Abstract
Glycans of human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) play pivotal roles in modulating virus-target cell interactions. We have previously reported that, whereas SIVmac239 is pathogenic, its deglycosylated essentially nonpathogenic mutant (Δ5G) serves as a live-attenuated vaccine, although both replicate similarly during primary infection. These findings prompted us to determine whether such a polarized clinical outcome was due to differences in the immune tissues targeted by these viruses, where functionally and phenotypically different memory CD4+ T cells reside. The results showed that Δ5G replicates in secondary lymphoid tissue (SLT) at 1- to 2-log-lower levels than SIVmac239, whereas SIVmac239-infected but not Δ5G-infected animals deplete CXCR3+ CCR5+ transitional memory (TrM) CD4+ T cells. An early robust Δ5G replication was localized to small intestinal tissue, especially the lamina propria (effector site) rather than isolated lymphoid follicles (inductive site) and was associated with the induction and depletion of CCR6+ CXCR3− CCR5+ effector memory CD4+ T cells. These results suggest that differential glycosylation of Env dictates the type of tissue-resident CD4+ T cells that are targeted, which leads to pathogenic infection of TrM-Th1 cells in SLT and nonpathogenic infection of Th17 cells in the small intestine, respectively.
INTRODUCTION
Heavily glycosylated viral spikes are the hallmark of both human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) and have been reasoned to serve as a shield to fend off host immune responses, leading to the failure to induce protective responses and promote a persistent chronic infection that erodes the host immune system, eventually leading to AIDS in HIV-infected individuals (5, 18, 19, 26, 49, 61) and SIV-infected macaques (6, 29, 41). However, the precise mechanism(s) underlying the pathogenic events caused by HIV/SIV infection and, in particular, the role of the Env glycans remains to be elucidated.
Details of the viral spike composed of a trimer of the gp120 (surface protein) and gp41 (transmembrane protein) heterodimer have been elucidated by using a combination of X-ray crystallography and electron microscopy, and in their unligated form, the viral spikes are covered with massive amounts of host origin N-linked and O-linked glycans designated as “glycan shields” (18, 19). The number of potential N-linked glycosylation sites (PNGs) of primate lentivirus gp120 is conserved and range from 20 to 30. The number and position of the PNGs differ among isolates such as HIV-1 subtypes, HIV-2 subtypes, SIVsmm, SIVagm, and SIVcpz (63), and are thought to be associated with a wide variety of functions, which include evading host humoral responses (5, 49, 61), tropism for cells required for dissemination of founder/early-transmitting viruses (15, 31), infection of cells within different anatomical sites of specific tissues, and other viral properties such as fitness (17, 43).
Our laboratory has been studying the properties, functions, and roles of the glycans with a focus on their potential role in conferring pathogenicity using a SIV-macaque AIDS model. SIVmac239 has a total of 23 PNGs within gp120 (32, 40, 41). All of the PNGs except for amino acid (aa) 247 are conjugated with N-glycans. Of the remaining 22 N-glycans, while those at aa 278, 284, 295, and 316 are considered essential, the remaining 18 are considered nonessential based on levels of virus replication in a CD4+ T cell line (32). To study the in vivo role of N glycosylation of gp120 with regard to viral replication and pathogenesis, we created a series of recombinant SIVmac239-mutants in which select PNGs were mutated to reduce glycosylation (32). Among these mutants was a quintuple deglycosylation mutant termed Δ5G consisting of nonsynonymous (Asn-to-Gln) mutations of five nonessential PNGs at aa 79, 146, 171, 460, and 479 which was shown to replicate in vitro to similar levels and kinetics as the wt SIVmac239 in primary in vitro-activated rhesus macaque CD4+ T cells. Studies were thus carried out to compare and contrast the in vivo characteristics of Δ5G to that of SIVmac239. Consistent with the in vitro studies, Δ5G showed the same replication kinetics and peak viral loads as SIVmac239 during acute primary infection of rhesus macaques (29). However, whereas the SIVmac239 showed variable but high sustained plasma viral loads (VL) during the chronic-phase, the Δ5G-infected animals showed relatively rapid decreases to nearly undetectable plasma viremia for > 10 years and near-sterile protection against homologous challenge virus. These results demonstrate that despite a similar level of viral replication during the primary infection, antiviral host response in Δ5G-infected animals successfully contain the infection similar to viruses that cause a robust but controllable acute infection such as flu viruses. Essentially similar in vitro and in vivo properties were also observed in animals infected with two other quintuple- and one triple-deglycosylation mutants, that share potent live-attenuated vaccine properties, including protection against challenge with homologous SIVmac239 and significant protective effects against challenge with heterologous SIVsmE543-3 (52).
A role for glycans in conferring conformational masking of neutralizing epitopes or receptor binding sites was reasoned to contribute to the evasion of HIV-1/SIV against humoral and especially neutralizing antibodies (NAb) (18). Differences in the humoral immune responses elicited by the Δ5G- and SIVmac239-infected animals were noted. Whereas the SIVmac239-infected animals rarely developed NAb, Δ5G-infected animals showed variable NAb response, including one animal that did not elicit any detectable NAb (50). Since 100% of the Δ5G-infected animals showed spontaneous viral containment, it was reasoned that NAb alone could not account for the live attenuated phenotype of Δ5G SIV. However, it cannot be excluded that humoral response played a role in the divergent outcomes between the two SIV infections, since Δ5G-infected animals elicited Δ5G-specific antibodies recognizing epitopes in V1/V2 of gp120 earlier than NAb (50). Since HIV/SIV preferentially infects and depletes virus-specific CD4+ T cells (7, 44, 62), we hypothesized that one reason for the polarized clinical outcome noted with SIVmac239 compared to Δ5G could be due to differences in the subsets of CD4+ T cells and/or the tissues in which these viruses replicate. Accordingly, we examined the cell lineage and anatomic distribution of SIV infection and the potential mechanisms for the differences in immunopathology typically seen in SIVmac239- but not Δ5G-infected animals during primary infection. In addition, the objective was to attempt to define the potential mechanism that led to complete and partial protection against not only homologous but also heterologous viral challenge, respectively, during chronic infection.
The CD4+ CCR5+ T cell subset is the primary target for HIV/SIV during the primary infection (4, 24) and are particularly abundant within the gastrointestinal tissues (GIT), such as the lamina propria (LP). Thus, these tissues have been considered a primary target for HIV/SIV replication, together with secondary lymphoid tissues (SLT) (21, 23, 53, 54). However, CD4+ CCR5+ T cells in SLT and GIT are functionally and phenotypically different, exemplified by the fact that the memory cells residing in the SLT immune inductive sites such as the mesenteric lymph nodes (LN) are distinct from those residing in the effector sites such as the LP (56). The impact of HIV/SIV infection and consequent depletion of these CD4+ T cell subsets on pathogenesis and their role in the induction of protective host response continues to be refined. Thus, CD4+ T cells are quite heterogeneous and are currently classified into functionally different subsets, such as Th1, Th2, Th17, Treg, and Tfh cells, based either on the spectrum of cytokines and/or intracellular transcription factors (34). The expression of select chemokine receptors helps distinguish Th17 cells as those that express CCR6 (10, 12) and pre-Th1 cells that express CXCR3 (42), each of which play a crucial role in the regulation of immune response in different compartments of lymphoid tissues (16, 34, 42). Recent studies have also demonstrated differences in the relative susceptibilities of these CD4+ T cells subsets to HIV (10) and preferential infection and depletion of Th17 cells and α4β7hi-expressing CD4+ T cells in HIV-1 and SIV infections (3, 39). Accordingly, we hypothesized that differences in the outcome of infection with SIVmac239 and Δ5G could be secondary to differential targeting of subsets of CD4+ T cells particularly during primary infection.
In the present study, we show that an apparent paucity of SIV infection in secondary lymphoid tissues in the animals infected with a deglycosylated mutant Δ5G accounts for its apathogenicity. In contrast, a fully N-glycosylated SIVmac239 is required for productive infection and a selective depletion of the CD4+ cell subset (transitional memory-Th1) residing within the SLT inductive tissues. We also show that an intense viral replication that is confined to the small intestinal tissues during acute infection in the Δ5G-infected animals is accompanied by the depletion of the Th17 CD4+ T cell subset, which might contribute to the induction of protective immune responses.
MATERIALS AND METHODS
Viruses.
Δ5G was derived by site-directed mutagenesis of an SIVmac239 infectious DNA clone in which the asparagine residues for the N glycosylation sites at aa 79, 146, 171, 460, and 479 in gp120 were replaced with glutamine residues (29). The stocks of SIVmac239 and Δ5G were prepared by DNA transfection of the respective proviral DNAs into 293T cells. These virus stocks were propagated in phytohemagglutinin-stimulated peripheral blood mononuclear cells (PBMCs) from rhesus macaques as previously described (28, 52). Nucleotide sequence of Δ5G was confirmed that no mutation occurred in env gene during the preparation of the virus stock. Since suboptimal nucleotides in SIVmac239 were reported (2), we examined the corresponding mutations in Δ5G and detected no mutation except the one in the Lys-tRNA primary binding site in the 5′ long terminal repeat.
Animals and ethics statement.
Juvenile rhesus macaques originating from Burma were used following negative results of screening for SIV, simian T-cell lymphotropic virus, herpes B virus, and type D retrovirus infection prior to study inception. All animals were housed in individual cages and maintained according to the rules and guidelines for experimental animal welfare as outlined by the National Institute of Infectious Diseases and National Institute of Biomedical Innovation (Japan). The full details of the study were approved by the National Institute of Infectious Diseases Institutional Animal Care and Use Committee in accordance with the recommendations of the Weatherall report. Early endpoints were adopted including frequent monitoring of viral loads and immunological parameters, and humane euthanasia was conducted once any manifestation of clinical AIDS or signs of fatal disease were noted.
Infection and collection of tissue specimens.
A total of 16 rhesus macaques were used for the present study. Seven animals each group were intravenously inoculated with 300 50% tissue culture infective doses of the deglycosylation mutant Δ5G and SIVmac239, respectively. Two uninfected animals served as controls. To examine SIV replication in tissues and the impact on the immune system during primary infection, blood samples were collected at regular time intervals until necropsy, namely, at 3, 7, 9, 12, 14, and 21 days postinfection (p.i.). Tissues for routine immunohistological examination and tissues for detailed analyses of SIV infection which included a variety of lymphoid tissues (the axillary, cervical, hilar, iliac, inguinal, mesenteric, pancreatic, and submandibular LN, the tonsils, and the spleen) and intestinal tissues (entire small intestines and colon) were collected. Δ5G-infected animals were euthanized at different times postinfection: Mm0306 and Mm0401 at 9 days p.i., Mm0624 and Mm0625 at 12 days p.i., Mm0402 at 14 days p.i., and Mm0609 and Mm0611 at 21 days p.i. SIVmac239-infected animals were also euthanized at different times postinfection: Mm0403 at 7 days p.i., Mm0406 and Mm0519 at 9 days p.i., Mm0405 at 12 days p.i., Mm9212 at 14 days p.i., and Mm0610 and Mm0612 at 21 days p.i. Animals Mm0308 and Mm0407 were used as uninfected controls.
Preparation of immune cells from tissues.
Immune cells from intestinal jejunum, ileum, and colon were isolated by a method described previously (58) with some modifications. Briefly, excised pieces of tissue were treated with 5 mM EDTA and Hanks balanced salt solution to remove intestinal epithelial lymphocytes and then digested with collagenase and DNase I to isolate leukocytes in the LP. These leukocytes were purified by Percoll density gradient. Immune cells from the spleen and mesenteric LN were isolated as follows: the tissue was gently minced and then passed through a 19-gauge needle. The cells obtained were purified by using a Ficoll-Paque density gradient. Residual red blood cells were removed after treatment with NH4Cl solution. Isolated immune cells were cryopreserved in a liquid nitrogen storage apparatus.
Flow cytometry and cell sorting.
Peripheral blood or thawed cryopreserved immune cells prepared from the intestinal mucosa or lymphatic tissues were used for flow cytometric analysis. Phenotypic analyses of immune cells were performed using a FACSCalibur (BD) with fluorescein isothiocyanate (FITC), phycoerythrin (PE), PerCP5.5, and allophycocyanin (APC) as fluorescent probes for blood. FACS Aria (BD) was used with FITC/Alexa Fluor 488, PE, ECD, PerCP5.5, PE-Cy7, APC, Alexa Fluor 700, APC-Cy7, Pacific Blue, and AmCyan as 10 fluorescent probes for the tissue samples. Fluorochrome-conjugated monoclonal antibodies (MAbs) used were as follows: MAbs L200 (CD4), SP34-2 (CD3), SK1 (CD8), DX2 (CD95), L27 (CD20), 3A9 (CCR5), 1C6/CXCR3 (CXCR3), 11A9 (CCR6), and L243 (MHC-II DR) were obtained from BD. MAb 150503 (CCR7) and MAb CD28.2 (CD28) were obtained from R&D and Beckman Coulter, respectively. Flow data were analyzed using FlowJo software (v9.3.2; Tree Star). Stained cells were sorted on a FACS Aria.
Immunohistological detection of SIV-infected cells.
Collected tissues were fixed with 4% paraformaldehyde for 24 h and then embedded in paraffin. SIV-infected cells were detected by double immunostaining methods described previously (51). Briefly, SIV viral proteins were detected with antibodies against p27 Gag (rabbit polyclonal, provided by T. Sata) or Nef (MAb; FIT Biotech). T cells, B cells, and macrophages were detected with antibodies against CD3 (rabbit polyclonal antibody; Dako), CD20 (L26 MAb; Dako), and CD68 (KP1 MAb; Dako), respectively. Sections were incubated with mouse IgG and rabbit IgG and then incubated with a peroxidase-conjugated anti-rabbit IgG polymer (Envision; Dako) and a biotin-conjugated anti-mouse IgG (Dako). SIV proteins and cells were visualized with fluorescein tyramide (Molecular Probes) and Alexa 594-conjugated streptavidin (Molecular Probes).
Viral load measurements.
The level of SIV infection was monitored by measuring the plasma viral RNA load using a highly sensitive quantitative real-time reverse transcription-PCR (RT-PCR). Viral RNAs from plasma samples and PBMCs were isolated by using a MagNA PureCompact nucleic acid isolation kit (Roche Diagnostics). Real-time RT-PCR was performed using the QuantiTec probe RT-PCR kit (Qiagen) and an SDS7000 sequence detection system (Applied Biosystems). For viral DNA measurement, PBMCs or sorted cells from tissues were suspended in lysis buffer (10 mM Tris, 0.5% NP-40, 0.5% Tween20) with proteinase K (0.1 mg/ml), incubated at 55°C for 1 h, and then heat inactivated at 95°C for 5 min. Real-time PCR was performed by using the qPCR Mastermix Plus kit (Eurogentec) and the SDS7000 sequence detection system. To detect SIVmac239 gag, the following primers and probe sets were synthesized: SIVmac239-specific gag primers 5′-GCAGAGGAGGAAATTACCCAGTAC-3′ and 5′-CAATTTTACCCAGGCATTTAATGTT-3′ and probe 5′-FAM-TGTCCACCTGCCATTAAGTCCCGA-TAMRA-3′. The detection sensitivity of plasma viral RNA by this method was calculated to be 100 viral RNA copies per ml of plasma. Cell number was validated by detection of the cellular interleukin-4 (IL-4) sequence with the rhesus IL-4-specific primers 5′-TGTGCTCCGGCAGTTCTACA-3′ and 5′-CCGTTTCAGGAATCGGATCA-3′ and probe 5′-FAM-TGCACAGCAGTTCCACAGGCACAAG-TAMRA-3′.
Statistical analysis.
Statistical analysis was performed by the Mann-Whitney test by using GraphPad Prism 4.0 software, with P values as indicated on the figures. Differences were considered to be statistically significant when P values were <0.05.
RESULTS
No detectable depletion of CCR5+ CD4+ T cells in Δ5G infection during acute infection despite similar plasma viral loads compared to SIVmac239 infection.
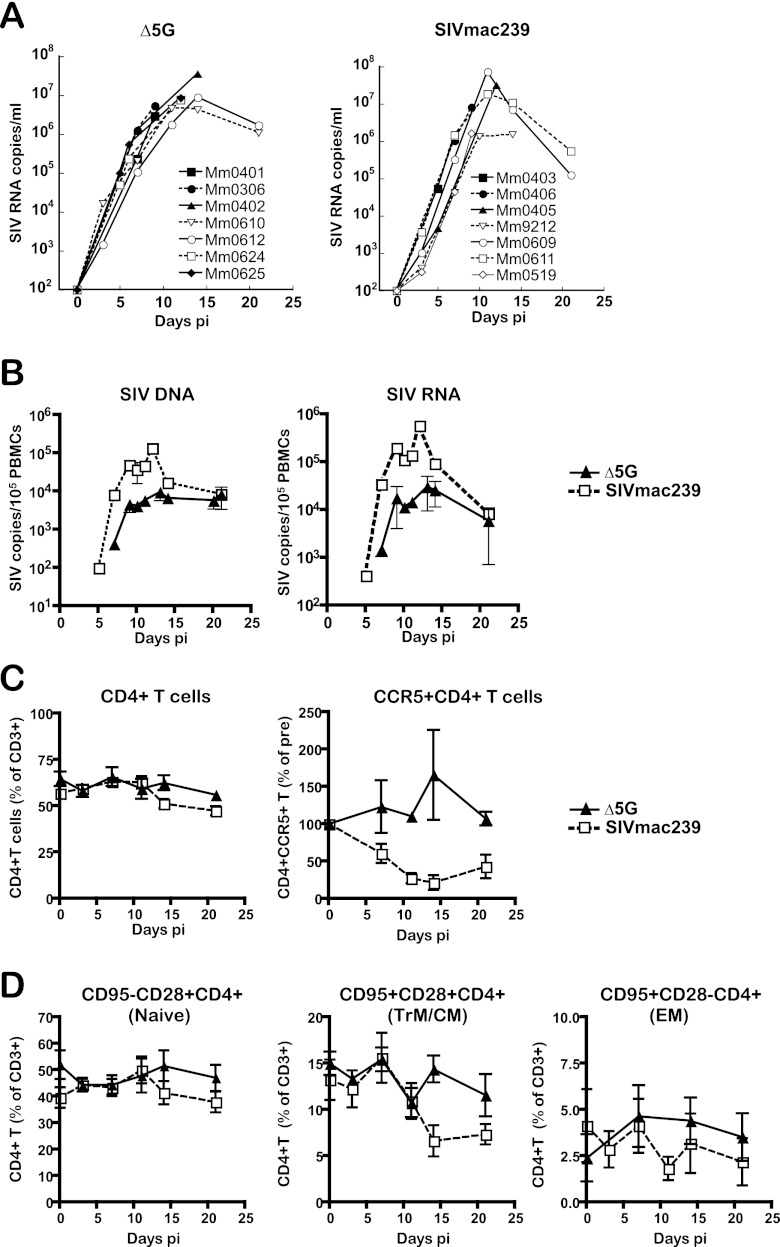
Fourteen rhesus macaques divided into two groups of seven animals each were intravenously infected with Δ5G or SIVmac239. Blood specimens were collected at 2- to 3-day intervals from Δ5G- and SIVmac239-infected animals until euthanasia at 9, 12, 14, or 21 days p.i. and at 7, 9, 12, 14, or 21 days p.i., respectively. Potential target tissues of SIV infection such as a variety of SLT, GIT, and others were collected at necropsy. Two uninfected macaques served as naive controls. First, we examined plasma VL by a quantitative real-time PCR. Consistent with our previous studies (29, 50), plasma VL of Δ5G-infected animals were essentially similar to those of SIVmac239-infected animals during the primary infection, with a peak plasma VL reaching over 107 copies/ml at about 12 days p.i. (Fig. 1A). In contrast to the plasma VL, PBMC cellular VL (SIV DNA and RNA) were significantly lower during acute infection with Δ5G than SIVmac239 (P < 0.01) (Fig. 1B). The mean value of peak PBMC SIV DNA load of Δ5G- and SIVmac239-infected animals measured at 12 to 13 days p.i. was 9.7 × 103 and 1.4 × 105copies/105 cells, respectively (ratio of 1:14). The mean values of the peak PBMC SIV RNA loads of Δ5G- and SIVmac239-infected animals at 12 to 13 days p.i. were 3.1 × 104 and 5.9 × 105 copies/105 cells, respectively (1:19).
Fig 1.
Viral loads and CD4+ T cell subsets in the peripheral blood. Seven animals each were infected intravenously with Δ5G and SIVmac239, respectively, and the blood specimens were examined for the following parameters. (A) Plasma viral loads. SIV RNA copies/ml in plasma from each animal were determined by real-time PCR with a probe and primers specific for Gag sequences of SIVmac239 (B) PBMC viral loads. SIV DNA and RNA copies in PBMCs were determined by real-time PCR with the same probe and primers as for the plasma vial loads. The results were plotted as mean values with the SEM. (C) Frequencies of CD4+ T cells and CD4+ CCR5+ T cells. The frequencies of CD4+ T cells and CD4+ CCR5+ T within gated populations of CD3+ T cells were examined by flow cytometry. The percentages of CD4+ T cells among the gated CD3+ T cells and the relative percentage to baseline (preinfection) for CD4+ CCR5+ T cells are shown with mean values with the SEM. (D) Naive, central memory (CM)/transitional memory (TrM), and effector memory (EM) CD4+ T cells. Naive (CD28+ CD95−), CM/TrM (CD28+ CD95+), and EM (CD28− CD95+) subsets in CD4+ T cells were examined by flow cytometry and the percentages (mean and SEM) within gated CD3+ T cells are shown.
Since the CCR5+ CD4+ T cells in PBMCs are preferentially depleted during primary infection of both rhesus macaques and humans infected with SIV and HIV-1, respectively (23, 24, 38, 57), we examined the frequencies of this subset. As seen in Fig. 1C, while the overall frequency of CD4+ T cells decreased only slightly, the levels of the CCR5+ CD4+ T cell subset in the SIVmac239-infected animals were preferentially decreased to ∼10% of the preinfection levels at 14 days p.i. In contrast, the frequencies of the same CCR5+ CD4+ T cell subset and CD4+ T cells in the Δ5G-infected animals did not decline up to 21 days p.i. (Fig. 1C). Next, we examined the frequencies of naive/memory subsets of CD4+ T cells such as CD95− CD28+ (naive), CD95− CD28+ (central memory and transitional memory [CM and TrM]) and CD95+ CD28− (effector memory [EM]) cells based on markers utilized for subset identification elsewhere (33). Whereas modest decreases were observed in naive and EM subsets of CD4+ T cells, major decreases were noted in the CM/TrM subset at 12 days p.i. and thereafter in SIVmac239-infected animals (Fig. 1D), but not in Δ5G-infected animals, which is similar to the data for the CCR5+ CD4+ T cell subset.
These results indicate that, although the plasma VL shows a similar SIV replication profile during primary infection between the two groups of animals, the >1-log-lower cellular VL in PBMCs and the preservation of CCR5+ and CM/TrM subsets of CD4+ T cells noted in the Δ5G-infected animals suggests that the two viruses most likely replicate/deplete in distinct subsets of cells within distinct tissues.
Differences in cellular VL in PBMCs correlate with different tissue localization of SIVmac239 compared to Δ5G.
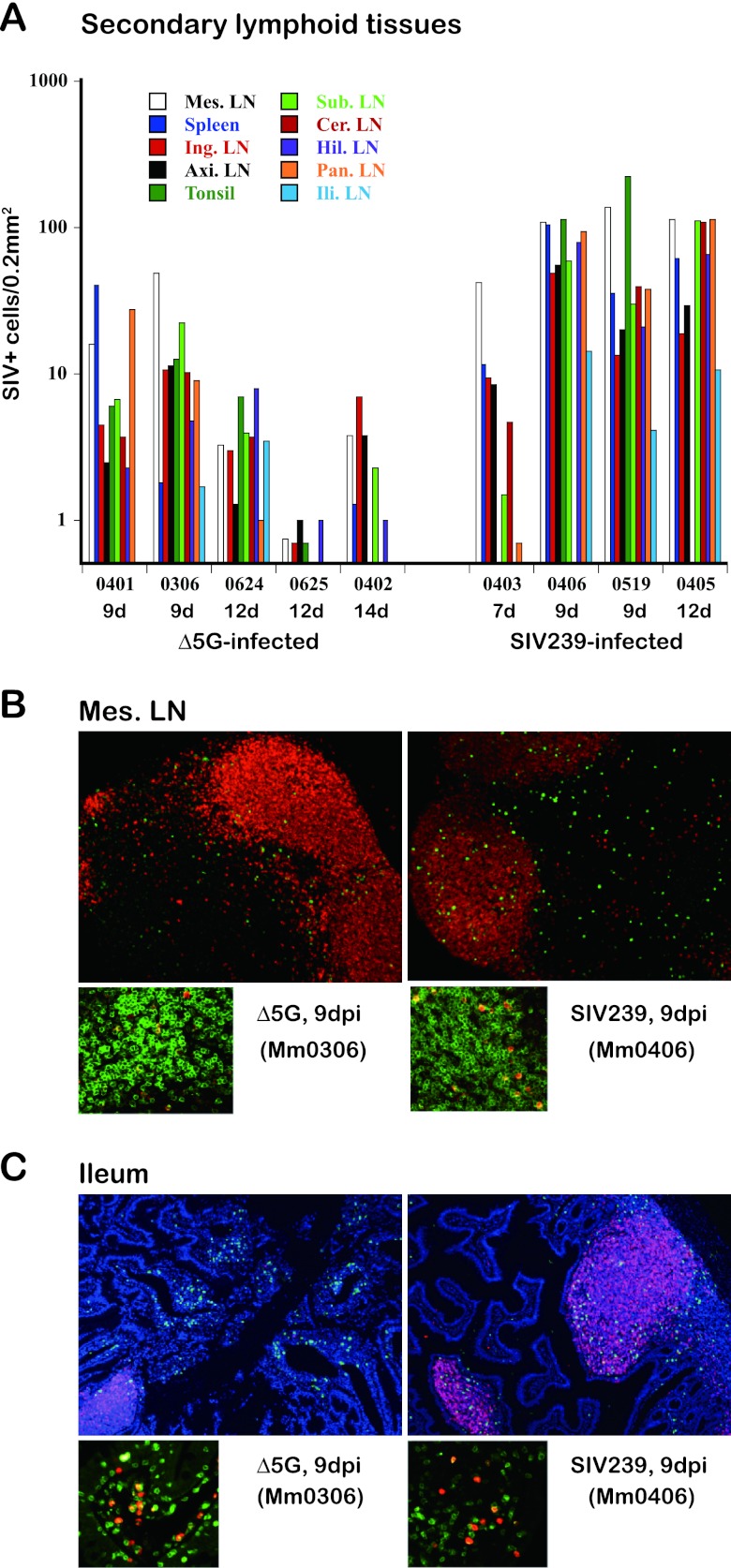
In efforts to identify the potential tissues that serve as the primary source for SIV replication, we examined the tissues that are typically reported to be associated with SIV infection (21, 23, 54). These tissues were individually collected at necropsy, and sections were examined immunohistologically for the frequency of SIV-infected cells. Since a vast majority of peripheral blood cells are derived from SLT, we first examined SIV infection in SLT such as the axillary, cervical, hilar, iliac, inguinal, mesenteric, pancreatic, and submandibular LN, the tonsils, and the spleen. As noted above, whereas SIV plasma VL were similar at 9 days p.i. in the two groups, these VL were subsequently higher at 12 and 14 days p.i. for SIVmac239-infected animals over Δ5G-infected animals. In contrast, analysis of all SLT tissue sites from day 9 until 14 p.i. showed markedly lower frequencies of SIV-infected cells in the Δ5G-infected versus SIVmac239-infected animals (Fig. 2A). Thus, by day 9 p.i. the levels of SIV+ cell density were ∼8-fold lower in Δ5G-infected versus SIVmac239-infected tissues, and by day 12 to 14 p.i. the levels had decreased 33-fold (Table 1). Thus, these results indicate that the differences in SIV infection of SLT between the two groups correlate with the differences seen in SIV levels in PBMCs.
Fig 2.
Immunohistological detection of SIV+ cells in secondary lymphoid and intestinal mucosal tissues. (A) SIV-infected cells in secondary lymphoid tissues. SIV-infected cells in the secondary lymphoid tissues (variety of LN and spleen) of Δ5G- or SIVmac239-infected animals were examined by immunohistochemical staining with anti-SIV antibodies against Gag or Nef. Total numbers of SIV+ cells detected in the tissue sections were plotted. Since very few SIV+ cells were detected in the given tissues from the infected animals at 21 days p.i., the results were not shown. (B and C) SIV-infected cells in the mesenteric LN and ileal tissues from the Δ5G- and SIVmac239-infected animals were examined, and the localization and identification of SIV+ cells were determined by double staining with cell-type-specific antibodies and SIV specific antibodies. (B) SIV+ cells in mesenteric LN. Most of the SIV+ cells (green) were localized at the boundary of the B cell (red) and T cell areas in the Δ5G-infected animal at 9 days p.i. (left). SIV+ cells were distributed in both B and T cell areas in tissues from the SIVmac239-infected animals (right). Most of the SIV+ cells (red) in Δ5G- and SIVmac239-infected animals were merged with the CD3+ T cells (green) (lower panels). (C) Localization of SIV+ cells in the ileum. Foci of SIV+ cells (green) were detected in the LP of the Δ5G-infected animals at 9 days p.i., whereas a few SIV+ cells were detected in the SLF where B cells (red) accumulated (left). In contrast, many SIV+ cells were found in the SLF of the SIVmac239-infected animals at 9 days p.i., whereas some, but not many, SIV+ cells were also detected in the LP (right). Most of the SIV+ cells (red) in the Δ5G- and SIVmac239-infected animals were merged with the CD3+ T cells (green) (lower panels).
Table 1.
SIV+ cells in secondary lymphatic tissues
| Animal no. | Virus | Days p.i. | No. of SIV+ cells/0.2 mm2a |
Median | Meanb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLN | Spleen | Tonsil | INLN | ALN | SLN | CLN | HLN | PLN | ILLN | |||||
| Mm0401 | Δ5G | 9 | 16.0 | 40.3 | 6.0 | 4.5 | 2.5 | 6.8 | 3.7 | 2.3 | 27.8 | 0 | 5.3 | 7.9 |
| Mm0306 | Δ5G | 9 | 49.0 | 1.8 | 12.7 | 10.6 | 11.5 | 22.2 | 10.3 | 4.8 | 9.0 | 1.7 | 10.5 | |
| Mm0624 | Δ5G | 12 | 3.3 | 0 | 7 | 3 | 1.3 | 4 | 3.7 | 8 | 1 | 3.5 | 3.4 | 1.8 |
| Mm0625 | Δ5G | 12 | 0.75 | 0 | 0.7 | 0.7 | 1 | 0 | NA | 1 | 0 | 0 | 0.7 | |
| Mm0402 | Δ5G | 14 | 3.8 | 1.3 | 0 | 7.0 | 3.8 | 2.3 | 0.5 | 1.0 | NA | 0.4 | 1.3 | |
| Mm0403 | SIVmac239 | 7 | 42.1 | 11.7 | 0 | 9.5 | 8.5 | 1.5 | 4.7 | 0.3 | 0.7 | 0 | 3.1 | |
| Mm0406 | SIVmac239 | 9 | 110.2 | 104.6 | 113.7 | 48.8 | 55.2 | 59.4 | NA | 78.8 | 93.8 | 14.3 | 78.8 | 59.3 |
| Mm0519 | SIVmac239 | 9 | 137.0 | 36.0 | 224.0 | 13.5 | 20.0 | 30.0 | 40.0 | 21.0 | 38.0 | 4.1 | 33.0 | |
| Mm0405 | SIVmac239 | 12 | 112.8 | 62.2 | NA | 18.8 | 29.4 | 110.8 | 108.3 | 66.2 | 114.5 | 10.8 | 66.2 | |
MLN, mesenteric lymph nodes; INLN, inguinal lymph nodes; ALN, axillary lymph nodes; SLN, submandibular lymph nodes; CLN, cervical lymph nodes; HLN, hilar lymph nodes; PLN, pancreatic lymph nodes; ILLN, iliac lymph nodes. NA, not assayed.
The means of animals Mm0401 and Mm0306 (7.9), Mm0624, Mm0625, and Mm0402 (1.8), and Mm0406, Mm0519, and Mm0405 (59.3) are indicated.
Abundance of SIV-infected cells within intestinal tissues of both Δ5G- and SIVmac239-infected animals but localized differently within the GIT (LP versus solitary lymphoid follicles).
The GIT has been shown to serve as a primary target tissue of HIV/SIV infection and replication during primary and chronic-infection (4, 21, 23, 24, 55). Accordingly, we examined SIV infection in the jejunum, ileum, and colon of each SIV-infected animal. Our initial analyses clearly indicated that the SIV+ cells were not evenly distributed in the tissues but instead accumulated as localized foci to areas such as the LP or solitary lymphoid follicles (SLF), prompting a need for detailed studies. The frequencies of SIV+ cells were thus enumerated within sectioned tissues prepared from the LP and SLF (Table 2). Levels of SIV+ cells in the ileum and jejunum peaked at 9 days p.i. and were comparable between the two groups, indicating that the intestinal tissue was indeed a primary site for productive SIV replication, a finding consistent with previous reports. However, SIV+ cells were differently compartmentalized at 9 days p.i. between Δ5G- and SIVmac239-infected animals. The majority of SIV+ cells were detected in the LP for Δ5G-infected animals and in the SLF for the SIVmac239-infected animals (Table 2 and Fig. 2C). Notably, the focus of SIV+ cells was not detected in LP of the jejunum and ileum from the Δ5G-infected animals at 12 and 14 days p.i. Differences in the distribution of SIV+ cells were also observed in sections of the colon tissue with massive SIV+ cells detected in only SIVmac239-infected animals but not in Δ5G-infected animals (Table 2). These results clearly showed that in addition to markedly lower frequencies of SIV+ cells in tissues of Δ5G-infected animals relative to SIVmac239-infected animals, whereas Δ5G and SIVmac239 infections were both targeted to the intestinal organs, they were targeted to distinct compartments.
Table 2.
SIV+ cells in intestinal tissuesa
| Animal (dpi) | Tissue | Lymphoid follicles |
Lamina propria |
||
|---|---|---|---|---|---|
| No. of focia | No. of SIV+ cells | No. of foci | No. of SIV+ cells | ||
| Δ5G-infected animals | |||||
| Mm0401 (9) | Jejunum | ND | <5 | ND | <5 |
| Ileum | 1 | 152 | 3 | 527 | |
| Colon | ND | <5 | ND | <5 | |
| Mm0306 (9) | Jejunum | 2 | 611 | 2 | 340 |
| Ileum | ND | <5 | 12 | 3,213 | |
| Colon | ND | <5 | ND | <5 | |
| Mm0624 (12) | Jejunum | 2 | 151 | ND | <5 |
| Ileum | 2 | 156 | ND | <5 | |
| Colon | 1 | 16 | ND | <5 | |
| Mm0625 (12) | Jejunum | ND | <5 | ND | <5 |
| Ileum | ND | <5 | ND | <5 | |
| Colon | ND | <5 | ND | <5 | |
| Mm0402 (14) | Jejunum | ND | <5 | ND | <5 |
| Ileum | 1 | 91 | ND | <5 | |
| Colon | ND | <5 | ND | <5 | |
| SIVmac239-infected animals | |||||
| Mm0403 (7) | Jejunum | ND | <5 | ND | <5 |
| Ileum | 10 | 1,348 | ND | <5 | |
| Colon | ND | <5 | ND | <5 | |
| Mm0406 (9) | Jejunum | ND | <5 | ND | <5 |
| Ileum | 9 | 3,081 | ND | <5 | |
| Colon | 8 | 2,816 | ND | <5 | |
| Mm0519 (12) | Jejunum | 2 | 488 | ND | <5 |
| Ileum | 5 | 2,166 | 2 | 131 | |
| Colon | 17 | 3,666 | ND | <5 | |
| Mm0405 (12) | Jejunum | ND | <5 | 3 | 541 |
| Ileum | 17 | 1,703 | 3 | 317 | |
| Colon | 14 | 1,259 | ND | <5 | |
SIV+ cells accumulated as a focus; thus, the numbers of foci detected in the lymphoid follicle or lamina propria are shown. dpi, days postinfection. ND, not detected.
Infection of CD4+ T cells is preferentially localized to the SLT for SIVmac239 infection and to the LP of the small intestine for Δ5G infection.
SIV-infected cells in the mesenteric LN and intestinal ileum tissue were identified using standard immunohistological techniques using antibodies specific for SIV (Gag or Nef), B cells (CD20), and T cells (CD3) (Fig. 2B and C). In addition to the quantitative differences in SIV infection in SLT described above, we also observed histological differences in the distribution of SIV+ cells in the mesenteric LN between the two groups. Thus, whereas many SIV+ cells (green) were detected in both the B cell region (red) and the T cell region (unstained) in SIVmac239-infected animals, SIV-infected cells were predominantly localized to the boundary between T and B cell areas in tissues from the Δ5G-infected animals (Fig. 2B, upper panels). The merging of the pictures (for SIV+ cells and for CD3+ cells) indicate that most SIV+ cells were CD3+, suggesting that CD4+ T cells are likely the primary target cells for both viruses (Fig. 2B, lower panels). However, whereas the SIV+ cells appeared primarily localized to the LP in ileum tissues of the Δ5G-infected animals (Fig. 2C, left), SIV+ cells in the ileum of SIVmac239-infected monkeys were predominantly detected in SLF, as identified by the finding of accumulated B cells (Fig. 2C, right). Irrespective of the different localizations, most of the SIV+ cells were CD4+ T cells, similar to the mesenteric LN (Fig. 2C, lower panels).
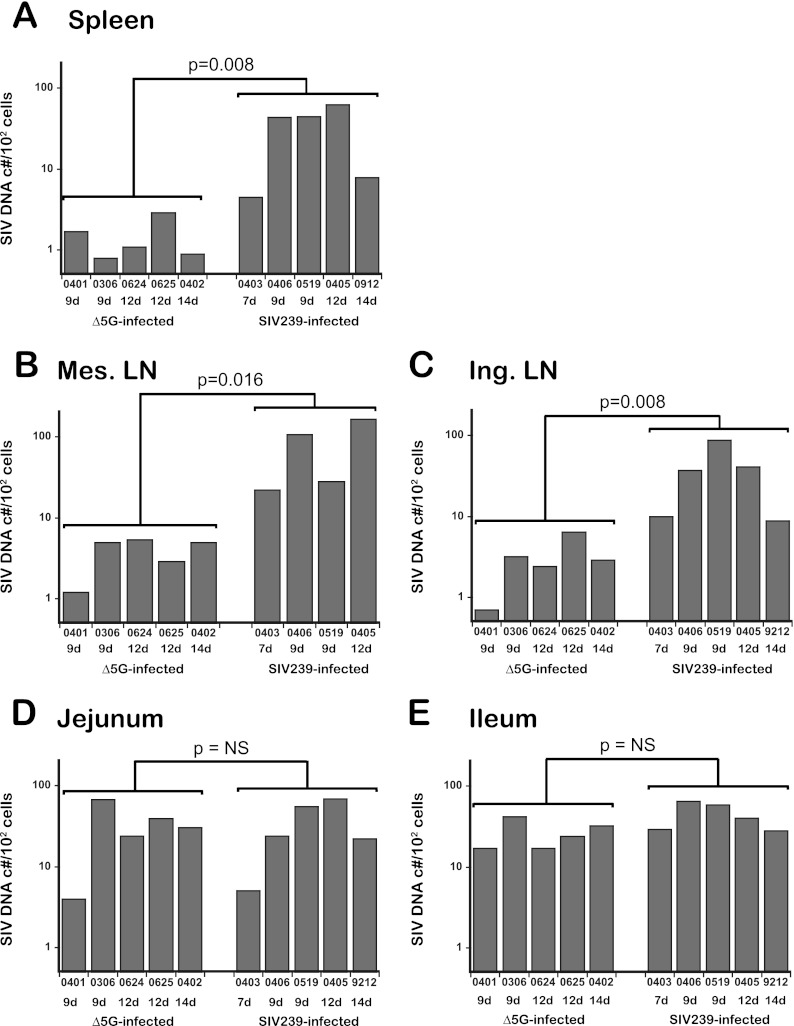
Since CD4+ T cells were the major infected cells in both Δ5G- and SIVmac239-infected animals (Fig. 2B and C), CD4+ T cells isolated from the tissues were sorted by flow cytometry, and the levels of SIV DNA were determined by a real-time PCR method. The cellular VL in spleen, mesenteric LN, and inguinal LN from Δ5G-infected animals during the primary infection (9 to 14 days p.i.) were statistically lower than those in similar tissues from the SIVmac239-infected animals (7 to 14 days p.i.) (Fig. 3A, B, and C). Thus, the means ± the standard errors of the mean (SEM) SIV DNA copies/102 cells in spleens from the Δ5G and SIVmac239 infections were 32 ± 11 and 1.5 ± 0.39, respectively. The levels in the mesenteric LN were 80 ± 34 and 3.9 ± 0.79 (mean ± the SEM) SIV DNA copies/102 cells for tissues from the Δ5G- and SIVmac239-infected animals, respectively. Finally, the mean ± the SEM SIV DNA copies/102 cells in inguinal LN for Δ5G- and SIVmac239-infected animals were 37 ± 14 and 3.1 ± 0.93, respectively. Thus, the ratios of the cellular VL for Δ5G versus SIVmac239-infected animals in the spleen, mesenteric LN, and inguinal LN tissues were approximately 1:22, 1:21, and 1:12, respectively, indicating that the differences in SIV infection in SLT were primarily due to differences in the frequencies of infected CD4+ T cells.
Fig 3.
SIV DNA loads in CD4+ T cells from intestinal and secondary lymphoid tissues. SIV DNA levels (SIV DNA copies/102 cells) in CD4+ T cells from the spleen (A), mesenteric LN (B), and inguinal LN (C) as representative secondary lymphoid tissues and from intestinal ileal (D) and jejunal (E) tissues from Δ5G- and SIVmac239-infected animals were determined by real-time PCR. The significance of differences in SIV DNA levels in CD4+ T cells between Δ5G- and SIVmac239-infected animals during the primary infection (7 to 14 days p.i. for SIVmac239-infected animals and 9 to 14 days p.i. for Δ5G-infected animals) was determined utilizing the unpaired Student t test. Differences were considered statistically significant when P values were <0.05. NS, not significant.
In the small intestinal tissues, the overall frequencies of SIV+ cells at day 9 p.i. were essentially similar for both Δ5G- and SIVmac239-infected animals (Table 2). Consistent with this observation, SIV DNA loads in CD4+ T cells in jejunum and ileum tissues were similar between the two groups (Fig. 3D and E), indicating that a robust SIV replication occurred in CD4+ T cells in these tissues from both groups at this early time point. However, as indicated in Fig. 2C, it is important to note that SIV-infected target cells were preferentially detected in LP for Δ5G- and SLF in SIVmac239-infected animals. Although LP is an effector site comprised predominantly of effector immune cells, SLF is an inductive site, consisting primarily of immune cells primed for an initiating adaptive immune response, also classified as SLT.
SIVmac239 infection preferentially depletes CXCR3+ CCR6− CCR5+ transitional memory (TrM) CD4+ T cells in SLT.
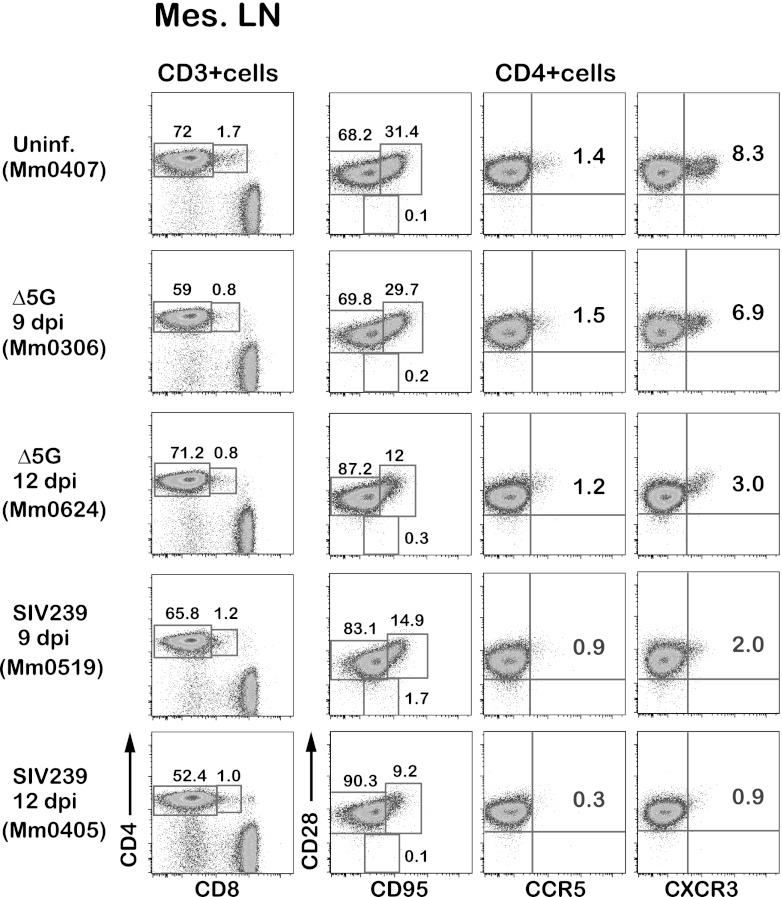
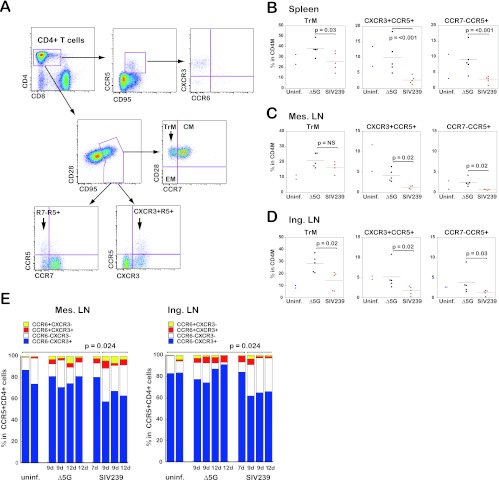
We found 1- to 2-log greater VL in CD4+ T cells in SLT of SIVmac239-infected animals compared to Δ5G-infected animals (Fig. 3A, B, and C). Since CCR5+ and CD28+ CD95+ (CM/TrM) subsets of CD4+ T cells were preferentially depleted in PBMCs during the primary infection with SIVmac239 but not Δ5G (Fig. 1C and D), we assumed that the depletion of these subsets should also occur in the SLT of the SIVmac239-infected animals but not those of Δ5G-infected animals. Accordingly, we analyzed phenotypes of CD4+ T cells in the spleen, mesenteric LN, and inguinal LN by polychromatic flow cytometry. As expected, the frequencies of CD28+ CD95+ and CCR5+ subsets in SIVmac239-infected animals at 9 and 12 days p.i. were lower than in Δ5G-infected and uninfected animals (Fig. 4). In addition, we noted a decrease in CXCR3+ CD4+ cells in SIVmac239-infected animals at 9 and 12 days p.i. Based on these results, we attempted to define phenotypes of CD4+ T cells by using cell surface markers that distinguish CM from TrM cells. Thus, the CD28+ CD95+ cells are divided into CCR7+ CD28+ (CM) and CCR7− CD28+ (TrM) cells (33) (Fig. 5A). Using such markers for classification, we examined the impact of SIV infection on CD4+ T cell subsets in isolated cells from SLT sites. Whereas no statistical differences between the two groups were observed in the frequencies of naive, CM, and EM subsets in CD3+ T cells (data not shown), the frequencies of TrM were significantly lower in the spleen and inguinal LN but not in the mesenteric LN from SIVmac239-infected animals as previously reported (33) (Fig. 5B, C, and D, left panels). Furthermore, we found a more marked difference in the depletion of CD4+ T subsets defined by the expression of the chemokine receptors such as CCR5, CXCR3, and CCR7 between the two groups. As seen in Fig. 5B, C, and D, middle panels, the numbers of CXCR3+ CCR5+ cells were significantly lower in the SIVmac239-infected animals than in Δ5G-infected animals in cells isolated from the spleen, inguinal LN, and mesenteric LN. Similar differences were observed in the CCR7− CCR5+ cells (Fig. 5B, C, and D, right panels). However, no significant difference was observed in the frequencies of CXCR3− CCR5+ cells and CCR7+ CCR5+ cells (data not shown). These results demonstrate that CXCR3+ CCR5+ TrM cells were preferentially depleted in the SIVmac239-infected animals but spared by the Δ5G infection.
Fig 4.
CD4+ T cell subsets in the mesenteric LN. Flow profiles of CD4+ T cell subsets from the mesenteric LN of Δ5G- and SIVmac239-infected animals at 9 and 12 days p.i. were examined compared to those from the uninfected animals. CD4+ T cells were gated into the naive/memory subsets by the expression of CD28/CD95, the CCR5+ cells by the expression of CD28/CCR5, and the CXCR3+ cells by the expression of CD28/CXCR3.
Fig 5.
CD4+ T cell subsets in spleen, mesenteric, and inguinal LN. (A) Gating strategies for CD4+ T cell subsets. (Top) CCR5+ CD4+ T cells were gated into four CXCR3/CCR6 subsets: CXCR3+ CCR6−, CXCR3− CCR6−, CXCR3+ CCR6+, and CXCR3− CCR6+ cell. (Middle) Gating for central memory (CM), transitional memory (TrM), and effector memory (EM) subsets. (Bottom left) Gating for CCR7− CCR5+ and CCR7− CCR5+ subsets. (Bottom right) Gating for CXCR3− CCR5+ and CXCR3+ CCR5+ subsets. The TrM, CXCR3+ CCR5+, and CCR7− CCR5+ subsets are indicated by arrows. (B, C, and D) Frequencies of the TrM, CXCR3+ CCR5+, and CCR7− CCR5+ subsets in CD4+ memory T cells from the spleen (B), mesenteric LN (C), and inguinal LN (D) were compared between Δ5G-infected animals at 9 to 14 days p.i. and SIVmac239-infected animals at 7 to 14 days p.i. The significance of differences was determined by using the unpaired Student t test. The frequencies in the uninfected animals are shown as references. The differences implicate background immune responses in the corresponding animal. (E) Percentages of four CXCR3/CCR6 subsets in CCR5+ CD4+ T cells from the mesenteric LN (left) and inguinal LN (right) of Δ5G- and SIVmac239-infected animals. The percentages of the CXCR3+ CCR6− cells from these tissues of the SIVmac239-infected animals at 9 to 12 days p.i. were significantly lower than those in the uninfected animals, Δ5G-infected animals, and SIVmac239-infected animals at 7 days p.i.
Several reports have previously documented that heterogeneity exists among CD4+ effector T cells based on the expression of cytokines, transcription factors, and chemokine receptors (34, 42). Thus, the cell surface expression of CCR6+, CCR4+, CXCR3+ CCR6+, and CXCR3+ CCR6− cells are defined as representing Th17, Th2, Th1/Th17, and Th1, respectively (1, 10, 12). Accordingly, we further examined the phenotype of the depleted CD4+ T cell subset in the SLT from SIVmac239-infected animals. As shown in Fig. 5A (upper panels), while the CXCR3+ CCR6− cells were predominant among CCR5+ CD4+ T cells in the mesenteric and inguinal LN from uninfected and Δ5G-infected animals, the frequencies of this subset were significantly (P = 0.024) lower in CCR5+ CD4+ cells in the animals that exhibited high levels of SIV infection in SLT (Fig. 5E) from the SIVmac239-infected animals at 9 and 12 days p.i. (Fig. 2A and 3A to C). These results demonstrate that SIVmac239 preferentially replicates and depletes CXCR3+ CCR6− CCR5+ TrM cells (TrM-Th1 cells) in the SLT during primary infection.
Emergence and depletion of the CXCR3− CCR6+ CCR5+ EM CD4+ T cell subset occur in small intestinal tissues from Δ5G-infected animals.
As seen in Fig. 2C, SIV-infected cells were localized to different areas within the LP and SLF of the intestinal ileum, jejunum, and colon from the two groups of animals (Table 2 and Fig. 2C). Such differences suggest that distinct subsets of CD4+ T cells were infected by the two viruses in vivo. In general, CD4+ T cells in the GIT contain higher proportions of CD4+ CD8+ T cells compared to those in the SLT (Fig. 4 and 6). Thus, we examined CD4+ T cell subsets among the CD4+ CD8+ T cells and CD4+ CD8− T cells, respectively (Fig. 6A). Although intestinal CD4+ CD8+ T cells are a major early target during primary SIV infection (57) and are highly activated memory cells (35), we found a markedly high frequency of CD95+ CD28− (EM) cells among the CD4+ CD8+ cells from the Δ5G-infected animals at 9 days p.i. compared to that of uninfected animals (Fig. 6A). Interestingly, this CD28− subset contained higher frequencies of CCR5+ cells than the uninfected controls, and a majority of the CCR5+ cells were CXCR3− CCR6+ (Fig. 6A). However, a majority of the CCR5+ subset among the CD4+ CD8− T cells were CXCR3+ CCR6− TrM (CCR7− CD28+ CD95+) cells similar to CCR5+ cells found in the SLT (Fig. 5A and 6A). Based on all of these findings, we found two different major CCR5+ CD4+ T cells in the ileal (Fig. 6A) and jejunal (data not shown) tissues consisting of CXCR3− CCR6+ EM and CXCR3+ CCR6− TrM within the CD4+ CD8+ T cells and CD4+ CD8− T cells, respectively.
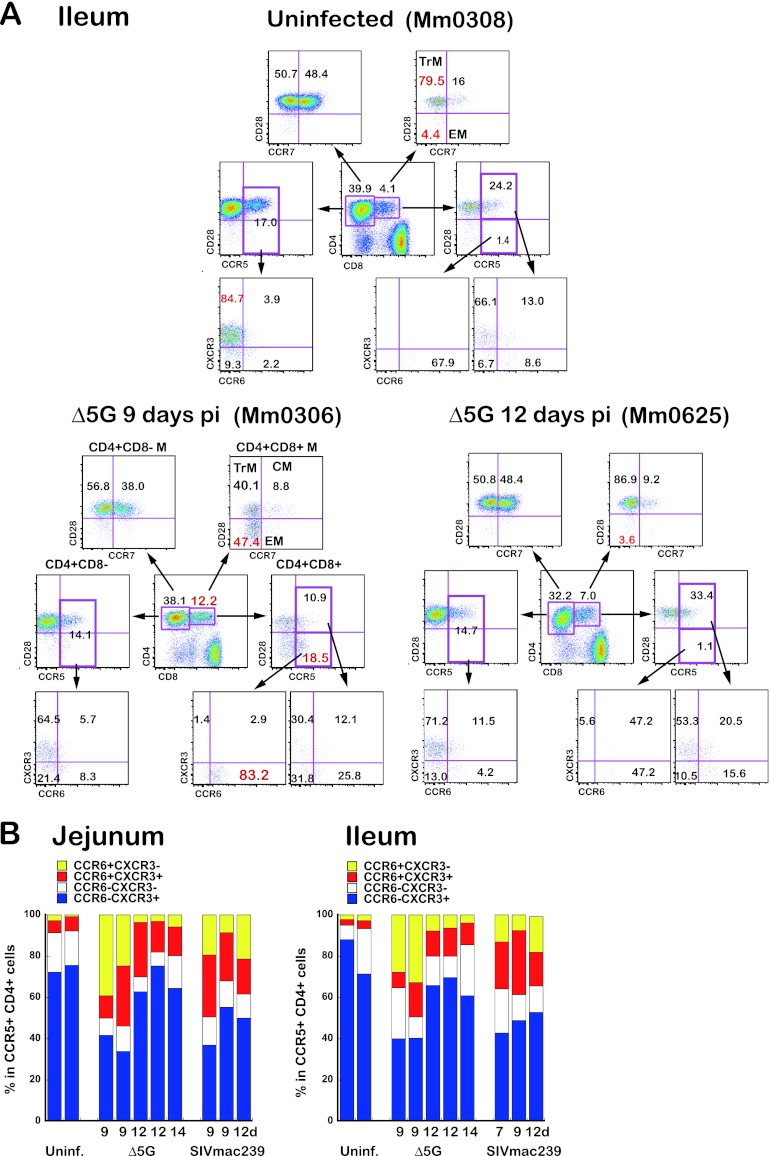
Fig 6.
CD4+ T cell subsets in the small intestinal tissues. (A) Flow profiles of CD4+ T cell subsets in the ileal tissues from an uninfected animal (upper panel), a Δ5G-infected animal at 9 days p.i. (lower left), and a Δ5G-infected animal at 12 days p.i. (lower right). The CD4+ CD8− T cells and CD4+ CD8+ T cells were gated as indicated. (Top) CM, TrM, and EM subsets. (Middle) CCR5+ TrM and CCR5+ EM subsets. (Bottom) CXCR3/CCR6 subsets in CCR5+ cells. (B) Frequencies of CXCR3/CCR6 subsets in CCR5+ CD4+ T cells from jejunal tissues (left) and ileal tissues (right) from uninfected animals, Δ5G-infected animals, and SIVmac239-infected animals. The flow analysis was based on the gating strategy shown in Fig. 5A (top profiles). Although the CXCR3+ CCR6− subset (blue) was predominant in uninfected animals, a remarkably higher CXCR3− CCR6+ subset (yellow) from these tissues of Δ5G-infected animals at 9 days p.i. was noted.
The significantly higher frequencies of EM T cells among the CD4+ CD8+ T cells in Δ5G-infected animals noted at 9 days p.i. relative to uninfected animals were transient and returned to the levels seen in the uninfected animals by day 12 p.i. (Fig. 6A) Interestingly, the differences in the frequencies of this CD4+ T cell subset were correlated with the differences in SIV infection in the intestinal tissues (Table 2). Thus, we assumed that the frequencies of CCR6+ CXCR3− CCR5+ cells might be related to the level of SIV infection in the tissues. Flow-based studies using the gating strategy shown in Fig. 5A (upper panels) were conducted to examine the frequencies of CCR6+ CXCR3−, CCR6+ CXCR3+, CCR6− CXCR3+, and CCR6− CXCR3− cells among the CCR5+ CD4+ T cells in single cells isolated from the ileal and jejunal tissues from the Δ5G- and SIVmac239-infected animals during the primary infection. Whereas the frequencies of CCR6+ CXCR3− cells were very low in the tissues from uninfected animals (<2%), the frequencies of this subset were significantly higher (20 to 40%) from tissues of the Δ5G-infected animals at 9 days p.i. (Fig. 6B) but decreased (3 to 8%) by 12 days p.i. In contrast, whereas the frequency of CCR6+ CXCR3− cells from SIVmac239-infected animals at 9 days p.i. was also higher (7 to 22%), this elevated frequency remained higher in tissues at 12 days p.i. relative to uninfected animals. The frequency of this subset was correlated with SIV infection. Several studies have shown that the CCR6+ CXCR3− CD4+ T cells comprise the Th17 subset of cells (1, 10, 12), which preferentially localize to the small intestine (8, 60), and that the Th17 cells are the major target of HIV infection cells in the GIT (3, 27). Taken together, we believe Δ5G preferentially replicates within Th17 cells that were mobilized by SIV infection to the LP of small intestinal tissues.
DISCUSSION
To our knowledge, there have been limited studies of the potential function and role of glycans of HIV/SIV during virus-cell interactions. One of the areas that has received considerable attention has been the role of glycans serving as shields as a mechanism by which HIV/SIV evades host humoral immune responses and contributes to the emergence of a variety of mutants with altered PNGs (18, 61). The results of the studies reported herein show that N glycosylation of gp120 may also distinguish pathogenic from nonpathogenic infection as noted for monkeys infected with SIVmac239 and Δ5G. We submit that one explanation for the findings of our studies is that these two viruses target distinct CD4+ T cell subsets, with SIVmac239 primarily targeting the TrM subset within SLT and the Δ5G primarily targeting the EM within the small intestine, respectively, during primary infection with polarized clinical outcomes. These data also correlate with the recent report by Paiardini et al. (36) whereby SIV infection is found predominantly in EM T cells in the asymptomatic natural infection of sooty mangabeys.
Since the discovery that the gastrointestinal tract serves as a primary target tissue for viral replication and depletion of CD4+ T cells after both HIV and SIV infection (54), studies have focused on the intestinal mucosal tissue to elucidate the mechanisms of pathogenesis that would help guide the formulation of vaccines and therapeutics (4, 24). It is becoming increasingly clear, however, that the intestinal tissue is complex, comprised of different compartments with diverse physiological and immune function. Some of these compartments include the LP (effector site) and Peyer's patches and SLF (inductive sites). What remains unclear is whether there are discrete and/or subtle differences in the way different viruses target each of these tissues and structures within these tissues. Recent data from our lab and others have highlighted the exclusion of CD8+ T cells from germinal centers within lymphoid follicles, while SIV signal was readily detected within germinal centers during SIVmac239 infection (13, 59), as seen for the same virus in Fig. 2C but not for Δ5G. It remains to be seen whether such different targeting results in distinct clinical outcome associated with differences on the local and general immune function. Of relevance to this line of thinking is the fact that, whereas considerable recent attention has been paid to the pathogenic consequences of both HIV/SIV infection in the GIT, a direct systematic comparison with SLT during acute infection has not been performed. The studies reported here precisely are aimed at delineating the potential differences in the infection of CD4+ T cells in the SLT versus GIT and the outcome of such differential targeting to the pathogenic events following SIV infection. This line of reasoning is partially supported by the finding that, whereas SIV infection of natural hosts during acute infection results in depletion of CD4+ T cells in the intestinal tissues, such marked pathology is not associated with progression to disease and/or of detectable immunodeficiency (9, 25, 37). A recent study has pointed out that an association exists between AIDS-free infection and low level of infection of CM CD4+ T cells (36). Inverse correlations between VL and frequencies of CM CD4+ T cells have also been reported in pathogenic SIV/macaque models (20, 22, 33). In addition, these studies suggest that it is likely that the immunological process such as chronic immune activation, rather than the magnitude of viral replication, that plays a key factor in the pathogenesis of HIV/SIV infection (9, 45–47).
Progressive decline in the number of TrM CD4+ T cells results in insufficient effector CD4+ T cells that is associated with onset of AIDS in chronic SIV infection (33, 47). Our study demonstrates that a 1- to 2-log lower acute viral load among the CD4+ T cells in SLT occurs during “nonpathogenic” Δ5G-infected animals compared to SIVmac239-infected animals. In addition, such lower levels of infection in the Δ5G-infected animals are associated with the preservation of CXCR3+ CCR5+ TrM CD4+ T cells, i.e., TrM-Th1 cells whose depletion is linked to an insufficient virus-specific cellular response leading to persistent viral replication in SIVmac239 infection (28, 51).
Robust levels of Δ5G replication during the primary infection were detected within the effector site (LP) of small intestine (Table 2 and Fig. 3), a finding consistent with studies of HIV-1 and SIV infection (21, 24). The phenotype of the cells targeted was the CCR6+ CXCR3− EM cell, consistent with the view that these are Th17 cells (12, 27). Since Δ5G infection to a large extent was nonpathogenic, the result of the studies reported here suggest that depletion of the targeted Th17 lineage cells is dispensable for the health of the animal at least during the acute phase of infection. Supporting this view is our finding that infection with a live-attenuated SIVmac239 (ΔNef) was shown to preferentially replicate within CD4+ T cells in the B cell region of the LN, where follicular helper CD4+ T cells reside, but not in the T cell region, where CM, TrM, and naive CD4+ T cells reside (51). Thus, SIVΔ5G shares an in vivo property with SIVΔnef. Whereas the underlying mechanism seems different, the two viral moieties, Nef proteins and glycosylated viral spikes, likely facilitate the productive infection of the relatively less susceptible CD4+ T cells in the SLT. Notably, since both Δ5G and ΔNef elicit potent protective immune responses in the infected animals that are sufficient to induce nearly 100% protection against high-dose virus challenges with homologous virus and are variable but quite effective against high doses of heterologous challenge viruses, the infection and targeting of EM CD4+ T cells for a short term (Fig. 6A) might allow the generation of highly effective antiviral host immune responses by the nondepleted and/or unaffected cell populations, as previously reported (29, 30, 52).
This study reveals that N glycans must play a role in the targeting of SIV to infection and replication to different subsets of CD4+ T cells, such as TrM-Th1 cells in SLT and Th17 cells in the intestinal tissues. The recent findings that N glycans in HIV-1 gp120 play a role in their selective tropism for α4β7 high CD4+ T cells and that these cells are the primary target cells for founder/early transmitting viruses (31) supports the findings of the studies reported herein. This view is also consistent with our findings that animals that were vaccinated with Δ5G who failed to control challenge with heterologous virus showed the rapid emergence of mutants with increased PNGs (52). These results underscore the important role of glycans that function to dictate tropism of the various HIV/SIV virus isolates for different target cells and the critical role that subsets of target cells may play in immune homeostasis. As mentioned above, each of the HIV/SIV isolates that have been studied to date have their own optimal number of PNGs in the Env. We submit that such glycans help define tropism for a distinct set of target cells that result in persistent viral replication in their host.
The gp120 of SIVmac239, which consists of 23 PNGs, replicates quite efficiently within the TrM CD4+ T cells located within the SLT and a reduction of five PNGs from this wt SIVmac239 causes a marked loss of replication. We believe that mechanisms by which N glycans of SIVmac239 but not those of Δ5G facilitate targeting and viral replication in the TrM CD4+ T cells in SLT is closely related to the pathogenesis of HIV/SIV infection. Several studies suggested that dendritic cells (DCs) might play a key role in the immune activation and productive infection of CD4+ T cells through immunological or virological synapses (14, 48). DCs expresses several c-type lectin receptors that might also function as the receptors for HIV/SIV, since viral glycans can bind to them. To support this hypothesis, a recent study demonstrated that, together with TLR8, a c-type lectin receptor DC-SIGN facilitated the productive infection of DC with HIV-1 (11). It is thus reasoned that SIVmac239 but not Δ5G might replicate in TrM CD4+ T cells through select lectin-like receptor-mediated infection of CD4+ T cells. We submit that there exists an important potential role of glycans in the pathogenesis of HIV/SIV infection and that much has yet to be learned on this subject.
The findings reported here demonstrate that glycans play a critical role in conferring tropism for a particular CD4+ T cell subset to SIV and that the tropism for TrM CD4+ T cells in SLT defines the pathogenicity of SIV. Understanding the mechanisms underlying methods by which glycans facilitate SIV/HIV to replicate in particular immune cells in different tissues might provide clues to discover key biological molecules and virus-host cell interactions required for the control of HIV infection.
ACKNOWLEDGMENTS
We thank Kayoko Ueda for excellent technical assistance. We acknowledge all of the animal care and veterinary staff at the Tsukuba Primate Research Center.
This study was conducted through the Cooperative Research Program in the Tsukuba Primate Research Center, National Institute of Biomedical Innovation. This study was supported by AIDS research grants from the Health Sciences Research Grants from the Ministry of Health, Labor, and Welfare in Japan and from the Ministry of Education, Culture, Sports, Science, and Technology in Japan.
Footnotes
Published ahead of print 20 June 2012
REFERENCES
- 1. Acosta-Rodriguez EV, et al. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646 [DOI] [PubMed] [Google Scholar]
- 2. Alexander L, Denekamp L, Czajak S, Desrosiers RC. 2001. Suboptimal nucleotides in the infectious, pathogenic simian immunodeficiency virus clone SIVmac239. J. Virol. 75:4019–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brenchley JM, et al. 2008. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112:2826–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brenchley JM, et al. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao J, et al. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808–9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chackerian B, Rudensey LM, Overbaugh J. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719–7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Douek DC, et al. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98 [DOI] [PubMed] [Google Scholar]
- 8. Esplugues E, et al. 2011. Control of TH17 cells occurs in the small intestine. Nature 475:514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gordon SN, et al. 2007. Severe depletion of mucosal CD4+ T cells in AIDS-free simian immunodeficiency virus-infected sooty mangabeys. J. Immunol. 179:3026–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gosselin A, et al. 2010. Peripheral blood CCR4+ CCR6+ and CXCR3+ CCR6+ CD4+ T cells are highly permissive to HIV-1 infection. J. Immunol. 184:1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gringhuis SI, et al. 2010. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat. Immunol. 11:419–426 [DOI] [PubMed] [Google Scholar]
- 12. Hirota K, et al. 2007. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J. Exp. Med. 204:2803–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hong JJ, Amancha PK, Rogers K, Ansari AA, Villinger F. 2012. Spatial alterations between CD4+ TFH, B, and CD8+ T cells during SIV infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J. Immunol. 188:3247–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keele BF, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim CH, et al. 2001. Rules of chemokine receptor association with T cell polarization in vivo. J. Clin. Invest. 108:1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong X, et al. 2008. The human immunodeficiency virus type 1 envelope confers higher rates of replicative fitness to perinatally transmitted viruses than to nontransmitted viruses. J. Virol. 82:11609–11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwong PD, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leonard CK, et al. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373–10382 [PubMed] [Google Scholar]
- 20. Letvin NL, et al. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science 312:1530–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Q, et al. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434:1148–1152 [DOI] [PubMed] [Google Scholar]
- 22. Mattapallil JJ, et al. 2006. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge. J. Exp. Med. 203:1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mattapallil JJ, et al. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434:1093–1097 [DOI] [PubMed] [Google Scholar]
- 24. Mehandru S, et al. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200:761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Milush JM, et al. 2007. Virally induced CD4+ T cell depletion is not sufficient to induce AIDS in a natural host. J. Immunol. 179:3047–3056 [DOI] [PubMed] [Google Scholar]
- 26. Montefiori DC, Robinson WE, Jr, Mitchell WM. 1988. Role of protein N glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 85:9248–9252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monteiro P, et al. 2011. Memory CCR6+ CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J. Immunol. 186:4618–4630 [DOI] [PubMed] [Google Scholar]
- 28. Mori K, et al. 2005. Influence of glycosylation on the efficacy of an Env-based vaccine against simian immunodeficiency virus SIVmac239 in a macaque AIDS model. J. Virol. 79:10386–10396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mori K, et al. 2001. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J. Virol. 75:4023–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mori K, et al. 2000. Suppression of acute viremia by short-term post-exposure prophylaxis of simian/human immunodeficiency virus SHIV-RT-infected monkeys with a novel reverse transcriptase inhibitor (GW420867) allows for development of potent antiviral immune responses resulting in efficient containment of infection. J. Virol. 74:5747–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nawaz F, et al. 2011. The genotype of early-transmitting HIV gp120s promotes α4β7 reactivity, revealing α4β7+/CD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 7:e1001301 doi:10.1371/journal.ppat.1001301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohgimoto S, et al. 1998. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J. Virol. 72:8365–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okoye A, et al. 2007. Progressive CD4+ central memory T cell decline results in CD4+ effector memory insufficiency and overt disease in chronic SIV infection. J. Exp. Med. 204:2171–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Shea JJ, Paul WE. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327:1098–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pahar B, Lackner AA, Veazey RS. 2006. Intestinal double-positive CD4+ CD8+ T cells are highly activated memory cells with an increased capacity to produce cytokines. Eur. J. Immunol. 36:583–592 [DOI] [PubMed] [Google Scholar]
- 36. Paiardini M, et al. 2011. Low levels of SIV infection in sooty mangabey central memory CD T cells are associated with limited CCR5 expression. Nat. Med. 17:830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pandrea IV, et al. 2007. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J. Immunol. 179:3035–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Picker LJ, et al. 2004. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J. Exp. Med. 200:1299–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raffatellu M, et al. 2008. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 14:421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Regier DA, Desrosiers RC. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 6:1221–1231 [DOI] [PubMed] [Google Scholar]
- 41. Reitter JN, Means RE, Desrosiers RC. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679–684 [DOI] [PubMed] [Google Scholar]
- 42. Rivino L, et al. 2004. Chemokine receptor expression identifies Pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J. Exp. Med. 200:725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salazar-Gonzalez JF, et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seth N, Kaufmann D, Lahey T, Rosenberg ES, Wucherpfennig KW. 2005. Expansion and contraction of HIV-specific CD4 T cells with short bursts of viremia, but physical loss of the majority of these cells with sustained viral replication. J. Immunol. 175:6948–6958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Silvestri G, et al. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441–452 [DOI] [PubMed] [Google Scholar]
- 46. Sodora DL, Silvestri G. 2008. Immune activation and AIDS pathogenesis. AIDS 22:439–446 [DOI] [PubMed] [Google Scholar]
- 47. Sopper S, et al. 2003. Impact of simian immunodeficiency virus (SIV) infection on lymphocyte numbers and T-cell turnover in different organs of rhesus monkeys. Blood 101:1213–1219 [DOI] [PubMed] [Google Scholar]
- 48. Sowinski S, et al. 2008. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10:211–219 [DOI] [PubMed] [Google Scholar]
- 49. Stamatatos L, Cheng-Mayer C. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840–7845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sugimoto C, et al. 2008. Impact of glycosylation on antigenicity of simian immunodeficiency virus SIV239: induction of rapid V1/V2-specific non-neutralizing antibody and delayed neutralizing antibody following infection with an attenuated deglycosylated mutant. J. Gen. Virol. 89:554–566 [DOI] [PubMed] [Google Scholar]
- 51. Sugimoto C, et al. 2003. nef gene is required for robust productive infection by simian immunodeficiency virus of T-cell-rich paracortex in lymph nodes. J. Virol. 77:4169–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sugimoto C, et al. 2010. Protection of macaques with diverse MHC genotypes against a heterologous SIV by vaccination with a deglycosylated live-attenuated SIV. PLoS One 5:e11678 doi:10.1371/journal.pone.0011678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tenner-Racz K, et al. 1998. The unenlarged lymph nodes of HIV-1-infected, asymptomatic patients with high CD4 T cell counts are sites for virus replication and CD4 T cell proliferation: the impact of highly active antiretroviral therapy. J. Exp. Med. 187:949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Veazey RS, et al. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427–431 [DOI] [PubMed] [Google Scholar]
- 55. Veazey RS, Lackner AA. 1998. The gastrointestinal tract and the pathogenesis of AIDS. AIDS 12(Suppl A):S35–S42 [PubMed] [Google Scholar]
- 56. Veazey RS, Lackner AA. 2004. Getting to the guts of HIV pathogenesis. J. Exp. Med. 200:697–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Veazey RS, et al. 2000. Dynamics of CCR5 expression by CD4+ T cells in lymphoid tissues during simian immunodeficiency virus infection. J. Virol. 74:11001–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Veazey RS, et al. 1997. Characterization of gut-associated lymphoid tissue (GALT) of normal rhesus macaques. Clin. Immunol. Immunopathol. 82:230–242 [DOI] [PubMed] [Google Scholar]
- 59. Vojnov L, et al. 2011. GagCM9-specific CD8+ T cells expressing limited public TCR clonotypes do not suppress SIV replication in vivo. PLoS One 6:e23515 doi:10.1371/journal.pone.0023515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang C, Kang SG, Lee J, Sun Z, Kim CH. 2009. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol. 2:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 62. Yue FY, et al. 2005. Preferential apoptosis of HIV-1-specific CD4+ T cells. J. Immunol. 174:2196–2204 [DOI] [PubMed] [Google Scholar]
- 63. Zhang M, et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]