Abstract
Metastable conformations of the gp120 and gp41 envelope glycoproteins of human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) must be maintained in the unliganded state of the envelope glycoprotein trimer. Binding of gp120 to the primary receptor, CD4, triggers the transition to an open conformation of the trimer, promoting interaction with the CCR5 chemokine receptor and ultimately leading to gp41-mediated virus-cell membrane fusion and entry. Topological layers in the gp120 inner domain contribute to gp120-trimer association in the unliganded state and to CD4 binding. Here we describe similarities and differences between HIV-1 and SIVmac gp120. In both viruses, the gp120 N/C termini and the inner domain β-sandwich and layer 2 support the noncovalent association of gp120 with the envelope glycoprotein trimer. Layer 1 of the SIVmac gp120 inner domain contributes more to trimer association than the corresponding region of HIV-1 gp120. On the other hand, layer 1 plays an important role in stabilizing the CD4-bound conformation of HIV-1 but not SIVmac gp120 and thus contributes to HIV-1 binding to CD4. In SIVmac, CD4 binding is instead enhanced by tryptophan 375, which fills the Phe 43 cavity of gp120. Activation of SIVmac by soluble CD4 is dependent on tryptophan 375 and on layer 1 residues that determine a tight association of gp120 with the trimer. Distinct biological requirements for CD4 usage have resulted in lineage-specific differences in the HIV-1 and SIV gp120 structures that modulate trimer association and CD4 binding.
INTRODUCTION
The primate immunodeficiency viruses (PIVs) include the human immunodeficiency viruses, HIV-1 and HIV-2, and the simian immunodeficiency viruses (SIVs). In nature, HIV-1 and HIV-2 infect humans, HIV-1-related SIVcpz viruses infect chimpanzees, and SIV variants infect African monkeys (6, 23, 25, 43, 44). Based on phylogenetic evidence, SIV variants form the primary reservoir, with HIV-1 and HIV-2 resulting from zoonotic cross-species transmissions within the last century or two (7, 24, 66). Clear lineage-specific genetic differences are observed between variants of SIV and also between these variants and their more recent human crossovers (4, 7, 24, 66). Humans infected with HIV-1 and HIV-2 and Asian macaques infected by certain SIVs frequently develop life-threatening immunodeficiency (AIDS) due to depletion of CD4-positive T lymphocytes (6, 11, 23, 32).
Entry of HIV-1 and SIV into the host cell is mediated by the viral envelope glycoproteins, which are derived by proteolytic cleavage of a trimeric, glycosylated gp160 envelope glycoprotein precursor (2, 64). The resulting mature envelope glycoproteins, the gp120 exterior envelope glycoprotein (SU) and the gp41 transmembrane envelope glycoprotein (TM), constitute a trimeric complex on the virion surface that is anchored by the gp41 membrane-spanning segments (8, 18, 71, 79). The gp120 exterior envelope glycoprotein is retained on the trimer via labile, noncovalent interactions with the gp41 ectodomain and perhaps with other gp120 protomers (21, 29, 75, 77). The gp120 glycoprotein is the most exposed element on the trimer and mediates binding to the viral receptors on the host cell. Binding to the initial receptor, CD4 (12, 36), triggers conformational changes in gp120 that promote its interaction with one of the chemokine receptors, usually CCR5 (1, 10, 13–15, 19, 70, 73). CD4 binding also induces conformational changes in the assembled HIV-1 envelope glycoprotein trimer that result in a more “open” configuration in which a helical heptad repeat (HR1) segment of the gp41 ectodomain is exposed (22, 28, 38, 45, 67). Further conformational changes lead to the formation of a gp41 six-helix bundle composed of the HR1 and HR2 heptad repeat regions, which provides the energy needed to fuse the viral and target cell membranes (8, 46, 71).
The movement of the HIV-1 and SIV envelope glycoprotein trimer from its unliganded state to the CD4-bound state must be carefully regulated. Premature triggering of the metastable envelope glycoprotein complex to downstream conformations results in functional inactivation (22, 27, 28, 33, 34, 38). Because detailed structural information about gp120 in the unliganded HIV-1 trimer is lacking, only a rudimentary understanding of the sequence of events initiated by CD4 binding exists. The X-ray crystal structure of an HIV-1 gp120 “core,” which lacks some of the gp120 hypervariable loops, in complex with CD4 has been solved (41, 54). In the CD4-bound state, the gp120 core consists of a gp41-interactive inner domain, an outer domain that faces outward on the assembled envelope glycoprotein trimer, and a minidomain called the bridging sheet. CD4 is thought to initially contact the outer domain of gp120 and to induce the formation of the bridging sheet, a four-stranded antiparallel β-sheet that interacts directly with CD4 (41). Although the gp120 inner domain does not contact CD4 (41, 54), conformational rearrangements within this domain decrease the off rate of CD4 binding and thus increase gp120-CD4 affinity (21). In the CD4-bound conformation, the HIV-1 gp120 inner domain consists of a β-sandwich that, along with the gp120 N and C termini, mediates the noncovalent interaction with gp41 (29, 77); three topological layers (layers 1, 2, and 3) in the inner domain emanate from this β-sandwich and were suggested to undergo rearrangement in the transition of gp120 from the unliganded to the CD4-bound state (54). The interaction of layer 1 and layer 2 within the HIV-1 gp120 inner domain contributes in an indirect manner to CD4 binding (21). Thus, gp120 refolds itself upon binding to CD4, with the inner and outer domains and bridging sheet brought into proximity at the interface with CD4. An approximately 150-Å3 cavity between HIV-1 gp120 and CD4 is bounded by Phe 43 of CD4, which makes numerous contacts with conserved gp120 residues critical for CD4 binding (41). The gp120 residues that abut this “Phe 43 cavity” are also highly conserved in HIV-1 strains, even though many of these residues do not contact CD4. Some of these cavity-lining residues contribute to an aromatic array that helps stabilize the CD4-bound conformation (21, 41, 54). Because changes in some inner domain residues in layer 2 result in shedding of gp120 from the envelope glycoprotein trimer, it has been suggested that CD4-induced conformational changes involving the inner domain layers might promote partial release of the restraints that keep gp120 and gp41 in their unliganded conformation (21, 63).
Although HIV-1 and SIV use CD4 as a primary receptor, differences exist between these PIV lineages. First, SIV is generally less dependent on CD4 for infection than HIV-1 and can efficiently infect cells with low levels of surface CD4 expression (17, 50, 51, 58–61, 65). The SIVsmm gp120 glycoproteins naturally have a tryptophan 375 residue that is predicted to fill the Phe 43 cavity (39) and may contribute to the lower dependence on CD4. In support of this hypothesis, substitution of tryptophan for serine 375, which is found in most HIV-1 strains (39), fills the Phe 43 cavity and allows HIV-1 gp120 to sample conformations closer to the CD4-bound state spontaneously (76). Second, when soluble CD4 (sCD4) or CD4-mimetic compounds trigger the CD4-bound state on virions that are distant from a target cell, the infectivity of HIV-1 decays with a half-life of 5 to 7 min at 37°C (27); by contrast, the sCD4-activated state of SIV is long-lived, allowing the virus to infect CCR5-expressing cells hours after binding sCD4 (65). Intriguingly, the half-life of the sCD4-activated state of HIV-1 was increased by changes in layer 1 of the inner domain (21). Moreover, the impact of alterations in layers 1 and 2 on CD4 binding could be diminished by replacement of serine 375 with tryptophan in HIV-1 gp120 (21). It is noteworthy that the disulfide loop in layer 1 of the gp120 inner domain is truncated in SIV relative to that of HIV-1 (see below). These observations led us to hypothesize that the interaction of layers 1 and 2 of the HIV-1 gp120 inner domain and the Phe 43 cavity-filling tryptophan 375 of SIV gp120 similarly serve to promote the CD4-bound conformation and thus facilitate CD4 binding. A corollary of this hypothesis is that the contribution of layers 1 and 2 to SIV gp120 interaction with CD4 will be less than that previously documented for HIV-1 gp120 (21). Here we tested this hypothesis by investigating the contributions of the gp120 inner-domain layers 1 and 2 and tryptophan 375 to the distinct biological properties of SIV. For these studies, we chose the molecularly cloned and well-characterized SIVmac239 (32, 35, 52) from the diverse group of SIVs. The SIVmac239 envelope glycoproteins exhibit a dependence on target cell CD4 levels similar to that of the HIV-1YU2 envelope glycoproteins (5), which were studied in the previous mutagenic analysis of the HIV-1 gp120 inner domain layers 1 and 2 (21). Both SIVmac239 and HIV-2, which cause AIDS in Asian monkeys and humans, respectively, were derived from SIVsmm of sooty mangabeys (11, 24, 31, 43, 44). Thus, SIVmac239, SIVsmm, and HIV-2 are members of the same lineage of PIVs (24, 31, 39, 66).
MATERIALS AND METHODS
Modeling full-length SIV gp120.
Modeling of the full-length SIVmac239 gp120 was done by using the X-ray crystal structure (PDB identifier 3JWD) of the ternary complex consisting of an HIV-1HXBc2 gp120 core with intact N and C termini, two-domain CD4, and the antigen-binding fragment of the human antibody 48d (54) as a template. Modeling was performed on the Discovery Studio software platform (Accelrys Software, Inc.). SIVmac239 gp120 glycoprotein modeling was based on protein sequence alignment with full-length HIV-1HXBc2 gp120, using the Modeler (version 9) program in the Accelrys software package. The gp120 V4 variable loop, consisting of the sequence WSTEGSNNT, was disordered in the X-ray crystal structure (54) and was added by modeling.
Cell lines.
The 293T human embryonic kidney, Cf2Th canine thymocyte (American Type Culture Collection), and TZM-bl cell lines (NIH AIDS Research and Reference Reagent Program) were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Sigma) and 100 μg/ml of penicillin-streptomycin (Mediatech, Inc.). Cf2Th cells stably expressing human CD4 and CCR5 (42) were grown in medium supplemented with 0.4 mg/ml of G418 (Invitrogen) and 0.15 mg/ml of hygromycin B (Roche Diagnostics). Cf2Th-CCR5 cells were grown in medium supplemented with 0.4 mg/ml of G418 (Invitrogen). The TZM-bl cell line is a HeLa cell line stably expressing high levels of CD4 and CCR5 and possessing an integrated copy of the luciferase gene under the control of the HIV-1 long terminal repeat (57).
Site-directed mutagenesis.
Mutations were introduced individually or in combination into the previously described vector expressing the SIVmac239 envelope glycoproteins (pSIVmac239) (47, 48). Site-directed mutagenesis was performed using the QuikChange II XL site-directed mutagenesis protocol (Stratagene). The presence of the desired mutations was determined by automated DNA sequencing. The numbering of the SIVmac239 and HIV-1 envelope glycoprotein amino acid residues is based on that of the prototypic HXBc2 strain of HIV-1, where 1 is the initial methionine (37).
Immunoprecipitation and CCR5 binding of envelope glycoproteins.
For pulse-labeling experiments, 3 × 105 293T cells were cotransfected by the calcium phosphate method with pLTR-Tat and the pSIVmac239 vector expressing the SIVmac239 envelope glycoproteins. One day after transfection, cells were metabolically labeled for 16 h with 100 μCi/ml [35S]methionine-cysteine (35S] protein labeling mix; Perkin-Elmer) in Dulbecco's modified Eagle's medium lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum. Cells were subsequently lysed in RIPA buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% NP-40, and 0.05% sodium dodecyl sulfate [SDS]). Precipitation of radiolabeled SIVmac239 envelope glycoproteins from cell lysates or medium was performed with a mixture of sera from SIV-infected macaques. Alternatively, medium containing radiolabeled envelope proteins was immunoprecipitated with increasing amounts of CD4-Ig for 1 h at 37°C in the presence of 50 μl of 10% protein A-Sepharose (Amersham Biosciences).
For CCR5 binding, normalized amounts of radiolabeled SIVmac239 gp120 envelope glycoproteins were incubated in the presence or absence of 200 nm of sCD4 for 1 h at 37°C. Subsequently, 2 × 106 Cf2Th-CCR5 cells were added for an additional 1 h at 37°C and washed twice with phosphate-buffered saline (PBS) prior to cell lysis in RIPA buffer. Cell lysates were immunoprecipitated with a mixture of sera from SIV-infected macaques. All samples were loaded on NuPAGE Novex Bis-Tris polyacrylamide gels (Invitrogen) and analyzed by autoradiography and by using a PhosphorImager (Molecular Dynamics). For CD4-Ig and CCR5 immunoprecipitations, samples were analyzed under nonreducing conditions as previously described (20).
Cell-based ELISA.
Detection of trimeric envelope glycoproteins at the surface of COS-1 cells was performed by cell-based ELISA, as described previously (27). Briefly, COS-1 cells were seeded in 96-well plates (2 × 104 cells per well) and transfected the next day with 0.15 μg of a plasmid expressing the envelope glycoproteins and 0.01 μg of a Tat-expressing plasmid per well using the standard polyethylenimine (PEI) (Polyscience Inc., Pennsylvania) transfection method. Two days later, cells were washed twice with blocking buffer (10 mg/ml nonfat dry milk, 1.8 mM CaCl2, 1 mM MgCl2, 25 mM Tris, pH 7.5, and 140 mM NaCl) and then incubated for 1 h at room temperature with a 1/2,000 dilution of heat-inactivated serum from SIV-infected monkeys (a kind gift from Norman Letvin, Beth Israel Deaconess Medical Center). A horseradish peroxidase-conjugated antibody specific for the Fc region of human IgG (Pierce) was then incubated with the samples for 45 min at room temperature. Cells were washed 5 times with blocking buffer and 5 times with washing buffer. Horseradish peroxidase (HRP) enzyme activity was determined after the addition of 30 μl per well of a 1:1 mix of Western Lightning oxidizing and luminol reagents (Perkin Elmer Life Sciences). Light emission was measured with an LB 941 TriStar luminometer (Berthold Technologies).
Recombinant luciferase viruses.
Recombinant viruses containing the firefly luciferase gene were produced by calcium phosphate transfection of 293T cells with the HIV-1 proviral vector pNL4.3 Env−Luc and the plasmid expressing the wild-type or mutant SIVmac239 envelope glycoproteins at a ratio of 2:1. Two days after transfection, the cell supernatants were harvested; the reverse transcriptase activities of all viruses were measured as described previously (62). The virus-containing supernatants were stored in aliquots at −80°C.
Infection by single-round luciferase viruses.
Cf2Th-CD4-CCR5 target cells were seeded at a density of 5 × 103 cells/well in 96-well luminometer-compatible tissue culture plates (Dynex) 24 h before infection. Recombinant viruses (10,000 reverse transcriptase units) in a final volume of 100 μl were then added to the target cells, followed by incubation for 48 h at 37°C; the medium was then removed from each well, and the cells were lysed by the addition of 30 μl of passive lysis buffer (Promega) and three freeze-thaw cycles. An EG&G Berthold microplate luminometer, LB 96V, was used to measure the luciferase activity of each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μl of 1 mM D-luciferin potassium salt (BD Pharmingen).
Cell-cell fusion.
To assess cell-to-cell fusion, 3 × 105 293T cells were cotransfected by the calcium phosphate method with an HIV-1 Tat-expressing plasmid, pLTR-Tat, and the pSIVmac239 vector expressing the SIVmac239 envelope glycoproteins. Two days after transfection, 3 × 104 293T cells were added to TZM-bl target cells that were seeded at a density of 3 × 104 cells/well in 96-well luminometer-compatible tissue culture plates (Dynex) 24 h before the assay. Cells were coincubated for 6 h at 37°C, after which they were lysed by the addition of 30 μl of passive lysis buffer (Promega) and three freeze-thaw cycles. Luciferase activity in each well was measured as described above.
RESULTS
Comparison of the HIV-1 and SIV gp120 inner domains.
We wished to investigate the function of the inner domain of SIVmac gp120. Because a structure of SIV gp120 with the complete inner domain is not available (9), we modeled the SIVmac239 gp120 envelope glycoprotein based on the X-ray crystal structure of the HIV-1 gp120 core with intact N and C termini, bound to soluble CD4 and a neutralizing antibody fragment (54). This X-ray crystallographic study revealed the structure of the complete HIV-1 gp120 inner domain (54) (Fig. 1A). A key component of the inner domain, a seven-stranded β-sandwich, serves as a point of departure for the gp120 N and C termini, which meander toward the viral membrane. The β-sandwich and the gp120 terminal strands have been implicated in the noncovalent association of gp120 with gp41 (21, 29, 77). Projecting from the β-sandwich toward the target cell membrane are three excursions that compose the rest of the inner domain. These excursions form three topological layers (“layers 1, 2, and 3”) that have been proposed to change conformation as gp120 undergoes the transition from the unliganded state to the CD4-bound state (54). Changes in layer 2 can decrease the stable association of HIV-1 gp120 with the envelope glycoprotein trimer and result in shedding of gp120 (21). The interaction of layers 1 and 2 in the HIV-1 gp120 inner domain strengthens gp120-CD4 binding by reducing the off rate (21).
Fig 1.
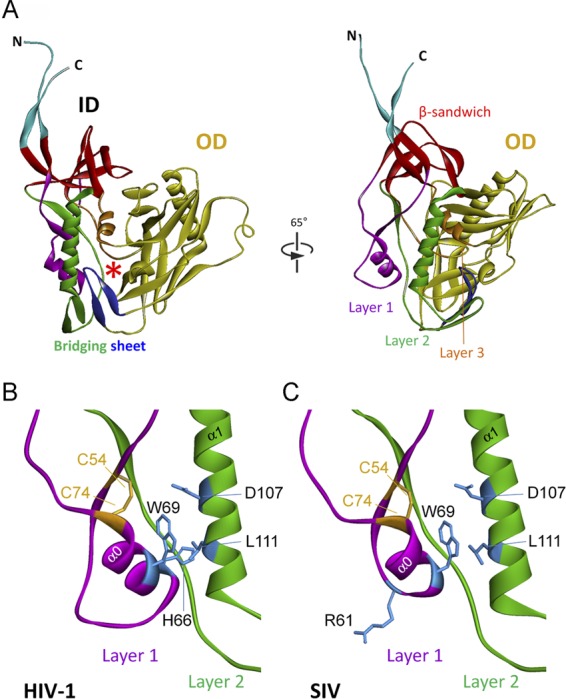
Structure of the inner domain of HIV-1 gp120 and modeled SIV gp120 in the CD4-bound conformation. (A) The structure of the HIV-1HXBc2 gp120 core with N/C termini in the CD4-bound state (54) is shown from the perspective of CD4 (left panel) and from the approximate perspective of the envelope glycoprotein trimer axis (right panel). The outer domain (OD) of gp120 is colored yellow. The N and C termini are colored cyan. The components of the gp120 inner domain (ID) are the β-sandwich (red) and three loop-like extensions: layer 1 (magenta), layer 2 (green), and layer 3 (orange). The β20-β21 strands of gp120 (blue) project from the outer domain and, in the CD4-bound conformation, compose two of the strands of the four-stranded bridging sheet. The other two strands of the bridging sheet are derived from the distal portion of layer 2. The location of the Phe 43 cavity is indicated by a red asterisk. A close-up view of the interactions of the inner domain layer 1 and layer 2 from HIV-1 gp120 (B) or a modeled SIV gp120 glycoprotein (C) is shown. The perspective in panels B and C is rotated approximately 25° in the counterclockwise direction around the Z axis from the perspective in the right part of panel A. The cysteine residues that form a disulfide bond in layer 1 are colored yellow, and some of the layer 1 and layer 2 residues implicated in layer 1-layer 2 interaction in HIV-1 gp120 (21) are colored blue. The α0 helix in layer 1 and the α1 helix in layer 2 are labeled.
The SIVmac239 gp120 primary amino acid sequence was aligned with that of HIV-1 gp120. The nine intrachain disulfide bonds in HIV-1 gp120 glycoproteins are conserved in all PIV gp120 glycoproteins, assisting the alignment of the HIV-1 and SIVmac239 gp120 sequences (37, 39). The alignment of the sequences of the inner domain (layer 1, layer 2, β-sandwich, and N and C termini) and Phe 43 cavity-lining residues relevant to this study is shown in Fig. 2. Layer 1 is truncated in the gp120 glycoproteins of HIV-2 and most monkey SIV strains compared with that of HIV-1/SIVcpz strains (Fig. 2 and 3). Notably, the only exceptions are the gp120s of the SIVgsn/SIVmon/SIVmus clade, which are thought to provide the env gene of the recombinant SIVcpz that gave rise to HIV-1 (4, 25, 66) (Fig. 3). Modeling suggests that the α0 helix in the SIVmac239 gp120 inner domain is shorter than that in HIV-1 gp120 (compare Fig. 1B and C). Interestingly, as observed for HIV-1 gp120, the conserved residues Trp 69, Asp 107, and Leu 111 are predicted to be located at the interface of layers 1 and 2 of SIVmac239 gp120 (compare Fig. 1B and C). However, a fourth residue, His 66, at the layer 1-layer 2 interface of HIV-1 gp120, lacks a counterpart in SIV gp120 as a result of the truncation of layer 1. Of note, HIV-1/SIVcpz gp120 glycoproteins, which invariably have a histidine at position 66, also have a proline at position 212 (Fig. 3); this proline makes stacking interactions with His 66 and helps to stabilize the CD4-bound conformation (21, 34, 54). Interestingly, the gp120 glycoproteins of the HIV-2/SIVsmm lineage, which includes SIVmac239, lack a counterpart of His 66 and also lack a proline residue at position 212; in the HIV-2/SIVsmm lineage, residue 212 is an alanine, serine, or threonine residue (39) (Fig. 2 and 3). Thus, in HIV-2/SIVsmm strains, fewer predicted layer 1-layer 2 interactions are available to promote/stabilize the CD4-bound conformation of gp120 than in HIV-1/SIVcpz strains.
Fig 2.
Alignment of HIV-1 and SIVmac239 gp120 regions. The primary amino acid sequences of specific regions of HIV-1 (accession number K03455), SIVcpz (accession number AY169968), and SIVmac239 (accession number M33262) are aligned. Sequence identity is indicated by a solid vertical line, and amino acid residues that are conserved are connected by a dotted vertical line. Gaps in the sequence are indicated by dashes. Residue numbering is based on that of the HXBc2 strain of HIV-1 (37).
Fig 3.
Evolution of gp120 changes in PIVs. The phylogenetic relationship of envelope glycoprotein sequences of the PIVs is shown, based on previous studies (4, 7, 66, 68). The evolutionary distances in this tree are approximate. The disulfide loop (DSL) length of layer 1 in the gp120 inner domain is shown in the first column. The identities of residues 66, 212, and 375 in gp120 are shown in the other columns. A dash indicates the absence of a corresponding residue in the gp120 glycoprotein of the indicated PIV.
The above analysis predicts that substantial differences exist between the inner domain layer 1 of HIV-1 and SIVmac239 gp120. Although the primary sequences of the other gp120 inner domain and N/C-terminal elements diverge between HIV-1 and HIV-2/SIVsmm lineages, no major insertions/deletions were observed in these regions of PIV envelope glycoproteins (Fig. 2).
CD4 binding depends upon the α3 helix of HIV-1 gp120, and the adjacent β16 strand flanks the Phe 43 cavity (41). The side chain of residue 375 is oriented toward the Phe 43 cavity, and thus large residues have the potential to occupy the cavity (76). Comparison of the gp120 β16 sequences from different PIVs indicates that HIV-2 and the SIVs of monkeys and apes have large residues (Trp, Met, Tyr, His, and Phe) at position 375 that likely fill the Phe 43 cavity (39) (Fig. 2 and 3). HIV-1 strains in the N and O groups also have these large residues at position 375 (Fig. 3). In contrast, the HIV-1 strains in group M, the major group of HIV-1, have the smaller Ser 375, which allows the existence of an “empty” Phe 43 cavity. Apparently, gp120 residue 375 has evolved from tryptophan in most monkey SIVs to other large residues (Met, His, Tyr, and Phe) in most ape SIVs and in group N and O HIV-1. Evolution of the group M HIV-1 from SIVcpzPtt (66) favored a serine residue at position 375. Based on the CD4-binding phenotypes of HIV-1 gp120 mutants with different substitutions at Ser 375 (76), the phylogenetic changes in residue 375 accompanying the evolution of the group M HIV-1 envelope glycoproteins would be expected to decrease the tendency to assume the CD4-bound conformation.
Effect of gp120 inner domain changes on processing and subunit association of the SIVmac239 envelope glycoproteins.
The contribution of the gp120 inner domain to the structure and function of the SIVmac239 envelope glycoproteins was evaluated by studying the phenotypes of a panel of mutant envelope glycoproteins (Table 1). Proteolytic processing of the envelope glycoprotein precursor and the noncovalent association of the gp120 exterior envelope glycoprotein with the gp41 transmembrane envelope glycoprotein in the trimer were evaluated. Some of the amino acid changes (W96A and L225A) in the inner domain β-sandwich severely affected precursor processing; these mutants were defective in multiple envelope glycoprotein functions, suggesting that they may experience folding problems.
TABLE 1.
Phenotypes of SIVmac239 gp120 mutantsa
| Mutation(s)b | Residue location | Association indexc | Processing indexd | Relative infectivitye | Cell-to-cell fusionf | CD4-Ig bindingg |
|---|---|---|---|---|---|---|
| None | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| V36A | N terminus | 0.26 | 0.98 | 0.46 | 1.29 | 1.88 |
| Y40A | N terminus | 0.81 | 1.20 | 0.48 | 0.76 | 1.30 |
| N47A | N terminus | 1.05 | 0.56 | 0.26 | 1.25 | 0.53 |
| L52A | Layer 1 | 0.41 | 0.70 | 0.04 | 0.97 | 0.54 |
| T56A | Layer 1 | 0.38 | 0.72 | 0.10 | 0.88 | 0.73 |
| R61A | Layer 1 | 0.28 | 0.82 | 0.23 | 0.67 | 1.03 |
| R61S | Layer 1 | 0.36 | 0.93 | <0.01 | 1.20 | 1.38 |
| D62A | Layer 1 | 0.12 | 0.49 | <0.01 | 0.88 | 1.17 |
| T63A | Layer 1 | 0.20 | 1.20 | 0.10 | 1.04 | 1.60 |
| W69L | Layer 1 | 0.34 | 0.58 | 0.02 | 0.89 | 0.96 |
| W69F | Layer 1 | 0.23 | 0.84 | 0.01 | 1.02 | 1.38 |
| T72A | Layer 1 | 0.38 | 0.80 | 0.11 | 1.14 | 0.83 |
| Q73A | Layer 1 | 0.27 | 0.67 | 0.03 | 1.09 | 0.82 |
| P76A | Layer 1 | 0.94 | 0.81 | <0.01 | 0.01 | 0.97 |
| N78A | Layer 1 | 0.96 | 0.89 | 0.01 | 0.08 | 0.67 |
| G79S | Layer 1 | 1.19 | 0.85 | 0.47 | 0.91 | 0.61 |
| E83A | β-Sandwich | 0.83 | 0.15 | 0.17 | 0.85 | NA |
| A85G | β-Sandwich | 1.14 | 0.93 | 0.33 | 0.60 | 0.54 |
| L86V | β-Sandwich | 0.93 | 1.15 | 1.09 | 0.99 | 1.23 |
| V89A | β-Sandwich | 0.74 | 1.34 | 0.42 | 0.95 | 1.53 |
| T90A | β-Sandwich | 0.65 | 0.48 | 0.27 | 0.80 | 0.47 |
| E91A | β-Sandwich | 0.75 | 4.52 | 0.77 | 0.82 | 1.24 |
| W96A | β-Sandwich | 0.37 | 0.07 | 0.02 | 0.01 | NA |
| Q103A | Layer 2 | 0.17 | 0.78 | <0.01 | 0.94 | 0.01 |
| D107A | Layer 2 | 0.48 | 0.88 | 0.01 | 0.87 | 0.46 |
| W109I | Layer 2 | 0.96 | 0.95 | 0.45 | 0.53 | 0.85 |
| L111A | Layer 2 | 0.56 | 0.83 | 0.07 | 0.85 | 0.49 |
| F112A | Layer 2 | 0.07 | 0.75 | 0.01 | 1.17 | 0.23 |
| I116A | Layer 2 | 0.17 | 1.04 | 0.07 | 1.98 | 1.02 |
| A212P | Layer 2 | 0.09 | 1.10 | 0.03 | ND | 1.32 |
| F215A | Layer 2 | 0.25 | 0.14 | <0.01 | 0.64 | 0.01 |
| R216A | Layer 2 | 0.62 | 0.30 | 0.02 | 0.01 | 0.23 |
| Y217A | Layer 2 | 0.58 | 0.10 | 0.01 | 0.47 | NA |
| P221A | Layer 2 | 0.42 | 0.57 | 0.06 | 0.18 | 0.46 |
| Y223A | β-Sandwich | 0.33 | 0.14 | 0.22 | 1.10 | NA |
| L225A | β-Sandwich | 0.52 | 0.04 | <0.01 | 0.21 | NA |
| L226A | β-Sandwich | 0.65 | 0.15 | 0.14 | 0.99 | NA |
| F233A | β-Sandwich | 0.53 | 0.06 | <0.01 | 0.01 | NA |
| W375S | Phe 43 cavity | 0.76 | 1.09 | 0.24 | 0.41 | 0.48 |
| K487A | C terminus | 0.34 | 0.08 | 0.03 | 1.07 | NA |
| V491A | C terminus | 0.37 | 0.26 | 0.44 | 1.28 | 0.22 |
| I494A | C terminus | 0.07 | 0.37 | 0.01 | 0.84 | 1.88 |
| L496A | C terminus | 0.14 | 0.59 | 0.28 | 1.17 | 1.52 |
| W69L/W375S | Layer 1/Phe 43 | 0.32 | 1.36 | 0.02 | 0.78 | 0.27 |
| D107A/W375S | Layer 2/Phe 43 | 0.44 | 1.47 | 0.03 | 0.97 | 0.37 |
| W109I/W375S | Layer 2/Phe 43 | 0.65 | 1.12 | 0.29 | 0.09 | 0.26 |
| L111A/W375S | Layer 2/Phe 43 | 0.87 | 1.58 | 0.03 | 0.58 | 0.27 |
Values presented in this table represent the means of data from at least two independent experiments, with experimental variation typically not more than 20% of the value reported.
The numbering of the SIVmac239 gp120 envelope glycoprotein amino acid residues is based on that of the prototypic HXBc2 strain of HIV-1, where 1 is the initial methionine (37). The mutations result in the substitution of the amino acid residue shown on the right for the amino acid residue shown on the left of the number; for example, W69L indicates a substitution of a leucine residue for the tryptophan residue at position 69.
The association index is a measure of the ability of the mutant gp120 molecule to remain associated with the envelope glycoprotein complex on the expressing cell relative to that of the wild-type envelope glycoproteins. The association index is calculated as follows: association index = ([mutant gp120]cell × [wild-type gp120]supernatant)/([mutant gp120]supernatant × [wild-type gp120]cell).
The processing index is a measure of the conversion of the mutant gp160 envelope glycoprotein precursor to mature gp120 relative to that of the wild-type envelope glycoproteins. The processing index was calculated by the following formula: processing index = ([total gp120]mutant × [gp160]wild-type)/([gp160]mutant × [total gp120]wild-type).
Relative infectivity was assessed by infecting Cf2Th-CD4/CCR5 cells with similar amounts of recombinant HIV-1 pseudotyped with wild-type and mutant SIVmac239 envelope glycoproteins, normalized according to reverse transcriptase (see Materials and Methods). The ratio of mutant/wild-type virus infectivity is reported.
Cell-cell fusion ability was assessed by coincubation for 6 h at 37°C of 293T cells expressing the SIVmac239 envelope glycoprotein variants with the reporter TZM-bl cells, as described in Materials and Methods. ND, not determined.
Normalized amounts of radiolabeled wild-type and mutant gp120 glycoproteins were incubated with 13 nM CD4-Ig for 1 h at 37°C. The immunoprecipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry. NA, not applicable, because the levels of gp120 secreted into the supernatant for these mutants were not sufficient to assess CD4 binding.
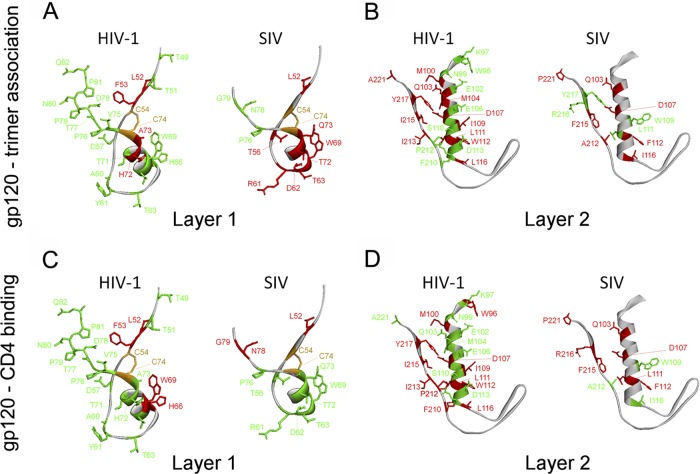
Several changes in the SIVmac239 gp120 inner domain decreased the association of gp120 with the envelope glycoprotein trimer (Fig. 4 and Table 1). Multiple changes (L52A, T56A, R61A, D62A, T63A, W69L, W69F, T72A, and Q73A) in layer 1 disrupted gp120-trimer association. These results were noteworthy, since relatively few changes in layer 1 of HIV-1 gp120 exerted effects on subunit association (21) (Fig. 5A). Alteration of several layer 2 residues affected the association of the SIVmac239 gp120 glycoprotein with the trimer, consistent with the phenotypes of layer 2 mutants of the HIV-1 gp120 glycoprotein (21) (Fig. 4 and 5B and Table 1). However, changes in particular layer 2 residues (Gln 103, Trp 109, and Ala 212) affected SIVmac239 gp120-trimer association differently than alteration of the equivalent residue in HIV-1 gp120 (21). Likewise, although several changes in the SIVmac239 gp120 inner domain β-sandwich and N/C termini resulted in significant shedding of gp120 from the trimer, the association of SIVmac239 gp120 with the trimer was less sensitive than the HIV-1 gp120-trimer association to changes in Tyr 40, Val 85, Ala/Leu 86, and Val 89. These observations suggest that layer 1 contributes significantly to the stability of the SIV envelope glycoprotein trimer, in contrast to layer 1 of HIV-1 gp120. Layer 2, the β-sandwich, and the N/C termini of both HIV-1 and SIV gp120 support gp120-trimer association, although the contribution of particular residues to this property appears to differ between these viruses.
Fig 4.
Precursor processing and gp120-trimer association of selected SIVmac239 envelope glycoprotein mutants. Cell lysates and supernatants (SN) of 35S-labeled cells transiently expressing the SIVmac239 wild-type and indicated mutant envelope glycoproteins were precipitated with serum from an SIV-infected macaque. The precipitated proteins were loaded onto NuPAGE Novex Bis-Tris polyacrylamide gels and analyzed by autoradiography and densitometry.
Fig 5.
Residues important for gp120-trimer association and CD4 binding. Layer 1 and Layer 2 of HIV-1 gp120 (54) and the modeled SIVmac239 gp120 are shown from the same perspective as those in Fig. 1, B and C. In panels A and B, the ribbon and side chain residues that were altered in this and a previous study (21) are colored according to the gp120-trimer association index (Red: association index < 0.5 and Green: association index ≥ 0.7). In panels C and D, the ribbon and side chain residues are colored according to CD4-Ig binding ability (Red: relative CD4-Ig binding ≤ 0.5 and Green: relative CD4-Ig binding > 0.5).
Interaction of mutant SIVmac239 envelope glycoproteins with CD4.
Alteration of the interface between layers 1 and 2 of the HIV-1 gp120 inner domain has been shown to decrease CD4 binding, revealing an important mechanism whereby HIV-1 gp120 regulates CD4-induced conformational rearrangement and achieves a reduction in the off rate (21, 34, 54). To investigate the contribution of the inner domain of SIV gp120 to CD4 binding, the CD4-binding abilities of the panel of SIVmac239 gp120 mutants were examined. Wild-type and mutant SIVmac239 envelope glycoproteins were transiently expressed in 293T cells, which were radiolabeled for 16 h. The amount of radiolabeled gp120 glycoproteins shed into the cell medium was normalized by immunoprecipitation with polyclonal serum from SIV-infected macaques before assessing the ability to bind CD4-Ig. Notably, none of the changes in the disulfide-bonded loop of the SIV gp120 layer 1 significantly decreased CD4-Ig binding (Fig. 5C, Fig. 6A and B, and Table 1). The W69L and W69F changes, which decrease CD4 binding of the HIV-1 gp120 glycoprotein (21), did not significantly diminish the interaction of SIVmac239 gp120 with CD4-Ig. Only two changes, E47A and L52A, which affect the layer 1 strand that is N-terminal to the disulfide loop, resulted in a decrease in CD4-Ig binding; the L52A change in the HIV-1 gp120 glycoprotein has been previously shown to decrease CD4-Ig binding (21). These results suggest that the disulfide-bonded loop in layer 1 of the SIV gp120 inner domain makes less of a contribution to CD4 binding than the corresponding region of HIV-1 gp120 (Fig. 5C).
Fig 6.
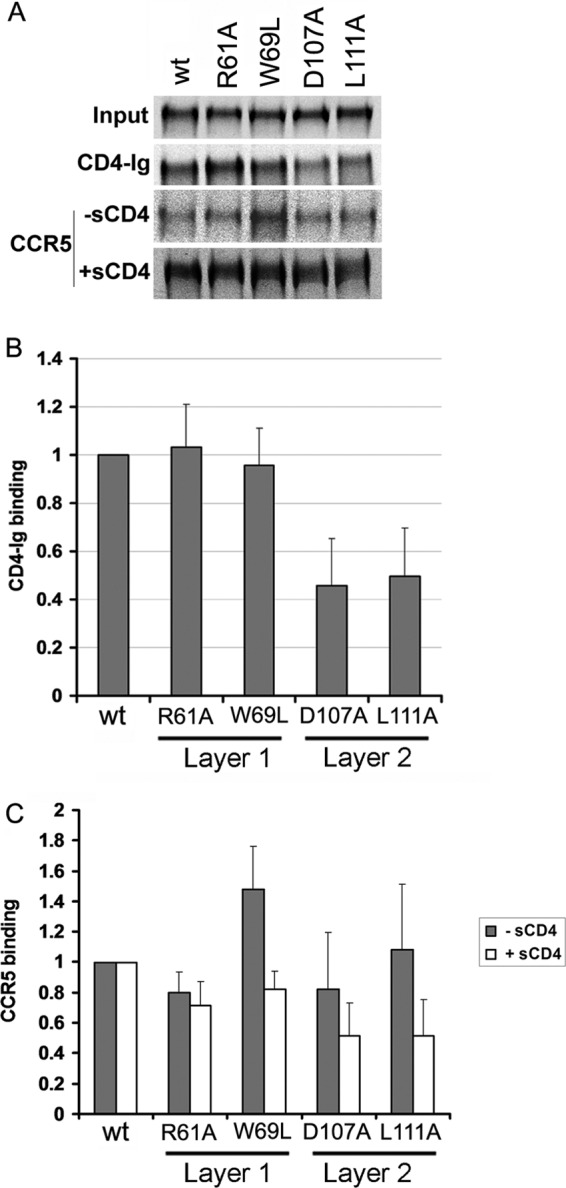
Binding of soluble SIVmac239 gp120 glycoproteins to CD4-Ig and CCR5. Normalized amounts of radiolabeled wild-type and mutant gp120 glycoproteins were incubated with 13 nM CD4-Ig for 1 h at 37°C (A and B). The precipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry. Representative results from at least five independent experiments are shown. (C) Similar amounts of radiolabeled gp120 glycoproteins were incubated in the absence (gray bars) or presence (white bars) of 200 nM sCD4 prior to addition to cells expressing CCR5. After 1 h at 37°C, the amount of bound mutant gp120 was determined and normalized to the observed amount of bound wild-type (wt) gp120. Incubation with sCD4 increased the binding of wt SIVmac239 gp120 to CCR5 by 8-fold. The data shown represent the means ± SEM of four independent experiments.
A previous study (21) showed that the phenotypic effects of the changes in His 66 and Trp 69 in layer 1 of the HIV-1 gp120 glycoprotein on CD4 binding were eliminated by the S375W change, which fills the Phe 43 cavity (76). We hypothesized that the presence of a tryptophan residue at position 375 in the SIVmac239 gp120 glycoprotein likewise diminished the potential contribution of layer 1-layer 2 interactions to CD4 binding. To test this hypothesis, we examined the CD4-binding ability of an SIVmac239 W375S mutant gp120 that, in some cases, had additional changes predicted to decrease layer 1-layer 2 interaction. The W375S change decreased the interaction of the SIVmac239 gp120 glycoprotein with CD4-Ig (Table 1; see also Fig. S1 in the supplemental material). In combination with the W375S alteration, the W69L change in layer 1 and the W109I change in layer 2 reduced CD4-Ig binding further; of note, neither the W69L change nor the W109I change significantly decreased CD4-Ig binding in the context of the wild-type SIVmac239 gp120 glycoprotein (Table 1). Thus, the presence of the Phe 43 cavity-filling tryptophan 375 residue diminishes the contribution of inner domain layer 1-layer 2 interactions to CD4 binding.
The majority of SIVmac239 gp120 inner domain changes that decreased CD4-Ig binding involved layer 2 residues. Changes in residues Asp 107, Leu 111, Trp/Phe 112, and Ile/Phe 215 decreased CD4 binding in both HIV-1 and SIVmac239 gp120 glycoproteins (Table 1) (21). The Q103A change decreased CD4-Ig binding of SIVmac239 gp120 but not that of HIV-1 gp120. Conversely, changes in Ile/Trp109, Leu/Ile 116, and Pro/Ala 212 decreased CD4 binding of HIV-1 gp120 but not that of SIVmac239 gp120. Thus, layer 2 changes can affect the CD4-binding affinity of both HIV-1 and SIVmac239 gp120 glycoproteins, but the contribution of individual layer 2 residues to CD4 binding may differ between the two viruses (Fig. 5D).
Abilities of mutant envelope glycoproteins to interact with CCR5.
A functionally critical consequence of CD4 binding to the HIV-1 and SIV gp120 glycoproteins is the induction of conformational changes that lead to the binding of the chemokine receptor CCR5 or CXCR4 (1, 10, 13–15, 19, 70, 74). We analyzed whether introducing changes in the interface between layers 1 and 2 in the SIVmac239 gp120 inner domain affected CCR5 recognition. Radiolabeled wild-type and mutant SIV gp120 glycoproteins were incubated in the presence or absence of sCD4 for 1 h at 37°C prior to incubation with cells expressing the CCR5 coreceptor for 1 h at 37°C. After washing and lysis of the cells, bound gp120 was detected by immunoprecipitation. Consistent with a modest effect of the D107A and L111A changes in layer 2 on CD4 binding, only slight decreases in CCR5 recognition were observed for these mutants in the presence of sCD4 (Fig. 6A and C).
Function of mutant SIV envelope glycoproteins.
The wild-type and mutant SIVmac239 envelope glycoproteins were assessed for the ability to mediate cell-cell fusion with cells expressing both CD4 and CCR5, as a measure of their functionality. With the exception of only 8 mutants (P76A, N78A, W96A, R216A, P221A, L225A, F233A, and W109I/W375S), the SIVmac239 envelope glycoprotein mutants mediated efficient cell-cell fusion (Fig. 7A and Table 1). Thus, in most cases, the introduction of the inner domain changes did not abolish the abilities of the SIV envelope glycoproteins to bind receptors and mediate membrane fusion.
Fig 7.
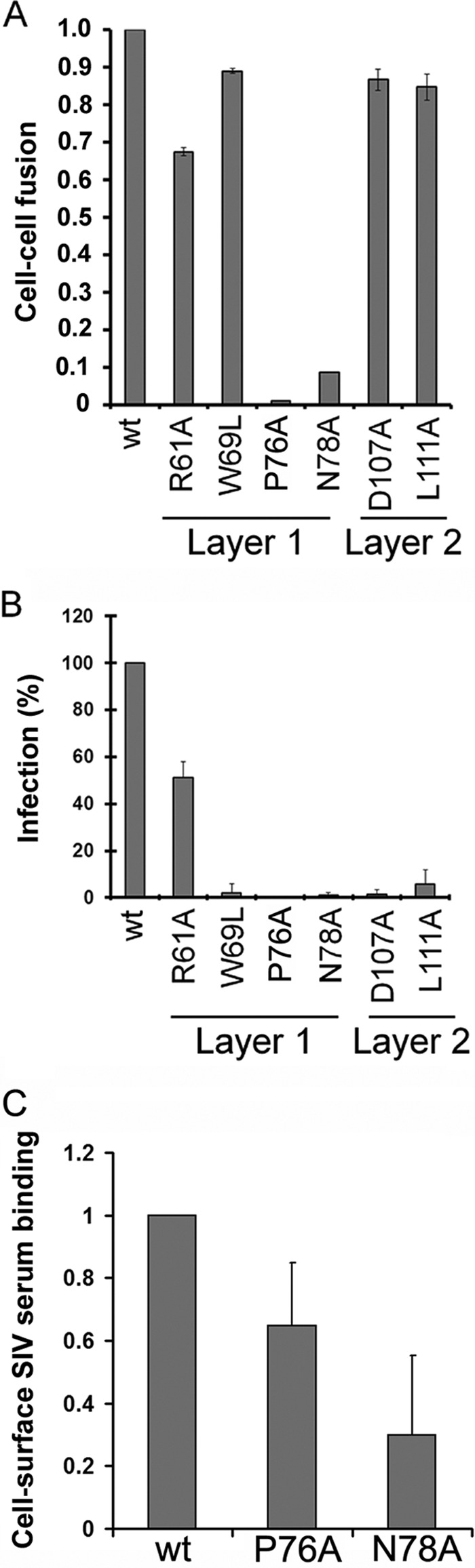
Functionality of SIVmac239 gp120 envelope glycoprotein variants. (A) Cell-cell fusion ability was assessed by coincubation of 293T cells expressing the envelope glycoprotein variants with the reporter TZM-bl cells for 6 h at 37°C. The activities of the mutant envelope glycoproteins were normalized to that of the wild-type (wt) envelope glycoproteins. The results shown represent the means ± SEM obtained from at least three independent experiments. (B) Relative infectivity was assessed by incubating Cf2Th-CD4/CCR5 cells with similar amounts of recombinant HIV-1 pseudotyped with wild-type and mutant SIVmac239 envelope glycoproteins, normalized according to reverse transcriptase. Data shown represent the means ± SEM obtained from at least three independent experiments. (C) The cell surface expression of the wt and fusion-defective SIVmac239 P76A and D78A envelope glycoproteins was measured by a cell-based ELISA. The results are normalized to the value obtained for the wt SIVmac239 envelope glycoproteins and represent the average for at least three independent experiments ± SD performed in quadruplicate.
We also evaluated the abilities of the panel of SIVmac239 envelope glycoprotein mutants to support the infection of cells expressing CD4 and CCR5. As expected, the mutants exhibiting decreased gp120-trimer association were significantly attenuated in the ability to mediate cell-free virus infection (Fig. 7B and Table 1). The majority of these mutant envelope glycoproteins mediated cell-cell fusion much more efficiently than cell-free virus infectivity. This phenotype has been previously observed for HIV-1 envelope glycoprotein mutants that exhibit decreased gp120-trimer association (21, 77); because the time between envelope glycoprotein synthesis and engagement of the target cell is much longer in the virion infectivity assay than in the cell-cell fusion assay, the former assay is more sensitive to decreases in the stability of gp120-trimer association.
Two of the SIVmac239 layer 1 mutants, P76A and N78A, did not mediate efficient cell-cell fusion or virus entry yet exhibited processing and association indices and CD4-Ig binding values similar to those of the wild-type SIVmac239 envelope glycoprotein. These mutants are capable of engaging the CD4 receptor, but they exhibit decreased cell surface expression that might contribute to their decreased functionality (Fig. 7C). The analogous mutants in the HIV-1 envelope glycoproteins were not as attenuated in cell-cell fusion activity (21).
SIV envelope glycoprotein determinants of sCD4 activation of entry.
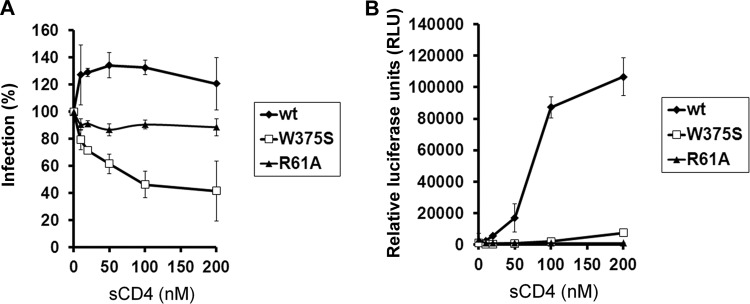
Unlike HIV-1 envelope glycoproteins, which undergo a relatively short-lived activation by sCD4 treatment (27), the SIVmac239 envelope glycoproteins assume a long-lived, activated state after sCD4 binding (3, 65). We examined whether the gp120 structural elements that differ between HIV-1 and SIV might explain these distinct responses to sCD4. Specifically, we tested whether changes in the Phe 43 cavity-filling residue, Trp 375, and layer 1 of the inner domain affected the response of viruses with the SIVmac239 envelope glycoproteins to sCD4. sCD4 activation of entry mediated by the wild-type SIVmac239 envelope glycoproteins was evident on CD4− CCR5+ target cells (Fig. 8B) and to a lesser extent on CD4+ CCR5+ target cells (Fig. 8A). The W375S envelope glycoproteins were minimally activated by sCD4 for infection of CD4− CCR5+ target cells (Fig. 8B) and were inhibited by sCD4 during infection of CD4+ CCR5+ cells (Fig. 8A). Soluble CD4 activation of infection of both CD4− CCR5+ and CD4+ CCR5+ cells was eliminated by the R61A change in layer 1 (Fig. 8A and B). Thus, sCD4 activation of the SIVmac239 envelope glycoproteins is dependent upon the Phe 43 cavity-filling Trp 375 residue and on elements in layer 1 of the inner domain.
Fig 8.
Effect of sCD4 on infection of CD4+ CCR5+ and CD4−CCR5+ cells. The amounts of recombinant luciferase-expressing HIV-1 pseudotyped with the wt and mutant SIVmac239 envelope glycoproteins were normalized according to reverse transcriptase. Viruses were incubated at 37°C with increasing concentrations of sCD4 (0 to 200 nM) for 1 h prior to addition of Cf2Th-CD4/CCR5 (A) or Cf2Th-CCR5 (B) target cells. Luciferase activity was measured after a 48-h incubation at 37°C. In panel A, the level of infection after incubation with sCD4 relative to the level observed for the same virus without sCD4 treatment is shown. In panel B, the absolute levels of infection in relative luciferase units are shown. The results represent the averages (± SD) from three independent experiments performed in triplicate.
DISCUSSION
HIV-1 and HIV-2/SIV share ancestry and many properties, including the ability to infect CD4+ CCR5+ T lymphocytes, monocytes/macrophages, and microglia. Low-resolution cryoelectron tomograms of HIV-1 and SIV virions indicate that the envelope glycoprotein trimers of these viruses exhibit a similar overall architecture (45, 72, 73, 78, 79, 80). A recent study has shown that the HIV-1 gp120 core has a propensity to assume the CD4-bound conformation spontaneously once the V1/V2 and V3 variable loops are removed (40). Thus, in the unliganded envelope glycoprotein complex, gp120 must retain a stable association with the trimer but must also be restrained from sampling downstream conformations, such as the CD4-bound state. Some of the HIV-1 gp120 elements (N/C termini, inner domain β-sandwich and layer 2, and V3 [in primary strains]) that contribute to the stable association of gp120 with the envelope glycoprotein trimer have been characterized (21, 30, 54, 75, 77). Here we have provided evidence that the SIVmac239 gp120 N/C termini and inner domain β-sandwich and layer 2 also contribute to gp120-trimer association. Thus, the global architecture of the envelope glycoprotein structures that govern gp120-trimer association is likely shared by HIV-1 and SIVmac239. Some differences exist in the trimer association phenotypes associated with changes in particular residues within these elements of HIV-1 and SIV gp120, suggesting that there may be subtle differences in the specific structural relationships between the envelope glycoproteins of these virus lineages.
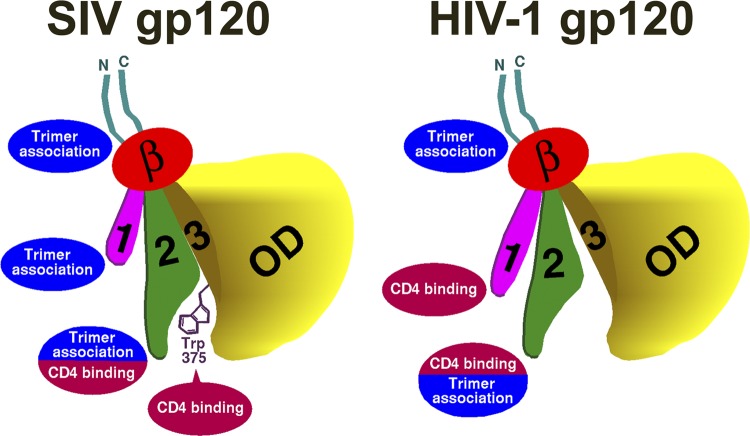
Our study highlights two structural elements that differ between the HIV-1 and SIV gp120 exterior envelope glycoproteins, i.e., layer 1 of the inner domain and the Phe 43 cavity-lining residue 375. We investigate the function of these elements in the SIVmac239 envelope glycoproteins, and we show that these elements interdependently determine phenotypes related to CD4 binding. Relative to layer 1 of HIV-1 gp120, the SIVmac239 gp120 layer 1 plays a more prominent role in maintaining gp120-trimer association and is minimally involved in promoting CD4 binding. HIV-2/SIVsmm gp120 glycoproteins, like those of most monkey SIVs, typically have a tryptophan residue at position 375 (39). In HIV-1, substitution of tryptophan for serine 375, which is well conserved in the major group of HIV-1, results in spontaneous sampling of a conformation closer to the CD4-bound state (76). The S375W HIV-1 mutant binds CD4 more efficiently and exhibits increased sensitivity to sCD4-induced shedding of gp120 and neutralization (21, 76). These observations suggested the hypothesis that relative to HIV-1 gp120, the HIV-2/SIVsmm gp120 glycoproteins, by virtue of the Phe 43 cavity-filling Trp 375, might naturally exhibit a greater propensity to sample the CD4-bound conformation. We found that a W375S change in the SIVmac239 envelope glycoproteins decreased CD4 binding and eliminated the enhancing effect of sCD4 on infection of CD4+ CCR5+ and CD4− CCR5+ target cells (3, 65). This sCD4-induced activation of SIVmac239 infection was also dependent on the integrity of gp120 layer 1. Moreover, some additional decrease in CD4 binding was associated with SIV gp120 layer 1 mutants in the context of the W375S, but not the wild-type, SIVmac239 envelope glycoproteins. The cooperative influence of both residue 375 and layer 1 in determining CD4 binding and its consequences is supported by the converse studies of the HIV-1 envelope glycoproteins. The H66A and W69L layer 1 changes decreased the CD4-binding affinity of HIV-1 gp120 but had no effect on CD4 binding of an S375W HIV-1 mutant (21). Moreover, changes in these HIV-1 layer 1 residues, particularly in combination with the S375W change, allowed the formation of a longer-lived activated intermediate upon sCD4 binding, closer to the sCD4-induced phenotype observed for SIV (21). Thus, the gp120 inner domain layer 1 and residue 375 exert reciprocal influences on the affinity of CD4 binding and the stability of the CD4-activated state.
These observations suggest that some differences between the SIVmac239 and HIV-1 gp120 subunits in the assembled envelope glycoprotein trimer exist, as summarized in Fig. 9. By virtue of the Phe 43 cavity-filling Trp 375, SIVmac239 gp120 has a strong propensity to assume the CD4-bound conformation. Since premature movement into the CD4-bound conformation can have negative consequences (gp120 shedding, exposure of neutralization epitopes, and rapidly decaying intermediate states), this propensity in SIVmac239 gp120 is compensated by the strong contribution of the inner domain layer 1 to the association of gp120 with the trimer. With Trp 375 filling the Phe 43 cavity, layer 1-layer 2 interactions make minimal contributions to CD4 binding. In contrast, in HIV-1 gp120, which lacks a Phe 43 cavity-filling residue at position 375, the propensity to assume the CD4-bound conformation is lower than that in SIVmac239 gp120. The association of HIV-1 gp120 with the unliganded trimer is adequately maintained by the N/C termini and the inner domain β-sandwich and layer 2. In HIV-1, therefore, the major function of layer 1 is to promote high-affinity CD4 binding by solidifying the interaction with layer 2 and decreasing the off rate of CD4.
Fig 9.
Summary of functional differences between HIV-1 and SIVmac239 gp120 glycoproteins. One of the three gp120 subunits of the SIVmac239 and HIV-1 envelope glycoprotein trimer is depicted. In the orientation shown, the trimer axis runs vertically on the left of each figure and the viral membrane is at the top of the figure. The gp120 N and C termini are colored cyan, the β-sandwich is red, layer 1 is magenta, layer 2 is green, layer 3 is ochre, and the outer domain (OD) is yellow. The Trp 375 side chain that fills the Phe 43 cavity of SIV gp120 is depicted. The major contributions of the gp120 N and C termini, the inner domain (the β-sandwich, layer 1, and layer 2), and the Phe 43 cavity to gp120-trimer association and/or CD4 binding are shown. Note that the contribution of the filled SIVmac239 gp120 Phe 43 cavity to CD4 binding is lacking in HIV-1 gp120; this lack is compensated by a contribution of the HIV-1 gp120 inner domain layer 1 to CD4 binding. In contrast, the inner domain layer 1 of SIVmac239 gp120 makes a larger contribution to gp120-trimer association than layer 1 of HIV-1 gp120.
The SIVmac239 envelope glycoproteins were chosen for this study because they exhibit a level of CD4 dependence comparable to that of the previously studied HIV-1YU2 envelope glycoproteins (5, 21). However, SIVmac239 is more CD4 dependent, more neutralization resistant, and less able to infect monkey macrophages than most other members of the SIVsmm clade. How generalizable are the results that we obtained with the SIVmac239 envelope glycoproteins? First, despite the similar levels of CD4 dependence of the SIVmac239 and HIV-1YU2 envelope glycoproteins, the phenotypes of the gp120 inner domain layer 1 mutants differed. Thus, layer 1 function in other members of the SIVsmm/HIV-2 clade, which are less dependent on CD4 than either SIVmac239 or HIV-1YU2 (5), is also expected to differ from that of HIV-1. Second, the SIVmac239 gp120 residues identified in this study as important for gp120-trimer association and CD4 binding are well conserved in all SIVsmm and HIV-2 strains. Therefore, all SIVsmm and HIV-2 gp120 envelope glycoproteins likely share common structural and functional inner domain elements. However, although the Phe 43 cavity-filling tryptophan 375 residue is found in the gp120 glycoproteins of HIV-2 and most monkey SIVs, differences in the inner domain layer 1 and layer 2 sequences occur among the distinct SIV lineages associated with different African monkey species. Additional studies will be required to determine the functional implications of these layer 1/layer 2 differences in SIV lineages other than the HIV-2/SIVsmm clade.
What was the sequence of events during PIV evolution that led to the observed changes in the gp120 inner domain and Phe 43 cavity? Based on our current understanding of PIV evolution (66) and the patterns observed in Fig. 3, we can suggest a model. The ancestral pool of indigenous SIVs likely had short inner domain layer 1 structures and a Phe 43 cavity filled with a tryptophan residue. At some point in time and for reasons that are unknown, the SIVgsn/SIVmus/SIVmon gp120 glycoproteins evolved longer disulfide loops in layer 1 without changing the Phe 43 cavity-filling tryptophan residue. The longer layer 1 sequences in the gp120 glycoproteins of SIVgsn/SIVmus/SIVmon are not likely to result in an extension of the α0 helix, in contrast to the situation in HIV-1 gp120 (54). This assertion is supported by the presence of Pro 65 and Gly 66 residues, both unfavorable for stable helix formation, in layer 1 of the SIVgsn/SIVmus/SIVmon gp120 inner domains. Nonetheless, some advantage resulted in fixation of the longer layer 1 in the gp120 glycoproteins of these SIVs. SIVgsn, SIVmus, and SIVmon infect three closely related Cercopithecus species (C. nictitans, C. cephus,, and C. mona, respectively), which are hunted by chimpanzees in central Africa. Chimpanzees are thought to have become infected with one of these SIVs, which, through a recombination event with a coinfecting SIVrcm, supplied the env gene to the SIVcpz precursor of HIV-1 (4, 66). With the evolution of SIVcpz, His 66 makes its first appearance in the elongated layer 1, allowing stacking interactions to occur with the preexisting Pro 212 in layer 2. Thus, by the time SIVcpz variants became indigenous in African chimpanzees, the ability of layer 1-layer 2 interactions to bolster CD4 binding was established. This may have relaxed the evolutionary pressure to maintain a tryptophan 375 residue in the Phe 43 cavity, as evidenced by the preference in SIVcpzPtt, SIVgor, and HIV-1 strains for residues other than tryptophan at position 375. Subsequent adaptation in humans further influenced the identity of residue 375. The less successful group N and group O HIV-1 strains retain the large residues at position 375 found in their respective chimpanzee and gorilla SIV ancestors. The extremely successful group M HIV-1 evolved an empty Phe 43 cavity. Thus, a series of sequential structural changes in the gp120 inner domain and Phe 43 cavity apparently provided the circumstances conducive to the evolution of a group M HIV-1 envelope glycoprotein well suited for transmission and spread in humans.
The different arrangements of the Phe 43 cavity and inner domain of HIV-1 and SIVsmm gp120 glycoproteins helps to explain why SIVsmm envelope glycoproteins are generally less dependent on CD4 for CCR5 interaction, are able to infect cells that express low levels of CD4, and can maintain a long-lived sCD4-activated state (17, 50, 58–61, 65). What biological necessities shaped the divergent evolution of these HIV-1 and SIV gp120 elements? The very low level of CD4 expression on the surface of tissue macrophages of monkeys (5, 51) may provide selective pressure on the SIV envelope glycoproteins to achieve and maintain the CD4-bound conformation after engaging CD4 at low stoichiometry. Indeed, the nearly undetectable levels of CD4 expression on monkey macrophages have been shown to represent the major factor limiting their infection by T cell-line-tropic SIVsmm strains and, notably, by macrophage-tropic HV-1 strains (5, 51). The ability of the latter viruses to infect primary human macrophages suggests that human macrophages targeted for HIV-1 infection must have higher levels of surface CD4 available for envelope glycoprotein interaction than primary macrophages from monkeys. Freed of the necessity to maintain the ability to infect cells with extremely low levels of CD4 expression, HIV-1 may have taken advantage of the benefits of avoiding the CD4-bound conformation. Transmitted/founder HIV-1 typically retains the ability to infect T lymphocytes but not monocytes/macrophages (53). Thus, HIV-1 tropism for cells with low CD4 levels, like monocytes/macrophages and microglia cells, may need to be acquired as the virus evolves in particular tissue compartments, such as the central nervous system (16, 26, 49, 55, 56, 69). An understanding of PIV adaptation to species-specific environments may assist efforts to intervene in virus transmission, spread within the infected host, and pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yvette McLaughlin and Elizabeth Carpelan for manuscript preparation. We thank Norman Letvin and Anna Gonzales for serum from SIV-infected macaques.
This work was supported by intramural funding by the VRC, NIAID, and by grants from the National Institutes of Health (AI24755, GM56550, and AI67854), by the International AIDS Vaccine Initiative, by the late William F. McCarty-Cooper (to J.S.), and by an amfAR Mathilde Krim Fellowship in Basic Biomedical Research (phase I, 107963-49-RKVA; phase II, 108092-50-RKVA), by a Canada Foundation for Innovation Program Leader grant (no. 29866), and by a CIHR operating grant, 257792, to A.F.
We have no conflicts of interest to report.
Footnotes
Published ahead of print 13 June 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alkhatib G, et al. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958 [DOI] [PubMed] [Google Scholar]
- 2. Allan JS, et al. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091–1094 [DOI] [PubMed] [Google Scholar]
- 3. Allan JS, et al. 1992. Strong association of simian immunodeficiency virus (SIVagm) envelope glycoprotein heterodimers: possible role in receptor-mediated activation. AIDS Res. Hum. Retroviruses 8:2011–2020 [DOI] [PubMed] [Google Scholar]
- 4. Bailes E, et al. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 5. Bannert N, Schenten D, Craig S, Sodroski J. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984–10993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barre-Sinoussi F, et al. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871 [DOI] [PubMed] [Google Scholar]
- 7. Bibollet-Ruche F, et al. 2004. New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a Cercopithecus monkey virus clade. J. Virol. 78:7748–7762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273 [DOI] [PubMed] [Google Scholar]
- 9. Chen B, et al. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841 [DOI] [PubMed] [Google Scholar]
- 10. Choe H, et al. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 11. Clavel F, et al. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233:343–346 [DOI] [PubMed] [Google Scholar]
- 12. Dalgleish AG, et al. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 13. Deng H, et al. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666 [DOI] [PubMed] [Google Scholar]
- 14. Doranz BJ, et al. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158 [DOI] [PubMed] [Google Scholar]
- 15. Dragic T, et al. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673 [DOI] [PubMed] [Google Scholar]
- 16. Dunfee RL, et al. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160–15165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edinger AL, et al. 1997. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc. Natl. Acad. Sci. U. S. A. 94:14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farzan M, et al. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 72:7620–7625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877 [DOI] [PubMed] [Google Scholar]
- 20. Finzi A, et al. 2010. Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J. Virol. Methods 168: 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finzi A, et al. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell 37:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Furuta RA, Wild CT, Weng Y, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276–279 [DOI] [PubMed] [Google Scholar]
- 23. Gallo RC, et al. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500–503 [DOI] [PubMed] [Google Scholar]
- 24. Gao F, et al. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature 358:495–499 [DOI] [PubMed] [Google Scholar]
- 25. Gao B, et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- 26. Gorry PR, et al. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haim H, et al. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360 doi:10.1371/journal.ppat.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He Y, et al. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 77:1666–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Helseth E, Olshevsky U, Furman C, Sodroski J. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65:2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Helseth E, et al. 1990. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J. Virol. 64:6314–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirsch VM, Olmsted RA, Murphey-Curb M, Purcell RH, Johnson PR. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389–392 [DOI] [PubMed] [Google Scholar]
- 32. Kanki PJ, et al. 1985. Serologic identification and characterization of a macaque T-lymphotropic retrovirus closely related to HTLV-III. Science 228:1199–1201 [DOI] [PubMed] [Google Scholar]
- 33. Kassa A, et al. 2009. Identification of a human immunodeficiency virus type 1 envelope glycoprotein variant resistant to cold inactivation. J. Virol. 83:4476–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kassa A, et al. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J. Virol. 83:8364–8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kestler HW, III, et al. 1988. Comparison of simian immunodeficiency virus isolates. Nature 331:619–622 [DOI] [PubMed] [Google Scholar]
- 36. Klatzmann D, et al. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768 [DOI] [PubMed] [Google Scholar]
- 37. Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. 1998. Numbering positions in HIV relative to HXB2CG, p 102–111 In Korber B, et al. (ed), Human retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 38. Koshiba T, Chan DC. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J. Biol. Chem. 278:7573–7579 [DOI] [PubMed] [Google Scholar]
- 39. Kuiken C, et al. 2008. HIV sequence compendium 2008. Los Alamos HIV sequence database. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 40. Kwon YD, et al. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A. 109:5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwong PD, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. LaBonte JA, Patel T, Hofmann W, Sodroski J. 2000. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J. Virol. 74:10690–10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Letvin NL, et al. 1985. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science 230:71–73 [DOI] [PubMed] [Google Scholar]
- 44. Letvin NL, King NW. 1990. Immunologic and pathologic manifestations of the infection of rhesus monkeys with simian immunodeficiency virus of macaques. J. Acquir. Immune Defic. Syndr. 3:1023–1040 [PubMed] [Google Scholar]
- 45. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075–1082 [DOI] [PubMed] [Google Scholar]
- 47. Marcon L, et al. 1997. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J. Virol. 71:2522–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marcon L, Sodroski J. 1997. High degree of sensitivity of the simian immunodeficiency virus (SIVmac) envelope glycoprotein subunit association to amino acid changes in the glycoprotein 41 ectodomain. AIDS Res. Hum. Retroviruses 13:441–447 [DOI] [PubMed] [Google Scholar]
- 49. Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F. 2006. HIV-1 tropism for the central nervous system: brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology 346:169–179 [DOI] [PubMed] [Google Scholar]
- 50. Martin KA, et al. 1997. CD4-independent binding of SIV gp120 to rhesus CCR5. Science 278:1470–1473 [DOI] [PubMed] [Google Scholar]
- 51. Mori K, Rosenzweig M, Desrosiers RC. 2000. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J. Virol. 74:10852–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Naidu YM, et al. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 62:4691–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ochsenbauer C, et al. 2012. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 86:2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pancera M, et al. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Peters PJ, et al. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J. Virol. 78:6915–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peters PJ, et al. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J. Virol. 80:6324–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pohlmann S, et al. 2004. Amino acid 324 in the simian immunodeficiency virus SIVmac V3 loop can confer CD4 independence and modulate the interaction with CCR5 and alternative coreceptors. J. Virol. 78:3223–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Puffer BA, Altamura LA, Pierson TC, Doms RW. 2004. Determinants within gp120 and gp41 contribute to CD4 independence of SIV Envs. Virology 327:16–25 [DOI] [PubMed] [Google Scholar]
- 60. Puffer BA, et al. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reeves JD, et al. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J. Virol. 73:7795–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360 [DOI] [PubMed] [Google Scholar]
- 63. Rits-Volloch S, Frey G, Harrison SC, Chen B. 2006. Restraining the conformation of HIV-1 gp120 by removing a flexible loop. EMBO J. 25:5026–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Robey WG, et al. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228:593–595 [DOI] [PubMed] [Google Scholar]
- 65. Schenten D, et al. 1999. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J. Virol. 73:5373–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1:a006841 doi:10.1101/cshperspect.a006841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Si Z, et al. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 101:5036–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Takehisa J, et al. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thomas ER, et al. 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360:105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Trkola A, et al. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187 [DOI] [PubMed] [Google Scholar]
- 71. Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430 [DOI] [PubMed] [Google Scholar]
- 72. White TA, et al. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6(12):e1001249 doi:10.1371/journal.ppat.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. White TA, et al. 2011. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J. Virol. 85:12114–12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu L, et al. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183 [DOI] [PubMed] [Google Scholar]
- 75. Xiang SH, et al. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 84:3147–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xiang SH, et al. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 76:9888–9899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang X, Mahony E, Holm GH, Kassa A, Sodroski J. 2003. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 313:117–125 [DOI] [PubMed] [Google Scholar]
- 78. Zanetti G, Briggs JA, Grünewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2(8):e83 doi:10.1371/journal.ppat.0020083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhu P, et al. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. U. S. A. 100:15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhu P, et al. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.