the efficacy of chemotherapeutic agents can be examined in conditionally immortalized primary cells from normal and tumor tissues (10, 35).
ACKNOWLEDGMENTS
This work was supported by grant R01-CA053371 from the National Cancer Institute awarded to R.S.
We thank the Proteomics and Metabolomics Shared Resource at the Georgetown University Lombardi Comprehensive Cancer Center (Washington, DC).
Footnotes
Published ahead of print 27 June 2012
REFERENCES
- 1. Allen-Hoffmann BL, et al. 2000. Norman growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J. Invest. Dermatol. 114:444–455 [DOI] [PubMed] [Google Scholar]
- 2. Arbeit JM, Munger K, Howley PM, Hanahan D. 1994. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J. Virol. 68:4358–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashrafi GH, Haghshenas M, Marchetti B, Campo MS. 2006. E5 protein of human papillomavirus 16 downregulates HLA class I and interacts with the heavy chain via its first hydrophobic domain. Int. J. Cancer 119:2105–2112 [DOI] [PubMed] [Google Scholar]
- 4. Baege AC, Berger A, Schlegel R, Veldman T, Schlegel R. 2002. Cervical epithelial cells transduced with the papillomavirus E6/E7 oncogenes maintain stable levels of oncoprotein expression but exhibit progressive, major increases in hTERT gene expression and telomerase activity. Am. J. Pathol. 160:1251–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker CC, et al. 1987. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 61:962–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bauer-Hofmann R, et al. 1996. Genomic cloning and characterization of the nonoccupied allele corresponding to the integration site of human papillomavirus type 16 DNA in the cervical cancer cell line SiHa. Virology 217:33–41 [DOI] [PubMed] [Google Scholar]
- 7. Bouvard V, Matlashewski G, Gu ZM, Storey A, Banks L. 1994. The human papillomavirus type 16 E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology 203:73–80 [DOI] [PubMed] [Google Scholar]
- 8. Boyer SN, Wazer DE, Band V. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620–4624 [PubMed] [Google Scholar]
- 9. Chang JL, et al. 2001. The expression of HPV-16 E5 protein in squamous neoplastic changes in the uterine cervix. J. Biomed. Sci. 8:206–213 [DOI] [PubMed] [Google Scholar]
- 10. Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. 2010. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest. 120:2619–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choo KB, et al. 1987. Presence of episomal and integrated human papillomavirus DNA sequences in cervical carcinoma. J. Med. Virol. 21:101–107 [DOI] [PubMed] [Google Scholar]
- 12. Clifford GM, Smith JS, Plummer M, Munoz N, Franceschi S. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer 88:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collins SI, et al. 2009. Disruption of the E2 gene is a common and early event in the natural history of cervical human papillomavirus infection: a longitudinal cohort study. Cancer Res. 69:3828–3832 [DOI] [PubMed] [Google Scholar]
- 14. Corden SA, Sant-Cassia LJ, Easton AJ, Morris AG. 1999. The integration of HPV-18 DNA in cervical carcinoma. Mol. Pathol. 52:275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crusius K, Auvinen E, Steuer B, Gaissert H, Alonso A. 1998. The human papillomavirus type 16 E5-protein modulates ligand-dependent activation of the EGF receptor family in the human epithelial cell line HaCaT. Exp. Cell Res. 241:76–83 [DOI] [PubMed] [Google Scholar]
- 16. Cullen AP, Reid R, Campion M, Lorincz AT. 1991. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J. Virol. 65:606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Disbrow GL, Sunitha I, Baker CC, Hanover J, Schlegel R. 2003. Codon optimization of the HPV-16 E5 gene enhances protein expression. Virology 311:105–114 [DOI] [PubMed] [Google Scholar]
- 18. Fehrmann F, Laimins LA. 2003. Human papillomaviruses: targeting differentiating epithelial cells for malignant transformation. Oncogene 22:5201–5207 [DOI] [PubMed] [Google Scholar]
- 19. Forsman A, Ruetschi U, Ekholm J, Rymo L. 2008. Identification of intracellular proteins associated with the EBV-encoded nuclear antigen 5 using an efficient TAP procedure and FT-ICR mass spectrometry. J. Proteome Res. 7:2309–2319 [DOI] [PubMed] [Google Scholar]
- 20. Genther Williams SM, et al. 2005. Requirement of epidermal growth factor receptor for hyperplasia induced by E5, a high-risk human papillomavirus oncogene. Cancer Res. 65:6534–6542 [DOI] [PubMed] [Google Scholar]
- 21. Gruener M, Bravo IG, Momburg F, Alonso A, Tomakidi P. 2007. The E5 protein of the human papillomavirus type 16 down-regulates HLA-I surface expression in calnexin-expressing but not in calnexin-deficient cells. Virol. J. 4:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hafner N, et al. 2008. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene 27:1610–1617 [DOI] [PubMed] [Google Scholar]
- 23. Huh KW, et al. 2005. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. U. S. A. 102:11492–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Isaacson Wechsler E, et al. 2012. Reconstruction of human papillomavirus type 16-mediated early-stage neoplasia implicates E6/E7 deregulation and the loss of contact inhibition in neoplastic progression. J. Virol. 86:6358–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jeon S, Allen-Hoffmann BL, Lambert PF. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69:2989–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kabsch K, et al. 2004. The HPV-16 E5 protein inhibits TRAIL- and FasL-mediated apoptosis in human keratinocyte raft cultures. Intervirology 47:48–56 [DOI] [PubMed] [Google Scholar]
- 27. Kell B, et al. 1994. Detection of E5 oncoprotein in human papillomavirus type 16-positive cervical scrapes using antibodies raised to synthetic peptides. J. Gen. Virol. 75:2451–2456 [DOI] [PubMed] [Google Scholar]
- 28. Krawczyk E, Hanover JA, Schlegel R, Suprynowicz FA. 2008. Karyopherin beta3: a new cellular target for the HPV-16 E5 oncoprotein. Biochem. Biophys. Res. Commun. 371:684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krawczyk E, et al. 2008. Koilocytosis: a cooperative interaction between the human papillomavirus E5 and E6 oncoproteins. Am. J. Pathol. 173:682–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 31. Lemaire R, et al. 2007. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. Proteome Res. 6:1295–1305 [DOI] [PubMed] [Google Scholar]
- 32. Lewis JK, Bendahmane M, Smith TJ, Beachy RN, Siuzdak G. 1998. Identification of viral mutants by mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 95:8596–8601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li M, Garcea RL. 1994. Identification of the threonine phosphorylation sites on the polyomavirus major capsid protein VP1: relationship to the activity of middle T antigen. J. Virol. 68:320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lim PS, et al. 1990. Distribution and specific identification of papillomavirus major capsid protein epitopes by immunocytochemistry and epitope scanning of synthetic peptides. J. Infect. Dis. 162:1263–1269 [DOI] [PubMed] [Google Scholar]
- 35. Liu X, et al. 2012. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 180:599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu X, Roberts J, Dakic A, Zhang Y, Schlegel R. 2008. HPV E7 contributes to the telomerase activity of immortalized and tumorigenic cells and augments E6-induced hTERT promoter function. Virology 375:611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsukura T, Koi S, Sugase K. 1989. Both episomal and integrated forms of human papillomavirus type 16 are involved in invasive cervical cancers. Virology 172:63–72 [DOI] [PubMed] [Google Scholar]
- 38. Mincheva A, Gissmann L, zur Hausen H. 1987. Chromosomal integration sites of human papillomavirus DNA in three cervical cancer cell lines mapped by in situ hybridization. Med. Microbiol. Immunol. 176:245–256 [DOI] [PubMed] [Google Scholar]
- 39. Morgenstern JP, Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Munger K, et al. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pater MM, Pater A. 1985. Human papillomavirus types 16 and 18 sequences in carcinoma cell lines of the cervix. Virology 145:313–318 [DOI] [PubMed] [Google Scholar]
- 42. Pear WS, Nolan GP, Scott ML, Baltimore D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 90:8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pim D, Collins M, Banks L. 1992. Human papillomavirus type 16 E5 gene stimulates the transforming activity of the epidermal growth factor receptor. Oncogene 7:27–32 [PubMed] [Google Scholar]
- 44. Pirami L, Giache V, Becciolini A. 1997. Analysis of HPV16, 18, 31, and 35 DNA in pre-invasive and invasive lesions of the uterine cervix. J. Clin. Pathol. 50:600–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Regan JA, Laimins LA. 2008. Bap31 is a novel target of the human papillomavirus E5 protein. J. Virol. 82:10042–10051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Riley RR, et al. 2003. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 63:4862–4871 [PubMed] [Google Scholar]
- 47. Rodriguez J, Gupta N, Smith RD, Pevzner PA. 2008. Does trypsin cut before proline? J. Proteome Res. 7:300–305 [DOI] [PubMed] [Google Scholar]
- 48. Romanczuk H, Howley PM. 1992. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl. Acad. Sci. U. S. A. 89:3159–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136 [DOI] [PubMed] [Google Scholar]
- 50. Sherman L, Alloul N, Golan I, Durst M, Baram A. 1992. Expression and splicing patterns of human papillomavirus type-16 mRNAs in pre-cancerous lesions and carcinomas of the cervix, in human keratinocytes immortalized by HPV 16, and in cell lines established from cervical cancers. Int. J. Cancer 50:356–364 [DOI] [PubMed] [Google Scholar]
- 51. Shirasawa H, et al. 1988. Transcriptional differences of the human papillomavirus type 16 genome between precancerous lesions and invasive carcinomas. J. Virol. 62:1022–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shirasawa H, Tomita Y, Sekiya S, Takamizawa H, Simizu B. 1987. Integration and transcription of human papillomavirus type 16 and 18 sequences in cell lines derived from cervical carcinomas. J. Gen. Virol. 68:583–591 [DOI] [PubMed] [Google Scholar]
- 53. Stoppler MC, Straight SW, Tsao G, Schlegel R, McCance DJ. 1996. The E5 gene of HPV-16 enhances keratinocyte immortalization by full-length DNA. Virology 223:251–254 [DOI] [PubMed] [Google Scholar]
- 54. Straight SW, Hinkle PM, Jewers RJ, McCance DJ. 1993. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J. Virol. 67:4521–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suprynowicz FA, Campo MS, Schlegel R. 2006. Biological activitites of papillomavirus E5 proteins, p 97–113 In Campo MS. (ed), Papillomavirus research: from natural history to vaccines and beyond. Caister Academic Press, Wymondham, United Kingdom [Google Scholar]
- 56. Suprynowicz FA, Disbrow GL, Simic V, Schlegel R. 2005. Are transforming properties of the bovine papillomavirus E5 protein shared by E5 from high-risk human papillomavirus type 16? Virology 332:102–113 [DOI] [PubMed] [Google Scholar]
- 57. Suprynowicz FA, et al. 2010. The human papillomavirus type 16 E5 oncoprotein inhibits epidermal growth factor trafficking independently of endosome acidification. J. Virol. 84:10619–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thomsen P, van Deurs B, Norrild B, Kayser L. 2000. The HPV-16 E5 oncogene inhibits endocytic trafficking. Oncogene 19:6023–6032 [DOI] [PubMed] [Google Scholar]
- 59. Thorland EC, Myers SL, Gostout BS, Smith DI. 2003. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene 22:1225–1237 [DOI] [PubMed] [Google Scholar]
- 60. Valle GF, Banks L. 1995. The human papillomavirus (HPV)-6 and HPV-16 E5 proteins co-operate with HPV-16 E7 in the transformation of primary rodent cells. J. Gen. Virol. 76:1239–1245 [DOI] [PubMed] [Google Scholar]
- 61. Veldman T, Horikawa I, Barrett JC, Schlegel R. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 75:4467–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Werness BA, Levine AJ, Howley PM. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76–79 [DOI] [PubMed] [Google Scholar]
- 63. Yee C, Krishnan-Hewlett I, Baker CC, Schlegel R, Howley PM. 1985. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am. J. Pathol. 119:361–366 [PMC free article] [PubMed] [Google Scholar]
- 64. zur Hausen H, de Villiers EM. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427–447 [DOI] [PubMed] [Google Scholar]