Abstract
The RNA export adaptor protein Rec, encoded for by the human endogenous retrovirus HERV-K/HML-2 elements, binds to the Rec responsive element (RcRE) located in the 3′ untranslated region of HERV-K/HML-2 transcripts. Binding allows the nucleocytoplasmic export of unspliced viral RNA, thereby overcoming host restriction. Chemical probing of the secondary structure of the RcRE corroborated the theory that the RcRE forms a complex folded structure with seven stem-loop regions. Laser-induced liquid beam ion desorption mass spectrometry revealed that Rec forms stable tetramers, which are further stabilized upon RNA binding. The RNA protein complex consists of three Rec tetramers, which bind to multiple sites on the RcRE—preferentially to purine-rich motifs—which represent several low-affinity binding sites. Mutated RcREs, with one to three purine-rich motifs deleted, were still bound and exported by Rec, indicating that the complex folded structure of the RcRE is important for Rec binding. This suggests a binding model where up to three Rec tetramers bind to the complex folded structure of the RcRE and the binding seems to be tightened by recognition of the purine-rich motifs.
INTRODUCTION
Approximately 8% of the human genome consists of retrovirus-like sequences (19). These human endogenous retroviruses (HERVs) were presumably acquired during the evolution by occasional infections of individual germ cells with exogenous viruses, followed by fixation of such “endogenized” retroviral elements in the respective population (for reviews, see references 5 and 24). Like exogenous retroviruses, HERVs can be grouped into families which often comprise several hundred or even several thousand members defined by closely related sequences and dispersed over the human chromosomes. One of these families is designated HERV-K/HML-2, abbreviated as HERV-K here. All HERVs identified thus far are noninfectious, and most members of the HERV-K family have accumulated mutations and deletions (24). However, some HERV-K elements encode for some or even all viral proteins enabling formation of virus-like particles, which were first observed in tumor cell lines of germ cells (23, 32). It is assumed that these proteins may provide protection against related exogenous viruses (31, 44). Intact HERV-K elements encode for the common viral genes gag, prt, pol, env, and rec. The later codes for the essential, accessory protein Rec, an RNA export adaptor protein, which has an important role in carcinogenesis (7, 9, 17). Transgenic Rec can cause testicular carcinoma in situ in mice (17), probably by binding to the androgen receptor and thus counteracting zinc finger regulators of germ stem cell pluripotency and spermatogenesis, as suggested by in vivo and in vitro reporter assays (7, 21).
Rec is responsible for the export of unspliced and incompletely spliced viral RNAs to the cytosol and mostly present in the nucleus (28, 43). It has an arginine-rich nuclear import signal, which is also used for RNA binding (27). It was proposed that Rec binds as an oligomer to its responsive element (RcRE), which is located in the U3 region of the 3′ untranslated region of HERV-K transcripts and exports RNA to the cytosol via a Crm1/RanGTP-dependent pathway (28, 46).
The exogenous human immunodeficiency virus (HIV) and human T-cell leukemia virus (HTLV) possess functional analogues of Rec (called Rev and Rex, respectively) (16, 25, 29, 35, 47). Rev (of HIV) binds to a complex folded Rev responsive element (RRE), which is located within the env gene and is excised during splicing events. Rev dimers bind to a specific stem-loop (IIB) within the RRE and export unspliced viral RNA as a multimer on the same Crm1-dependent pathway as Rec (10, 11, 13, 29, 30). The Rex protein of HTLV also exports unspliced viral RNA to the cytosol via a Crm1-dependent mechanism. It binds to a specific stem-loop within the Rex responsive element (RxRE), which is located within the 3′ long terminal repeats (8, 20, 22, 27, 47).
The sequence of a prototypic, 429-nucleotide-long, functional Rec responsive element (RcRE), pck30, has been identified (26). A secondary structure prediction of RcRE pck30 suggested a highly complex folding with seven stem-loops in the core and a 30-bp closing stem. Individual single point mutations within the RcRE did not severely impair the nuclear export of a Rev-dependent reporter construct (26). Interestingly, it was shown that Rev and Rex are able to mediate the export of RcRE, whereas Rec neither exports the RRE nor the RxRE (26).
This prompted us to analyze the binding of Rec on RcRE in more detail to evaluate the binding characteristics compared to Rev and Rex. We determined the secondary structure of RcRE by structural probing and mapped the Rec binding sites on the RcRE. We identified several GGAA motifs, which are important for Rec binding. Laser-induced liquid beam ion desorption mass spectrometry (LILBID-MS) revealed that Rec binds as a tetramer to RcRE pck30.
MATERIALS AND METHODS
Plasmid construction.
RcRE pck30 was amplified via PCR with the oligonucleotides Fwd_pck30uLTR21 (Purimex) and Rev_pck30uLTR21 (Purimex) using pBluescript RcREpck30 as a template. Fwd_pck30uLTR21 inserts an EcoRI restriction site and the T7-promoter, while Rev_pck30uLTR21 inserts an XbaI restriction site. The fragment was cloned into pSP64 (Promega), which carries an ampicillin resistance selection marker gene, creating pSP64_pck30. All sequences are given in Table S1 in the supplemental material.
Overlapping fragments (20 nucleotides [nt] in length) for small hairpin RNAs were ordered from Eurofins MWG (see Table S1 in the supplemental material). The fragments were amplified by overlap primer extension reaction, cleaved with EcoRI and XbaI (New England BioLabs), and inserted into pSP64. The in vitro transcription resulted in 40-nt small hairpin RNAs (GGU CGU ACA GCU UGU CGN NNN NNC GGC GAG CAG UAU GAU C) with different hexaloops (an N6 pyrimidine-rich loop [UUC CGU], a GGAA loop [AGG AAG], a purine-rich loop [AAG UAA], a AAGG loop [AAA GGG], and a GGA loop [AGG AUG]) and a hairpin with a GGAA motif within the stem (GGUCGUAUAUCUUGUCGUUCCGUCGGCGAGGAAUAUGAUC).
The mutations in RcRE pck30 were inserted into pSP64_pck30 by using a Phusion site-directed mutagenesis kit (New England BioLabs) according to the manufacturer instructions with primers ordered from Eurofins MWG (see Table S1 in the supplemental material). Several mutations of RcRE pck30 were created as follows: M1, A82U G83C; M2, G106U A107U A108C; M3, A125C G126C; M4, A168C G169C; M5, G270C A271U.
The mutated RcREs were amplified via PCR with the oligonucleotides Fwd_RcRE pck30 plus SacII and Rev_RcRE pck30 plus XhoI (Sigma-Aldrich; the sequences are listed in Table S1 in the supplemental material). Fwd_RcRE pck30 plus SacII inserts a SacII and Rev_RcRE pck30 plus a XhoI restriction site. The fragments were cloned into pHIVgag deltaU3 RIII, which carries an ampicillin resistance selection marker and the HIV gag gene sequence 5′ of the mutated RcREs. Successful molecular cloning of all constructs was confirmed by sequencing.
In vitro transcription.
HERV-K RcRE pck30 RNA and mutated versions, as well as small hairpin RNAs, were in vitro transcribed from a T7 promoter by runoff transcription using linearized plasmids. XbaI (New England BioLabs) was used for linearization. In vitro transcription was performed at 37°C overnight. The 1-ml reaction contained 100 μg of template, 25 mM magnesium acetate, 200 mM Tris-HCl (pH 8.0), 20 mM spermidine, 4 mM concentrations of each nucleotide, and 10 mg of homemade T7 polymerase (14). The pyrophosphate was pelleted by centrifugation, and a 0.5 volume of 0.5 M EDTA was added. After ethanol precipitation, the RNA was separated by gel electrophoresis (6% polyacrylamide, 8 M urea), detected by UV shadowing, and eluted in 300 mM sodium acetate (pH 6.5) at 4°C overnight. The supernatant was filtered using a 0.45-μm-pore-size filter (Sarstedt) and ethanol precipitated. The RNA was resuspended in H2O and stored at −20°C.
Labeling of RNA.
In vitro-transcribed RNA was dephosphorylated with alkaline phosphatase (Roche) at 50°C for 15 min and ligated to a Cy5-labeled DNA oligonucleotide [DNA-RNA-Hybrid Cy5-RNA_RcRE pck30: Cy5-d(AAA AAA AAA A) r(UUG AAA AA); Biospring] for in-line probing. The oligonucleotides were heated to 95°C for 5 min and annealed during slowly cooling to 30°C. Then, 10 μl of T4 RNA ligase 2 (New England BioLabs) was added, and the total reaction volume was adjusted to 50 μl, followed by incubation at 37°C for 2 to 3 h. The reaction was stopped by ethanol precipitation. Dephosphorylated RNA was radioactively labeled for electrophoretic mobility shift assays (EMSAs) using a 3-fold excess of [γ-32P]ATP (Hartmann Analytic) by polynucleotide kinase (Roche) at 37°C for 1 h.
Selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE).
A total of 8 pmol of RcRE pck30 RNA was denatured in 50 mM Tris-HCl (pH 7.4) and 20 mM KCl at 90°C for 2 min and folded during slow cooling to room temperature. Afterward, 1.2 mmol of benzoylcyanide (BzCN; Sigma-Aldrich) in dimethyl sulfoxide (DMSO) or DMSO absolute (control) was added, followed by incubation at room temperature for 5 min. Subsequently, ethanol precipitation was performed overnight. Cy5-labeled primers (middle-primer_SHAPE_pck30uLTR21 and 3′-primer _SHAPE_pck30uLTR21 [Biospring]; the sequences are listed in Table S1 in the supplemental material) were used for reverse transcription. The cDNA synthesis was performed using SuperScript III (Invitrogen) with 4 pmol of RNA at 50°C for 1 h. The reaction was stopped by heat inactivation at 70°C for 15 min, and 6 μl of alkaline loading buffer (300 mM NaOH, 6 mM EDTA, 40% [vol/vol] sucrose, 0.2% [wt/vol] croceine orange) was added, followed by incubation at 90°C for 3 min. DNA fragments were separated using gel electrophoresis (12% polyacrylamide, 8 M urea; ALFexpress [Amersham]). SHAPE analysis was performed at least six times for RcRE pck30 RNA. To assign the SHAPE signals, Sanger sequencing was performed with 0.1 μM RcRE pck30, 500 mM primer (see the SHAPE primers), 0.5 mM deoxynucleoside triphosphates (dNTPs), and 0.13 mM ddNTP (Roche).
The analysis was performed in absence or presence of 3 mM MgCl2 and in the presence of Rec protein (3.2, 6.3, and 12.7 μM Rec in 10 mM Tris-HCl [pH 7.5], repeated at least twice with each concentration).
In-line probing.
In-line probing was performed as previously described (41). In detail, 3 pmol of Cy5-labeled folded RNA was incubated in 50 mM Tris, 20 mM MgCl2, and 100 mM KCl (pH 8.3) at room temperature for 24 to 72 h in a total volume of 10 μl. Thirty pmol of Rec protein (in 10 mM Tris-HCl [pH 7.5]) was added to the reaction for analysis of protein binding sites on RcRE pck30 RNA. The cleavage pattern was analyzed by gel electrophoresis (12% polyacrylamide, 8 M urea). All experiments were repeated at least twice. RNase T1 cleavage and a hydrolysis ladder were performed to assign the cleavage products. For the hydrolysis ladder, 3.6 pmol of Cy5-labeled RNA was incubated in 250 mM sodium carbonate at 90°C for 1 to 2 min. The reaction was stopped by the addition of a 0.5 volume of 1 M sodium acetate. For the RNase T1 cleavage, 3 pmol of Cy5-labeled RNA in 25 mM sodium citrate (pH 5.0) was incubated with 1 μl of RNase T1 (New England BioLabs) at 55°C for 5 min in a total volume of 10 μl.
EMSA.
For the EMSA analyses, 1 to 10 fmol of [γ-32P]ATP-labeled RNA in H2O was heated at 55 to 80°C for 5 min and then cooled on ice to fold the RNA. The RNA was incubated in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 25 mM NaCl, and 25 μg of yeast tRNA/ml for the EMSA. Increasing concentrations of Rec (0 to 20 μM in 10 mM Tris-HCl [pH 7.5]) were added, and the samples were incubated at room temperature for 30 min. The reaction was stopped by the addition of 89 mM Tris, 89 mM boric acid, and 5% glycerol and subsequently analyzed by gel electrophoresis (1 to 3% agarose in 89 mM Tris–89 mM boric acid, 70 V). The gel shifts were performed at least twice, the bands were integrated, and the arithmetic mean was calculated. The KD (equilibrium dissociation constant) was estimated by fitting the arithmetic mean to Hill function: y = (Bmax·xn)/(KDn + xn).
LILBID-MS.
The mass spectra were recorded using the previously described laser-induced liquid beam ion desorption mass spectrometry (LILBID-MS) (18, 36, 37). In brief, tiny droplets of native solution (∼65 pl) were injected into a high vacuum, where they were irradiated by mid-infrared laser pulses. During the subsequent explosion, the analyte molecules were set free and analyzed by a time-of-flight mass spectrometer.
In vitro-transcribed RNA was denatured in H2O at 90°C for 3 min, folded during cooling on ice, and diluted in buffer to denoted concentrations. Rec was added, and the reaction was incubated at room temperature for 30 min. The spectra for Rec were recorded in 10 mM Tris-HCl (pH 7.5) for 0.5 μM RcRE pck30 in complex with 2 μM Rec in 5 mM Tris-HCl (pH 7.5), 5 mM NH4HCO3, and 1 mM MgCl2. The spectrum of 460 nM RcRE pck30 in complex with 180 nM Rec was recorded in 10 mM Tris-HCl (pH 7.5) and 1 mM MgCl2. Portions (5 μl) of the respective samples were loaded into the spectrometer.
Export assay.
Export assays were performed to analyze the function of the different RcRE mutants in tissue culture cells. A total of 250 ng of p24 HIV capsid reporter constructs carrying different RcRE variants (pHIVgag_RCRE, pHIVgag_M1, pHIVgag_M2, pHIVgag_M3, pHIVgag_M4, pHIVgag_M5, pHIVgag_M2&M5, pHIVgag_M3&M5, and pHIVgag_M3&M4&M5) were transfected together with 100 ng of Rec plasmid encoding the viral export adapter HERV-K Rec (26–28). The Rec-dependent RNA export activity can be determined by measuring the amount of the reporter protein translated from the exported transcripts. Transient transfections into HeLa Tat cells (42) were performed in 24-well plates (Greiner; 0.2 million cells per well) for HIV p24 quantification. Transfections were carried out with GeneJuice transfection reagent (Novagen) according to the user manual. At 24 h after transfection, the cells were lysed in 200 μl of passive lysis buffer (Promega) containing 1/10 of lysis buffer provided in detection kit (ZeptoMetrix). The lysates were cleared by centrifugation. Quantification of HIV p24 was carried out using RETROtek HIV-1 p24 Antigene ELISA 2.0 (ZeptoMetrix) with 15 μl of lysate according to the instructions. All transfections were performed in triplicates and normalized to the protein content. Within each set of experiments, the HIV Gag expression level mediated by the RcRE sequence of the pcK30 LTR and Rec was set as 100%.
RESULTS
Analysis of the secondary structure of the RcRE pck30 by SHAPE and in-line probing.
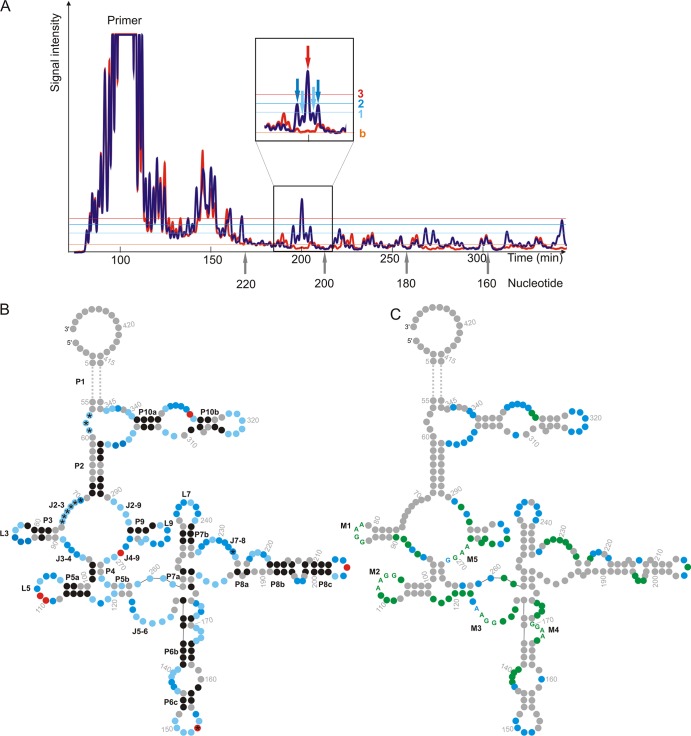
In vitro-transcribed RcRE pck30 was treated with benzoyl cyanide (BzCN) for SHAPE analysis according to the method of Weeks (34, 38). BzCN builds an ester with ribose, a modification that interferes with the processivity of the reverse transcriptase detectable by gel electrophoresis. The degree of modification depends on the flexibility of nucleotides, thus indicating whether the nucleotide is located in a helical part or a more flexible loop region. Two Cy5-labeled primers were used to specifically start reverse transcription. One primer anneals close to the 3′ end of the RcRE pck30 RNA and the second anneals in the middle of the sequence, allowing complete analysis of RcRE pck30 between nt 67 and 381 of the 429-nt RcRE pck30. The SHAPE reactivities were classified to three categories (strong, medium, and weak signals) according to the reference peak of the spectrum. Always the same peak of the spectra was used as a reference. A representative spectrum including the reference peak is shown in Fig. 1. Nucleotides that gave rise to peaks below the threshold for weak signals were assumed to be in double-stranded regions. The SHAPE reactivities were aligned to the secondary structure prediction of RcRE pck30 (26) and, based on this, a secondary structure model was developed, which is displayed in Fig. 1 with the different sensitivities of the nucleotides reflected in the color depth (Fig. 1B). The analysis confirmed the existence of seven stem-loop regions and a 50 nt closing stem P1.
Fig 1.
Analysis of the secondary structure of RcRE pcK30 with SHAPE and in-line probing. (A) Section of a representative spectrum for SHAPE (blue) and control reactivities (orange). The inlet shows the pattern of nt 207 to 203, which were used as reference for each SHAPE analysis (light blue arrow, 1; dark blue arrow, 2; red arrow, 3). The light blue, dark blue, and red lines, respectively, indicate signals set as 1 (weak), 2 (medium), or 3 (strong). The baseline is shown in orange (indicated by “b”). (B) Secondary structure of RcRE pck30 with highlighted signal intensities for SHAPE where the 50-nt closing stem (P1) is omitted (replaced by dotted line). Stems (P), junctions (J), and loops (L) are indicated. The color depth represents the intensity of the signals (black, nucleotide in double strand; light blue, weak signal for nucleotide in single strand; dark blue, medium strong signal for nucleotide in single strand; red, strong signal for nucleotide in single strand; and gray, nucleotide could not be resolved). Nucleotides marked with an asterisk (*) were analyzed with in-line probing only, because the 5′ end could not be resolved with SHAPE. (C) Secondary structure of RcRE pck30 with marked nucleotides that showed prominent changes in the probing pattern in the presence of Rec and therefore are potential Rec binding sites (marked in green) and marked nucleotides that are stabilized with magnesium ions (marked in blue). The purine-rich motifs inside the potential Rec binding sites are given as letters and are indicated as M1 to M5.
The strong SHAPE reactivities without magnesium indicated a flexible structure with a large unfolded region at junction J2-3, J3-4, J4-9, and J2-9 and also at J5-6. In addition, all terminal loops L3 to L10 and several internal loops within P6, P8, and P10 were accessible. The majority of all SHAPE reactivities decreased in the presence of magnesium ions, indicating magnesium-dependent folding of the structure (displayed in Fig. 1C). The most prominent changes were observed within P10 and the terminal loops L6, L7, and L9. In addition, the internal loops of P8, junctions J3-4 and J2-9, showed reduced sensitivity, indicating tertiary interactions of these regions.
In addition, we performed in-line probing to complement structural data obtained by SHAPE. Here the secondary structure is analyzed by self cleavage of RNA. This is triggered by a nucleophilic attack of the 2′ OH group of ribose on the neighboring 3′ phosphate. The reaction only takes place when the 2′ OH group is in-line to the phosphate group, which occurs for flexible nucleotides only.
We used a shortened construct of RcRE pck30 (nt 45 to 354) missing the closing stem P1 for in-line probing. The RNA was Cy5 labeled and incubated at room temperature for 32, 52, or 72 h. The fragmentation pattern was analyzed by gel electrophoresis. The signals were classified to three categories (strong, medium, and weak signals) and incorporated into the color code of the structure shown in Fig. 1B. The in-line probing data corroborated the SHAPE reactivity information. In addition, the first 80 nt were only accessible by in-line probing. Hereby, junction J2-3 was solved, and the loop regions of P3 to P8 were verified. Taken together, the structural probing confirms in parts the predicted secondary structure and shows that RcRE pck30 folds into a highly structured RNA.
Mapping of binding sites of Rec on the RcRE pck30.
SHAPE and in-line probing were performed in the presence of Rec to determine the binding sites of Rec on the RcRE pck30 RNA. All nucleotides that showed prominent changes in the probing pattern are displayed in Fig. 1C.
A multitude of signals decreased in the presence of the protein, indicating that Rec might contact the RNA at different sites. The most prominent changes were located in the central region of the structure, especially the terminal loops L3, L5, and L9 and junctions J3-4, J4-9, J5-6, and J7-8. Interestingly, very often purine-rich stretches with a preference for GGAA or AAGG are protected by Rec (marked as M1 to M5 in Fig. 1C), indicating that these might be potential binding motifs for Rec.
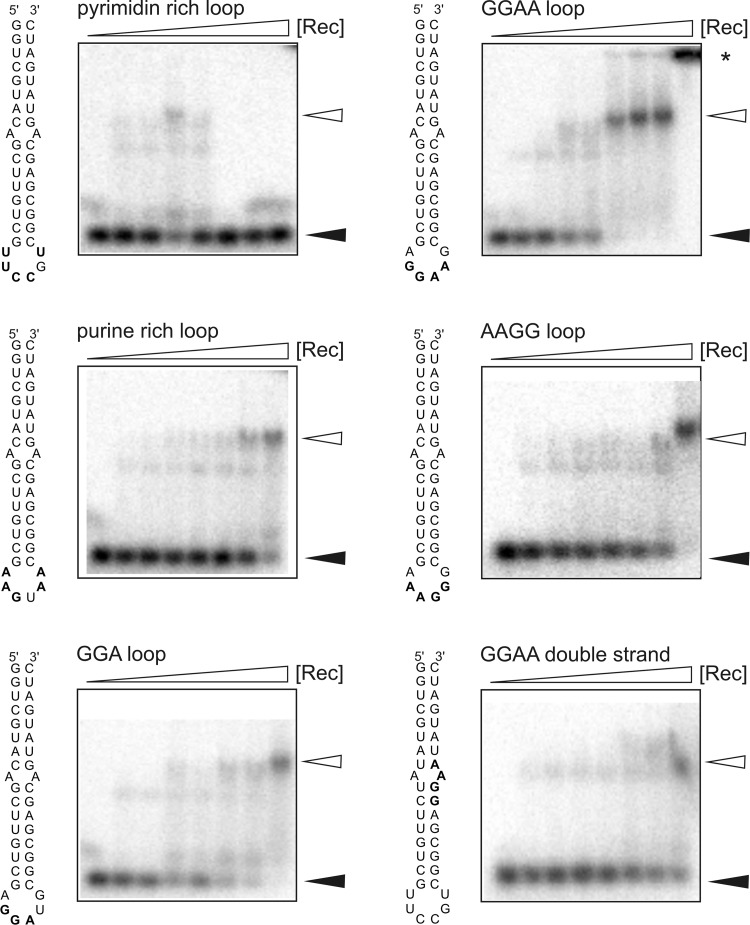
We performed EMSAs to prove this hypothesis. We designed 40-nt hairpin RNAs with different purine-rich motifs presented in terminal hexaloops (AGGAAG, AGGAUG, AAAGGG, and AAGUAA) and a pyrimidine-rich control construct (UUCCGU). In addition, a construct with a GGAA motif within the stem was designed (Fig. 2).
Fig 2.
EMSA results for small hairpin RNAs. Portions (10 fmol) of radiolabeled RNA were incubated with increasing concentrations of Rec (0, 0.8, 1.2, 1.6, 2.0, 2.4, 2.8, and 3.2 μM). The reaction was performed in the presence of 25 μg tRNA/ml to prevent nonspecific RNA binding to Rec. Complex formation was analyzed using 3% agarose gels. Filled and open arrowheads correspond to free and shifted RNA, respectively. RNA retained in the loading pocket is indicated by a star.
We observed clear binding of Rec to the hairpin containing the GGAA motif (AGGAAG hexaloop) with an estimated KD of ∼2.5 ± 0.06 μM (Fig. 2). The fit indicated a highly cooperative binding. The binding of Rec to the constructs with AGGAUG, AAAGGG, or AAGUAA loops were slightly weaker than to the hairpin with the AGGAAG loop. No binding could be observed for the construct with the pyrimidine-rich loop. The RNA with a GGAA motif in the stem showed only marginal binding to Rec (Fig. 2). These data indicate that there is specific binding of Rec to purine-rich loops, especially those with GGAA motifs.
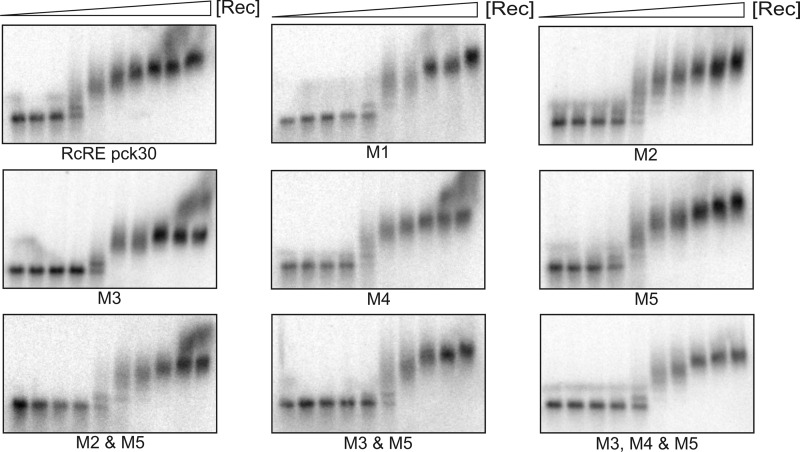
In contrast to the small hairpin constructs, the complete RcRE pck30 bound Rec in the low nanomolar range with an estimated KD of ∼ 45 ± 1.6 nM (Fig. 3; Table 1). The RNA shifted at different heights and showed highly cooperative binding, thus the KD could only be roughly estimated. The differences in binding between RcRE pck30 and the small hairpins indicate that although there is a favored binding of Rec to GGAA motifs, the complex folded structure of the RcRE pck30 is necessary for high-affinity binding. To confirm this, we created a set of RcRE pck30 variants in which different GGAA and AAGG motifs were mutated as follows: M1, the AAGG motif of L3 was mutated to AUCG; M2, the GGAA motif of L5 was mutated to GUUC; M3, the AAGG motif of J5-6 was mutated to ACCG; M4, the AAGG motif in the internal loop of P6 was mutated to ACCG; and M5, the GGAA motif of J4-9 was mutated to GCUA. In addition, M2/M5 and M3/M5 double mutants and an M3/M4/M5 triple mutant were created. The motifs are highlighted in Fig. 1C.
Fig 3.
EMSA of RcRE pck30 mutants. Portions (10 fmol) of radiolabeled RNA were incubated with increasing concentrations of Rec (0, 0.02, 0.04, 0.06, 0.08, 0.12, 0.16, 0.32, 0.56, and 2 μM), and complex formation was analyzed using 1% agarose gels. The location of the mutations is shown in Fig. 1. M1, the AAGG motif was mutated to AUCG; M2, the GGAA motif was mutated to GUUC; M3, the AAGG motif was mutated to ACCG; M4, the AAGG motif was mutated to ACCG; M5, the GGAA motif was mutated to GCUA. Calculated KD values are given in Table 1.
Table 1.
CalculatedKD values for Hill fita
| Construct | Mean KD (nM) ± SD |
|---|---|
| RcRE pck30 | 45.25 ± 1.59 |
| M1 | 88.15 ± 1.89 |
| M2 | 80.43 ± 0.19 |
| M3 | 98.03 ± 2.28 |
| M4 | 92.23 ± 1.66 |
| M5 | 148.90 ± 0.86 |
| M2 and M5 | 89.64 ± 4.60 |
| M3 and M5 | 117.01 ± 1.00 |
| M3, M4, and M5 | 99.75 ± 1.01 |
The calculated KD values for RcRE pck30 and its mutations are shown.The arithmetic mean of at least two EMSAs was fitted to the Hill equation (see Materials and Methods), and the standard deviation was calculated.Representative shifts of each mutation are shown in Fig. 3.
Using EMSA, a reduced level of binding for all mutations was observed (Fig. 3). All mutations of one motif had comparable effects on Rec binding. The KD values for the single mutations slightly increased to values ∼90 nM compared to 45 nM for RcRE pck30 (Table 1). The highest KD was observed for M5 with 150 nM, pointing to this motif as especially important for Rec binding. As soon as two or more motifs are destroyed the binding is further impaired. Double mutations showed a KD of 117 nM. For the triple mutation, the KD was comparable to that for the double mutations.
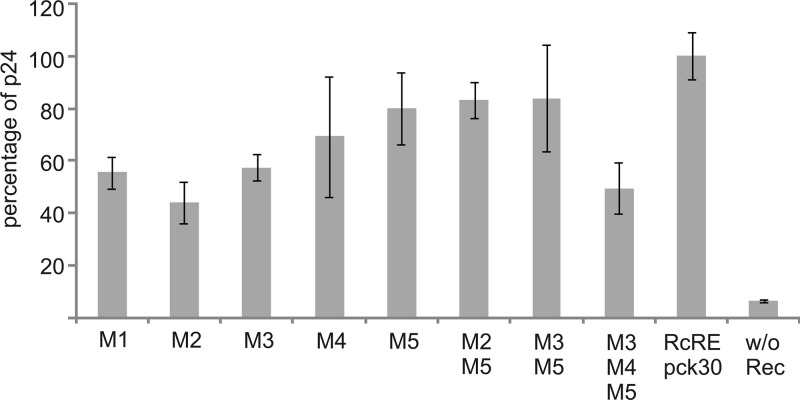
Export studies support these data. The export of the RcRE was impaired for all single mutations with the greatest impact of M2 (Fig. 4). Here, export efficacies were reduced to 45% of wild-type RcRE pck30. The triple mutation clearly showed an additive effect (reduction to 50%, which is not so prominent for the double mutations). The data clearly show the importance of the identified purine-rich motifs for functionality in cell culture.
Fig 4.
Export assay of RcRE pck30 mutants. The HIV Gag precursor was determined in cell lysates after cotransfection of the indicated pHIVgag_RcRE pck30 mutant vector into HeLa Tat cells with the effector plasmid pRec. The results of one representative triplicate are shown. Error bars indicate the standard deviations.
Taken together, we identified several Rec binding sites on the RNA; however, single mutations lead to small effects only. Therefore, we assume that the GGAA and AAGG motifs contribute to the binding of Rec, but the main part of the Rec binding is mediated through the structure of the RcRE pck30.
Analysis of multimerization of Rec bound to RcRE pck30.
EMSA of RcRE pck30 resulted in bands at different heights, indicating that several Rec proteins may bind to RcRE pck30. We performed LILBID-MS to characterize the stoichiometry of Rec binding. LILBID is a soft MS method for the analysis of noncovalent bound complexes (36). The fluid sample is irradiated by different laser strengths, and the decay of the complex is observed.
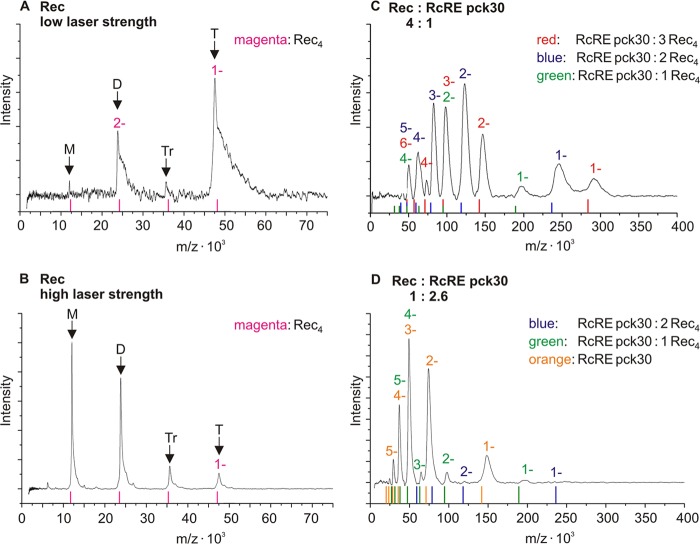
LILBID spectra of Rec were recorded at low laser strength and are displayed in Fig. 5A. The spectra show the mass per charge plotted against the laser intensity. A prominent peak at 47 kDa emerged, which corresponds to a Rec tetramer. A smaller fraction of dimers could be observed, whereas the concentration of Rec monomers and trimers was negligible. The tetramers can be dissociated with high laser strength, which is indicated by prominent peaks for Rec monomers and dimers in Fig. 5B.
Fig 5.
LILBID spectra of Rec in the absence or presence of RcRE pck30. The signal intensity is plotted against mass per charge. For each complex, ions with different charges could be detected. The charges of these species are indicated by 1 to 6 in the complex-specific color. Thus, some peaks overlay because the mass per charge can be the same for different complexes with differing charges. (A and B) Spectra of 10 μM Rec with low (A) and high (B) laser strength. The expected masses of oligomers are indicated by magenta lines and arrows (monomer [M], 11.8 kDa; dimer [D], 23.6 kDa; trimer [Tr], 35.4 kDa; tetramer [T], 47.2 kDa). With increasing laser strength, the Rec tetramers fall apart. (C) Spectrum of 0.5 μM RcRE pck30 in complex with 2 μM Rec recorded with high laser strength. Peaks composed of complexes of RcRE pck30 with one Rec tetramer are marked with green charges (RcRE pck30, 141.6 kDa; complex, ∼190 kDa). For peaks of complexes composed of RcRE pck30 and two or three Rec tetramers, the charge is marked in blue and red, respectively (RcRE pck30: 2 Rec4, ∼236 kDa; RcRE pck30: 3 Rec4, ∼283 kDa). The observed complexes differ by one Rec tetramer. (D) Spectrum of 460 nM RcRE pck30 in complex with 180 nM Rec recorded with high laser strength. Peaks for RcRE pck30 are marked with an orange charge. For peaks composed of complexes of RcRE pck30 with one or two Rec tetramers, the charges are marked in green and blue, respectively.
We then recorded Rec in complex with RcRE pck30 (Fig. 5C and D), and large complexes with up to three Rec tetramers bound to one RcRE pck30 appeared. This is shown by the peaks arising for the complexes with one, two, and three Rec tetramers bound to RcRE pck30 (indicated as RcRE pck30; 1 Rec4 [green], 2 Rec4 [blue], and 3 Rec4 [red] [Fig. 5C]). The smallest complexes observed here consisted of one Rec tetramer and one RcRE pck30. These complexes were disassembled with the applied laser strength, but there were always Rec tetramers leaving the RNA complexes. This can be concluded because there are no peaks for complexes consisting of less than one RcRE pck30 and one Rec tetramer at high laser strength (Fig. 5C and D).
When RNA was supplied in excess, we exclusively observed peaks for the complex of one RcRE pck30 molecule bound to at least one Rec tetramer, as well as peaks for unbound RcRE pck30 (marked in green and orange in Fig. 5D). This indicates that there was free RNA rather than complexes smaller than Rec tetramers bound to RcRE pck30. Therefore, the Rec tetramers seemed to be highly stabilized by binding to RcRE pck30 because Rec monomers or dimers have only been observed in the absence of RNA (Fig. 5B).
Taken together, Rec protein binds as tetramer to the RcRE and becomes stabilized upon RNA binding. Based on the results of the shift assay, we assume that there might be a binding of at least three Rec tetramers to RcRE pck30, which is also supported by the LILBID data.
DISCUSSION
We have shown in the current report that RcRE pck30 builds a complex folded secondary structure with seven different stem-loops and a highly flexible central region. Both SHAPE and in-line probing coincided with each other and confirmed large parts of the predicted two-dimensional structure (26). The main difference is that the probed structure is more flexible than the predicted one. Mainly junctions J3-4, J2-9, J4-9, and J5-6 seem to be flexible, while they are predicted to be in a double strand.
Rec binding to the RNA occurs in a rather complex binding pocket instead of using a defined binding element with binding sites in loops L3, L5, and L9 and junctions J3-4, J5-6, and J7-8. These regions have already been proposed to be important for RNA export, underlining the importance of Rec binding to this region as a crucial step for the export of unspliced viral RNA from the nucleus (26).
Nearly all sites protected by Rec contain purine-rich loops, often with GGAA motifs. Within the HERV-K family members, all five GGAA/AAGG motifs of RcRE pck30 are conserved (26). We have proven the direct binding by EMSA using small purine-rich hairpin RNAs. The assay clearly showed that purine-rich, but not pyrimidine-rich motifs are bound. Rec recognized the small hairpins as ∼50-fold weaker than the complete RcRE pck30, indicating that a correct presentation of the purine-rich motifs by the correctly folded RcRE pck30 is important. This is further supported by the mutational analysis of GGAA motifs within the context of the complete RcRE pck30 by which the mutation of one to three motifs lead to a stepwise decrease in binding. This was confirmed by export studies. The export efficacy was affected by the mutations as well. The triple mutant export efficacy was decreased to 50% of wild type. The effect of the mutations on the export seemed to be stronger than on the binding to Rec. Therefore, we assume that the mutations may destabilize the export complex. The purine-rich motifs in the center of the RcRE pck30, such as M2, M3, and M5, might be the most important ones for Rec binding because their single mutations showed the strongest effect, as well as did the triple mutant. The export was not inhibited completely by the mutations, indicating that the complex folded structure of the RcRE pck30 is important for both Rec binding and export.
The binding of purine-rich RNA sequences was already described for a bacterial regulatory protein called CsrA. The protein binds to the untranslated region of mRNA, thereby regulating their translation (45). It has been shown that CsrA recognizes a conserved GGA motif within the loop of a hairpin structure. Also, the sequence and the structure are important for CsrA binding (15).
Another protein that recognizes purine-rich RNA sequences is TRAP (1, 3). This protein regulates the translation and transcription of the trp operon in Gram-positive bacteria such as Bacillus subtilis (4, 33, 39). Here, TRAP multimers bind to GAG repeats on RNA by wrapping the RNA around the protein (1, 3). It has been shown that mainly lysines and arginines mediate the binding of TRAP to purine-rich motifs (2). Rec seems to use its arginine-rich motif to bind to RcRE pck30 as well (27). Therefore, binding comparable to purine-rich motifs is imaginable, although it is unlikely that the RcRE is wrapped around Rec because of its complex folded structure.
Rev of HIV is a functional analogue of Rec (16, 25, 29). It has been shown that Rev binds preferentially to the stem-loop IIB within its responsive element (RRE) but oligomerizes on a well-defined multiple stem-loop structure (6, 10, 11, 13, 30). No binding of Rev to RRE was observed once stem-loop IIB was deleted (10, 30). In contrast to Rec, Rev shows no sequence specific binding to its responsive element (30). Rev binds even when the sequence of the stem-loop IIB is completely changed by maintaining the secondary structure, indicating that the structure seems to be much more important for Rev binding than the sequence (30). It was shown that Rec's export efficiency is also affected when the structure of the RcRE is destroyed (26).
Rev binds with three dimers to its responsive RNA element, as shown in the recently solved structure of the Rev-RRE complex (13). A sequential binding of Rev to one RRE was proposed with the multimeric assembly nucleated by the high-affinity binding of Rev (12, 13, 40). The binding of additional Rev dimers occur to already bound Rev proteins and adjacent sites on the RNA with less affinity (13, 40). This seems to be comparable to our Rec binding model with three tetramers binding to the complex folded structure of the RcRE pck30 with several low-affinity binding sites.
The Rec protein forms stable tetramers, as demonstrated by LILBID-MS. However, up to three tetramers were bound to one RNA, which was also supported by EMSA. It also binds as tetramer to RcRE pck30 even when the RNA was in excess. It seems that unbound RNA is preferred over splitting Rec tetramers. Within the RNA protein complex the tetramers could not be ruptured into monomers any more. Therefore, we propose that the binding of RcRE pck30 might induce a conformational change in Rec that tightens the interaction between the subunits. This is again comparable to Rev binding because the RRE seems to control the oligomeric state of Rev as well (12).
In summary, we have shown that Rec binds to and thereby stabilizes the complex folded structure of RcRE pck30. It forms tetramers, which are highly stabilized by RNA binding, and up to three Rec tetramers bind to one RcRE pck30. We assume that Rec binds to the complex folded structure of RcRE pck30 predominantly. This binding might be tightened by the binding of Rec to purine-rich motifs in single-stranded regions, especially GGAA motifs with a sequential binding model comparable to the one for Rev (13, 40). In this model, initially one Rec tetramer may bind with high affinity to the complex folded central region of RcRE pck30 containing a GGAA and an AAGG motif. In the following steps, the cooperative binding of additional Rec tetramers might occur.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ute Scheffer and Michael Goebel for their support with the SHAPE measurements and the Cy5 labeling of RNA and for providing the ALFexpress. We are very grateful to Oliver Weichenrieder who kindly provided the T7 polymerase. We thank Matilda Oke for carefully reading the manuscript.
This study was supported by the Aventis Foundation (B.S.) and the Deutsche Forschungsgemeinschaft (Cluster of Excellence Macromolecular Complexes [B.S., J.H., and B.B.] and GRK 1172 [J.S.L.]).
J.S.L. performed experiments for SHAPE, in-line probing and EMSA, analyzed data, and wrote the manuscript. N.V.F. performed export studies and analyzed these data. J.H. performed the LILBID-MS and analyzed these data. A.W. performed preliminary experiments. R.L. gave the impulse for this study and designed experiments. B.B. designed experiments, and B.S. designed experiments and edited the manuscript.
Footnotes
Published ahead of print 13 June 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Antson AA, et al. 1994. 11-fold symmetry of the trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis determined by X-ray analysis. J. Mol. Biol. 244:1–5 [DOI] [PubMed] [Google Scholar]
- 2. Antson AA, et al. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401:235–242 [DOI] [PubMed] [Google Scholar]
- 3. Antson AA, et al. 1995. The structure of trp RNA-binding attenuation protein. Nature 374:693–700 [DOI] [PubMed] [Google Scholar]
- 4. Babitzke P, Yanofsky C. 1993. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc. Natl. Acad. Sci. U. S. A. 90:133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 101(Suppl 2):14572–14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel DP, Zapp ML, Green MR, Szostak JW. 1991. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell 67:529–536 [DOI] [PubMed] [Google Scholar]
- 7. Boese A, et al. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328–4336 [DOI] [PubMed] [Google Scholar]
- 8. Bogerd HP, Tiley LS, Cullen BR. 1992. Specific binding of the human T-cell leukemia virus type I Rex protein to a short RNA sequence located within the Rex-response element. J. Virol. 66:7572–7575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buscher K, et al. 2006. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec, and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 16:223–234 [DOI] [PubMed] [Google Scholar]
- 10. Cook KS, et al. 1991. Characterization of HIV-1 Rev protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 19:1577–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daly TJ, Cook KS, Gray GS, Maione TE, Rusche JR. 1989. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 342:816–819 [DOI] [PubMed] [Google Scholar]
- 12. Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD. 2010. HIV Rev response element (RRE) directs assembly of the Rev homo-oligomer into discrete asymmetric complexes. Proc. Natl. Acad. Sci. U. S. A. 107:12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daugherty MD, Liu B, Frankel AD. 2010. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 17:1337–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 81:2035–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dubey AK, et al. 2003. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 185:4450–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Felber BK, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis GN. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. U. S. A. 86:1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galli UM, et al. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223–3228 [DOI] [PubMed] [Google Scholar]
- 18. Hoffmann J, Schmidt TL, Heckel A, Brutschy B. 2009. Probing the limits of liquid droplet laser desorption mass spectrometry in the analysis of oligonucleotides and nucleic acids. Rapid Commun. Mass Spectrom. 23:2176–2180 [DOI] [PubMed] [Google Scholar]
- 19. Hughes JF, Coffin JM. 2001. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 29:487–489 [DOI] [PubMed] [Google Scholar]
- 20. Inoue J, Yoshida M, Seiki M. 1987. Transcriptional (P40x) and posttranscriptional (P27x-Iii) regulators are required for the expression and replication of human T-cell leukemia virus type I genes. Proc. Natl. Acad. Sci. U. S. A. 84:3653–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaufmann S, et al. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J. Gen. Virol. 91:1494–1502 [DOI] [PubMed] [Google Scholar]
- 22. Kiyokawa T, Kawaguchi T, Seiki M, Yoshida M. 1985. Association of the Px gene product of human T-cell leukemia virus type I with nucleus. Virology 147:462–465 [DOI] [PubMed] [Google Scholar]
- 23. Lower R, et al. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. U. S. A. 90:4480–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lower R, Lower J, Kurth R. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 93:5177–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J. 1995. Identification of a Rev.-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Magin-Lachmann C, et al. 2001. Rec (formerly Corf) function requires interaction with a complex, folded RNA structure within its responsive element rather than binding to a discrete specific binding site. J. Virol. 75:10359–10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Magin C, Hesse J, Lower J, Lower R. 2000. Corf, the Rev/Rex homologue of HTDV/HERV-K, encodes an arginine-rich nuclear localization signal that exerts a trans-dominant phenotype when mutated. Virology 274:11–16 [DOI] [PubMed] [Google Scholar]
- 28. Magin C, Lower R, Lower J. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. 1989. The HIV-1 Rev transactivator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254–257 [DOI] [PubMed] [Google Scholar]
- 30. Malim MH, et al. 1990. HIV-1 structural gene expression requires binding of the Rev transactivator to its RNA target sequence. Cell 60:675–683 [DOI] [PubMed] [Google Scholar]
- 31. Mayer J, et al. 2004. Human endogenous retrovirus HERV-K(HML-2) proviruses with Rec protein coding capacity and transcriptional activity. Virology 322:190–198 [DOI] [PubMed] [Google Scholar]
- 32. Mayer J, et al. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21:257–258 [DOI] [PubMed] [Google Scholar]
- 33. Merino E, Babitzke P, Yanofsky C. 1995. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 177:6362–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127:4223–4231 [DOI] [PubMed] [Google Scholar]
- 35. Mertz JA, Simper MS, Lozano MM, Payne SM, Dudley JP. 2005. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J. Virol. 79:14737–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgner N, Barth HD, Brutschy B. 2006. A new way to detect noncovalently bonded complexes of biomolecules from liquid micro-droplets by laser mass spectrometry. Aust. J. Chem. 59:109–114 [Google Scholar]
- 37. Morgner N, Kleinschroth T, Barth HD, Ludwig B, Brutschy B. 2007. A novel approach to analyze membrane proteins by laser mass spectrometry: from protein subunits to the integral complex. J. Am. Soc. Mass Spectrom. 18:1429–1438 [DOI] [PubMed] [Google Scholar]
- 38. Mortimer SA, Weeks KM. 2008. Time-resolved RNA SHAPE chemistry. J. Am. Chem. Soc. 130:16178–16180 [DOI] [PubMed] [Google Scholar]
- 39. Otridge J, Gollnick P. 1993. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 90:128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pond SJ, Ridgeway WK, Robertson R, Wang J, Millar DP. 2009. HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc. Natl. Acad. Sci. U. S. A. 106:1404–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Regulski EE, Breaker RR. 2008. In-line probing analysis of riboswitches. Methods Mol. Biol. 419:53–67 [DOI] [PubMed] [Google Scholar]
- 42. Schwartz S, Felber BK, Benko DM, Fenyo EM, Pavlakis GN. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh S, Kaye S, Gore ME, McClure MO, Bunker CB. 2009. The role of human endogenous retroviruses in melanoma. Br. J. Dermatol. 161:1225–1231 [DOI] [PubMed] [Google Scholar]
- 44. Tonjes RR, et al. 1996. HERV-K: the biologically most active human endogenous retrovirus family. J. Acquir. Immune Defic. Syndr Hum. Retrovirol. 13(Suppl 1):S261–S267 [DOI] [PubMed] [Google Scholar]
- 45. Wei BL, et al. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245–256 [DOI] [PubMed] [Google Scholar]
- 46. Yang J, et al. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. U. S. A. 96:13404–13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Younis I, Green PL. 2005. The human T-cell leukemia virus Rex protein. Front. Biosci. 10:431–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.