Abstract
Adeno-associated virus (AAV) has attracted considerable interest as a vector for gene therapy owing its lack of pathogenicity and the wealth of available serotypes with distinct tissue tropisms. One of the most promising isolates for vector development, based on its superior gene transfer efficiency to the liver in small animals compared to AAV type 2 (AAV2), is AAV8. Comparison of the in vivo gene transduction of rAAV2 and rAAV8 in mice showed that single amino acid exchanges in the 3-fold protrusions of AAV8 in the surface loops comprised of residues 581 to 584 and 589 to 592 to the corresponding amino acids of AAV2 and vice versa had a strong influence on transduction efficiency and tissue tropism. Surprisingly, not only did conversion of AAV8 to AAV2 cap sequences increase the transduction efficiency and change tissue tropism but so did the reciprocal conversion of AAV2 to AAV8. Insertion of new peptide motifs at position 590 in AAV8 also enabled retargeting of AAV8 capsids to specific tissues, suggesting that these sequences can interact with receptors on the cell surface. However, a neutralizing monoclonal antibody that binds to amino acids 588QQNTA592 of AAV8 does not prevent cell binding and virus uptake, indicating that this region is not necessary for receptor binding but rather that the antibody interferes with an essential step of postattachment processing in which the 3-fold protrusion is also involved. This study supports a multifunctional role of the 3-fold region of AAV capsids in the infection process.
INTRODUCTION
Adeno-associated virus (AAV) vectors belong to the most frequently used viral vectors in current gene therapy applications. They combine several advantageous features, including a good safety profile, stable long-term gene expression in several tissues, the ability to transduce dividing and nondividing cells, and physicochemical stability (11, 66). AAV vectors show generally a low innate immunity, as well as low efficiency to transduce professional antigen-presenting cells (87), although recent studies warrant a more differentiated view of the immune response to AAV-mediated gene transfer (8, 22, 23, 26, 29, 32, 33, 37, 50, 65, 84, 88). Humoral immune responses are generated and memory CD8+ T-cell responses have been observed in clinical trials (36, 40, 58). To circumvent these issues, many different AAV capsid variants have been isolated from nonhuman primates (6, 14, 16, 41, 61, 63, 82), which exhibit enhanced transduction of certain tissues and potentially a lower seroprevalence and diminished preexisting capsid immune responses. In addition, different types of capsid modifications have been explored for improving tissue targeting and transduction efficiency (10, 39, 59, 70).
Among the AAV serotypes characterized thus far, AAV type 8 (AAV8) vectors showed an outstanding liver tropism in mice compared to AAV2, the most extensively studied serotype (12, 15, 27, 45). This trend is also true in skeletal muscle (35, 60, 76), cardiac tissue (53, 68, 76), pancreas (72), and glioblastoma (21), and specialized cells in brain and retina are preferentially transduced by AAV8 (5, 9). In many animal models, AAV8 has already proven to provide an enormous therapeutic potential (7, 18, 38, 47, 54, 55, 62), although translation of the exceptional performance of AAV8 to higher primates has been challenging (25, 74, 75). Application of AAV8 vectors in a clinical trial has recently provided successful proof of concept for expression of the factor IX gene in human liver (48). These observations have stimulated an interest in understanding the different vector performances of AAV2 and AAV8 at a mechanistic level and relating it to structural features of the AAV capsids. The crystal structure of the AAV8 capsid revealed significant differences to AAV2 within the BC and GH loops between the β-strands of the core eight-stranded (βB-βI) β-barrel (46). Of particular interest, these regions play a crucial role in AAV2-mediated gene transduction and antibody recognition (28, 34, 52, 80, 81). A reduced amount of basic residues was observed for AAV8 at the mapped AAV2 heparan sulfate proteoglycan (HSPG) binding region, which is consistent with the non-heparin-binding phenotype of AAV8 (46). A primary attachment receptor for AAV8 has not yet been reported. The 37/67-kDa laminin receptor (LamR) has been suggested to act as a coreceptor of AAV8, with the binding site mapped to two protein domains including amino acids (aa) 491 to 547 and aa 593 to 623 on the AAV capsid exterior (1). However, AAV2 can also use this receptor for cell entry (1), although with lower affinity. Therefore, binding to LamR may only partially explain the different transduction efficiencies of AAV8 and AAV2. In vitro experiments showed that serine proteases cathepsin B and L bind and cleave AAV8 and AAV2 in slightly different patterns to prime the AAV capsids for subsequent nuclear uncoating (2). AAV8 capsids appear to be cleaved more efficiently and quicker than AAV2 capsids, a finding that is consistent with a study demonstrating a faster uncoating rate of AAV8 (69). Altogether, the slower rate of uncoating vector genomes is likely a limiting step of AAV2 transduction compared to AAV8.
Recently, other studies have reported differences between AAV8 and AAV2 that may contribute to the different gene transduction in vivo (43, 64). In a protein microarray-based protein binding assay, AAV8 capsids bound a number of kinases that were not bound by AAV2 (43). However, the influence of these different interaction partners on gene transduction was not assayed. In another report, domain swap experiments showed that crucial differences between the liver transduction efficiencies of AAV2 and AAV8 are located in the GH loop, between aa 432 and 639 of AAV8, which form the surface of the 3-fold protrusions (64). Another study identified a peptide within this region, aa 585 to 590 of AAV8, as conferring its muscle transduction properties to AAV2 (3). These observations suggest that residues within the 3-fold protrusions or adjacent to it are critical structures involved in AAV transduction, given that they also contain an HS binding site and an integrin binding site in AAV2 (4, 28, 52).
The aim of the present study was to map the capsid protein sequences of AAV8 involved in tissue tropism and superior gene transduction compared to AAV2. Knowledge of such capsid protein positions should provide the basis for the insertion of vector targeting peptide sequences. By conversion of specific sequence motifs in the GH loop of AAV8 to AAV2 and vice versa, we have identified two very closely located sequences that strongly influence gene transfer specificity and efficiency of AAV8 and AAV2. Modification of this region by single amino acid substitutions not only affected the transduction efficiency and tissue tropism of AAV8 but also that of AAV2. The AAV8 capsid proteins tolerate peptide insertions at one of these sites for retargeting AAV8 gene transduction in vivo. Most surprisingly, however, specific blocking of one of the motifs by a neutralizing antibody did not prevent vector binding to the host cell but inhibited gene transduction at a postbinding step. This strongly suggests that this region of the AAV8 3-fold protrusion determines receptor binding and is also involved in gene transduction at a postattachment processing step.
MATERIALS AND METHODS
Animals, cell lines, and cell culture.
Six- to nine-week-old, female NMRI mice and male B6 mice were used for the experiments. Mice were purchased from Charles River Wiga (Sulzfeld, Germany) and kept according to the guidelines of the German Cancer Research Center. HEK293T (56) and HepG2 cell lines were maintained in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C in 5% CO2. HEK293T cells were transfected at a confluence of 70%. Primary murine hepatocytes were isolated by liver perfusion as previously described (73). About 4 × 106 primary murine hepatocytes were added to a collagen-coated 10-cm dish (collagen I-coated plates; Nunc, Langenselbold, Germany); after 4 h, adhesion medium (Williams E medium [Sigma-Aldrich, Munich, Germany], 10% FCS, 1% penicillin-streptomycin, 2 mM glutamine, insulin [0.01 mg/ml], 100 nM dexamethasone) was exchanged with the culture medium (Williams E medium [Sigma-Aldrich], 10% FCS, 1% penicillin-streptomycin, 2 mM glutamine). After 24 h, the primary hepatocytes were used for in vitro selection.
Plasmids and site-directed mutagenesis.
Vector plasmid pTRUF2CMV-Luc is a recombinant AAV2 plasmid expressing a luciferase reporter gene under the control of a cytomegalovirus (CMV) promoter and is flanked by inverted terminal repeats (90). Plasmid pDGΔVP expresses all essential Ad helper proteins and the AAV Rep proteins but not the VP proteins (13). The p5E18-VD2/8 helper construct (16) supports the synthesis of AAV2 Rep proteins and AAV8 VP proteins, and pBSΔTR18 provides AAV2 Rep and VP proteins (78). Both plasmids served as templates for site-directed mutagenesis reactions. Mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene, Amsterdam, Netherlands) according to the manufacturer's instructions. For domain swap exchanges between p5E18-VD2/8 and pBSΔTR18, three sets of mutations had to be produced. For the Eco47III insertion into pBSΔTR18, the primers I-f-AAG (5′-GGAAAACAGCAAGCGCTGGAATCCCG-3′) and I-r-CTT (5′-CGGGATTCCAGCGCTTGCTGTTTTCC-3′) were designed. For the MluI insertion, the primers III-f-ACGC (5′-GCCAGCAACGCGTATCAAAGACATCTGC-3′) and III-r-GCGT (5′-GCAGATGTCTTTGATACGCGTTGCTGGC-3′) were used. Finally, for the removal of one HindIII site, the primers II-f-CGA (5′-CGATATCGAGCTTATCGATACCG-3′) and II-r-TCG (5′-CGGTATCGAGCTTATCGATACCG-3′) were used. Subsequently, five domain swaps between AAV8 and AAV2 could be performed.
Single amino acid exchanges between AAV2 and AAV8 were generated using the primer combinations listed in Tables 1 and 2. The complete fragments with domain or single residue swaps were sequenced to exclude additional mutations.
TABLE 1.
Single amino acid exchanges from AAV2 into the cap gene of AAV8 (p5E18-VD2/8)
| Region | Primer | Sequence (5′–3′) |
|---|---|---|
| 1 | QQR-for/rev | GGCACGGCAAATCAGCAGCGTCTGGGCTTCAGCC/GGCTGAAGCCCAGACGCTGCTGATTTGCCGTGCC |
| 2 | DMR-for/rev | GGTGGGCCTAATGATATGAGAAATCAGGCAAAGAAC/GTTCTTTGCCTGATTTCTCATATCATTAGGCCCACC |
| 3 | ERT-for/rev | GGCAAACAAAATGCTGAGAGAACCAATGCGGATTACAGC/GCTGTAATCCGCATTGGTTCTCTCAGCATTTTGTTTGCC |
| 4 | I-for/rev | CAGAGACAATGCGGATATCAGCGATGTCATGCTC/GAGCATGACATCGCTGATATCCGCATTGTCTCTG |
| 5 | SVAT-for/rev | GAGGAATACGGTTCCGTGGCAACTAACTTGCAGCAGC/GCTGCTGCAAGTTAGTTGCCACGGAACCGTATTCCTC |
| 6 | GNRQ-for/rev | GATAACTTGCAGCAGGGAAACCGGCAACCTCAAATTGG/CCAATTTGAGGTTGCCGGTTTCCCTGCTGCAAGTTATC |
| 7 | ATGD-for/rev | CACGGCTCCTGCAACTGGAGATGTCAACAGCCAGGGG/CCCCTGGCTGTTGACATCTCCAGTTGCAGGAGCCGTG |
TABLE 2.
Single amino acid exchanges from AAV8 into the cap gene of AAV2 (pBSΔTR18)
| Region | Primer | Sequence (5′–3′) |
|---|---|---|
| 1 | TQTL-for/rev | CCAAGTGGAACCACCAATACGCAAACTCTTGGGTTTTCTCAGGC/GCCTGAGAAAACCCAAGAGTCTGCGTATTGGTGGTTCCACTTGG |
| 2 | NTMAN-for/rev | CTCAGGCCGGAGCGAATACAATGGCCAATCAGTCTAGGAACTG/CCAGTTCCTAGACTGATTGGCCATTGTATTCGCTCCGGCCTGAG |
| 4 | DYSD-for/rev | CTCAGAGAAAACAAATGTGGACTACAGCGATGTCATGATTACAG/CTGTAATGATGATCGCTGTAGTCCACATTTGTTTTCTCTGAGC |
| 5 | IVAD-for/rev | GGAGCAGTATGGTATTGTAGCTGACAACCTCCAGAGAGG/CCTCTCTGGAGGTTGTCAGCTACAATACCATACTGCTCC |
| 6 | QQNTA-for/rev | CCAACCTCCAGCAACAAAACACAGCGGCAGCTACCGC/GCGGTAGCTGCCGCTGTGTTTTGTTGCTGGAGGTTGG |
| 7 | QIGTV-for/rev | GGCAACAGACAAGCACAGATCGGAACTGTCAACACACAAGGC/GCCTTTGTGTTACAGTTCCGATCTGTGCTTGTCTGTTGCC |
Plasmids and peptide insertions.
For the insertion of peptides into the AAV8 VP proteins, a SfiI binding site was generated (42) by synthesis of a 743-bp fragment (GeneArt, Regensburg, Germany). It was cloned as a XcmI1277-EcoRV2020 fragment into plasmid p5E18-VD2/8. Four peptides were chosen for insertion into the SfiI binding site: PSVSPRP and VNSTRLP (85), ASSLNIA (86), and GQHPRPG (Y. Ying, unpublished data). The oligonucleotides encoding the respective peptides were designed and synthesized by MWG Biotech (Ebersberg, Germany) with a SfiI linker for vector plasmid insertion. To convert oligonucleotides into double-stranded DNA (dsDNA), 2 μg of each forward and reverse oligonucleotide was mixed with 40 μl of annealing buffer (10 mM Tris-HCl [pH 8.5],150 mM NaCl) and annealed under one cycle of 5 min at 95°C, 20 min at 76°C, and 20 min at 37°C in a PCR thermal cycler. The backbone plasmid, p5E18-VD2/8+SfiI, was digested with SfiI to insert the annealed dsDNA in the presence of 1 U of T4 DNA ligase (Roche, Mannheim, Germany) and 10× ligation buffer (Roche). A final volume of 20 to 30 μl was kept overnight at 12°C. Plasmids were sequenced (GATC Biotech, Constance, Germany) to verify the insertion of the correct oligonucleotides.
Generation of an AAV8 random peptide display library.
To produce an rAAV8 library backbone plasmid, the plasmid pE18-VD2/8+SfiI was restricted by EcoRV and HindIII and cloned into pMT187-XX2 (85) to obtain pMT182-XX2/8. After ligation had been checked in a sequence analysis, pMT182-XX2/8 was restricted with XbaI to ligate rep and cap gene (plus stuffer) into pSSV9 (received from J. Samulski, Chapel Hill, NC). The pLib588-92+ITRs plasmid was used for production of the AAV8 peptide display library, which was carried out as described previously (42, 77).
In vitro selection by the AAV8 peptide display library.
For in vitro selection, 2 × 106 primary murine hepatocytes, were seeded out in culture dishes 1 day prior to infection. At 70% confluence, the cells were infected with the random AAV8 display peptide library at a multiplicity of infection (MOI) of 10,000 viral genomes (vg) per cell for the first screening round. After 4 to 6 h of incubation at 37°C, the cells were washed with phosphate-buffered saline (PBS; 18.4 mM Na2HPO4, 10.9 mM KH2PO4, 125 mM NaCl), superinfected with Ad5 at 50 PFU/cell, and incubated for 3 to 5 days at 37°C until 50% of the cells showed a cytopathic effect. The cells were then harvested, pelleted by centrifugation at 1,000 × g for 10 min, and resuspended in 1 ml of PBS. A 200-μl aliquot of cell suspension was removed to extract viral DNA for further analysis. The remaining suspension was pelleted by additional centrifugation and resuspended in 1 ml of lysis buffer. Replicated rAAV particles were harvested from cell lysates after three freeze-thaw cycles, and the numbers of viral genomes were determined. For each subsequent selection round, preselected viruses recovered from the preceding screening round were added to target cells at reduced MOIs.
PCR amplification and sequencing of selected clones.
Isolated viral DNA from harvested cells of selection rounds 3 and 4 served as templates for PCR amplification. AAV genomic DNA comprising the oligonucleotide library insert region of the cap gene was amplified by PCR using the TopoLib8-f primer (5′-CTGGCATCGCTATGGCAACACAC-3′) and the TopoLib8-r primer (5′-GGATCTGAGGCGGAGGATGTTTC-3′). The PCR product was purified and subcloned into the plasmid pCR2.1 using a TOPO-TA cloning kit (Invitrogen, Karlsruhe, Germany). After 24 h, white colonies were selected, and 32 clones were sent for sequencing for every third and fourth selection round (GATC Biotech) using the M13 primer 5′-AGGAAACAGCTATGACCATG-3′. According to the isolated peptides, primers were designed and inserted into the AAV8 backbone plasmid p5E18-VD2/8+SfiI as described above.
Cell transfection and vector production.
A triple transfection (1:1:1) was carried out using calcium phosphate precipitation. For serotypes, as well as the vector mutant, HEK293T cells were seeded out and transfected 24 h later. Per plate, 50 μg of DNA was resuspended in 1.125 ml of sterile Braun H2O, mixed with 125 μl of CaCl2 (Sigma, St. Louis, MO), and added slowly to 1.25 ml of 2× Hanks balanced salt solution (280 mM NaCl, 50 mM HEPES, 1.5 mM Na2HPO4, 10 mM KCl, 12 mM glucose [pH 7.05]), with constant shaking. After 1 min of incubation, the mix was added to 7.5 ml of medium and added to the cells. After 48 h at 37°C, 5% CO2, the cells were harvested, washed with PBS, centrifuged at 200 × g for 15 min, and stored at −80°C until additional purification.
Purification and titration of rAAV vector stocks.
A 15-ml portion of lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.5]) was added to the cell pellets, followed by five rounds of freeze-thawing, 30 min of Benzonase (Sigma) treatment (50 U/ml) at 37°C, and centrifugation for 15 min at 3,000 × g. By an iodixanol step gradient, rAAV particles were purified from the crude cell lysate (89) by a centrifugation step at 50,000 × g at 4°C for 2 h. Viral particles were aspirated from the 40% iodixanol phase and deep-frozen at 20°C. Quantization of rAAV genomic particles was assayed by quantitative real-time PCR adapted from previously described methods (71). After alkaline lysis of the rAAV particles, genomes were subjected to the TaqMan Universal Master mix, including the primers for-5′-TGCCCAGTACATGACCTTATGG -3′ and rev-5′-GAAATCCCCGTGAGTCAAACC-3′ and the probe 6-FAM-AGTCATCGCTATTACCATGG-MGB, and analyzed under standard quantitative real-time PCR conditions (Applied Biosystems, Inc., Foster City, CA).
Heparin binding analysis.
The heparin binding of rAAV8 and rAAV2 vectors was analyzed by chromatography of 1011 genome-containing vector particles on heparin agarose (Sigma-Aldrich). After loading of the vector, the columns were washed twice with PBS, and heparin-bound vector particles were eluted with PBS containing 1 M sodium chloride. Input, flowthrough, wash, and elution fractions were collected for the purification of viral DNA using a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Isolated viral DNA from each fraction was immobilized onto a GeneScreen Plus nylon membrane (Perkin-Elmer, Rodgau, Germany) and analyzed by DNA dot blotting with a CMV-specific radioactive 32P-labeled probe.
AAV in vivo application and analysis.
NMRI mice were injected intravenously (i.v.) with 1011 vg-containing particles. Groups of at least three mice were injected per vector. One month after injection, animals were intraperitoneally injected with d-luciferin (Synchem, Felsberg, Germany) and imaged with an IVIS imager system (IVIS 100; Xenogen, Illinois) for 5 min, 10 min after injection. Mice were sacrificed, and organs were extracted.
Luciferase expression analysis.
For the in vivo expression analysis, excised organs of an injected animal were shock frozen in liquid nitrogen and stored at −80°C until further analysis. Before expression analysis could be performed, 1 μl of reporter lysis buffer (Promega GmbH, Mannheim, Germany) was added per mg of organ, and the organ samples were homogenized and left for 10 min at room temperature. Samples were centrifuged at 10,000 × g for 10 min, and the supernatant was transferred into a new tube. Expression analysis with a luminometer was performed using 50 μl of sample plus 100 μl of luciferin, and emitted photons were measured for 20 s to determine the relative light units (RLU) per organ. Protein amounts were determined by the standard protocol of the NanoOrange technology (Invitrogen).
Genomic DNA extraction and analysis.
The peqGOLD tissue DNA minikit (Peqlab Biotechnology, Erlangen, Germany) was a rapid method to isolate up to 30 μg of genomic DNA from mouse tissue. Tissue samples were cut into small pieces (30 mg in all), and genomic DNA was extracted according to the manufacturer's protocols. Quantitative reverse transcription-PCR was performed to determine viral vector genomes and complete genome amounts. Probes against CMV and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; TaqMan, Applied Biosystems), respectively, were used.
Analysis of viral protein expression.
Western blot analysis was performed with equal amounts of genome containing particles according to standard methods (Harlow and Lane 1988). Monoclonal antibodies were applied as previously described (79).
Molecular modeling.
The three-dimensional illustration of the AAV8 capsid trimer was visualized by Visual Molecular Dynamics (VMD 1.8.7.) (24) based on the coordinates of the AAV8 structure (Protein Data Bank [PDB] accession no. 2QAO).
Statistical data analysis.
Statistical analysis was obtained by using an unpaired Student t test. The presented values are means of at least three independent measurements plus the corresponding standard deviations. P values of <0.05 were considered to be significant.
Analysis of the influence of antibody ADK8 on rAAV8 gene transduction.
The influence of ADK8 on rAAV8-mediated cell transduction was analyzed by incubation of the vector with HepG2 cells for 4 h, followed by analysis of the luciferase transgene expression. On the day before the experiment, 5 × 103 cells were seeded per well of a 96-well plate, followed by incubation at 37°C under appropriate conditions. Per well, 1.25 × 108 rAAV-8 vector particles (representing an MOI of 25,000 vector genomes per cell) were preincubated at 37°C with or without the addition of 10 ng of ADK8 antibody for 1 h at 37°C, before the vector solution was added to the cells, followed by incubation for 4 h. Thereafter, vector solution was replaced by medium. At 24 h postinfection, the cells were harvested, and the luciferase transgene expression was analyzed using a luciferase assay system (Promega).
The influence of ADK8 on the cell binding of rAAV8 was analyzed by incubation of the vector with HepG2 cells at 4°C, followed by quantification by real-time PCR. One day before the experiment, 5 × 105 cells were seeded per well of a six-well plate, followed by incubation at 37°C under appropriate conditions. Per well, 1.25 × 1010 rAAV8 vector particles (representing an MOI of 25,000 vector genomes per cell) were preincubated at 37°C for 1 h with or without the addition of 100 ng of ADK8 antibody. For the binding experiment, the cells were cooled down to 4°C for 20 min to stop all endocytic processes prior to the addition of the precooled rAAV8 vector solution in a total volume of 500 μl of culture medium containing no FCS. After incubation for 1 h at 4°C under minimal agitation (∼10 rpm), the rAAV vectors present either in the supernatant or attached to the cells were recovered, and the viral DNA was purified using a Blood & Tissue kit (Qiagen). Purified DNA was then quantified by real-time PCR using a probe directed against the CMV promoter.
RESULTS
VP protein domain exchange between AAV2 and AAV8 identifies transduction determinant.
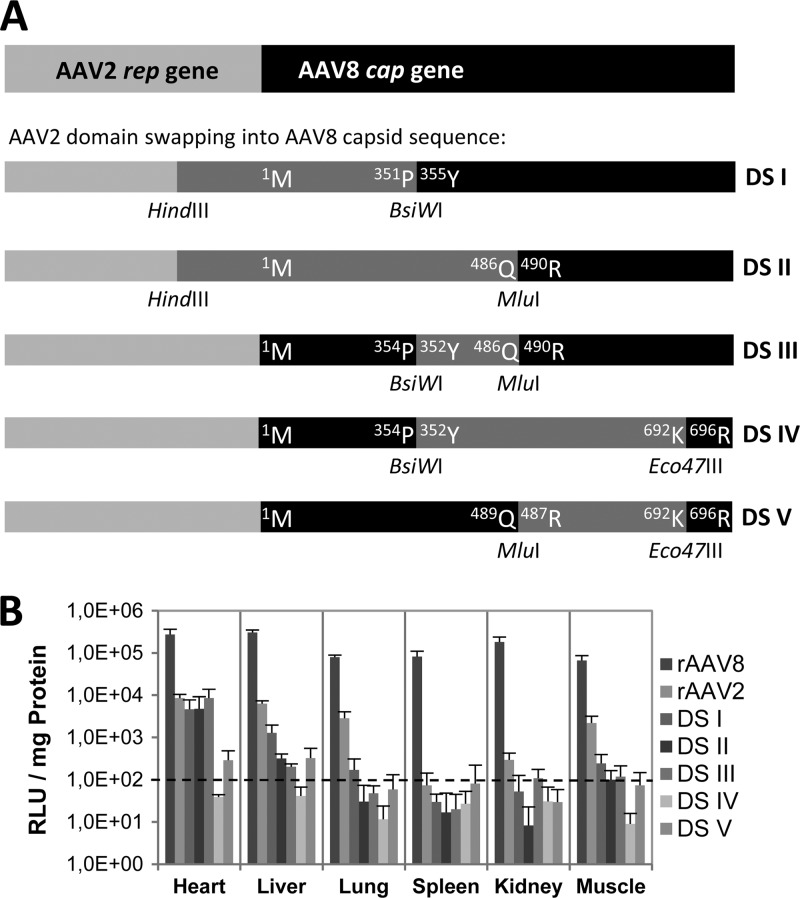
Even though AAV2 and AAV8 share an 82% capsid amino acid sequence identity, the VP protein sequences responsible for their unequal in vivo gene transduction efficiency have not been identified. A first comparative study between AAV2 and AAV8 demonstrated that within the VP1 encoding sequence, amino acids within the GH loop (between the βG and βH strands of the core β-barrel) play a critical role for mouse liver gene transduction efficiency (64). In a comparable approach five different VP domains were transferred from AAV2 into AAV8 to generate chimeric AAV packaging vectors (Fig. 1A). All domain swap (DS) mutant vectors showed strongly reduced transduction efficiency compared to rAAV8, although the packaging efficiency and vector yields were equivalent for these vectors (data not shown). The in vivo performance of several domain swap mutant vectors was even lower than that of wild-type (wt) rAAV2, especially in liver, lung, and muscle tissues, indicating that AAV2 and AAV8 VP protein domains influence each other. The mutant vector, containing a complete swap of the GH loop of AAV2 inserted in AAV8, demonstrated by far the lowest transduction efficiencies (DS IV in Fig. 1B), confirming the observations of Shen et al. (64). These observations provided a basis for a more detailed sequence analysis of this region.
Fig 1.
AAV2 capsid protein domain insertions into the AAV8 capsid. (A) Schematic overview of the cloning strategy for domain swaps cloned into packaging helper plasmid P5E18-VD2/8 (16). The rep2 gene and cap8 gene are indicated. Domain swaps I to V were produced by insertion of parts of the AAV2 genome (dark gray boxes) into AAV8. Restriction sites for MluI and Eco47III were introduced in the AAV2 genome by site-directed mutagenesis. Amino acids of the VP proteins of the AAV2 and AAV8 at the boundaries are indicated within the boxes. (B) Reporter gene expression analysis of domain swap capsid mutants (DS I to V) in comparison to rAAV2 and rAAV8 in different mouse tissues. Seven groups of NMRI mice (n = 5) were i.v. injected with 1011 vg-containing particles and dissected 1 month after injection. Luciferase transgene expression per mg of protein was determined for heart, liver, lung, spleen, kidney, and muscle tissues. Bars indicate the standard deviations.
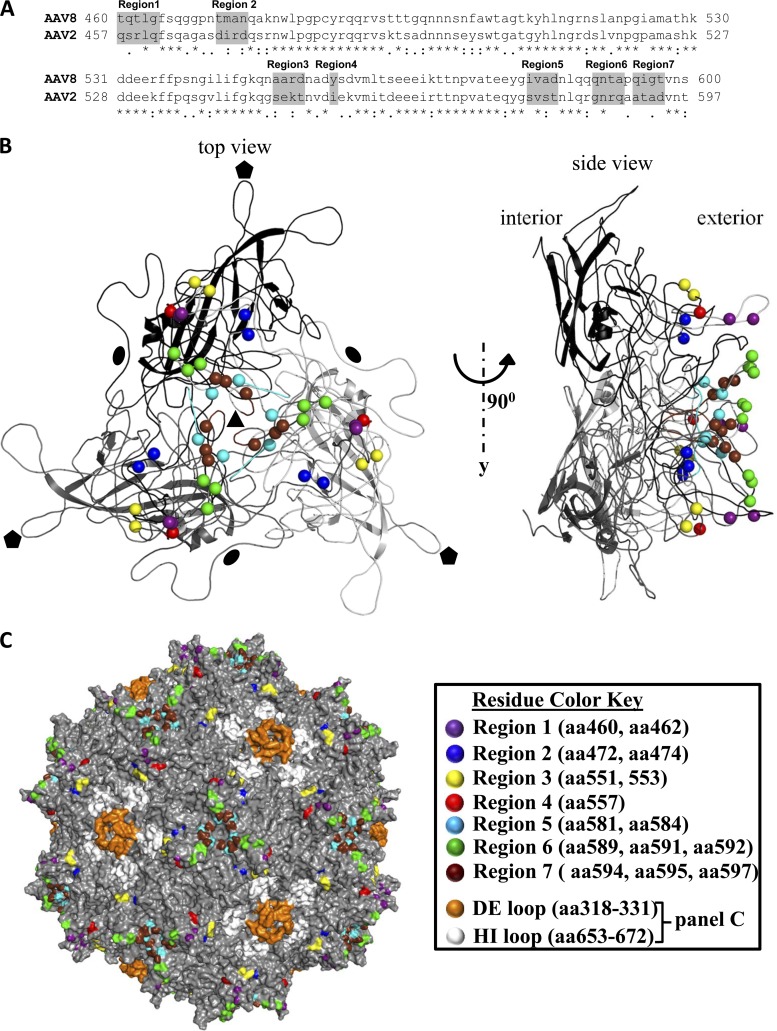
Comparison of amino acid sequences of the GH loops of AAV2 and AAV8 shows seven regions containing nonconserved amino acids (Fig. 2A). In a trimer of AAV8 VPs (Fig. 2B), region 1 is located close to the top of the protrusions near a previously defined AAV2/AAV8 variable region IV (VRIV) (46). Region 2 is located in a surface loop at the base of the protrusions surrounding the icosahedral 3-fold axes and is adjacent to VRI in a symmetry related VP monomer. Regions 3 and 4 are located in VRVII on the outer surface of the protrusions facing the 2-fold axes. Regions 5 to 7 are located on the surface of the protrusions facing the 3-fold axis with region 6 on the wall and regions 5 and 7 at the base, with all three located in AAV VRVIII. Based upon this information, vectors were generated in which the nonconserved amino acids in each region were converted from AAV8 to AAV2 (in the AAV8 capsid) and vice versa. Vector stocks were produced and analyzed for luciferase reporter gene expression in different tissues at 4 weeks after tail vein injection of mice. Vector yields of wt rAAV2, wt rAAV8, and mutants were equivalent.
Fig 2.
Sequence and structural analysis of AAV2 and AAV8 capsid protein sequences comprising loop IV. (A) Sequence alignment indicating identical residues (*), conserved substitutions (:), and semiconserved substitutions (.). Gray boxes illustrate the seven regions (regions 1 to 7) containing nonconserved amino acids that were mutated. (B) Top-down and side views (90° angle rotated) of an AAV8 capsid VP trimer (black, dark gray, and light gray, generated from the coordinates of its crystal structure (PDB accession no. 2QA0 [46]), shown as a ribbon diagram with the positions of nonconserved amino acids in regions 1 to 7 indicated by spheres colored according to the key on the right-hand side of panel C. The approximate positions of the icosahedral 2-, 3-, and 5-fold axes are depicted by the filled ovals, triangles, and pentagons, respectively, in the image on the left. (C) The capsid surface of AAV2 shown in gray and viewed down an icosahedral 3-fold axis with the positions of the seven regions of nonconserved aa colored as in panel B. The HI loop residues (between the β-H and β-I strands of the core eight-stranded β-barrel) which lie on the floor of the depression surrounding the icosahedral 5-fold axes are colored white; the 5-fold symmetry related DE loops (between the β-D and β-E strands), which form the channel at the icosahedral 5-fold axes, are colored brown.
Single amino acid residue swaps of nonconserved amino acids within the GH loop of AAV2 and AAV8 pinpoints tissue specific transduction regions.
Figure 3A and C show the capsid amino acid sequences of AAV8 (aa 460 to 597) and AAV2 (aa 458 to 595) with the amino acid substitutions in regions 1 to 7 indicated by capital letters and arrowheads. Transgene expression was measured for six different tissue types of mutants (8→2 and 2→8) in comparison to nonmutated rAAV8 and rAAV2 (Fig. 3B and D). The results demonstrated that the mutations have an impact on transduction efficiency and tissue specificity. Most surprisingly, the chimera 8→2 GnRQ (region 6), which partially reconstituted HS binding on the AAV8 capsid (Fig. 3E), resulted in a strong increase (>10-fold) in the transduction of heart tissue and a slight increase in the transduction of liver tissue compared to rAAV8 (Fig. 3B). The transduction of lung, spleen, and muscle tissue was equivalent to wt rAAV8. Similarly, the very closely located mutant 8→2 SvaT (region 5) also transduced heart and liver tissue very robustly and better (>10-fold) than wt AAV8 in heart tissue; however, its transduction of the other tissues was reduced compared to wt AAV8, indicating that modification of this site influenced tissue tropism in a different manner. Most of the other mutants (in regions 1, 3, 4, and 7) showed intermediary transduction efficiencies between AAV8 and AAV2 without selective effects on tissue tropism and transduction efficiency, suggesting a nonspecific reduction of transduction performance in total. The region 2 exchange (8→2 DmR; region 2) resulted in a vector that transduced all tested tissues very similarly to AAV2. This result shows that conversion of these two amino acids of AAV8 to those of AAV2 completely reversed the higher transduction efficiency of AAV8 without affecting the tissue tropism.
Fig 3.
Transduction analysis of AAV vectors with amino acid exchanges from AAV8 to AAV2 and from AAV2 to AAV8. (A) Part of the AAV8 capsid protein amino acid sequence with the seven regions containing nonconserved amino acids between AAV2 and AAV8 demarked by large letters. Amino acids converted from AAV8 into AAV2 are marked by arrowheads. (B) In vivo reporter gene expression analysis of AAV8 capsid mutants in comparison to rAAV2 and rAAV8. Nine groups of mice (n = 3) were i.v. injected with 1011 vg-containing particles. Mice experiments were performed twice with two independent vector productions. Animals were dissected 1 month after injection. Luciferase transgene expression per mg of protein was determined in heart, liver, lung, spleen, kidney, and muscle tissue. **, P < 0.01 (significantly increased or decreased transgene expression compared to rAAV8). (C) Excerpt of the AAV2 capsid protein amino acid sequence with regions containing nonconserved amino acids between AAV2 and AAV8 demarked by large letters. Amino acids converted from AAV2 into AAV8 are marked by arrowheads. (D) In vivo reporter gene expression analysis of AAV2 capsid mutants in comparison to rAAV2 and rAAV8. Eight groups of mice (n = 4) were i.v. injected with 1011 vg-containing particles. Mouse experiments were performed twice, with two independent vector productions. Animals were dissected 1 month after injection. Luciferase transgene expression per mg of protein was determined for heart, liver, lung, spleen, kidney, and muscle tissues. *, P < 0.05; **, P < 0.01 (significantly increased or decreased transgene expression compared to rAAV2). ns, not significant. (E) The heparin binding of rAAV2, rAAV8, and the domain swap mutants rAAV8/region 2 and rAAV8/region 6 was analyzed by chromatography on heparin agarose. Isolated viral DNA from input, flowthrough (FT), wash, and elution fractions was analyzed by DNA dot blotting with a CMV-specific radioactive probe.
Reverse mutants of regions 1, 2 and 4 (2→ 8) (mutant region 3 showed no difference to wt AAV2 and is not discussed further) showed a heterogeneous picture, including conservation of transduction efficiency in heart (regions 1 and 2) and lung (region 1) tissues, but a reduction of gene transgene expression in the other tissues compared to wt rAAV2 (Fig. 3C and D). Reversion of region 2, which decreased transgene expression of rAAV8 to the level of wt rAAV2 vectors in all tissues when the AAV2 amino acids were introduced (Fig. 3B), did not improve transgene expression of rAAV2. This indicates that the phenotype of this sequence in AAV8 requires the adjacent residues in the AAV8 capsid to lead to improved gene transduction. Mutant 2→8 QIGTv (region 7) showed also decreased gene expression in most tissue types and was not able to improve AAV2 performance. Unexpectedly, the amino acids exchanged in region 5 (2→8 IvAD) increased rAAV2 transduction efficiency in heart (>10-fold) and skeletal muscle (∼3-fold) tissues and kept it at the level of wt rAAV2 in the lungs, kidneys, and spleen. This means that transduction rates obtained with amino acids swaps at this position in both directions, AAV8 to AAV2 (8→2 SvaT vector) or AAV2 to AAV8 (2→8 IvAD), improved gene transduction. Conversion of region 6 (2→8 QQnTA) slightly increased transduction of heart tissue (∼4-fold), was neutral with respect to the transduction of spleen and skeletal muscle tissues, but significantly decreased the transduction of liver and lung tissues (>10-fold). This observation is in line with a recent report by Asokan et al. (3) for a nearly identical AAV2 mutant that showed improved heart tropism associated with reduced liver tropism. This mutation destroyed the HSPG binding motif in AAV2, which has been previously shown to decrease liver but not heart tissue transduction (28). Taken together, analysis of exchanges of nonconserved amino acids in the GH loop of AAV8 and AAV2 point to a selective and, in some instances, a strong influence of amino acids between positions 581 and 592 (regions 5 and 6) on gene transduction in heart and liver tissues. Unexpectedly, exchange in both directions induced in some tissues an improvement of transduction. All other mutations led to general reduction of gene transduction without selectivity and were not able to restore the overall high transduction efficiency of AAV8 in AAV2. We next sought to determine whether peptide insertions in region 6 of AAV8 would allow further manipulation of efficiency and specificity of transgene expression in different tissues.
Validation of a peptide insertion site within the AAV8 capsid.
An insertion site for displaying new peptides on the AAV8 capsid was designed at position 590 in the 3-fold protrusion with a SfiI restriction site inserted into the packaging plasmid p5E18-VD2/8 (16) (Fig. 4A). Two new amino acids (G and A) flanking the peptide insertions were introduced. In addition, several amino acids between positions 588 and 593 were mutated. Without peptide insertion, the modified helper plasmid could not generate functional AAV8 capsids, while with peptide insertions the vector yields were similar to wt rAAV8 vectors (data not shown). Four peptide sequences from previously derived AAV2 library selections (85, 86) were inserted into the AAV8 based capsids (AAV8-VNSTRLP, AAV8-ASSLNIA, AAV8-PSVSPRP, and AAV8-GQHPRPG) (Fig. 4B). Reporter gene expression in different tissues, 1 month after i.v.-injected particles (1011 vg/animal) showed that the insertion of two peptide sequences, ASSLNIA and GQHPRPG, resulted in a change in the transduction profile compared to rAAV8 (Fig. 4C). The transduction efficiency was slightly increased in heart tissue (4-fold, respectively 2-fold), while it was largely reduced in other tissues (10- to 100-fold), except for GQHPRPG, which showed nearly unchanged transgene expression in lung tissue. The other two peptides had only a negative effect on gene transduction. The results show that insertion of peptide sequences at this site can influence rAAV8 tissue targeting. Consequently, a random peptide display library was inserted into the AAV8 capsid at this position.
Fig 4.
AAV8 capsid retargeting by peptide insertions. (A) Concept of an AAV8 insertion site to insert AAV2 library selected peptides into the AAV8 capsid sequence. Stuffer sequences with SfiI restriction sites were generated and inserted into AAV8. In addition, aa 588 and 590 were exchanged from AAV8 to AAV2 because of previous mutant evaluations. (B) Helper plasmid p5E18-VD2/8 into which four different oligonucleotides (coding for 7 aa in length) were inserted into the SfiI restriction site to produce peptide-displaying AAV8 capsid mutants. (C) In vivo reporter gene expression analysis of four peptide-displaying AAV8 capsid mutants in comparison to rAAV2 and rAAV8 was performed. Six groups of mice (n = 3) were i.v. injected with 1011 vg-containing particles. Mouse experiments were performed twice, with two independent vector productions. Animals were dissected 1 month after injection. Luciferase transgene expression per mg of protein was determined for heart, liver, lung, spleen, kidney, and muscle tissues. *, P < 0.05; **, P < 0.01 (significantly increased or decreased transgene expression compared to rAAV8).
Generation, selection, and validation of an AAV8 random peptide display library.
A preparation of an AAV8 random peptide display library inserted at position 590 consisting of 7 × 1012 vg was generated. In vitro selections were performed on primary murine hepatocytes. After four selection rounds, several peptide sequences were enriched (Fig. 5A). The most highly enriched peptide sequence, SEGLKNL, was inserted into the AAV8 packaging helper plasmid after position 590, and chimeric vectors were produced by a triple transfection of HEK293T cells. The chimeras could efficiently be packaged and purified by iodixanol step gradients for gene transduction analysis.
Fig 5.
In vivo reporter gene expression analysis of AAV8 capsid mutants containing peptides enriched by selection of an AAV8 random peptide display library on primary hepatocytes. (A) Three peptide sequences enriched by in vitro selection of an AAV8 random peptide display library on primary hepatocytes. (B) IVIS images of mice analyzed 1 month after i.v. injection with rAAV8 or with the AAV8 capsid mutant displaying the most enriched peptide SEGLKNL (1011 vg). Images were taken 15 min after intraperitoneal substrate injection. (C) In vivo reporter gene expression analysis of the SEGLKNL peptide displaying AAV8 capsid mutant in comparison to rAAV8. Two groups of mice (n = 3) were i.v. injected with 1011 vg-containing particles. Mouse experiments were performed twice, with two independent vector productions. Animals were dissected 1 month after injection. Luciferase transgene expression per mg of protein was determined for heart, liver, lung, spleen, kidney, and muscle tissues.
In vivo imaging showed that the insertion of peptide sequence SEGLKNL triggered detargeting away from nonhepatic tissue (Fig. 5B). Quantification of reporter gene expression for the vector displaying peptide SEGLKNL in several tissues revealed only a 7-fold-lower transgene expression in the liver compared to AAV8 but a dramatic reduction in most other organs (>100-fold in the lungs, spleen, and kidneys), close to the detection level. In heart and skeletal muscle tissue, an ∼50-fold-reduced gene expression could be detected. In conclusion, the in vitro selection of an AAV8 peptide display library on primary hepatocytes gave rise to a peptide sequence SEGLKNL, which provided significant liver tropism in the mouse with only a weak reduction of liver transduction efficiency compared to rAAV8. This implies detargeting from native tropism by peptide insertions at the 3-fold protrusions of the AAV8 capsid. Thus, the question arises as to whether AAV8 VP protein sequences in this structurally prominent domain are involved in cellular receptor binding.
Binding of a monoclonal antibody to the 3-fold protrusions of AAV8 neutralizes infection at the postbinding level.
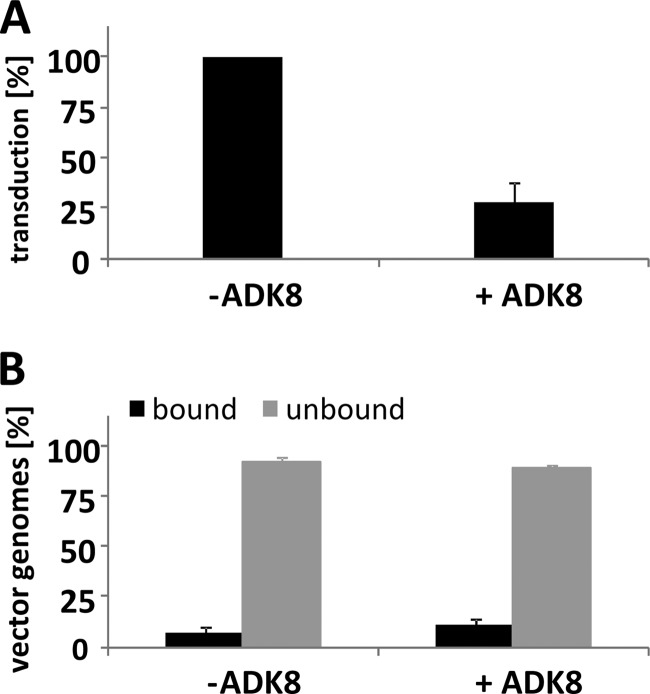
We have recently isolated and characterized an AAV8-specific monoclonal antibody, ADK8 (67), which efficiently neutralizes AAV8 gene transduction in vitro (Fig. 6A) and also in vivo in mice (20a). ADK8 binds to the sequence 588QQNTA592 located at the 3-fold protrusions of the AAV8 capsid as shown by cryo-electron microscopy and image reconstruction of the AAV8-ADK8 complex structure and analysis of capsid mutants (20a). The sequence represents region 6 and comprises the position where selected peptides and the peptide display library were inserted (see Fig. 4A and B). Mutation of this sequence or insertion of peptides within this sequence had a strong impact on in vivo gene transfer efficiency and tissue tropism as shown above and prevented binding of ADK8 to AAV8 capsids (20a). To test whether the binding of ADK8 prevents cell attachment, we analyzed rAAV8 cell binding by quantitative real-time PCR in the presence or absence of ADK8. As shown in Fig. 6B, preincubation of AAV8 vectors with ADK8 did not prevent binding of rAAV8 to HepG2 cells. This result was confirmed by an indirect immune fluorescence assay using HeLa cells (20a). Virus uptake, measured by shifting the temperature after binding at 4 to 37°C for 2 h, followed by trypsin treatment, indicated a weak reduction in the amount of internalized vector in the presence of the ADK8 antibody and a strong impairment of intracellular trafficking toward the nucleus (20a). This means that the binding of neutralizing antibody ADK8 to the sequence 588QQNTA592 (region 6) located at the inner shoulder of the 3-fold protrusions of AAV8 interfered with a postattachment process in AAV8 gene transduction.
Fig 6.
Neutralization of AAV8 gene transduction with monoclonal antibody ADK8. (A) Reporter gene transduction of HepG2 cells infected with an MOI of 25 × 103 vg/cell of a luciferase expressing AAV8 vector and inhibition of transduction by application of 10 ng of ADK8 antibody per 5 × 103 cells. (B) Detection of bound and unbound vector genomes after incubation of HepG2 cells with an MOI of 25 × 103 vg/cell in the absence or presence of 100 ng of ADK8 antibody per 5 × 103 cells. Shown are the mean values of three independent experiments. Bars indicate standard deviations.
Ratios of transgene expression to vector genome uptake level correlate with the transduction efficiency.
It has been reported that superior transduction of liver tissue by rAAV8 vectors compared to rAAV2 vectors is mainly due to more efficient uncoating (69). The contribution of postentry processing in gene transduction should be reflected by the ratio of transgene expression to the amount of accumulated genomes in a given tissue. To calculate such ratios, we determined the amount of vector genomes in heart tissue after tail vein injection of wt and mutant vectors and related the information obtained to the respective reporter gene expression. Heart tissue transduction with rAAV8 gave a 10-fold-higher expression-to-genome ratio than transduction with rAAV2 (Fig. 7), confirming the observation made for liver transduction that rAAV8 is more efficiently processed after cell uptake (69). Introduction of AAV2 amino acids into AAV8 at regions 1, 3, 4, and 7 (8→2 QqR; 8→2 ErT; 8→2 I; 8→2 ATgD) showed a similar expression/genome ratio as did rAAV2, indicating that these mutants are impaired in intracellular processing compared to wt rAAV8 (Fig. 7). Exchange of AAV8 region 2 (8→2 DmR) by AAV2 amino acids, which are located at the base of the 3-fold protrusion facing the 2-fold axes at a VP-VP interface, resulted in a very low ratio of transgene expression to genome uptake, suggesting that this region is most likely critical for intracellular conformational changes of the capsid leading to genome release and gene expression (Fig. 7). The two mutants that improved heart transduction of rAAV8 vectors after incorporation of AAV2 sequences of regions 5 or 6 (8→2 SvaT; 8→2 GnRQ) showed a clearly increased expression/genome ratio (Fig. 7). This suggests that their improved transduction efficiency is, at least partially, due to improved postentry processing.
Fig 7.
Correlation of viral vector genomes and transgene expression in heart tissue. By quantitative real-time PCR, the numbers of viral genomes per μg of total DNA of capsid mutants with amino acid substitutions from AAV8 to AAV2, as well as of AAV2 and AAV8, were determined in heart tissue after i.v. injection. Organs of six animals were analyzed in the experiment. The total amount of DNA was determined by using GAPDH. The ratios between gene expression (RLU/mg of protein) and viral genomes (viral genomes/μg of DNA) for heart tissue were determined.
DISCUSSION
Viral capsids are multifunctional protein assemblies that encapsidate, protect, and deliver the viral genome to the host cell. Relating structural elements of capsids to functions in the infection pathway is important for understanding AAV biology. Recombinant AAV8 and AAV2 show striking differences in the efficiency of transducing mouse tissues after systemic application. Domain swap experiments indicated that amino acid residues within the GH loop, which forms a significant component of the 3-fold protrusions, is mostly responsible for the transduction differences (64) (Fig. 1). This loop contains five structurally variable regions VRIV, VRV, VPII, VRVII, and VRVIII (as defined in reference 20) and seven clusters of nonconserved amino acids between AAV8 and AAV2 (designated regions 1 to 7 here). Single amino acid exchanges in these clusters, which did not affect capsid assembly or genome packaging, resulted in mutants with different gene transduction phenotypes. They are grouped according to their efficiency and selectivity of transduction.
Exchange of regions 1, 2, 3, 4, and 7 did not lead to transduction improvement or enhanced selectivity, nor did any of the mutants convert the low transduction efficiency of AAV2 to the high efficiency of AAV8. Replacement of region 2 in AAV8 by AAV2 amino acids converted the high transduction efficiency of AAV8 to the low efficiency of AAV2, but the reciprocal substitution did not restore the high transduction phenotype of AAV8. Strong inhibition of transduction of some tissues by AAV2, chimeric in regions 2 and 4, was not combined with improved transduction in other tissues. We therefore conclude that this part of the capsid structure does not harbor structural elements which provide the superior transduction properties of AAV8 compared to AAV2 and do not determine tissue tropism in a positive way. Interestingly, with the exception of region 7, these regions are all located on the outer wall of the 3-fold protrusions facing the 2-fold and 5-fold symmetry axes. The negative influence on gene transduction of substituting these regions may indicate that they perturb the interplay of chimeric VP amino acids and capsid regions with each other and/or with cellular proteins.
Surprisingly, introduction of amino acids of AAV2 into regions 5 and 6 of AAV8 improved the already high gene transduction efficiency of AAV8 in the heart and liver (Fig. 3A and B). Mutation of region 6 (QNTA to GNRQ) also maintained the level of transduction by rAAV8 in all other tissues. Mutation of region 5 (IVAD to SVAT), however, enhanced gene transduction only in the heart and reduced the transduction of lung, spleen, kidney, and skeletal muscle, indicating tissue selectivity and a mechanistically different influence on rAAV8 transduction. Both sites, region 5 and region 6, are located at the base and close to the top, respectively, of the protrusions facing the 3-fold axis. Region 6 contains in AAV2 R588 and R585, which have been shown to be involved in primary receptor binding and tissue tropism (28, 30, 31, 51, 52, 83). It is not known whether this region is also involved in cell binding and tissue tropism in AAV8. The laminin receptor-binding domains mapped to aa 492 to 557 and aa 593 to 623 of AAV8 do not overlap with this region (1), although the second stretch of residues is adjacent to the region 6 residues. There is a nearly symmetrical increase in heart transduction when regions 5 and 6 of AAV2 were replaced by the sequences of AAV8. With respect to region 6, this means that not only did conversion of QNTA in AAV8 to GNRQ enhance the transduction of the heart and liver, but also conversion of GNRQ to QNTA in AAV2 enhanced AAV2 transduction of the heart. This result excludes the interpretation that the corresponding sequences in AAV8 and AAV2 permit transduction by direct binding to the same receptor. The chimeric capsids could, however, recognize different receptors on the same cells or on different cell types within the same tissue. The same conclusion must be drawn for region 5, and in both cases the binding of different receptors may, by chance, enhance transduction to similar levels. On the other side, both results can also be interpreted by assuming that region 5 and 6 sequences cooperate with other receptor binding sites in the AAV capsids, which then permit cell entry.
By converting QNTA to GNRQ (region 6), a weak HS binding motif is created in AAV8 (Fig. 3E), which may enhance the retention period of the viral particle on the cell surface, thereby increasing the likelihood for interaction with a secondary receptor required for cell entry. This may explain why the transduction of all tissues tested is high. In the reciprocal conversion of GNRQ to QNTA in AAV2, the HS binding motif is partially destroyed. A previous report showed that disruption of the HS binding motif in AAV2 impaired transduction of most mouse tissues but still allowed good transduction of heart tissue (28), suggesting a second receptor binding site on the AAV2 capsid, which is different from the HS binding domain. Replacement of part of the HS binding domain, region 6, of AAV2 by sequences of different AAV serotypes also identified QNTA from AAV8 as a promising substitution (3). It showed that replacement of region 6 by sequences of other AAV serotypes did not increase AAV2 transduction implying that the sequence QNTA contains information which is either recognized by a receptor or used in another capsid protein interaction process. Influences of these sequences on postentry processing can also not be excluded and are even suggested by the ratio of transgene expression to genomes accumulated in heart tissue. An unexpected aspect of this detailed sequence replacement analysis is the observation that modifications of the very closely located sequences in the capsid (regions 5, 6, and 7) have strikingly different influences on gene transduction in different tissues. In particular, regions 5 and 7 are located at nearly overlapping positions, but chimeras in region 7 did not show any improved gene transduction in contrast to region 5. Thus far, we do not have an explanation for this observation.
Due to the observation that domain swaps in region 5 and 6 resulted in improved gene transduction by rAAV8 and rAAV2, these regions represented potential insertion sites for peptide sequences in AAV8 for tissue targeting, as has been demonstrated for AAV2 (19, 39, 42, 44, 57). Insertion of targeting peptides in region 6 clearly influenced transduction efficiency and tissue tropism of rAAV8 (Fig. 4). Although several peptides inserted in AAV8 capsids generally reduced transduction in all tissues, other peptides clearly induced selective transduction of heart or liver. These different transduction profiles indicate that there is specific sequence information required at this position, which was also concluded from the AAV2 to AAV8 sequence replacement experiments discussed above. The most obvious interpretation is that the peptides provide binding to an uptake receptor, either directly or by enhancing the interaction with a secondary receptor-binding domain located on another part of the capsid.
Antibodies can be used to characterize receptor-ligand interactions by blocking the interaction through competitive binding to the ligand or the receptor. Monoclonal antibody ADK8 binds to the sequence 588QQNTA592 on the AAV8 capsid corresponding exactly to region 6 in AAV8 (20a). If this sequence is the only one involved in receptor binding by AAV8, the antibody should compete for binding of the virus to the cell. Although ADK8 neutralizes rAAV8 gene transduction, it does not block the binding of AAV8 to cells (Fig. 6). This result argues for the interpretation that region 6 is not (or not alone) involved in receptor binding in AAV8 and that antibody binding inhibits a postbinding processing step. Also expression/genome ratios suggest an influence of regions 5 and 6 on postbinding processes. The mechanism by which the 3-fold protrusions could influence postbinding events remains a matter of speculation. In this regard it should be mentioned that a stem-like connection between packaged DNA and the inner capsid surface below the 3-fold symmetry axes has been observed (17, 49). Such a connection could mediate interactions with the 3-fold protrusions occurring outside the capsid to genome-capsid interactions inside the capsid and influence genome release. Alternatively, selection of an entry pathway mediated by the 3-fold protrusions may indirectly promote efficiency of postentry processing.
Overall, the data presented here suggest that modification of regions 5 and 6, which strongly influence efficiency and selectivity of AAV gene transduction in vivo, requires specific sequence information. There are several levels at which gene transduction in vivo could be influenced. These include blood clearance, penetration of the endothelial cell layer, sequestration at the extracellular matrix, cell binding and entry, and intracellular processing. It is unlikely that sequence-specific information is required for escape from trapping in a nonproductive state, e.g., at the extracellular matrix or opsonization by blood components. The need for specific sequence information for transduction improvement and retargeting rather suggests a gain of function. A gain-of-function mutation could lead to improved endothelial cell layer penetration, cell binding, cell entry, or postentry processing. The different assays conducted here point to a role of the 3-fold protrusions in both cell surface interaction and postentry processing; however, other activities cannot be excluded. Future studies are required to elucidate the role of this prominent structural domain of the viral capsid in AAV gene transduction.
ACKNOWLEDGMENTS
We thank A. Sacher and R. Popa-Wagner for valuable help.
This project was funded in part by grant KL516/7-1 from the Deutsche Forschungsgemeinschaft (to J.A.K.) and by National Institutes of Health grants R21 AI072341 and R01 GM082946 (to M.A.-M.).
Footnotes
Published ahead of print 20 June 2012
REFERENCES
- 1. Akache B, et al. 2006. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 80:9831–9836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akache B, et al. 2007. A two-hybrid screen identifies cathepsins B and L as uncoating factors for adeno-associated viruses 2 and 8. Mol. Ther. 15:330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asokan A, et al. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat. Biotechnol. 28:79–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asokan A, Hamra JB, Govindasamy L, Agbandje-McKenna M, Samulski RJ. 2006. Adeno-associated virus type 2 contains an integrin α5β1 binding domain essential for viral cell entry. J. Virol. 80:8961–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Auricchio A. 2011. Fighting blindness with adeno-associated virus serotype 8. Hum. Gene Ther. 22:1169–1170 [DOI] [PubMed] [Google Scholar]
- 6. Bantel-Schaal U, Delius H, Schmidt R, Hausen Hzur. 1999. Human adeno-associated virus type 5 is only distantly related to other known primate helper-dependent parvoviruses. J. Virol. 73:939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barbon CM, et al. 2005. AAV8-mediated hepatic expression of acid sphingomyelinase corrects the metabolic defect in the visceral organs of a mouse model of Niemann-Pick disease. Mol. Ther. 12:431–440 [DOI] [PubMed] [Google Scholar]
- 8. Breous E, Somanathan S, Bell P, Wilson JM. 2011. Inflammation promotes the loss of adeno-associated virus-mediated transgene expression in mouse liver. Gastroenterology 141:348–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Broekman ML, Comer LA, Hyman BT, Sena-Esteves M. 2006. Adeno-associated virus vectors serotyped with AAV8 capsid are more efficient than AAV-1 or -2 serotypes for widespread gene delivery to the neonatal mouse brain. Neuroscience 138:501–510 [DOI] [PubMed] [Google Scholar]
- 10. Choi VW, McCarty DM, Samulski RJ. 2005. AAV hybrid serotypes: improved vectors for gene delivery. Curr. Gene Ther. 5:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daya S, Berns KI. 2008. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 21:583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Denby L, Nicklin SA, Baker AH. 2005. Adeno-associated virus (AAV)-7 and -8 poorly transduce vascular endothelial cells and are sensitive to proteasomal degradation. Gene Ther. 12:1534–1538 [DOI] [PubMed] [Google Scholar]
- 13. Dubielzig R, King JA, Weger S, Kern A, Kleinschmidt JA. 1999. Adeno-associated virus type 2 protein interactions: formation of pre- encapsidation complexes. J. Virol. 73:8989–8998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao G, et al. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao G, Vandenberghe LH, Wilson JM. 2005. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 5:285–297 [DOI] [PubMed] [Google Scholar]
- 16. Gao GP, et al. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:11854–11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerlach B, Kleinschmidt JA, Bottcher B. 2011. Conformational changes in adeno-associated virus type 1 induced by genome packaging. J. Mol. Biol. 409:427–438 [DOI] [PubMed] [Google Scholar]
- 18. Ghosh A, et al. 2006. Long-term correction of murine glycogen storage disease type Ia by recombinant adeno-associated virus-1-mediated gene transfer. Gene Ther. 13:321–329 [DOI] [PubMed] [Google Scholar]
- 19. Girod A, et al. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 5:1052–1056 [DOI] [PubMed] [Google Scholar]
- 20. Govindasamy L, et al. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 80:11556–11570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a. Gurda BL, Raupp C, Popa-Wagner R, Naumer M, Olson NH, Ng R, McKenna R, Baker TS, Kleinschmidt JA, Agbandje-McKenna M. 16 May 2012. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J. Virol. doi:10.1128/JVI.00218-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harding TC, et al. 2006. Enhanced gene transfer efficiency in the murine striatum and an orthotopic glioblastoma tumor model, using AAV-7- and AAV-8-pseudotyped vectors. Hum. Gene Ther. 17:807–820 [DOI] [PubMed] [Google Scholar]
- 22. Hoffman BE, et al. 2011. Nonredundant roles of IL-10 and TGF-beta in suppression of immune responses to hepatic AAV-factor IX gene transfer. Mol. Ther. 19:1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hosel M, et al. 2012. Toll-like receptor 2-mediated innate immune response in human nonparenchymal liver cells toward adeno-associated viral vectors. Hepatology 55:287–297 [DOI] [PubMed] [Google Scholar]
- 24. Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 8:27–28 [DOI] [PubMed] [Google Scholar]
- 25. Hurlbut GD, et al. 2010. Preexisting immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Mol. Ther. 18:1983–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jayandharan GR, et al. 2011. Activation of the NF-κB pathway by adeno-associated virus (AAV) vectors and its implications in immune response and gene therapy. Proc. Natl. Acad. Sci. U. S. A. 108:3743–3748 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Jiang H, et al. 2006. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood 108:107–115 [DOI] [PubMed] [Google Scholar]
- 28. Kern A, et al. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laredj LN, Beard P. 2011. Adeno-associated virus activates an innate immune response in normal human cells but not in osteosarcoma cells. J. Virol. 85:13133–13143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lerch TF, Chapman MS. 2012. Identification of the heparin binding site on adeno-associated virus serotype 3B (AAV-3B). Virology 423:6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levy HC, et al. 2009. Heparin binding induces conformational changes in adeno-associated virus serotype 2. J. Struct. Biol. 165:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, et al. 2011. Capsid-specific T-cell responses to natural infections with adeno-associated viruses in humans differ from those of nonhuman primates. Mol. Ther. 19:2021–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H, et al. 2011. Adeno-associated virus vectors serotype 2 induce prolonged proliferation of capsid-specific CD8+ T cells in mice. Mol. Ther. 19:536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lochrie MA, et al. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 80:821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Louboutin JP, Wang L, Wilson JM. 2005. Gene transfer into skeletal muscle using novel AAV serotypes. J. Gene Med. 7:442–451 [DOI] [PubMed] [Google Scholar]
- 36. Manno CS, et al. 2006. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 12:342–347 [DOI] [PubMed] [Google Scholar]
- 37. Martino AT, et al. 2011. The genome of self-complementary adeno-associated viral vectors increases Toll-like receptor 9-dependent innate immune responses in the liver. Blood 117:6459–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McEachern KA, et al. 2006. AAV8-mediated expression of glucocerebrosidase ameliorates the storage pathology in the visceral organs of a mouse model of Gaucher disease. J. Gene Med. 8:719–729 [DOI] [PubMed] [Google Scholar]
- 39. Michelfelder S, Trepel M. 2009. Adeno-associated viral vectors and their redirection to cell-type specific receptors. Adv. Genet. 67:29–60 [DOI] [PubMed] [Google Scholar]
- 40. Mingozzi F, et al. 2007. CD8+ T-cell responses to adeno-associated virus capsid in humans. Nat. Med. 13:419–422 [DOI] [PubMed] [Google Scholar]
- 41. Mori S, Wang L, Takeuchi T, Kanda T. 2004. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology 330:375–383 [DOI] [PubMed] [Google Scholar]
- 42. Muller OJ, et al. 2003. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 21:1040–1046 [DOI] [PubMed] [Google Scholar]
- 43. Murphy SL, Bhagwat A, Edmonson S, Zhou S, High KA. 2008. High-throughput screening and biophysical interrogation of hepatotropic AAV. Mol. Ther. 16:1960–1967 [DOI] [PubMed] [Google Scholar]
- 44. Muzyczka N, Warrington KH., Jr 2005. Custom adeno-associated virus capsids: the next generation of recombinant vectors with novel tropism. Hum. Gene Ther. 16:408–416 [DOI] [PubMed] [Google Scholar]
- 45. Nakai H, et al. 2005. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J. Virol. 79:3606–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nam HJ, et al. 2007. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 81:12260–12271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nathwani AC, et al. 2011. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol. Ther. 19:876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nathwani AC, et al. 2011. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 365:2357–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ng R, et al. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J. Virol. 84:12945–12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nietupski JB, et al. 2011. Systemic administration of AAV8-alpha-galactosidase A induces humoral tolerance in nonhuman primates despite low hepatic expression. Mol. Ther. 19:1999–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. O'Donnell J, Taylor KA, Chapman MS. 2009. Adeno-associated virus-2 and its primary cellular receptor–cryo-EM structure of a heparin complex. Virology 385:434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pacak CA, et al. 2006. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ. Res. 99:e3–e9 [DOI] [PubMed] [Google Scholar]
- 54. Passini MA, et al. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 120:1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Paulk NK, et al. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology 51:1200–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pear WS, Nolan GP, Scott ML, Baltimore D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 90:8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Perabo L, et al. 2003. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol. Ther. 8:151–157 [DOI] [PubMed] [Google Scholar]
- 58. Pien GC, et al. 2009. Capsid antigen presentation flags human hepatocytes for destruction after transduction by adeno-associated viral vectors. J. Clin. Invest. 119:1688–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pulicherla N, et al. 2011. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol. Ther. 19:1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Qiao C, et al. 2009. Hydrodynamic limb vein injection of adeno-associated virus serotype 8 vector carrying canine myostatin propeptide gene into normal dogs enhances muscle growth. Hum. Gene Ther. 20:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rutledge EA, Halbert CL, Russell DW. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sarkar R, et al. 2006. Long-term efficacy of adeno-associated virus serotypes 8 and 9 in hemophilia a dogs and mice. Hum. Gene Ther. 17:427–439 [DOI] [PubMed] [Google Scholar]
- 63. Schmidt M, et al. 2008. Adeno-associated virus type 12 (AAV12): a novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J. Virol. 82:1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shen X, Storm T, Kay MA. 2007. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol. Ther. 15:1955–1962 [DOI] [PubMed] [Google Scholar]
- 65. Shin O, Kim SJ, Lee WI, Kim JY, Lee H. 2008. Effective transduction by self-complementary adeno-associated viruses of human dendritic cells with no alteration of their natural characteristics. J. Gene Med. 10:762–769 [DOI] [PubMed] [Google Scholar]
- 66. Snyder RO, Francis J. 2005. Adeno-associated viral vectors for clinical gene transfer studies. Curr. Gene Ther. 5:311–321 [DOI] [PubMed] [Google Scholar]
- 67. Sonntag F, et al. 2011. The assembly-activating protein (AAP) promotes capsid assembly of different AAV serotypes. J. Virol. 85:12686–12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun B, et al. 2005. Efficacy of an adeno-associated virus 8-pseudotyped vector in glycogen storage disease type II. Mol. Ther. 11:57–65 [DOI] [PubMed] [Google Scholar]
- 69. Thomas CE, Storm TA, Huang Z, Kay MA. 2004. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 78:3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vandenberghe LH, Wilson JM, Gao G. 2009. Tailoring the AAV vector capsid for gene therapy. Gene Ther. 16:311–319 [DOI] [PubMed] [Google Scholar]
- 71. Veldwijk MR, et al. 2002. Development and optimization of a real-time quantitative PCR-based method for the titration of AAV-2 vector stocks. Mol. Ther. 6:272–278 [DOI] [PubMed] [Google Scholar]
- 72. Wang AY, Peng PD, Ehrhardt A, Storm TA, Kay MA. 2004. Comparison of adenoviral and adeno-associated viral vectors for pancreatic gene delivery in vivo. Hum. Gene Ther. 15:405–413 [DOI] [PubMed] [Google Scholar]
- 73. Wang H, et al. 1998. Post-isolation inducible nitric oxide synthase gene expression due to collagenase buffer perfusion and characterization of the gene regulation in primary cultured murine hepatocytes. J. Biochem. 124:892–899 [DOI] [PubMed] [Google Scholar]
- 74. Wang L, et al. 2011. AAV8-mediated hepatic gene transfer in infant rhesus monkeys (Macaca mulatta). Mol. Ther. 19:2012–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang L, et al. 2011. Impact of preexisting immunity on gene transfer to nonhuman primate liver with adeno-associated virus 8 vectors. Hum. Gene Ther. 22:1389–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang Z, et al. 2005. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat. Biotechnol. 23:321–328 [DOI] [PubMed] [Google Scholar]
- 77. Waterkamp DA, Muller OJ, Ying Y, Trepel M, Kleinschmidt JA. 2006. Isolation of targeted AAV2 vectors from novel virus display libraries. J. Gene Med. 8:1307–1319 [DOI] [PubMed] [Google Scholar]
- 78. Weger S, Wistuba A, Grimm D, Kleinschmidt JA. 1997. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep. proteins. J. Virol. 71:8437–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wistuba A, Weger S, Kern A, Kleinschmidt JA. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wobus CE, et al. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wu P, et al. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xiao W, et al. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xie Q, et al. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405–10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xu D, Walker CM. 2011. Continuous CD8 T-cell priming by dendritic cell cross-presentation of persistent antigen following adeno-associated virus-mediated gene delivery. J. Virol. 85:12083–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ying Y, et al. 2010. Heart-targeted adeno-associated viral vectors selected by in vivo biopanning of a random viral display peptide library. Gene Ther. 17:980–990 [DOI] [PubMed] [Google Scholar]
- 86. Yu CY, et al. 2009. A muscle-targeting peptide displayed on AAV2 improves muscle tropism on systemic delivery. Gene Ther. 16:953–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zaiss AK, Muruve DA. 2008. Immunity to adeno-associated virus vectors in animals and humans: a continued challenge. Gene Ther. 15:808–816 [DOI] [PubMed] [Google Scholar]
- 88. Zhu J, Huang X, Yang Y. 2009. The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. J. Clin. Invest. 119:2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zolotukhin S, et al. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973–985 [DOI] [PubMed] [Google Scholar]
- 90. Zolotukhin S, Potter M, Hauswirth WW, Guy J, Muzyczka N. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]