Abstract
Replication of the human herpesvirus Epstein-Barr virus drastically impairs cellular protein synthesis. This shutoff phenotype results from mRNA degradation upon expression of the early lytic-phase protein BGLF5. Interestingly, BGLF5 is the viral DNase, or alkaline exonuclease, homologues of which are present throughout the herpesvirus family. During productive infection, this DNase is essential for processing and packaging of the viral genome. In contrast to this widely conserved DNase activity, shutoff is only mediated by the alkaline exonucleases of the subfamily of gammaherpesviruses. Here, we show that BGLF5 can degrade mRNAs of both cellular and viral origin, irrespective of polyadenylation. Furthermore, shutoff by BGLF5 induces nuclear relocalization of the cytosolic poly(A) binding protein. Guided by the recently resolved BGLF5 structure, mutants were generated and analyzed for functional consequences on DNase and shutoff activities. On the one hand, a point mutation destroying DNase activity also blocks RNase function, implying that both activities share a catalytic site. On the other hand, other mutations are more selective, having a more pronounced effect on either DNA degradation or shutoff. The latter results are indicative of an oligonucleotide-binding site that is partially shared by DNA and RNA. For this, the flexible “bridge” that crosses the active-site canyon of BGLF5 appears to contribute to the interaction with RNA substrates. These findings extend our understanding of the molecular basis for the shutoff function of BGLF5 that is conserved in gammaherpesviruses but not in alpha- and betaherpesviruses.
INTRODUCTION
Herpesviruses are large DNA viruses that cause lifelong infections in their host. After primary infection, herpesviruses enter a stage of latency during which few viral proteins are expressed. For transmission to another host, a lytic infection is required, resulting in the generation of viral progeny. In cells productively infected with either alpha- or gammaherpesviruses, an almost complete block in host protein synthesis is observed (20, 44, 46). This shutoff activity contributes to immune evasion by inhibiting the synthesis of proteins involved in antigen presentation and pathogen recognition (44, 48, 50). Furthermore, blocking host protein synthesis could provide an advantage to viral replication since it is anticipated to devote the cellular ribosomes to the synthesis of viral proteins.
For alphaherpesviruses, such as herpes simplex virus 1 (HSV-1) and HSV-2 and bovine herpesvirus type 1, shutoff is mediated by the virion host shutoff protein (vhs) (22, 25, 30, 33, 34, 38, 41). In contrast, shutoff upon productive infection with the gammaherpesviruses Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus (KSHV), and murine gammaherpesvirus 68 (MHV68) is mediated by the alkaline exonucleases BGLF5 (44), SOX (20), and muSOX (9), respectively. These alkaline exonucleases (AEs), which were originally identified as viral DNases, have both exo- and endonuclease activities toward DNA and are conserved throughout the herpesvirus family. This conservation in all herpesviruses is likely linked to the DNase activity that is essential for packaging of the viral genome. In contrast to their universal role in packaging, shutoff activity exerted by AE proteins is only observed for the gammaherpesvirus subfamily. Possibly, the AEs of gammaherpesviruses attained shutoff activity relatively late during evolution, i.e., after separation of the gammaherpesvirus subfamily from the alpha- and betaherpesvirus subfamilies. Alternatively, shutoff activity might have been lost by the AEs of alpha- and betaherpesviruses.
Despite the shutoff proteins encoded by alpha- and gammaherpesviruses being unrelated, they all act through mRNA degradation. The alphaherpesvirus HSV-encoded vhs protein, which was identified decades ago as a mediator of host shutoff, has intrinsic RNase activity (12, 53) and associates with eukaryotic translation initiation factor 4H (eIF4H) and eIF4F (16, 17, 39). Interaction of vhs with these translation initiation factors is likely to account for the specific targeting of mRNAs for degradation. For the gammaherpesviruses, the molecular basis underlying AE-mediated shutoff is just emerging. Interestingly, AE proteins of both EBV and KSHV have recently been shown to exert RNase activity in vitro (2, 5). The SOX protein of KSHV additionally affects the cytosolic poly(A) binding protein (PABPC). Under steady-state conditions, PABPC associates with both eIF4G and the poly(A) tail of mRNAs in the cytoplasm, thereby inducing the circularization of mRNAs. This facilitates translation initiation and hampers mRNA degradation (23). Furthermore, interaction of PABPC with mRNAs retains PABPC in the cytoplasm (32). Upon expression of KSHV SOX and subsequent mRNA depletion from the cytoplasm, PABPC is relocalized to the nucleus (32, 35), where it induces hyperadenylation and nuclear retention of mRNAs (31, 35). Thus far, it is unknown whether this also occurs for EBV BGLF5.
Recently, the crystal structures of the EBV BGLF5 (5) and KSHV SOX (2, 10) proteins were solved. These structures reveal a high degree of similarity between the viral proteins and lambda exonuclease, all belonging to the D-(D/E)XK superfamily of nucleases. Despite this progress, the molecular relationship between the DNase and RNase activities of gammaherpesvirus AE proteins remains not fully resolved. Likewise, the mechanism for the selective shutoff activity of AE proteins in gammaherpesviruses has not yet been elucidated. To address these issues and to obtain more detailed insight into the molecular mechanism(s) of BGLF5 function, we examined mutants of this EBV protein for their DNase and shutoff activities.
MATERIALS AND METHODS
Sequence analysis.
The sequences of different gammaherpesvirus AE homologues have been retrieved at the National Center for Biotechnology Information using BLAST (1) and aligned with CLUSTAL W (47). Sequence identities are >39%. Sequence conservation (identities) has been mapped onto a full model of the EBV nuclease structure (5) using ESPript (24). Using the HHV6 sequence as reference, betaherpesvirus sequences can be aligned with an overall sequence conservation of >30%. A model of the betaherpesvirus exonuclease based on the template KSHV exonuclease (10) (pdb entry 3fhd) and the sequence of HHV6 has been built using SWISS-MODEL (4) and has been used for the analysis of conserved residues. Alphaherpesvirus exonuclease sequences were aligned with the one of HSV-1 as reference (sequence identities above 35%). Because of the longer sequence of HSV-1 AE compared to EBV BGLF5 or KSHV SOX, we avoided to model the HSV-1 AE structure and only used the sequence alignment for analysis. A list of the sequences used is given in Table 1.
Table 1.
Sequences used in the analysis of the sequence conservation of herpesvirus exonucleases
| Virus gi identificationa | Description |
|---|---|
| Gammaherpesvirus | |
| gi|119691 | Epstein-Barr virus strain B95-8 |
| gi|82503243 | Epstein-Barr virus strain B95-8/Raji |
| gi|123845633 | Epstein-Barr virus strain GD1 derived from NPC |
| gi|139424516 | Epstein-Barr virus strain AG876 |
| gi|228537 | Epstein-Barr virus strain P3HR1 |
| gi|51518056 | Macacine herpesvirus 4 |
| gi|24943119 | Callitrichine herpesvirus 3 |
| gi|20453824 | Porcine lymphotropic herpesvirus 1 |
| gi|27452874 | Porcine lymphotropic herpesvirus 2 |
| gi|30348540 | Saimiriine herpesvirus 2 |
| gi|9625993 | Saimiriine herpesvirus 2 |
| gi|27452835 | Porcine lymphotropic herpesvirus 3 |
| gi|10140958 | Alcelaphine herpesvirus 1 |
| gi|9631229 | Ateline herpesvirus 3 |
| gi|9628040 | Equid herpesvirus 2 |
| gi|9629578 | Murid herpesvirus 4 |
| gi|83642873 | Ovine herpesvirus 2 |
| gi|78127870 | Ovine herpesvirus 2 |
| gi|262285082 | Wood mouse herpesvirus |
| gi|134038663 | Wood mouse herpesvirus |
| gi|13095614 | Bovine herpesvirus 4 |
| gi|2246484 | Human herpesvirus 8 |
| gi|1718290 | Human herpesvirus 8 type M |
| gi|139472831 | Human herpesvirus 8 |
| gi|46519394 | Macaca fuscata rhadinovirus |
| gi|18653844 | Macacine herpesvirus 5 |
| gi|7330027 | Rhesus monkey rhadinovirus H26-95 |
| gi|321496589 | Rodent herpesvirus Peru |
| Betaherpesvirus | |
| gi|9633139 | Human herpesvirus 6 |
| gi|51874292 | Human herpesvirus 7 |
| gi|190887040 | Muromegalovirus C4A |
| gi|51556591 | Macacine herpesvirus 3 |
| gi|9845385 | Murid herpesvirus 2 |
| gi|20026688 | Panine herpesvirus 2 |
| gi|259016230 | Human herpesvirus 5 strain AD169 |
| gi|213159230 | Caviid herpesvirus 2 |
| Alphaherpesvirus | |
| gi|330077 | Human herpesvirus 1 |
| gi|9629281 | Human herpesvirus 2 |
| gi|28804647 | Cercopithecine herpesvirus 1 |
| gi|83722580 | Papiine herpesvirus 2 |
| gi|312162843 | Saimiriine herpesvirus 1 |
| gi|165911567 | Anatid herpesvirus 1 |
| gi|50313291 | Equid herpesvirus 1 |
| gi|9629778 | Equid herpesvirus 4 |
| gi|216905903 | Equid herpesvirus 9 |
| gi|255683203 | Anatid herpesvirus 1 |
| gi|270339487 | Felid herpesvirus 1 |
| gi|11095844 | Meleagrid herpesvirus 1 |
| gi|125745063 | Gallid herpesvirus 2 |
| gi|66866009 | Human herpesvirus 3 |
| gi|57790979 | Gallid herpesvirus 1 |
| gi|10834881 | Gallid herpesvirus 3 |
| gi|51557529 | Suid herpesvirus 1 |
| gi|38638245 | Psittacid herpesvirus 1 |
| gi|296316049 | Bovine herpesvirus 5 |
| gi|145685896 | Anatid herpesvirus 1 |
| gi|9629863 | Bovine herpesvirus 1 |
| gi|13242441 | Cercopithecine herpesvirus 9 |
| gi|52551080 | Fibropapilloma-associated turtle herpesvirus |
The “gi” identifiers refer to the protein database at the National Center for Biotechnology Information.
Constructs.
The EBV BGLF5 gene with a C-terminal influenza virus hemagglutinin (HA) tag (BGLF5) was cloned into the pcDEF and pcDNA3-IRES-nlsGFP vectors as described previously (44). The pcDNA3 vector contains an internal ribosomal entry site (IRES) immediately downstream of the inserted gene-of-interest to allow coexpression of the marker gene green fluorescent protein (GFP [51]; kindly provided by E. Reits, Academic Medical Center, Amsterdam, Netherlands).
The BGLF5 mutants D203S, P158A, P158S, and P158G were constructed using a BGLF5-HA-encoding plasmid as a template for site-directed mutagenesis. The following primers were used: GAT GGC ATT TTT GGG GTG TCT CTA AGC TTG TGC GTC AAT GTG GAG TCA CAG GG (sD203S), CCC TGT GAC TCC ACA TTG ACG CAC AAG CTT AGA GAC ACC CCA AAA ATG CCA TC (asD203S), AAT CAC TAC TTT GGG GGC GCC GTG GCC TTT GGC CTG CGG (sP158A), CCG CAG GCC AAA GGC CAC GGC GCC CCC AAA GTA GTG ATT TG (asP158A), AAT CAC TAC TTT GGG GGA TCC GTG GCC TTT GGC CTG CGG (sP158S), CCG CAG GCC AAA GGC CAC GGA TCC CCC AAA GTA GTG ATT TG (asP158S), AAT CAC TAC TTT GGG GGA GGT GTG GCC TTT GGC CTG CGG (sP158G), and CCG CAG GCC AAA GGC CAC ACC TCC CCC AAA GTA GTG ATT TG (asP158G). The resulting mutant BGLF5 genes were cloned into the pcDNA3-IRES-nlsGFP and pcDEF vectors.
The BGLF5 mutants K231M and G305C were generated and cloned into the pcDNA3-IRES-nlsGFP vector as described previously (55) (kindly provided by J. Zuo and M. Rowe, University of Birmingham, Birmingham, United Kingdom), and were additionally cloned into the pcDEF vector. The pcDEF HSV-1 HA-AE and pcDEF KSHV HA-SOX constructs were described before (18) (kindly provided by B. Glaunsinger, University of California, Berkeley, CA). In addition, the HSV-1 HA-AE gene was cloned into the pcDNA3-IRES-nlsGFP vector. The pcDNA3-GFP (44), pcDNA3 HLA-A2.1 (28), and pcDNA3 EBV gp42-4Met (42) constructs were described before, and pMaxGFP was obtained from Lonza.
Recombinant BGLF5 protein.
Large quantities of recombinant proteins of wild-type (wt) and D203S mutant BGLF5 were produced using a baculovirus-based expression system, as described previously (5).
In vitro transcription and translation.
As a source of small amounts of (mutant) BGLF5 protein and other AEs, pcDNA3-IRES-nlsGFP-derived vectors were linearized and used for in vitro transcription with T7 polymerase (Promega) according to the manufacturer's instructions. Transcripts were translated at 30°C for 90 min in rabbit reticulocyte lysate (Promega) according to the manufacturer's instructions. To visualize the proteins produced, in vitro translation products were separated by SDS-PAGE and detected by Western blotting (see below).
Cell lines and transient transfection.
Human embryonic kidney 293T cells (293T cells) were maintained in complete culture medium consisting of Dulbecco modified Eagle medium (DMEM; Lonza) supplemented with 10% fetal calf serum (FCS; Lonza), 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Cells were transiently transfected with the above-mentioned constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
DNase assay.
In vitro translated AE protein products were incubated with linearized plasmids in IVT-Mg2+ buffer (5 mM MgCl2, 50 mM Tris [pH 8.8], 0.1 mg of bovine serum albumin [BSA]/ml, and 1.6% β-mercaptoethanol) at 37°C to allow digestion of the DNA. Alternatively, recombinant BGLF5 wild-type and D203S protein (see above) was incubated with linearized or circular plasmids in Mg2+ buffer (250 mM NaCl, 20 mM Tris [pH 7.5], and 10 mM MgCl2) at 37°C. At the indicated time points, samples of the degradation reaction were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
Antibodies.
The following antibodies were used: mouse monoclonal antibody (MAb) 311H, directed against BGLF5 (49) (kindly provided by J. Chen, National Taiwan University, Taipei, Taiwan); mouse MAb 10E10, directed against PABPC (Santa Cruz Biotechnology); mouse MAb H68.4, recognizing transferrin receptor (TfR; Roche Diagnostics); mouse MAb W6/32 (3), recognizing β2m-associated HLA class I molecules; rat MAb 3F10, recognizing an HA epitope that is used as a protein tag (Roche Diagnostics); and rabbit serum k120, specific for BGLF5 (13) (kindly provided by J. Middeldorp, Free University Medical Center, Amsterdam, Netherlands).
Western blot analysis.
SDS-PAGE and immunoblotting were performed as reported earlier (26). In brief, cell lysates were generated as follows: cells were lysed using 0.5% NP-40 buffer [0.5% NP-40, 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 10 μM leupeptin, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF)] and subsequently centrifuged to obtain postnuclear lysates. For Western blot analysis, cell lysates or in vitro translation products (see above) were boiled in sample buffer (0.05 M Tris [pH 8.0], 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.025% bromophenol blue), separated by SDS-PAGE, and transferred to polyvinylidene difluoride membranes (GE Healthcare). Proteins of interest were detected by incubating the membranes with specific antibodies (see above), followed by horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch Laboratories). Antibody binding was visualized using enhanced chemiluminescence (ECL; GE Healthcare).
Flow cytometry.
For cell surface detection of HLA class I molecules, cells were stained with MAb W6/32, washed, and stained with secondary goat anti-mouse PE antibodies (Dako) at 4°C (antibodies were diluted in phosphate-buffered saline [PBS] with 0.5% BSA and 0.02% sodium azide). For intracellular detection of the AE proteins, the cells were fixed using fix buffer (PBS with 3.7% formaldehyde) for 10 min at room temperature, washed, and permeabilized with perm buffer (PBS with 0.5% saponin and 2% FCS) for 10 min at room temperature. Subsequently, the cells were stained with specific primary antibodies, washed, and stained with secondary donkey anti-mouse APC antibodies (eBioscience) at 4°C (antibodies were diluted in perm buffer). HLA class I, BGLF5, and GFP levels were measured on a FACSCalibur and FACSCanto II flow cytometer using CellQuest Pro and FACSDiva software, respectively (all BD Biosciences) and analyzed using FlowJo (Tree Star) software.
Immunofluorescence.
Transfected 293T cells were grown overnight on microscope slides (Lab-Tek II chamber slide system; Thermo Scientific) and fixed in 3% paraformaldehyde-PBS for 30 min at 37°C. After permeabilization with perm buffer (1% Triton X-100 and 0.1% sodium citrate in PBS) for 5 min and blocking with 5% BSA in PBS for 60 min, the cells were stained with primary antibodies for 60 min. Subsequently, the cells were washed and stained during 60 min with fluorescently labeled secondary antibodies (Jackson Immunoresearch Laboratories) and DAPI (4′,6′-diamidino-2-phenylindole) to visualize nuclei and embedded with Mowiol (Brunschwig Chemie). All incubation steps were performed at room temperature unless stated otherwise. Cells were visualized with a fluorescence microscope (EVOS fl, Westburg).
RNase assay.
As a source of RNA, pcDNA3-derived vectors were linearized before or after the polyadenylation consensus sequence and used for in vitro transcription with T7 polymerase (Promega). In vitro transcripts were incubated with recombinant BGLF5 protein (see above) in Mn2+ buffer (250 mM NaCl, 20 mM Tris [pH 7.5], and 10 mM MnCl2) at 37°C to allow digestion of the RNA. At the indicated time points, samples of the degradation reaction were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
RESULTS
Modeling and sequence alignments point to a role for the “bridge” in the shutoff activity of EBV BGLF5.
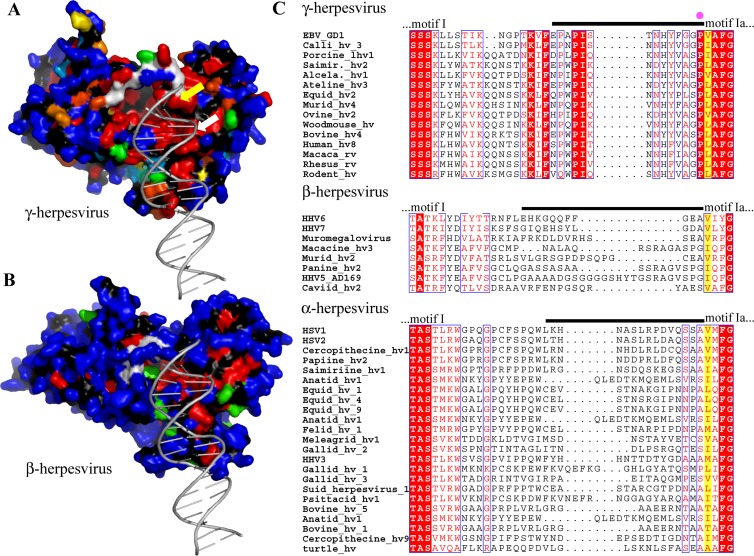
Selective functional conservation of shutoff activity by the gammaherpesvirus AE proteins could be controlled by amino acid residues that are conserved in the AE proteins of gammaherpesviruses, but not of alpha- and betaherpesviruses. To address this, we have aligned the amino acid sequences of the AE proteins encoded by 19 different gammaherpesviruses (see Fig. S1 in the supplemental material). The residue conservation profile was then mapped onto the tertiary structure of the EBV AE protein BGLF5 (Fig. 1A). We observed that conserved residues map principally to the region involved in catalysis and to the surface involved in contact with double-stranded DNA (dsDNA), as well as to the “bridge” that crosses the active site (Fig. 1A). To a lesser extent, residues on top of the nuclease domain are conserved (Fig. 1A, top right of the structure). There is no significant residue conservation on the bottom of the protein (data not shown).
Fig 1.
Structure and amino acid sequence comparisons for herpesvirus exonucleases. (A) Sequence conservation among 28 AE sequences of gammaherpesviruses (see Table 1) mapped onto the surface of the EBV nuclease BGLF5. Conservation of the side chains decreases from red (strictly conserved) over orange, yellow and green to blue (variable residues). Main chain atoms are colored in black, the ones of the bridge are colored in white. The position of a dsDNA substrate is derived from the KSHV dsDNA-SOX complex (2). The position of V159, the equivalent of a residue involved in the processivity of lambda exonuclease (54), is indicated by a yellow arrow, the white arrow indicates the K231 residue mutated in the present study. (B) Analysis of the sequence conservation within 8 betaherpesvirus exonucleases (see Table 1) based on a theoretical model of their structure. Colors are as defined in panel A. (C) Sequence conservation in the region of the bridge, which is indicated with a black line above the amino acid sequence alignments. A yellow background marks the residues corresponding to the amino acid essential for exonuclease activity in lambda exonuclease (54). The pink dot marks the position of EBV P158. hv, herpesvirus; rv, rhadinovirus.
Next, the AE protein sequences of these gammaherpesviruses were compared to those of 8 betaherpesviruses and 23 alphaherpesviruses, with the latter two only exerting DNase and lacking shutoff activity. The dual function of the gammaherpesvirus exonucleases, having both DNase and RNase activities, is probably reflected by their higher degree of conservation (39% for gammaherpesviruses versus 30 and 35% for betaherpesviruses and alphaherpesviruses, respectively). Within each herpesvirus subfamily (alpha-, beta-, and gammaherpesviruses), similar sequence divergences are observed (36). The difference in conservation between the three subfamilies is most pronounced in some areas, including the “bridge” region of the AE proteins, which appears to have adopted a disproportionately large number of insertions/deletions and substitutions during evolution of alpha- and betaherpesviruses versus gammaherpesviruses (Fig. 1C). In contrast, the areas likely involved in DNase function present sequence conservation among all herpesvirus AE proteins.
In conclusion, amino acids of the bridge (Fig. 1) show a high degree of conservation among the gammaherpesvirus AE proteins but not for alpha- and betaherpesvirus AE proteins. This subfamily-specific conservation of the AE protein “bridge” in the gammaherpesviruses is indicative of an additional function, possibly related to shutoff.
Selective mutagenesis of BGLF5.
To obtain more insight into the mechanism of shutoff by the gammaherpesvirus AEs, we integrated various assays to probe the different effector functions of EBV BGLF5 and mutants thereof. As a measure of DNase activity, we investigated the ability of in vitro synthesized (mutant) BGLF5 to degrade linearized DNA (55) (see Fig. 3). Shutoff activity was measured in transiently transfected cells by (i) downregulation of GFP reporter protein levels by (mutant) BGLF5 (55) (see Fig. 4) and (ii) nuclear relocalization of PABPC, used earlier to assess SOX shutoff activity (35) (see Fig. 5).
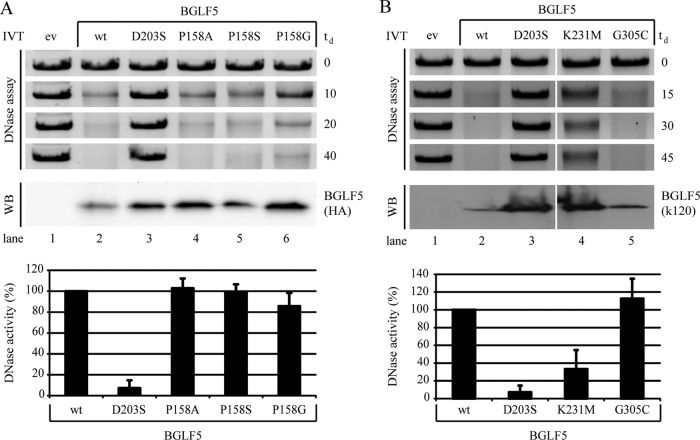
Fig 3.
Point mutations within EBV BGLF5 differentially affect DNase activity. (A and B) Linearized pcDNA3 DNA was incubated with the indicated (mutant) BGLF5 proteins synthesized as in vitro translation products. Samples of pcDNA3 substrate taken before and at the indicated time points during the degradation reaction (td, time of degradation in minutes) were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. To visualize the AE proteins as added to the enzyme reaction mixtures, 5-μl portions of the in vitro translation products were separated by SDS-PAGE and analyzed by Western blotting (WB) using either antibodies specific for the HA tag added to the C terminus of BGLF5 (3F10) (A) or with a BGLF5-specific polyclonal rabbit serum (k120) (B). For quantification of the DNase assays, the DNA amounts present after the degradation reaction were corrected for the corresponding empty vector control samples. Subsequently, the DNase activity of the BGLF5 proteins was determined with the activity of wild-type BGLF5 set at 100%. The standard deviations are represented by the error bars. All proteins were analyzed in at least four independent experiments, except for the K231M and G305C proteins that were analyzed in duplicate. ev, empty vector; wt, wild type.
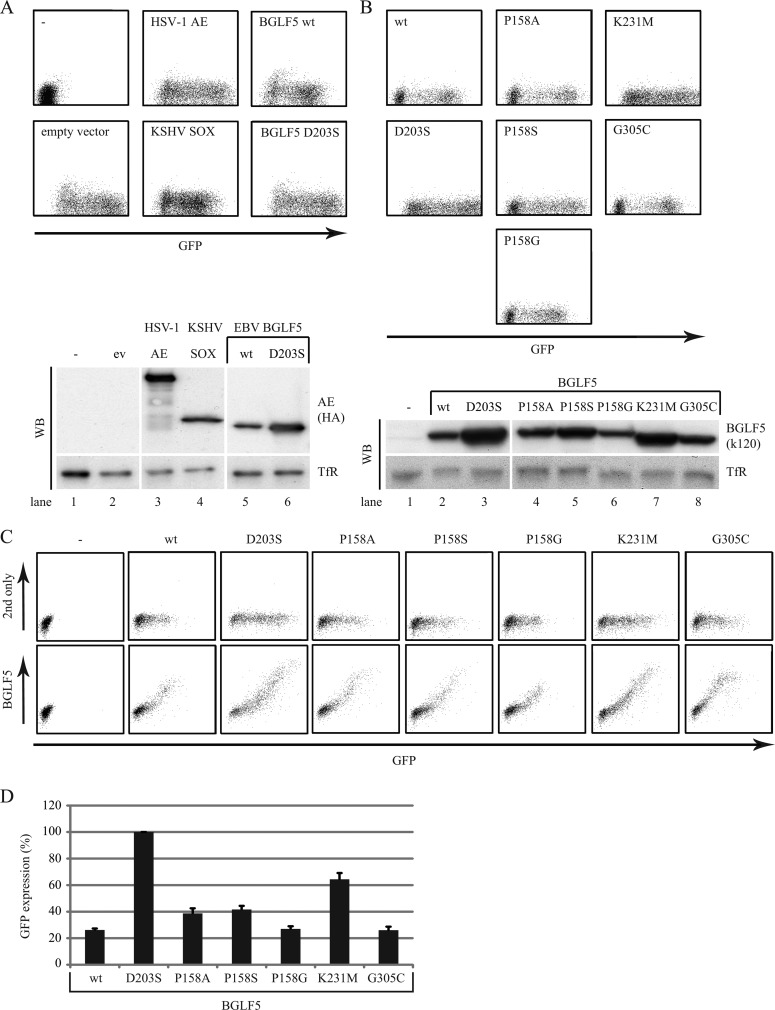
Fig 4.
Point mutations within BGLF5 differentially affect shutoff activity. (A and B) 293T cells were cotransfected with the reporter plasmid pcDNA3-IRES-nlsGFP and a pcDEF vector encoding HSV-1 AE, KSHV SOX, wild-type EBV BGLF5 (wt), or the indicated BGLF5 mutants (at a 1:4 ratio). At 48 h after transfection, the GFP intensity was analyzed by flow cytometry as a measure of the shutoff activity. Cellular expression of the various AE proteins was assessed by Western blot analysis with antibodies specific for the HA tag (3F10) (A) or BGLF5 (k120) (B). Transferrin receptor (TfR)-specific MAb H68.4 was taken along as a loading control. For the flow cytometric data, the results of one representative experiment out of five independent experiments are shown. For the Western blots, the results of one representative experiment out of two independent experiments are shown. (C) 293T cells were transfected with a pcDNA3-IRES-nlsGFP vector encoding wild-type BGLF5 (wt) or the indicated BGLF5 mutants. Nontransfected cells were taken along as a control. At 48 h after transfection, the cells were fixed, permeabilized, and stained for intracellular expression of BGLF5 (MAb 311H) and analyzed by flow cytometry. The results of one representative experiment out of two independent experiments are shown. (D) Quantification of flow cytometric data. The geometric mean fluorescence intensities for GFP within the BGLF5-positive cells depicted in panel C was determined. Subsequently, the values were set at 100% for the BGLF5 D203S catalytic site mutant. The standard deviations are represented by the error bars. All samples were analyzed in duplicate.
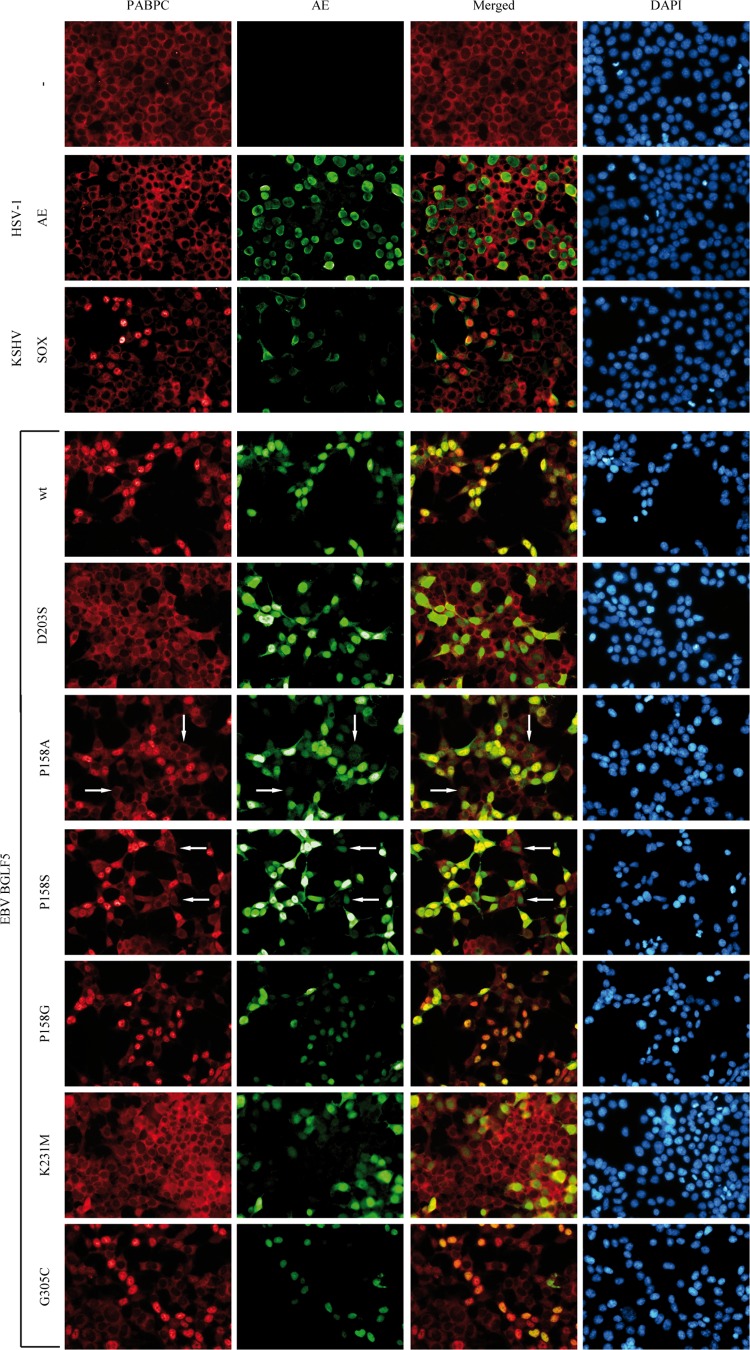
Fig 5.
EBV BGLF5 induces nuclear relocalization of PABPC, an activity that is correlated with shutoff. 293T cells were transfected with pcDEF plasmids encoding the individual herpesvirus AE proteins as in Fig. 4A and B. After 24 h of transfection, the cells were subjected to triple-label immunofluorescence analysis: PABPC expression was visualized by PABPC-specific antibodies (MAb 10E10 and anti-mouse-Cy3; left panels), AE expression was visualized with HA-specific antibodies (MAb 3F10 and anti-rat Alexa 488; second panels for negative control, HSV-1 AE and KSHV SOX samples) or with BGLF5-specific antibodies (k120 polyclonal antisera and anti-rabbit Alexa 488; second panels for [mutant] BGLF5 samples), and cells were counterstained with DAPI to identify nuclei (right panels). The white arrows indicate cells with low levels of the P158A and P158S mutant proteins. The KSHV SOX, BGLF5 wt, and D203S proteins were analyzed in triplicate; the other proteins were analyzed in duplicate.
Using these assays, we studied several mutants of BGLF5 (Fig. 2 and see Fig. S1 in the supplemental material). First, the catalytically inactive mutant D203S was included in our assays as a control, since this mutation is known to block both DNase and RNase activities (5). Second, guided by the structural model of BGLF5 and the sequence alignments of the herpesvirus AE proteins (Fig. 1), we mutated the proline residue at amino acid position 158 of BGLF5. This P158 residue is located at the C-terminal side of the bridge that crosses the active site of BGLF5 (5) (Fig. 1C, pink dot) and caps the α-helix that contains motif Ia. This residue is conserved in AE proteins of gammaherpesviruses but not of alpha- and betaherpesviruses (Fig. 1C). Because of its position in the tertiary structure, the P158 residue may restrict the flexibility of the bridge in BGLF5 and its homologues in gammaherpesviruses. This residue was substituted by an alanine, serine, or glycine residue, amino acid substitutions that are expected to have progressively greater effect on the flexibility of the bridge or to affect the terminus of the α-helix. We noted that all three residues naturally occur in the AE proteins encoded by alpha- and betaherpesviruses, although none is conserved in a large subset of these viruses (Fig. 1C). Third, an earlier random mutagenesis screen yielded a BGLF5 mutant containing two amino acid substitutions, K231M and G305C, that appeared to have lost shutoff activity while retaining DNase activity (55). The individual K231M substitution was concluded to be responsible for this genetic separation. However, from the position of the K231 residue in the structure and sequence alignments, no obvious explanation for a selective block in shutoff or retention of DNase activity occurred to us, since it localizes to motif III. Therefore, the individual K231M and G305C mutations were also included in our analysis.
Fig 2.
Location of the studied mutations within the EBV BGLF5 protein. Within the structural model of the EBV nuclease, the amino acid substitutions under study are colored as follows: P158 mutants in dark blue, D203S mutant in purple, K231M mutant in yellow, and G305C mutant in red. The exact context and conservation of the mutations is shown in Fig. S1 in the supplemental material. The flexible bridge is indicated by an arrow. (PDB entry 2W4B.)
Mutations affecting the conserved motifs of BGLF5 abolish DNase activity, whereas those in the bridge do not.
The effects of the mutations within the BGLF5 protein on DNase activity were examined by incubation of linearized DNA plasmid with in vitro synthesized BGLF5 protein. Western blot analysis verified that all BGLF5 proteins were synthesized (Fig. 3A and B). Subsequently, samples were taken at various time points during the DNase reaction and resolved by agarose gel electrophoresis. In the presence of wild-type (wt) BGLF5, linear DNA was completely digested within 40 min (Fig. 3A, lane 2). This DNase activity is comparable to that exerted by the AE protein of the alphaherpesvirus HSV-1 (data not shown). In contrast, no degradation was observed for samples incubated with the catalytic site mutant BGLF5 D203S (Fig. 3A, lane 3) (5). Analysis of the P158 “bridge” mutants revealed that the capacity to degrade DNA was comparable for the alanine and serine mutants and the wild-type protein at all time points (Fig. 3A, lanes 2, 4, and 5). Surprisingly, substitution of P158 by a glycine residue affected DNase activity, although the effect was less dramatic than the complete loss of DNase activity observed for the D203S mutant (Fig. 3A, lanes 2, 3, and 6).
We observed a drastically reduced ability of the BGLF5 K231M protein to digest linearized DNA plasmids compared to wild-type BGLF5 (Fig. 3B, lanes 2 and 4), as opposed to earlier data (55). However, unlike the complete inactivation of BGLF5 by the D203S mutation, the DNase activity was not entirely blocked by the K231M mutation (Fig. 3B, compare lanes 3 and 4), and the residual activity could thus account for the discrepancy between the present study and previous results (55). The BGLF5 G305C mutant degraded the linearized plasmid as efficiently as wild-type BGLF5 (Fig. 3B, lanes 2 and 5).
In summary, the proline 158 residue of EBV BGLF5 can be mutated into an alanine or serine residue without consequences for DNase activity. Surprisingly, replacement of P158 by a glycine residue partially impaired DNase activity. Mutation of the K231 residue into a methionine resulted in a substantial drop in DNase activity.
Shutoff function of BGLF5 is affected both by mutations leading to loss of DNase activity and, more selectively, by “bridge” mutations.
The shutoff activity of the AEs was examined using two approaches, namely, by flow cytometry- and by immunofluorescence-based assays.
First, 293T cells were cotransfected to express the reporter protein GFP and the alphaherpesvirus HSV-1 AE protein, or the gammaherpesvirus AE proteins KSHV SOX and EBV (wild-type and mutant) BGLF5. Two days after transfection, the shutoff activity was measured by determining the reduction in cellular GFP levels using flow cytometry.
In control cells cotransfected with GFP and an empty vector, high levels of GFP were observed (Fig. 4A, lower left panel). A vector encoding the HSV-1 AE protein, reported to lack shutoff activity, was taken along as a control. Expression of HSV-1 AE did not result in a major reduction in the expression of GFP (Fig. 4A, upper middle panel). In contrast, cells cotransfected with GFP and either wild-type SOX or BGLF5 had markedly reduced levels of GFP (Fig. 4A, lower middle and upper right panels). The BGLF5 D203S mutant did not profoundly diminish GFP expression (Fig. 4A, lower right panel), in support of this protein having lost its shutoff activity. Thus, this flow cytometry-based assay reflects the shutoff activity exerted by AE proteins of the subfamily of gammaherpesviruses.
Next, mutants of EBV BGLF5 were compared to the wild-type protein (Fig. 4B). Expression of the three P158 “bridge” mutants caused a reduction in GFP levels, but the degree of shutoff varied. The alanine and serine mutants were impaired in their shutoff activity compared to the wild-type protein, albeit less so than the D203S mutant (Fig. 4B, left and middle panels). Interestingly, substitution of the proline 158 residue by a glycine retained or even slightly enhanced shutoff activity (Fig. 4B, compare the lower middle and upper left panels). The K231M “motif III” mutation inhibited the ability of BGLF5 to block GFP expression (Fig. 4B, lower left and upper right panels). In contrast, shutoff activity was not affected by the G305C mutation (Fig. 4B, lower right panel).
Proper cellular expression of the constructs was verified by Western blotting with antibodies specific for BGLF5 or for the HA tag added to the herpesvirus AEs (Fig. 4A and B). Interestingly, expression levels of the AE proteins were negatively correlated with their shutoff activity. The highest protein band intensities were detected for HSV-1 AE and the catalytically inactive EBV BGLF5 D203S mutant (Fig. 4A, lanes 3 and 6). In contrast, the AE proteins capable of RNA degradation, BGLF5 and SOX (Fig. 4A, lanes 4 and 5), appeared to also reduce their own protein expression. The flow cytometric data were in line with the Western blot results in that the BGLF5 mutants that had (partly) lost shutoff activity were expressed to the highest levels (Fig. 4B, D203S, P158A, P158S, and K231M [lanes 3, 4, 5, and 7, respectively]). Conveniently, this strengthens the results regarding the shutoff-defective mutants: no reduction in GFP levels was observed, even though these proteins were expressed at higher levels.
To avoid influences of transfection efficiency, additional transfections were performed using a single plasmid encoding both (wild-type or mutant) BGLF5 and GFP separated by an IRES; coexpression of both proteins was confirmed by intracellular detection of BGLF5 in the GFP-positive population (Fig. 4C, lower panel). The results obtained using this experimental setup (Fig. 4C) were in good agreement with the data described above for the two-plasmid transfections (Fig. 4B) and further supported reproducibility of the shutoff phenotypes of the BGLF5 mutants (quantified in Fig. 4D).
Taken together, both amino acid residues important for DNase activity (D203 and K231), as well as a “bridge” amino acid (P158), are involved in BGLF5-mediated shutoff. The shutoff activity of the gammaherpesvirus AE proteins hampers their protein expression.
Shutoff-competent EBV BGLF5 induces nuclear relocalization of PABPC.
Our second approach to evaluate shutoff activity exerted by AE variants relied on the nuclear relocalization of PABPC. Previously, Lee et al. demonstrated that cellular SOX expression induced relocalization of PABPC to the nucleus, and this correlated with shutoff activity mediated by SOX (35). To assess whether BGLF5 has a similar effect on the distribution of PABPC, localization of endogenous PABPC was determined by immunofluorescence assays using 293T cells transfected to transiently express the shutoff-defective alphaherpesvirus HSV-1 AE protein or the shutoff-competent gammaherpesvirus KSHV SOX and EBV BGLF5 proteins. Paraformaldehyde-fixed, detergent-permeabilized cells were stained using antibodies specific for PABPC and for the viral nucleases. DAPI staining visualized the nuclei.
In control cells, PABPC resided exclusively in the cytoplasm (Fig. 5, upper-row panels). Whereas PABPC was still restricted to the cytoplasm of HSV-1 AE-expressing cells, nuclear relocalization of PABPC clearly occurred in cells that expressed KSHV SOX (Fig. 5, compare the second- and third-row panels), in agreement with published results (35). We also show here that BGLF5 induced nuclear relocalization of PABPC (Fig. 5, fourth-row panels), further supporting that BGLF5 and SOX use the same strategy to effectuate shutoff. The cytoplasmic localization of PABPC in cells expressing the catalytically inactive BGLF5 D203S protein (Fig. 5, fifth-row panels) is in line with its loss of shutoff activity.
Next, effects of the selected BGLF5 mutants on shutoff activity were evaluated using this immunofluorescence-based assay. Overall, the P158 and G305C mutants induced nuclear relocalization of PABPC comparable to wild-type BGLF5, whereas the K231M mutant did not (Fig. 5, sixth- to tenth-row panels). Interestingly, PABPC relocalization was not observed in cells with low levels of the “bridge” mutant P158A and P158S proteins (Fig. 5, sixth- and seventh-row panels, indicated by white arrow), suggesting that a certain threshold of BGLF5-mediated shutoff activity is required to mediate PABPC relocalization. These conclusions were supported by experiments with cells transiently transfected to coexpress exogenous flag-tagged PABPC and the viral nucleases (data not shown).
Altogether, these results show that EBV BGLF5 induces the nuclear relocalization of PABPC, as has been reported for KSHV SOX (35). The capacity of the mutant BGLF5 proteins to relocalize PABPC to the nucleus correlates with their ability to reduce GFP expression and, therefore, shutoff (Table 2).
Table 2.
Summary of the activities of the BGLF5 protein and its mutants
| BGLF5 | Motif | In vitro DNase activity | Reduction GFP levels | Relocalization PABPC | In vitro RNase activity |
|---|---|---|---|---|---|
| Wild type | ++ | ++ | ++ | + | |
| D203S | Catalytic site/II | − | − | − | − |
| P158A | Ia | ++ | + | + | NTa |
| P158S | Ia | ++ | + | + | NT |
| P158G | Ia | + | ++ | ++ | NT |
| K231M | III | +/− | +/− | − | NT |
| G305C | ++ | ++ | ++ | NT |
NT, not tested.
Specificity of the nuclease activities of BGLF5.
Production of relatively large quantities of purified baculovirus-expressed recombinant BGLF5 (wild type and D203S) allowed us to investigate the nuclease activities of the EBV protein in vitro. The specificity of the viral DNase activity was examined by incubation of linear and circular DNA with either recombinant wild-type or D203S mutant BGLF5. Both linear and circular DNA were digested by wild-type BGLF5, but linear DNA was degraded more efficiently (Fig. 6A, lanes 2 to 5), indicating that the exonuclease activity of BGLF5 is more potent than its endonuclease activity. Recombinant BGLF5 protein with the catalytic site mutation (D203S) did not induce degradation of linear DNA (Fig. 6A, lanes 6 to 9). Surprisingly, incubation of circular DNA with increasing amounts of the D203S mutant resulted in elevated levels of nicked plasmid (Fig. 6A, lanes 6 to 9 and bar graph), suggesting that some endonuclease activity is retained by BGLF5 D203S.
Fig 6.
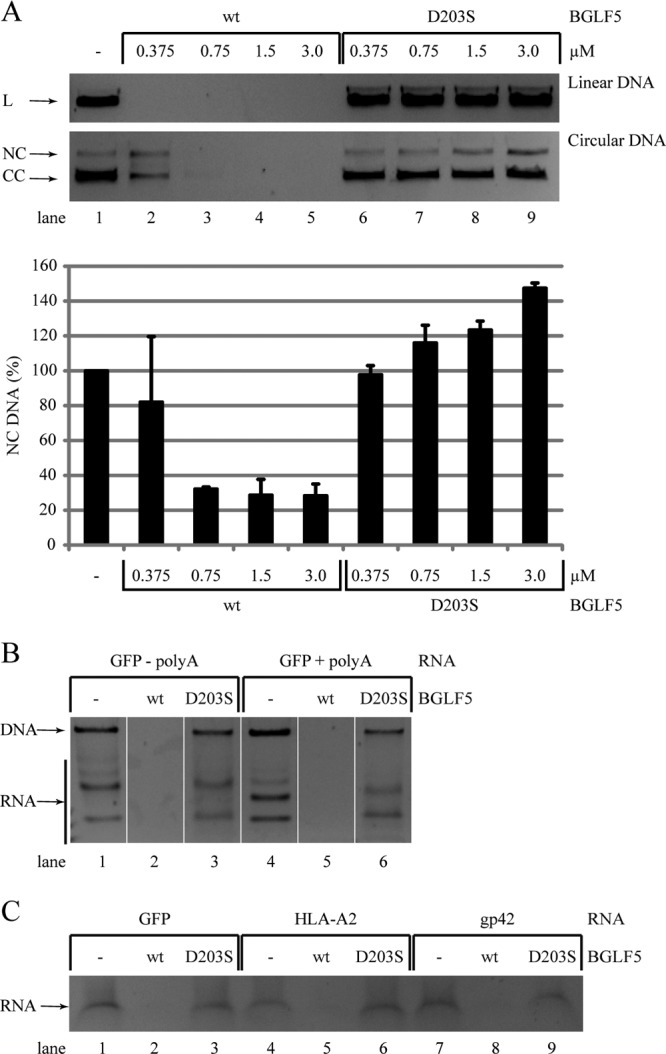
Specificities of EBV BGLF5-mediated DNA and RNA degradation. (A) To assess DNase activity, linear and circular pMaxGFP plasmids (0.05 pmol of DNA) were incubated with increasing concentrations of wt or D203S mutant EBV BGLF5 recombinant proteins in the presence of 5 mM Mg2+. After 2 h at 37°C, samples were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. The results of one representative experiment out of four independent experiments are shown. For quantification of the DNase assays, the NC DNA amounts present after the degradation reaction were corrected for the control samples (set at 100%). The standard deviations are represented by error bars. The quantification is based on two replicate experiments. L, linear DNA; CC, closed circle; NC, nicked circle. (B and C) The specificity of the RNase activity was studied by incubating wt or D203S mutant EBV BGLF5 recombinant proteins with in vitro-transcribed RNA in the presence of 10 mM Mn2+. (B) GFP RNA was synthesized from a plasmid linearized before (−polyA) or after (+polyA) the polyadenylation consensus sequence. (C) RNAs for a foreign protein (GFP), a cellular protein (HLA-A2), or a viral protein (EBV gp42) were used. After degradation periods of 30 min (B) or 6 h (C) at 37°C, the samples were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. DNA and RNA bands were identified according to their susceptibility to digestion with an RNase A/T1 mix (Ambion) (data not shown). Using this approach, the GFP (+polyA) and cellular mRNAs were analyzed once, the GFP (−polyA) and viral mRNAs were analyzed in at least two independent experiments.
Recently, we found that BGLF5 exerts intrinsic RNase activity, resulting in degradation of mRNA, but not of tRNA (5). This provided a mechanistic basis for BGLF5 blocking the synthesis of cellular proteins, most of which are translated from poly(A)-tailed mRNAs (44). In the present study, the specificity of this RNase activity was further investigated by incubating various RNA substrates with recombinant BGLF5 proteins. The requirement of mRNA polyadenylation for BGLF5-mediated degradation was analyzed using in vitro transcription of a GFP-encoding plasmid linearized before or after the polyadenylation consensus sequence. These experiments showed that, in addition to poly(A)-tailed mRNAs, mRNAs without a poly(A) tail are also degraded by BGLF5 (Fig. 6B, lanes 2 and 5). No RNase activity was observed for the control protein BGLF5 D203S (Fig. 6B, lanes 3 and 6). To examine whether BGLF5 specifically digests host-derived mRNA, recombinant BGLF5 was incubated with RNA encoding proteins of exogenous (GFP), cellular (HLA-A2), or viral (EBV gp42) origin. These different RNA substrates were all completely digested by wild-type BGLF5, but not by the catalytically inactive D203S mutant (Fig. 6C). Our results may thus imply that BGLF5 is incapable of discriminating host from viral RNAs.
In conclusion, BGLF5 has both endo- and exonuclease activities toward DNA substrates. Whereas the D203S mutation obstructs the exonuclease activity of BGLF5 toward DNA, some endonuclease activity is preserved. The RNase activity of BGLF5 is completely abolished by the D203S mutation. This intrinsic RNase activity does not appear to be influenced by polyadenylation or the source of the transcripts.
Shutoff activity of BGLF5 is required for immune evasion.
During EBV infection, shutoff by BGLF5 reduces cell surface HLA class I levels, thereby hampering recognition by CD8+ T cells (44, 55). We here investigated the effect of mutations within the BGLF5 protein on immune evasion. To this end, we examined HLA class I expression at the surface of 293T cells that were transiently transfected to express AE proteins. Transfected cells were identified by coexpression of GFP and compared to the nontransfected GFP-negative population.
Expression of wild-type BGLF5 induced a marked downregulation of surface HLA class I compared to a control protein (GFP) or the shutoff-defective alphaherpesvirus HSV AE protein (Fig. 7, compare panel 3 to panels 1 and 2). Of the BGLF5 mutants, shutoff-defective D203S and shutoff-impaired K231M have lost their capacity to interfere with HLA class I antigen presentation (Fig. 7, panels 4 and 8). For the “bridge” mutants, their ability to reduce HLA class I display correlated with their shutoff activity: HLA class I downregulation was less pronounced for the P158A and especially the P158S mutant compared to the P158G mutant (Fig. 7, panels 5 to 7). Expression of the shutoff-competent G305C mutant similarly reduced cell surface HLA class I levels as did wild-type BGLF5 (Fig. 7, panels 3 and 9).
Fig 7.
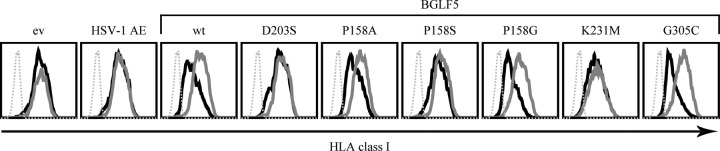
HLA class I downregulation by BGLF5 mutants depends on shutoff activity. 293T cells were transfected with a pcDNA3-IRES-nlsGFP vector encoding HSV-1 AE, wild-type EBV BGLF5, or the indicated BGLF5 mutants. As a control, the empty vector was transfected. At 48 h after transfection, the cells were stained for cell surface expression of HLA class I (MAb W6/32) and analyzed by flow cytometry. Dashed gray line, secondary antibody only; solid gray line, GFP-negative cells; solid black line, GFP-positive cells. The results of one representative experiment out of two independent experiments are shown. ev, empty vector; wt, wild type.
In conclusion, we found that the shutoff activity of BGLF5, rather than its DNase activity, correlates with surface HLA class I downregulation (Table 2). These data support the notion that BGLF5-mediated immune evasion relies on its shutoff activity.
DISCUSSION
In this study, we sought to identify elements that confer the additional shutoff activity onto AEs of the gammaherpesviruses, a trait complementing the DNase function shared by their counterparts in all herpesvirus family members.
We hypothesized that the additional constraint of a shutoff activity might explain the higher degree of sequence conservation observed among the gammaherpesvirus exonucleases. This conservation involved the enzyme's active site, the region involved in dsDNA binding, and, unexpectedly, the bridge that crosses the active-site cleft, the latter being poorly conserved in the homologous proteins of the other herpesvirus subfamilies (Fig. 1). Within the homologous bridge of lambda exonuclease, one residue has very recently been identified to directly participate in exonuclease processivity (54) (indicated in yellow in Fig. 1C); its counterpart in BGLF5 might play a similar role. Other conserved bridge residues might have a role in shutoff, for instance through interaction with RNA substrates. Indeed, mutagenesis of BGLF5 proline 158, located in the bridge base, differentially affects DNase and shutoff activities: substitution by an alanine (P158A) or serine (P158S) selectively hampers shutoff function, whereas replacement by a glycine (P158G) slightly reduces DNase activity (Table 2). Our results are in line with the reduced shutoff activity and preserved DNase activity observed upon mutation of the corresponding amino acid of KSHV SOX (P176S) (18). These combined results point toward a role for the bridge region in the shutoff function of gammaherpesvirus AE proteins.
The DNase and RNase activities of gammaherpesvirus exonucleases share their catalytic site, as supported by the observation that mutation of BGLF5 aspartic acid 203 (D203S) impaired both activities. Interestingly, this mutation blocked exonuclease, but not endonuclease activity toward DNA. A similar mutation in HSV AE (D340E) likewise impaired its exonuclease but not its endonuclease activity (21). These combined data suggest the endonuclease activity of the viral AE proteins to be mechanistically distinct from their exonuclease activity.
Previously, the DNase and shutoff activities of BGLF5 and (mu)SOX were found to be genetically separable (18, 43, 55). In the present study, complete separation of the two activities was never observed, although mutations in BGLF5 were found (P158, see above) that more selectively affected either its ability to degrade DNA or to inhibit host protein synthesis (Table 2).
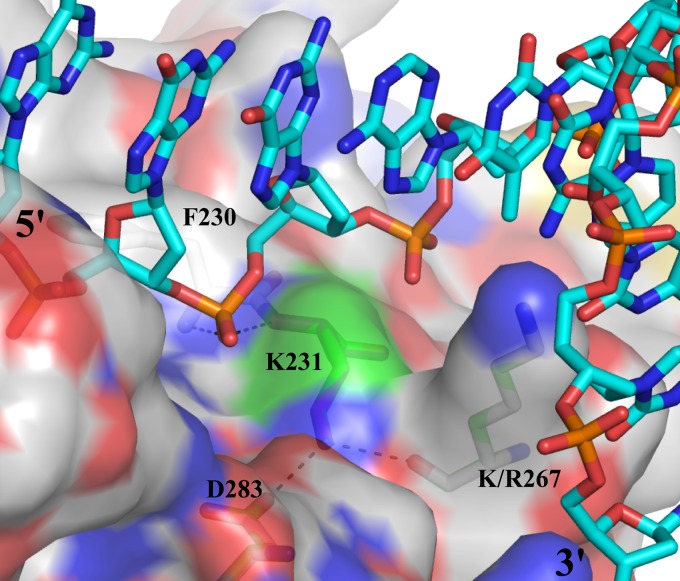
As deduced from the structure of DNA-bound KSHV SOX (2), EBV K231 is likely to form part of the DNA-binding surface (Fig. 8) through hydrogen bonds with D283 and the amide of R267; the neighboring amide nitrogen atoms of K231 and F230 form hydrogen bonds to a backbone phosphate of the dsDNA. Mutation of K231 to M causes loss of the hydrogen bonds, resulting in a different organization of the structure around the mutated residue and thereby providing a mechanistic basis for abolishment of nuclease activity of mutant BGLF5, both on DNA and RNA. The latter implicates that BGLF5-bound single-stranded RNA adopts a similar conformation as one of the DNA strands, at least for the first five bases. The apparent discrepancy between a prior report (55) and the present study related to K231M effects on DNase activity has most likely resulted from variation in DNA/BGLF5 ratios and incubation times during the DNase assay.
Fig 8.
Role of the EBV BGLF5 residue K231. View of the KSHV SOX structure in complex with dsDNA (2). Only one of the DNA strands is shown (cyan carbon atoms). The protein (white carbon atoms) is represented with a semitransparent surface with some key residues indicated. Residue K231 is shown with green carbon atoms. Residues are labeled with the numbers corresponding to EBV BGLF5. These residues are conserved between BGLF5 and SOX with the exception of an R/K substitution. Dotted lines show hydrogen bonds.
Expression of KSHV SOX (35) and EBV BGLF5 (the present study) results in nuclear relocalization of cytosolic PABPC, a protein involved in mRNA stabilization, and this correlates with the additional shutoff function of the gammaherpesvirus AE proteins. Recent data implicate that the nuclear accumulation of PABPC is resulting from virus-induced mRNA depletion from the cytoplasm (32). Interestingly, cellular expression of PABPC is altered upon infection by members of all herpesvirus subfamilies. The alphaherpesvirus HSV causes nuclear accumulation of PABPC in infected cells (11, 45), similar to the gammaherpesviruses. PABPC relocalization by HSV, however, occurs as a consequence of the combined expression of both ICP27, inhibiting mRNA splicing, and the vhs protein (11, 31). The betaherpesvirus human cytomegalovirus rather induces increased PABPC protein levels, which accumulate in the cytosol of infected cells (40, 52).
Recombinant gammaherpesvirus AE proteins exert RNase activity in vitro (2, 5), but it is not yet clear whether this is sufficient to mechanistically explain shutoff. The alphaherpesvirus HSV AE can also degrade RNA substrates in vitro but lacks shutoff activity in cells (27, 29). The strictly nuclear localization of HSV AE—in contrast to the additional cytoplasmic presence of gammaherpesvirus nucleases—could preclude the degradation of cytoplasmic mRNAs. However, mutation of HSV AE causing partially cytoplasmic localization did not restore shutoff (9). Besides being present in the cytoplasm, interaction with cellular cofactor(s) might increase RNA turnover activity of the gammaherpesvirus AE proteins in cells. The absence of gammaherpesvirus-specific conserved surface areas on the exonuclease would speak against the involvement of cofactor(s). However, most mutations found in shutoff-defective SOX variants map to the surface of the protein instead of the conserved catalytic site (10, 18). Thus, the contribution of cofactor binding to gammaherpesvirus AE-mediated shutoff remains to be assessed. Interestingly, in this context, Covarrubias et al. recently reported SOX to cleave mRNAs internally before their degradation by the cellular exonuclease Xrn1, indicating that gammaherpesviral AE-mediated shutoff is a two-step process (8).
In the present study, we showed that the BGLF5 protein can degrade mRNAs of cellular and viral origin in vitro (Fig. 6). Although additional studies are required to determine the full specificity spectrum of BGLF5-mediated mRNA degradation, data thus far imply broad reactivity of virus-induced shutoff. For instance, during EBV infection, BGLF5 reduces the mRNA and protein levels of several viral gene products, which is reverted in cells infected with deletion mutant EBV (14, 15). HSV vhs not only degrades cellular mRNAs but also viral mRNAs, facilitating the transition between the immediate early, early, and late phases of infection (33, 37, 38, 41). Some selectivity is likely, as a small subset of cellular transcripts, including interleukin-6, escapes KSHV SOX-induced degradation (6, 7, 19).
Combining the current and published findings, we propose the following model: BGLF5's exonuclease activity degrades both linear DNA and RNA using the same catalytic site, albeit that ion requirements and efficiencies differ for the two substrates. BGLF5's endonuclease activity seems to involve a somewhat different mechanism. Whereas the capacity of AE proteins to bind DNA is conserved throughout the herpesvirus family, additional binding of RNA substrates has probably been acquired during evolution by the gammaherpesviruses. Determinants for RNA recognition may include lineage-specific residues located in the flexible bridge sequence located between motifs I and Ia of the gammaherpesvirus AE proteins (5) (Fig. 1). BGLF5-mediated mRNA degradation blocks de novo protein synthesis and relocalizes PABPC. Within the nucleus, PABPC causes hyperadenylation and retention of nuclear mRNA molecules, thereby augmenting the shutoff phenotype initiated by BGLF5. Interestingly, the protein levels of BGLF5 (mutants) inversely correlate with shutoff activity, indicating that BGLF5 also degrades its own messenger.
Blocking protein synthesis may benefit herpesvirus infection at different levels. First, in the absence of host protein synthesis, cellular ribosomes will be committed to the synthesis of viral proteins. Second, by controlling the transition between immediate early, early, and late protein synthesis, the virus can progress efficiently between the different phases of productive infection. Recently, Richner et al. demonstrated a role for shutoff during latency establishment using an in vivo model of MHV68 infection (43). Finally, host shutoff contributes to immune evasion by preventing the synthesis of proteins involved in antiviral immunity. For example, EBV BGLF5 reduces expression of HLA molecules, thereby hampering antigen presentation to T cells (44, 55). Likewise, KSHV SOX and HSV vhs impair T cell activation (48, 55). Recently, BGLF5 was also found to downregulate Toll-like receptor 9 (50), an important pattern recognition receptor that detects viral dsDNA.
In conclusion, by orchestrating viral and cellular gene expression and thwarting both innate and adaptive immunity, BGLF5 aids the efficient synthesis of viral progeny in the absence of immune recognition, thereby facilitating the spread of EBV to new hosts.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Dutch Cancer Foundation (grant RUL/UU 2005–3259 to D.H., M.E.R., and E.J.H.J.W.), The Netherlands Scientific Organization (NWO Vidi 917.76.330 to M.E.R.), the ANR-MIME-2006 program of the French government (to W.P.B.), and the cluster 10 Infectiology of the Région Rhône-Alpes (to W.P.B.).
We gratefully acknowledge Jaap Middeldorp (Free University Medical Center, Amsterdam, Netherlands), Britt Glaunsinger (University of California, Berkeley, CA), Jen-Yang Chen (National Taiwan University, Taipei, Taiwan), Hans van Leeuwen and Sjaak van Voorden (both at Leiden University Medical Center, Leiden, Netherlands), Jianmin Zuo and Martin Rowe (both at University of Birmingham, Birmingham, United Kingdom), Eric Reits (Academic Medical Center, Amsterdam, Netherlands), and Janneke Peeters (University Medical Center Utrecht, Utrecht, Netherlands) for helpful advice and technical assistance and for generously sharing reagents and constructs. We thank Tracey Barret (Birkbeck College, London, United Kingdom) for providing coordinates for the SOX-DNA complex.
The authors declare that they have no conflicts of interest.
Footnotes
Published ahead of print 13 June 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagneris C, Briggs LC, Savva R, Ebrahimi B, Barrett TE. 2011. Crystal structure of a KSHV-SOX-DNA complex: insights into the molecular mechanisms underlying DNase activity and host shutoff. Nucleic Acids Res. 39:5744–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnstable CJ, et al. 1978. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell 14:9–20 [DOI] [PubMed] [Google Scholar]
- 4. Benkert P, Biasini M, Schwede T. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buisson M, et al. 2009. A bridge crosses the active-site canyon of the Epstein-Barr virus nuclease with DNase and RNase activities. J. Mol. Biol. 391:717–728 [DOI] [PubMed] [Google Scholar]
- 6. Chandriani S, Ganem D. 2007. Host transcript accumulation during lytic KSHV infection reveals several classes of host responses. PLoS One 2:e811 doi:10.1371/journal.pone.0000811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clyde K, Glaunsinger BA. 2011. Deep sequencing reveals direct targets of gammaherpesvirus-induced mRNA decay and suggests that multiple mechanisms govern cellular transcript escape. PLoS One 6:e19655 doi:10.1371/journal.pone.0019655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Covarrubias S, et al. 2011. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog. 7:e1002339 doi:10.1371/journal.ppat.1002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Covarrubias S, Richner JM, Clyde K, Lee YJ, Glaunsinger BA. 2009. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J. Virol. 83:9554–9566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dahlroth SL, Gurmu D, Haas J, Erlandsen H, Nordlund P. 2009. Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus. FEBS J. 276:6636–6645 [DOI] [PubMed] [Google Scholar]
- 11. Dobrikova E, Shveygert M, Walters R, Gromeier M. 2010. Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. J. Virol. 84:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Everly DN, Jr, Feng P, Mian IS, Read GS. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 76:8560–8571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fachiroh J, et al. 2004. Molecular diversity of Epstein-Barr virus IgG and IgA antibody responses in nasopharyngeal carcinoma: a comparison of Indonesian, Chinese, and European subjects. J. Infect. Dis. 190:53–62 [DOI] [PubMed] [Google Scholar]
- 14. Feederle R, Bannert H, Lips H, Muller-Lantzsch N, Delecluse HJ. 2009. The Epstein-Barr virus alkaline exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 83:4952–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feederle R, Mehl-Lautscham AM, Bannert H, Delecluse HJ. 2009. The Epstein-Barr virus protein kinase BGLF4 and the exonuclease BGLF5 have opposite effects on the regulation of viral protein production. J. Virol. 83:10877–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng P, Everly DN, Jr, Read GS. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272–10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng P, Everly DN, Jr, Read GS. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 79:9651–9664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glaunsinger B, Chavez L, Ganem D. 2005. The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J. Virol. 79:7396–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glaunsinger B, Ganem D. 2004. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. J. Exp. Med. 200:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glaunsinger B, Ganem D. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13:713–723 [DOI] [PubMed] [Google Scholar]
- 21. Goldstein JN, Weller SK. 1998. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology 244:442–457 [DOI] [PubMed] [Google Scholar]
- 22. Gopinath RS, Ambagala AP, Hinkley S, Srikumaran S. 2002. Effects of virion host shut-off activity of bovine herpesvirus 1 on MHC class I expression. Viral Immunol. 15:595–608 [DOI] [PubMed] [Google Scholar]
- 23. Gorgoni B, Gray NK. 2004. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief. Funct. Genomic Proteomic 3:125–141 [DOI] [PubMed] [Google Scholar]
- 24. Gouet P, Courcelle E, Stuart DI, Metoz F. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308 [DOI] [PubMed] [Google Scholar]
- 25. Hinkley S, Ambagala AP, Jones CJ, Srikumaran S. 2000. A vhs-like activity of bovine herpesvirus-1. Arch. Virol. 145:2027–2046 [DOI] [PubMed] [Google Scholar]
- 26. Horst D, et al. 2009. Specific targeting of the EBV lytic phase protein BNLF2a to the transporter associated with antigen processing results in impairment of HLA class I-restricted antigen presentation. J. Immunol. 182:2313–2324 [DOI] [PubMed] [Google Scholar]
- 27. Kehm E, Goksu M, Bayer S, Knopf CW. 1998. Herpes simplex virus type 1 DNase: functional analysis of the enzyme expressed by recombinant baculovirus. Intervirology 41:110–119 [DOI] [PubMed] [Google Scholar]
- 28. Kikkert M, et al. 2001. Ubiquitination is essential for human cytomegalovirus US11-mediated dislocation of MHC class I molecules from the endoplasmic reticulum to the cytosol. Biochem. J. 358:369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knopf CW, Weisshart K. 1990. Comparison of exonucleolytic activities of herpes simplex virus type-1 DNA polymerase and DNase. Eur. J. Biochem. 191:263–273 [DOI] [PubMed] [Google Scholar]
- 30. Koppers-Lalic D, et al. 2001. The UL41-encoded virion host shutoff (vhs) protein and vhs-independent mechanisms are responsible for down-regulation of MHC class I molecules by bovine herpesvirus 1. J. Gen. Virol. 82:2071–2081 [DOI] [PubMed] [Google Scholar]
- 31. Kumar GR, Glaunsinger BA. 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol. Cell. Biol. 30:4996–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar GR, Shum L, Glaunsinger BA. 2011. Importin α-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol. Cell. Biol. 31:3113–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kwong AD, Frenkel N. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. U. S. A. 84:1926–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwong AD, Kruper JA, Frenkel N. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee YJ, Glaunsinger BA. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 7:e1000107 doi:10.1371/journal.pbio.1000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McGeoch DJ, Rixon FJ, Davison AJ. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90–104 [DOI] [PubMed] [Google Scholar]
- 37. Oroskar AA, Read GS. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oroskar AA, Read GS. 1987. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J. Virol. 61:604–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Page HG, Read GS. 2010. The virion host shutoff endonuclease (UL41) of herpes simplex virus interacts with the cellular cap-binding complex eIF4F. J. Virol. 84:6886–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perez C, McKinney C, Chulunbaatar U, Mohr I. 2011. Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J. Virol. 85:156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Read GS, Frenkel N. 1983. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate early) viral polypeptides. J. Virol. 46:498–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ressing ME, et al. 2005. Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion. J. Virol. 79:841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Richner JM, et al. 2011. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLoS Pathog. 7:e1002150 doi:10.1371/journal.ppat.1002150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rowe M, et al. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. U. S. A. 104:3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salaun C, et al. 2010. Poly(A)-binding protein 1 partially relocalizes to the nucleus during herpes simplex virus type 1 infection in an ICP27-independent manner and does not inhibit virus replication. J. Virol. 84:8539–8548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smiley JR. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tigges MA, Leng S, Johnson DC, Burke RL. 1996. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J. Immunol. 156:3901–3910 [PubMed] [Google Scholar]
- 49. Tsai CH, et al. 1997. Characterization of monoclonal antibodies to the Zta and DNase proteins of Epstein-Barr virus. J. Biomed. Sci. 4:69–77 [DOI] [PubMed] [Google Scholar]
- 50. van Gent M, et al. 2011. EBV lytic-phase protein BGLF5 contributes to TLR9 downregulation during productive infection. J. Immunol. 186:1694–1702 [DOI] [PubMed] [Google Scholar]
- 51. van Lith M, et al. 2001. Regulation of MHC class II antigen presentation by sorting of recycling HLA-DM/DO and class II within the multivesicular body. J. Immunol. 167:884–892 [DOI] [PubMed] [Google Scholar]
- 52. Walsh D, Perez C, Notary J, Mohr I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 79:8057–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zelus BD, Stewart RS, Ross J. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 70:2411–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang J, McCabe KA, Bell CE. 2011. Crystal structures of λ exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl. Acad. Sci. U. S. A. 108:11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zuo J, et al. 2008. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J. Virol. 82:2385–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.