Abstract
Recent genetic studies suggested that viral nonstructural (NS) proteins play important roles in morphogenesis of flaviviruses, particularly hepatitis C virus (HCV). Adaptive and compensatory mutations occurring in different NS proteins were demonstrated to promote HCV production in cell culture. However, the underlying molecular mechanism of NS proteins in HCV morphogenesis is poorly understood. We have isolated a cell culture-adapted HCV of genotype 2a (JFH1) which grew to an infectious titer 3 orders of magnitude higher than that of wild-type virus. Sequence analysis identified a total of 16 amino acid mutations in core (C), E1, NS2, NS3, NS5A, and NS5B, with the majority of mutations clustered in NS5A. Reverse genetic analysis of these mutations individually or in different combinations demonstrated that amino acid mutations in NS2 and NS5A markedly enhanced HCV production. Additionally, mutations in C, E1, NS3, and NS5B synergistically promoted HCV production in the background of NS2 and NS5A mutations. Adaptive mutations in NS5A domains I, II, and III independently enhanced HCV production, suggesting that all three domains of NS5A are important for HCV morphogenesis. More importantly, adaptive mutations greatly enhanced physical interactions among HCV structural and NS proteins, as determined by studies with coimmunoprecipitation and mammalian two-hybrid assays. Collectively, these findings demonstrate that adaptive mutations can enhance specific protein-protein interactions among viral structural and NS proteins and therefore promote the assembly of infectious HCV particles.
INTRODUCTION
Hepatitis C virus (HCV) is a major cause of chronic liver diseases, affecting approximately 170 million people worldwide (39). The vast majority of acutely HCV-infected individuals become chronic carriers who are at higher risk for developing cirrhosis and hepatocellular carcinoma (32). HCV is the only member of the Hepacivirus genus in the Flaviviridae family (30). It is an enveloped RNA virus containing a single-stranded and positive-sense RNA genome. The genomic RNA is composed of a single open reading frame (ORF) and untranslated regions (UTR) at both the 5′ and 3′ ends (4, 20). The highly conserved cis-acting RNA elements within the 5′ and 3′ UTRs are important for mediating initiation of HCV polyprotein translation and controlling viral RNA replication (8, 21, 23). The ORF encodes a single polypeptide precursor which is proteolytically cleaved by cellular peptidases and viral proteases into individual mature HCV proteins in the order C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (21). It has been demonstrated by numerous studies that HCV RNA replication occurs in the membrane-bound replication complex consisting of HCV RNA and the NS3 to NS5B proteins, as well as cellular factors (7, 21, 25). The newly synthesized HCV proteins and genomic RNA are assembled to form progeny virus particles, which egress through secretory pathways. Although a great deal of progress has been made with respect to the importance of viral and cellular factors in the HCV life cycle, the underlying molecular mechanisms of HCV assembly, maturation, and egression remain poorly understood.
Viral nonstructural (NS) proteins of RNA viruses are known to play central roles in viral RNA replication (21). Strikingly, recent genetic studies suggested that viral NS proteins are also important for the assembly and/or production of flaviviruses (26). It was previously found that single amino acid mutations in the NS2A and NS3 proteins of the yellow fever virus and Kunjin virus resulted in abrogation of virus production but did not affect viral RNA replication, suggesting their importance in virus assembly and/or production (19, 22, 28). In the case of HCV, both NS2 and p7 are essential for HCV assembly and/or production although dispensable for viral RNA replication (15, 17, 36). Similarly, compensatory mutations that enhanced the production of the intragenotypic and intergenotypic HCV chimeras were found in the E1, p7, NS2, and NS3 proteins (1, 11, 40). Additionally, core protein mutants defective in HCV production could be rescued by compensatory mutations occurring in p7 and NS2 (27). Furthermore, NS5A is implicated in HCV assembly by the finding that a single phosphorylation site in domain III of NS5A is critically important for HCV production but not viral RNA replication (37). Collectively, these findings suggest that viral NS proteins are genetically important for HCV assembly and/or production. However, the molecular basis underlying the importance of viral NS proteins in HCV assembly is still poorly understood.
Several previous studies have demonstrated that adaptive mutations occurring in viral structural and NS proteins remarkably promoted the production of infectious HCV (6, 10, 13, 18, 29, 31, 41). Through serial passage of HCV in cell culture, we have also isolated an adapted HCV (JFH1/Ad16) which grew to 107 focus-forming units (FFU)/ml, 3 orders of magnitude higher (1,000-fold) than growth of wild-type virus. In the present study, we have determined the importance of the adapted amino acid mutations occurring in structural and NS proteins in HCV production when analyzed individually or in different combinations. Interestingly, mutations in the structural and NS proteins were found to synergistically promote HCV production. Mutations in the three different NS5A domains (I, II, and III) independently enhanced HCV production, suggesting that they all are important for HCV production. More importantly, we found that adaptive mutations remarkably enhanced protein-protein interactions among viral structural and NS proteins. Taken together, our findings demonstrate for the first time that adaptive mutations enhance specific protein-protein interactions among viral structural and NS proteins and therefore promote the formation and/or production of infectious HCV particles.
MATERIALS AND METHODS
Cell lines and cell culture.
Human hepatoma cell lines (Huh-7) that stably produce infectious HCV of genotype 2a (JFH1) were previously described (2). The Huh-7.5 cell line was kindly provided by Charles M. Rice (Rockefeller University). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), nonessential amino acids, penicillin, and streptomycin (Invitrogen).
Antibodies.
The HCV NS3-specific monoclonal antibody (MAb) was described previously (2). HCV NS2 polyclonal antibody (PAb), NS2 MAb (6H6), and NS5A MAb (9E10) were kindly provided by Charles M. Rice. The β-actin MAb and horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Sigma and Pierce, respectively. HCV core MAb and E1 MAb (E1A) were provided by Jake Liang (NIDDK/NIH).
Amplification of HCV cDNA by RT-PCR.
HCV virion RNA (vRNA) was extracted with TRIzol-LS reagent (Invitrogen) and was converted to cDNA by reverse transcription-PCR (RT-PCR) using a Superscript One-Step RT-PCR kit (Invitrogen) according to the manufacturer's instructions. PCR primers of 17 to 26 nucleotides (nt) in length were used to amplify eight fragments of HCV cDNA (nucleotides 78 to 1551, 1328 to 2999, 2936 to 4122, 4085 to 5334, 5274 to 6091, 5883 to 7509, 7415 to 8820, and 8765 to 9481). The resulting RT-PCR DNA products were sequenced at Northwestern University Biotechnology Laboratory (NUBL) (Chicago, IL).
Plasmid DNA construction.
The DNA vector pSGR/JFH1-FL/AR/pA, which contains the full-length cDNA of the JFH1 HCV RNA and a hepatitis delta virus antigenomic ribozyme at the immediate 3′ end, followed by a simian virus 40 poly(A) sequence, was described previously (2). The BamHI site at nucleotide 6006 of the JFH1 HCV cDNA (pSGR/JFH1-FL/AR/pA) was removed by introducing silent mutations using a two-step PCR method. The resulting DNA was designated pSGR/JFH1-FL/AR/pA/dBamHI6006. Specific adaptive mutations of C/E1, NS2, NS3, NS5A, and NS5B were introduced by replacing corresponding regions of wild-type DNA with RT-PCR cDNA fragments between AgeI and BsiWI (C/E1), NotI and SpeI (NS2), NotI and NsiI (NS3), BamHI and BsrGI (NS5A), and SfiI and EcoRV (NS5B), respectively. The resultant cDNA constructs were confirmed by DNA sequence analysis (NUBL, Chicago, IL). A mammalian two-hybrid system (Clontech) used for determining specific HCV protein-protein interactions in the cell was described previously (5). Each HCV protein was fused with the Saccharomyces cerevisiae Gal4 DNA-binding domain (Gal4-BD) and the activation domain of the herpes simplex virus (HSV) VP16, respectively. The cDNA of each HCV-specific gene was amplified by PCR using wild-type and adapted HCV cDNAs as templates, respectively, and synthetic oligonucleotides as primers (available on request). PCR DNA fragments were cut with the restriction enzymes EcoRI and XbaI and inserted into the similarly digested pM and pVP16 vectors, respectively. In order for detection of the E1 protein by an existing monoclonal antibody (12), the amino acid substitutions T197S, S199G, S200L, and M202H were introduced into E1 by overlapping PCR amplification using synthetic oligonucleotide primers E1A4/1 (5′-CAGGTGAAGAATAGCAGTGGCCTCTACCATGTGACCAATGACTGC-3′), E1A4/2 (5′-GCAGTCATTGGTCACATGGTAGAGGCCACTGCTATTCTTCACCTG-3′), 5′UTR/97 (5′-CTAGCCATGGCGTTAGTA-3′), and 2a/BsiWI-R (5′-CCTCGGGGACGCGCATC-3′). The PCR DNA fragment was digested with the restriction enzymes AgeI and BsiWI and cloned into JFH1 cDNA vectors, which were similarly cut by both AgeI and BsiWI, as previously described (12). The E1 protein containing these substitutions can be recognized by E1-specific MAb A4 (12).
In vitro transcription of HCV RNA and production of infectious HCV.
Wild-type and mutant plasmid DNAs were linearized with XbaI and were used for generation of infectious HCV RNA by transcription using a T7 RiboMax large-scale RNA production system (Promega). T7 transcripts of the HCV RNA genome were purified by passing them through Qiagen RNA isolation columns. To produce infectious HCV, HCV RNAs were transfected into Huh-7.5 cells using DMRIE-C reagent (Invitrogen), following the manufacturer's instructions. At 72 h posttransfection (p.t.), Huh-7.5 cells were either lysed for detection of HCV proteins or used for isolation of total RNAs and preparation of intracellular HCV. The levels of HCV replication in the RNA-transfected cells were determined by measuring the NS3 protein and positive-stranded RNA. The levels of HCV production were determined by measuring the infectivity and titers of infectious HCV in cell culture supernatants of the RNA-transfected Huh-7.5 cells.
HCV infection.
HCV derived from our previously described stable Huh-7 cell line was serially passaged by infection of Huh-7.5 cells more than 60 times (2). For determination of HCV protein and/or RNA levels, Huh-7.5 cells in 12-well (for protein detection) or 6-well (for viral RNA quantification) cell culture plates were infected with HCV at 37°C for 3 h. The HCV-infected cells were washed twice with phosphate-buffered saline (PBS) and then incubated with DMEM containing 10% FBS for 3 days. The HCV-infected cells in 12-well plates were lysed and used for detection of the NS3 protein by Western blotting. Total RNAs of the HCV-infected cells in 6-well plates were extracted with TRIzol reagent and used for determination of positive-strand HCV RNA by RNase protection assay (RPA).
Western blot analysis.
Twenty-five micrograms of total protein in cell lysates was loaded into a 10% sodium dodecyl sulfate-polyacrylamide gel, followed by electrophoresis. Separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane. HCV proteins were detected by Western blotting using specific monoclonal antibodies, as described previously (2). The β-actin protein was used as an internal control.
RNA extraction and quantification by RPA.
Total RNAs were extracted with TRIzol reagent (Invitrogen) from the HCV RNA-transfected or HCV-infected Huh-7.5 cells. The levels of positive-strand HCV RNA were determined by RPA, as described in our previous work (2). After digestion with RNase A/T1, RNA products were analyzed in a 6% polyacrylamide–7.7 M urea gel, visualized by autoradiography, and quantified with PhosphorImager analysis (2).
HCV vRNA extraction and quantification by qRT-PCR.
The vRNAs of extracellular and intracellular HCV particles were extracted using TRIzol LS reagent (Invitrogen). The levels of HCV vRNA were determined by quantitative RT-PCR (qRT-PCR) using a Superscript III Platinum One-Step qRT-PCR kit (Invitrogen). Oligonucleotides 2aF (5′-AGCCATGGCGTTAGTATGAGTGTC-3′) and 2aR (5′-ACAAGGCCTTTCGCAACCCAA-3′), complementary to sequences within the 5′ UTR, were used as primers. The probe (5′-AAACCCACTCTATGCCCGGCCATTT-3) containing a 5′ TAMRA and 3′ 6-carboxyfluorescein (FAM) was synthesized by Integrated DNA Technologies (IDT).
Determination of infectious HCV titers by IFA.
Infectious HCV titers were determined by limiting dilution using IFA and an NS3-specific MAb, as described previously (2, 5). Briefly, HCV in cell culture supernatants was serially diluted (10×) and used to infect naive Huh-7.5 cells in 96-well plates with glass bottoms. At 3 days postinfection (p.i.), the number of FFU/ml was determined by immunofluorescence assay (IFA).
co-IP.
Coimmunoprecipitation (co-IP) experiments were carried out using cell lysate of the HCV RNA-transfected cells and protein G-conjugated agarose beads, following the manufacturer's instructions (Pierce). Briefly, normal mouse IgG or HCV protein-specific MAbs were incubated with protein G-conjugated agarose beads at room temperature for 5 h. The unbound antibodies were removed by washing with 1× PBS three times. The antibody-bound agarose beads were then incubated with 300 μl of cell lysate at 4°C overnight. Upon co-IP, HCV C, E1, NS2, NS3, and NS5A were detected by Western blotting using specific antibodies.
Mammalian two-hybrid assay.
The mammalian two-hybrid assay for HCV protein-protein interaction was similar to the one used for our previous studies (5). Briefly, Huh-7.5 cells were seeded at 2 × 105 cells/well in 12-well cell culture plates overnight. Both the pM and pVP16 vectors (0.25 μg each), expressing individual HCV fusion proteins, were cotransfected with 0.5 μg of the reporter plasmid pFR-Luc (Stratagene) and 0.05 μg of the control vector phRL-SV40 (Promega) into Huh-7.5 cells using DMRIE-C reagent according to the manufacturer's instruction (Invitrogen). At 48 h p.t., firefly and Renilla luciferase activities were determined using dual-luciferase assay kits (Promega). Both pM and pVP16 vectors without HCV genes were used as negative controls. phRL-SV40 expressing a Renilla luciferase was used as an internal control to normalize DNA transfection efficiency.
Statistical analysis.
Student's t test was used for statistical analysis of the data. A P value of <0.05 was considered significant, and a P value of <0.01 was considered highly significant.
Nucleotide sequence accession number.
The GenBank accession number of the sequence for this adaptive HCV is JX014307.
RESULTS
A cell culture-adapted HCV variant with a higher infectious titer.
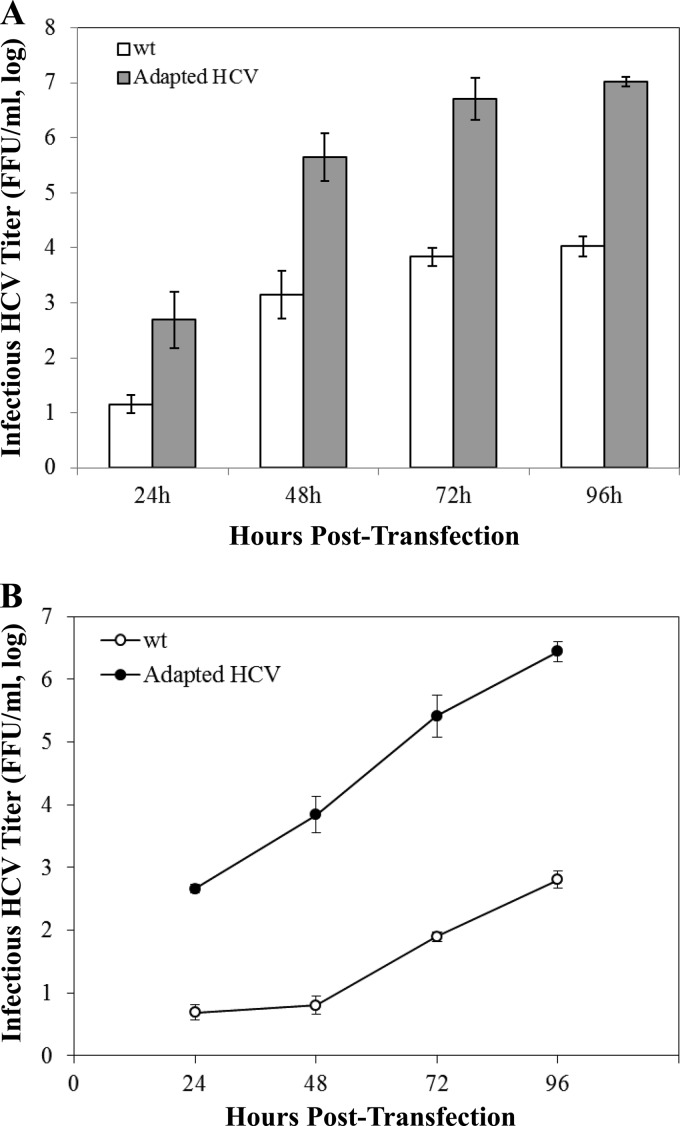
We had previously developed stable Huh-7 cell lines that robustly produce infectious HCV (2). However, the infectious titer of wild-type HCV produced in cultured cells is extremely low, typically at 104 FFU/ml. To select virus variants with higher infectious titers, HCV derived from a stable HCV-producing Huh-7 cell line (2) was adapted in naive Huh-7.5 cells by serial passage. Through numerous passages of HCV in Huh-7.5 cells, we identified an HCV variant which grew to a titer 3 orders of magnitude (1,000-fold) higher than that of wild-type virus when quantified at various time points (48, 72, and 96 h) after transfection with infectious HCV RNAs (Fig. 1A). The enhanced growth of the cell culture-adapted HCV variant was confirmed by infectious titers significantly higher than those of wild-type virus at different time points after infection with the same multiplicity of infection (MOI) of viruses (Fig. 1B). The availability of the HCV variant with a higher infectious titer made it possible to perform biochemical and genetic studies of HCV morphogenesis and egression.
Fig 1.
Growth comparison of the wild type (wt) and the cell culture-adapted HCV variant. (A) Cell culture-adapted HCV variant with an infectious titer higher than that of wild-type virus. Huh-7.5 cells were transfected with 2 μg of wild-type or adapted HCV RNAs. The supernatants were collected at 24, 48, 72, and 96 h posttransfection. The infectious HCV titers in the supernatant were determined by serial dilution and IFA. Results shown represents means ± standard deviations for three different experiments. (B) Growth curves of the wild-type and cell culture-adapted HCVs. Huh-7.5 cells in 24-well plates were infected with wild-type and cell culture-adapted HCV at a multiplicity of infection (MOI) of 0.2. The infectious HCV titers (FFU/ml) at different time points (24, 48, 72, and 96 h) postinfection were determined by serial dilution and immunostaining of the HCV-infected cells by IFA as described in Materials and Methods.
Determination and characterization of adaptive amino acid mutations.
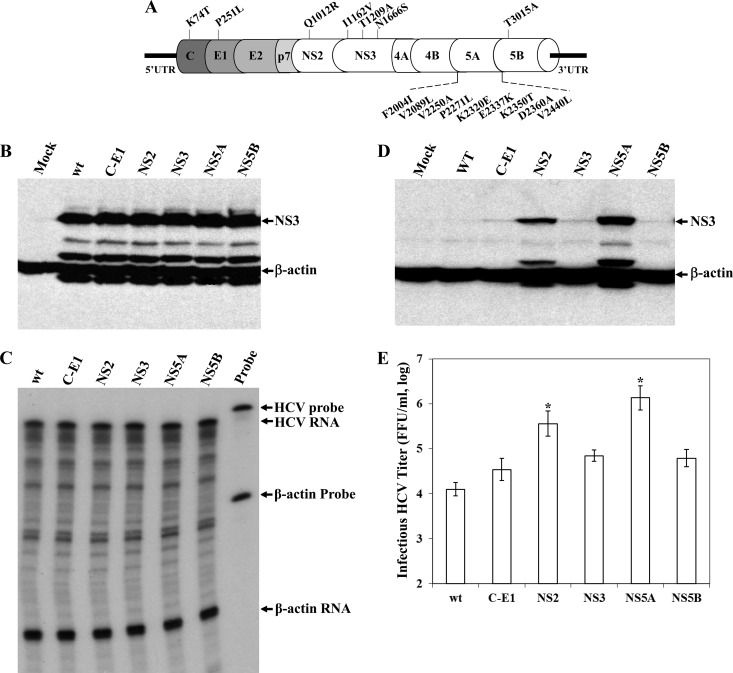
To identify cell culture-adaptive mutations, the HCV vRNA was extracted and its cDNA was amplified by RT-PCR. Upon DNA sequence analysis, a total of 16 amino acid mutations were identified in C (K74T), E1 (P251L), NS2 (Q1012R), NS3 (I1162V, T1209A, and N1666S), NS5A (F2004I, V2089L, V2250A, P2271L, K2320E, E2337K, K2350T, D2360A, and V2440L), and NS5B (T3015A), with most mutations clustered in NS5A (Fig. 2A). Additionally, there are 17 synonymous nucleotide mutations in the coding regions of C (A622G, C637A, and C655T), E1 (T1415C), E2 (A1888G, A1990T, C2068T, T2119C, A2251G, and G2257C), p7 (T2705C), NS3 (C4747T), NS4B (T5917C), NS5A (C6595T and G7468A), and NS5B (C7867G and A9211G). Interestingly, this HCV variant also contains the adaptive mutations Q1012R (NS2) and V2440L (NS5A), which were previously shown by others to promote HCV production (18, 31). Other adaptive mutations found in C, E1, NS3, NS5A, and NS5B are novel ones of our HCV variant. To determine the importance of adaptive mutations in the enhancement of HCV assembly and/or production, we carried out reverse genetics studies of mutations in individual HCV proteins. Adaptive mutations in different viral proteins were introduced back into wild-type HCV cDNA (Fig. 2A). Purified T7 transcripts of HCV RNAs were transfected into Huh-7.5 cells. At 72 h p.t., the cell culture supernatants were collected for the determination of infectious HCV titers and the levels of HCV vRNAs, while the levels of the HCV NS3 protein and positive-stranded RNA in the RNA-transfected cells were determined by Western blotting and RPA, respectively. None of the adaptive mutations affected viral RNA replication, as shown by similar levels of the NS3 protein (Fig. 2B) and positive-stranded HCV RNA by RPA (Fig. 2C) and qRT-PCR (data not shown) in the RNA-transfected Huh-7.5 cells. The effects of adaptive mutations on HCV production were determined by both HCV infectivity assay and infectious titers of HCV secreted into the cell culture supernatants. HCV infectivity was determined by measuring the levels of the NS3 protein in naive Huh-7.5 cells infected with the supernatants of the HCV RNA-transfected cells. Adaptive mutations in C-E1, NS3, and NS5B individually increased the production of infectious HCV titers by 3- to 5-fold compared to levels for wild-type virus (Fig. 2E). However, adaptive mutations in NS2 and NS5A significantly enhanced HCV production compared to that of wild-type virus, as shown by much higher levels of the NS3 protein than were seen for wild-type HCV, and adaptive mutations occurred in C/E1, NS3, and NS5B (Fig. 2D). Likewise, adaptive mutations in NS2 and NS5A remarkably increased infectious HCV titers, by 29- and 109-fold, respectively (Fig. 2E). Consistent with previous findings (18, 31), these results demonstrate that NS2 and NS5A play important roles in HCV morphogenesis.
Fig 2.
Effects of adaptive mutations in individual viral proteins on the production of infectious HCV. (A) Distribution of adapted amino acid mutations in HCV proteins. Amino acids are numbered based on the HCV polyprotein. For instance, K74T is a lysine-to-threonine mutation at amino acid residue 74 located in the C protein. (B) Detection of HCV NS3 protein by Western blotting. HCV RNAs (2 μg each) containing adapted mutations in individual HCV proteins (indicated on the top) were transfected into Huh-7.5 cells in 6-well plates. At 72 h p.t., supernatants were collected for the determination of infectious HCV titers (E). The RNA-transfected cells were lysed in a RIPA buffer. The NS3 protein was detected by Western blotting using an NS3-specific monoclonal antibody and Pierce ECL kits (Thermo Scientific). (C) Determination of the positive-stranded HCV RNA by RPA. The HCV RNA-transfected Huh-7.5 cells described for panel B were used for extraction of total RNA with TRIzol reagent (Invitrogen). The levels of positive-stranded HCV RNA were determined by RPA using a radiolabeled RNA probe containing the negative-stranded 3′ UTR, as previously described. The levels of β-actin mRNA were used as internal controls. (D) Determination of HCV infectivity by detecting NS3 in the infected naive Huh-7.5 cells. The culture supernatants of the RNA-transfected cells were used to infect naive Huh-7.5 cells. At 72 h p.i., the levels of the NS3 protein were measured by Western blotting using an NS3 MAb and Pierce ECL kits (Thermo Scientific). (E) Quantification of infectious HCV titers by limiting dilution. The infectious HCV in the supernatant (B) was serially diluted. The focus-forming units were determined by immunostaining of NS3-posive cells by IFA. The mean values and standard deviations from three independent experiments are calculated and plotted. The enhancement of infectious HCV titers by adaptive mutations in NS2 and NS5A is statistically significant (P < 0.05).
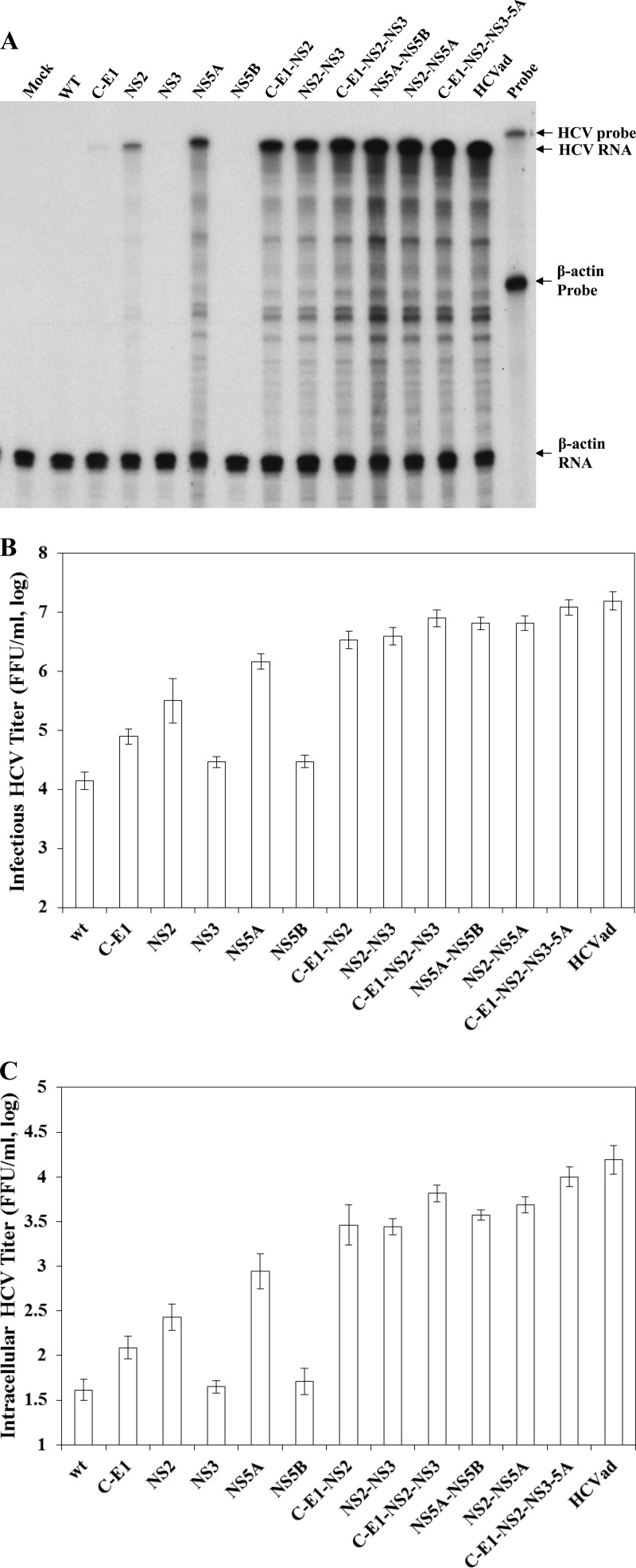
Synergistic enhancement of HCV production by adaptive mutations in viral structural and NS proteins.
To determine cooperative effects of adaptive mutations in structural and NS proteins on HCV assembly/production, we generated various recombinant viruses with adaptive mutations in different combinations. Like mutations in individual viral proteins (Fig. 2B and C), mutations in different combinations did not affect viral RNA replication based on similar levels of positive-stranded HCV RNA in the RNA-transfected cells as determined by RPA and qRT-PCR (data not shown). However, adaptive mutations in different combinations remarkably enhanced production of infectious HCV (Fig. 3). The levels of positive-stranded HCV RNAs were significantly increased in the cells infected with viruses containing adaptive mutations in NS2, NS5A, C-E1-NS2, NS2-NS3, C-E1-NS2-NS3, NS2/NS5A, NS2-NS5B, and C-E1-NS2-NS3-5A (Fig. 3A). More importantly, the adaptive mutations in C/E1, NS3, and NS5B synergistically enhanced the production of HCV containing NS2 or NS5A mutations, although these mutations per se only modestly increased HCV production, by about 3- to 5-fold (Fig. 3A). Adaptive mutations in C/E1, NS3, or NS5B further enhanced the titers of infectious HCV containing the NS2 or NS5A mutations by more than 10-fold (Fig. 3B).
Fig 3.
Effects of adaptive mutations in different combinations on HCV production. Infectious HCV RNAs containing adaptive mutations in HCV proteins (indicated on the top) were transfected into Huh-7.5 cells using the DMRIE-C reagent following the manufacturer's instructions. At 72 h p.t., total cellular RNAs were extracted with TRIzol reagent, while the cell culture supernatants were collected and used for the determination of HCV infectivity in the supernatants. Upon infection with HCV in the supernatants, the total RNAs in the infected Huh-7.5 cells were extracted using TRIzol reagent. The levels of HCV positive-strand RNA in the HCV-infected naive Huh-7.5 cells were determined by RPA (A). Both infectious HCV titers in the medium (B) and in the cells (C) were determined by serial dilution and IFA, as described previously. Mean values and standard deviations shown in the graph are derived from three independent experiments. The enhancement of infectious HCV production by combination of adaptive mutations in C-E1, NS3, and NS5B with that in NS2 or those in NS5A was statistically significant (P < 0.05) compared with the NS2 or NS5A mutations per se or highly significant (P < 0.01) compared with wild-type virus or adaptive mutations in C-E1, NS3, and NS5B alone.
To further determine whether adaptive mutations are important for HCV assembly and/or egression, the levels of intracellular HCV were determined by serial dilution and IFA. Intracellular HCV particles were prepared by repeated freezing and thawing of the HCV RNA-transfected cells, followed by centrifugation to remove any debris, as previously described (3, 5, 14). Similar to secreted HCV in the supernatants (Fig. 3B), the adaptive mutations in NS2 and NS5A significantly increased intracellular HCV titers by 7- and 24-fold (Fig. 3C). More significantly, the promotion of HCV assembly by adaptive mutations in NS2 and NS5A was synergistically enhanced by mutations in C/E1, NS3, and NS5B (Fig. 3C).
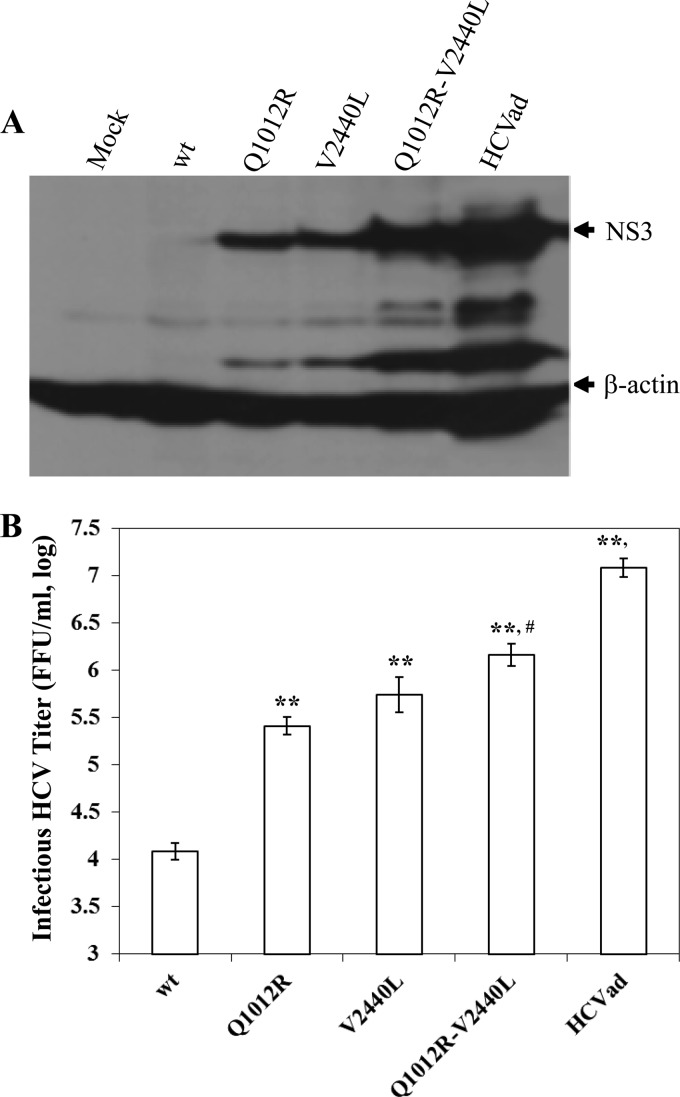
To further assess the synergistic cooperation of NS2 and NS5A with other viral proteins, we constructed infectious viruses containing Q1012R (NS2), V2440L (NS5A) which were previously shown to individually promote HCV production (18, 31), or both. The V2440L mutation in NS5A appeared to promote HCV production to a higher level than the Q1012R mutation in NS2 despite statistical insignificance, as shown by the level of the NS3 protein (Fig. 4A) and infectious HCV titers (Fig. 4B). However, both Q1012R and V2440L additively enhanced HCV production, as determined by increasing levels of infectivity (Fig. 4A) and infectious HCV titer in the supernatants (Fig. 4B). More importantly, mutations in the C/E1, NS3, and NS5B co-opted the Q1012R (NS2) and V2440L (NS5A) mutations in the enhancement of HCV production (Fig. 4). Taken together, these findings demonstrate that adaptive mutations in viral structural and NS proteins synergistically promoted HCV assembly, probably through increased physical protein-protein interactions in the cell.
Fig 4.
Effect of the double mutations Q1012R (NS2) and V2440L (NS5A) on the production of infectious HCV. HCV RNAs containing the indicated mutations were transfected into Huh-7.5 cells in 6-well plates. At 72 h p.t., supernatants were collected for the determination of HCV infectivity (A) or infectious HCV titers (B). (A) Determination of HCV infectivity by detecting NS3 in the infected naive Huh-7.5 cells. The culture supernatants of RNA-transfected cells were used to infect naive Huh-7.5 cells. At 72 h p.i., the levels of NS3 protein were measured by Western blotting using an NS3 MAb and Pierce ECL kits (Thermo Scientific). (B) Quantification of infectious HCV titers by limiting dilution. The infectious HCV in the supernatants was serially diluted. The focus-forming units were determined by immunostaining of NS3-positive cells by IFA. The mean values plus standard deviations from three independent experiments are calculated and plotted. The differences between wild-type and mutant viruses are highly significant (**, P < 0.01). The titer of infectious HCV containing the Q1012R V2440L double mutations was significantly higher than that with the single mutation Q1012R (#, P < 0.05) but significantly lower than that of the adapted HCV containing all mutations (§, P < 0.05).
Effects of different NS5A mutations on HCV production.
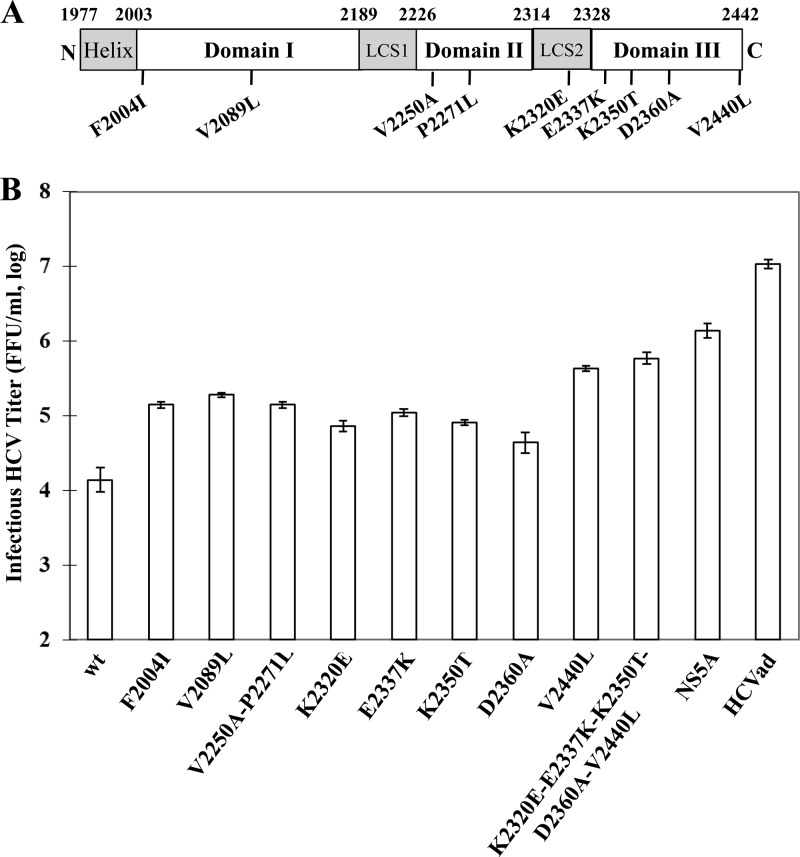
It was previously shown that NS5A domain III is important for production of infectious HCV (1, 37). The majority of adaptive mutations of the HCV variant described here are located in NS5A, including the mutations F2004I and V2089L in domain I, V2250A and P2271L in domain II, K2320E at the junction between domains I and II, and E2337K, K2350T, D2360A, and V2440L in domain III (Fig. 5A). To further determine the importance of different domains of NS5A in HCV assembly, we carried out reverse genetic analysis of the adaptive NS5A mutations individually and in combination. Interestingly, adaptive mutations in both domains I (F2004I and V2089L) and II (V2250A/P2271L) of NS5A promoted HCV production by more than 10-fold (Fig. 5). The mutations E2337K, K2350T, and D2360A in NS5A domain IIII increased HCV production by 8-, 6-, and 3-fold, respectively. The most critical mutation is V2440L in NS5A domain III, which enhanced HCV production by 31-fold (Fig. 5B, V2440L). This mutation was previously shown to suppress the cleavage between NS5A and NS5B (18). However, the combination of all NS5A mutations increased the titer of infectious HCV by 100-fold (Fig. 5B, NS5A). These results suggest that all three domains (I, II, and III) of NS5A play important roles in HCV assembly and/or production, particularly the V2440L mutation in NS5A domain III, consistent with previous findings that domain III of NS5A is critical to HCV production (18, 37).
Fig 5.
Effects of different NS5A mutations on HCV production. (A) Schematic distribution of adaptive mutations in NS5A. The three domains of NS5A are based on previous work by others (1, 38). (B) Effects of adaptive amino acid mutations in NS5A on HCV production. Infectious HCV RNAs containing adaptive amino acid mutations in NS5A individually or in different combinations, which are shown at the bottom, were transfected into Huh-7.5 cells, as described in Materials and Methods. At 72 h p.t., the supernatants were collected for the determination of infectious HCV titers by serial dilution and IFA. The enhancement of HCV production by individual NS5A adaptive mutations, except for D2360A, is either statistically significant (P < 0.05) or highly significant (P < 0.01; V2440L, K2320E-E2337K-K2350T-D2360A-V2440L, and NS5A).
Enhancement of viral protein-protein interactions by adaptive mutations.
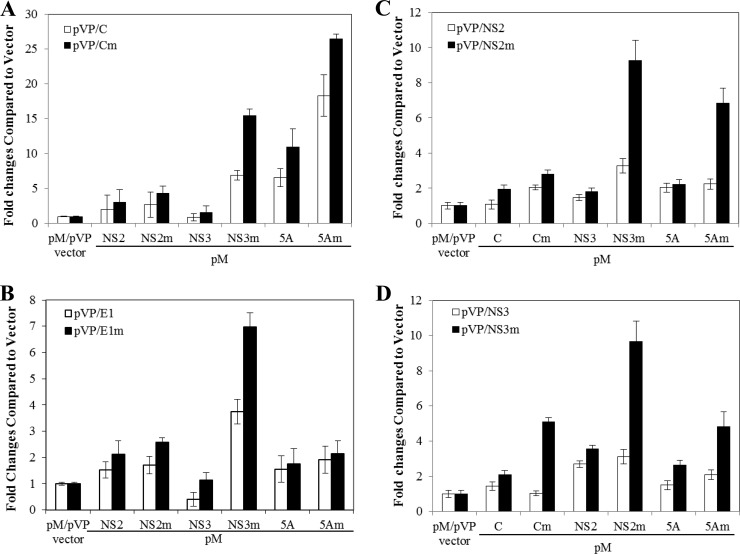
The question arose whether adaptive mutations enhance physical interactions among viral structural and NS proteins and therefore promote HCV assembly. To examine this possibility, we initially used a mammalian two-hybrid system to determine the effects of adaptive mutations on specific protein-protein interactions in the cell. Both wild-type and adapted mutant forms of C, E1, NS2, NS3, and NS5A were expressed as fusion proteins with either the DNA-binding domain of the yeast Gal4 protein or the transcriptional activation domain of the HSV VP16 protein, as described in our earlier work (5). HCV protein-protein interactions would result in transcriptional activation of the firefly luciferase gene downstream of the Gal4-binding sites. Thus, the degree of HCV protein-protein interactions could be determined by directly measuring firefly luciferase activity in a quantitative manner (Fig. 6). This mammalian two-hybrid assay revealed that wild-type C/NS2 and C/NS3 (Fig. 6A and C), E1/NS2 and E1/NS5A (Fig. 6B), and NS2/NS3 (Fig. 6C and D) resulted in only 2- to 3-fold higher levels of luciferase activity than the pM/pVP16 vector control. The coexpression of wild-type C and NS5A increased the luciferase activity by about 7-fold, consistent with previous findings that HCV C and NS5A interact with each other (9, 24, 34). However, the adaptive mutations K74T (C) and Q1012R (NS2) remarkably enhanced the physical interactions of C and NS2 with NS3 and NS5A (Fig. 6A and C). The coexpression of adaptive mutation-containing C/NS3 and C/NS5A resulted in 15- and 26-fold higher luciferase activity (Fig. 6A). Also, the adaptive mutation P251L (E1) significantly increased the E1 and NS3 interaction by 7-fold (Fig. 6B). Similarly, the adaptive mutations of NS3 significantly increased its interactions with adaptive mutation-containing C (5-fold), NS2 (10-fold), and NS5A (5-fold) (Fig. 6D). These results suggest that the adaptive mutations in different viral proteins enhanced protein-protein interactions.
Fig 6.
Determination of HCV protein-protein interactions by a mammalian two-hybrid system. Huh-7.5 cells in 12-well plates were cotransfected with 0.5 μg of pFR-Luc, 0.05 μg of phRL-SV40 (the Renilla luciferase as an internal control), 0.25 μg of the vector pM, expressing the fusion protein between Gal4-BD and each HCV protein, and 0.25 μg of the vector pVP16, expressing each HCV protein using the DMIRE-C reagent (Invitrogen). At 48 h p.t., the levels of firefly and Renilla luciferase activities were determined using dual-luciferase assay kits (Promega). The firefly luciferase activities (fold) relative to that of the vector (pM/pVP16) control, which is considered 1, are calculated and plotted upon normalization with Renilla luciferase activity. The numbers (fold) in the vertical axis are mean values with standard deviations derived from three independent experiments.
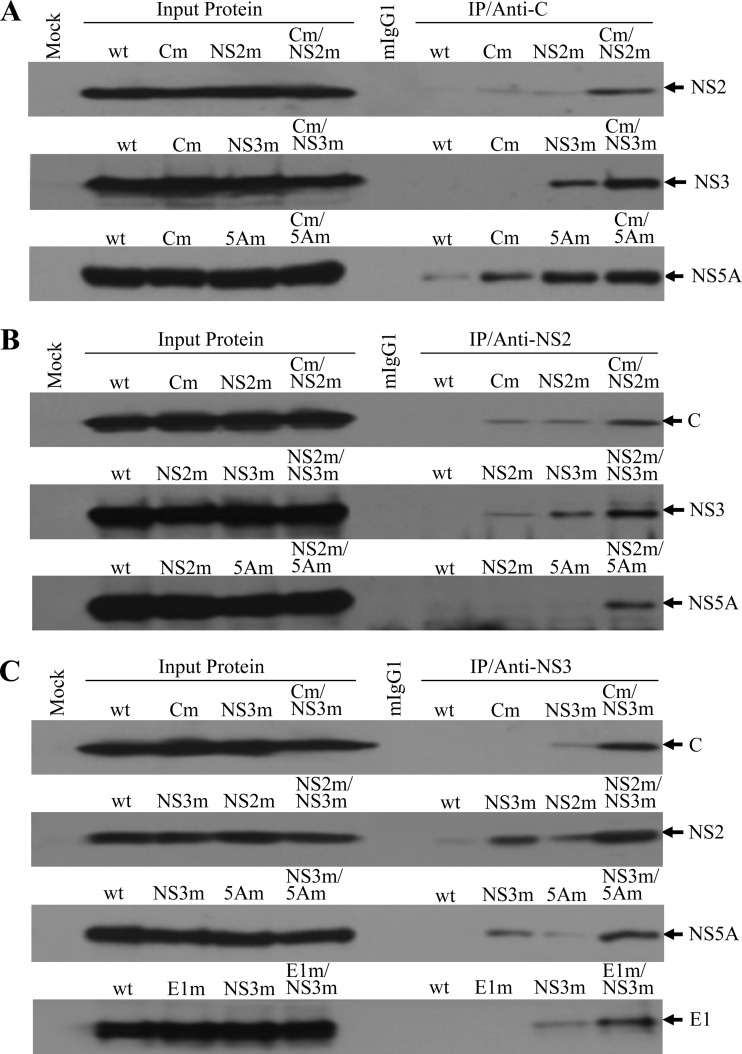
To confirm the enhancement of viral protein-protein interactions by adaptive mutations, coimmunoprecipitation (co-IP) experiments were subsequently carried out according to the method described in Materials and Methods and also our previous study (5). Huh-7.5 cells were transfected with infectious HCV RNAs containing mutations in the C, NS2, NS3, and NS5A genes individually or in different combinations. At 72 h p.t., cell lysate was prepared from the HCV RNA-transfected cells and was used for co-IP experiments. Monoclonal antibodies specific to HCV C (Fig. 7A), NS2 (Fig. 7B), or NS3 (Fig. 7C) were used to precipitate viral protein complexes. Upon co-IP, HCV C, E1, NS2, NS3, and NS5A were detected by Western blotting using specific antibodies, respectively. In the absence of adaptive amino acid mutations (wild type), the C-specific monoclonal antibody (anti-C) did not precipitate detectable amounts of HCV NS2 and NS3, although it brought down a small amount of NS5A (Fig. 7A, wt). Interestingly, the adaptive mutations present in individual C (Cm), NS2 (NS2m), NS3 (NS3m), and NS5A (5Am) slightly enhanced interactions of these proteins as shown by increased amounts of the NS2, NS3, and NS5A proteins precipitated by anti-C antibody (Fig. 7A, Cm, NS2m, NS3m, and 5Am). Strikingly, different combinations of adapted mutations present in C, NS2, NS3, and NS5A significantly increased physical interactions among these viral proteins (Fig. 7A, Cm/NS2m, Cm/NS3m, and Cm/5Am). These results were confirmed by co-IP experiments using NS2 (Fig. 7B) and NS3 (Fig. 7C) monoclonal antibodies. Both NS2 and NS3 monoclonal antibodies did not bring down significant amounts of the wild-type C, NS2, NS3, and NS5A proteins (Fig. 7B and Fig. 7C, wt). However, anti-NS2 and anti-NS3 precipitated significantly larger amounts of HCV C, NS2, NS3, and NS5A proteins containing adapted mutations when present in different combinations (Fig. 7B, Cm/NS2m, NS2m/NS3m, and NS2m/5Am; and Fig. 7C, Cm/NS3m, NS2m/NS3m, and NS3m/5Am). Additionally, the adaptive mutation in E1 significantly increased its interaction with mutant NS3 (Fig. 7C). Once again, these results suggest that the adapted mutations in C, E1, NS2, NS3, and NS5A promoted physical interactions among these viral proteins. Collectively, these findings suggest that cell culture-adapted mutations enhanced physical interactions between the structural protein C/E1 and NS proteins and among such NS proteins as NS2, NS3, and NS5A. The enhancement of the HCV protein-protein interactions by adaptive mutations likely contributes to the promotion of infectious HCV morphogenesis.
Fig 7.
HCV protein-protein interactions determined by co-IP. (A) Co-IP of HCV proteins using a C-specific monoclonal antibody. The wild-type and mutant JFH1 RNAs were transfected into Huh-7.5 cells as described in Materials and Methods. At 72 h p.t., the RNA-transfected cells were lysed in a RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM sodium chloride, 1% Nonidet P-40, and 0.5% sodium deoxycholate). HCV proteins were coprecipitated using an HCV C-specific MAb (anti-C), which was bound to protein G-conjugated agarose beads. Upon co-IP, HCV NS2, NS3, and NS5A were detected by Western blotting using specific antibodies. (B) Co-IP of HCV proteins by an NS2 monoclonal antibody. The co-IP experiments were done in the same way as for panel A except that an NS2 MAb (anti-NS2) was used for IP. Upon co-IP, HCV C, NS3 and NS5A were detected by Western blotting. (C) Co-IP of HCV proteins by an NS3 monoclonal antibody. The co-IP experiments were carried out by incubating the cell lysates with an NS3-specific MAb (anti-NS3). Upon co-IP, HCV C, E1, NS2, and NS5A were detected by Western blotting using specific antibodies. The input lysate is shown on the left side, and IP with C-, NS2-, and NS3-specific monoclonal antibodies is on the right side. HCV proteins detected by Western blotting are highlighted on the right. “Mock” indicates Huh-7.5 cells without HCV proteins. A negative control for IP with a normal mouse IgG1 is indicated by mIgG1. The lower-case m stands for HCV proteins containing adaptive amino acid mutations.
DISCUSSION
In this study, we have characterized a cell culture-adapted HCV variant of genotype 2a (JFH1) and determined the importance and mechanism of action of the adaptive amino acid mutations in the promotion of HCV assembly/morphogenesis in cell culture. Several previous studies found that cell culture-adapted HCVs of genotypes 1a, 1b, 2a, and 3a all grew to infectious titers 2 to 3 orders of magnitude higher than those of wild-type viruses. Sequence analysis and reverse genetics studies identified various amino acid mutations occurring in C (K78E), E1 (I372V and I374T), E2 (N417S, G451R, or N534K), p7 (N765D), NS2 (W879R or Q1012R), NS3 (M1290K or I1316V), NS4B (V1761L), NS5A (V2153A, L2175V, and V2440L), and NS5B (V2941M) individually or in different combinations (6, 10, 13, 18, 29, 31, 41). The Q1012R (NS2) mutation was previously found to coexist with the N765D (p7) mutation (31), while the V2440L (NS5A) mutation occurred together with mutations in C, E1, E2, and NS5B (18). However, these two mutations have not been previously found in the same adapted HCV. The adapted virus isolated in our lab contains both the Q1012R and V2440L mutations, two major determinants for the enhancement of HCV production, resulting in infectious titers 29- and 31-fold higher than those of wild-type virus, respectively (Fig. 2E and Fig. 5). The combination of these two mutations increased the titer of infectious HCV by about 100-fold (Fig. 4B). There are several other determinant mutations for HCV adaptation in cell culture, including the N417S, G451R, or N534K mutation in E2 (6, 31, 41) and the N765D mutation in p7 (18, 31). We have also made infectious viruses containing adaptive mutations in E2 or p7. In a parallel comparison with the mutations in E2 and p7, the adaptive mutations found in our HCV variant resulted in the highest infectious titer (J. Jiang and G. Luo, unpublished results). It will be interesting to determine whether combinations of the major adaptive mutations in E2, p7, NS2, and NS5A will result in an HCV with an even higher infectious titer. The availability of an HCV variant with the highest titer will greatly facilitate the structural determination of the HCV virion and the development of a potential inactivated HCV vaccine.
The characterization of our adapted HCV variant provided new evidence demonstrating the important roles of the cross-talk between HCV structural and NS proteins in virion assembly and morphogenesis. First, the adaptive mutations in C, E1, NS3, and NS5B per se did not significantly increase the production of infectious HCV but synergistically enhanced the promotion of the HCV assembly by the adaptive mutations in NS2 and NS5A, respectively (Fig. 3). This observation differs from the previous findings that the adaptive mutations in E1, E2, and NS5B had no significant effect on the enhancement of HCV production by the adaptive mutations in NS5A (18). Additionally, findings derived from our studies suggest that all three domains (I, II, and III) of NS5A are important for HCV assembly and/or morphogenesis. The adaptive mutations in NS5A domains I, II, and III were found to independently promote the production of infectious HCV by more than 10-fold, even though they did not modulate viral RNA replication (Fig. 5). It has been thought that NS5A domain III is a major determinant for HCV assembly and/or production (1, 37). A previous mutagenesis analysis of NS5A demonstrated that a single phosphorylation site in NS5A domain III is essential for the formation of infectious HCV particles without altering the efficiency of viral RNA replication (37). We have also examined the effects of adaptive mutations in domains I, II, and III on NS5A phosphorylation and did not detect significant differences in NS5A phosphorylation among the HCV variants containing adaptive mutations in NS5A domains I, II, and III (data not shown). Therefore, the enhancement of HCV production by the adaptive mutations in three different NS5A domains was most likely mediated by increased interactions among various viral proteins. This possibility is supported by the adaptive mutation-modulated enhancement of multiple interactions among HCV structural and NS proteins (Fig. 6 and Fig. 7). Further studies are warranted to determine whether the three NS5A domains mediate physical interactions with different viral proteins.
The underlying molecular mechanisms for the adaptive mutation-mediated enhancement of HCV morphogenesis have not been well understood. Similar to previous findings (6, 10, 13, 18, 29, 31, 41), the adaptive amino acid mutations did not significantly affect the efficiency of viral RNA replication, as suggested by similar levels of positive-stranded HCV RNA in the cells transfected with wild-type and mutant RNAs, which were quantified by both RPA and qRT-PCR methods (Fig. 2C and data not shown). Circumstantial evidence derived from several previous studies suggested that the adaptive mutations modulate HCV infectivity (6, 31, 41), stability (29), or polyprotein processing (18), depending on their locations in specific viral proteins. The adaptive mutations in E2 (N417S, G451R, and N534K) were found to primarily facilitate HCV infectivity (6, 31, 41), whereas the V2440L mutation in NS5A was shown to slow down cleavage at the junction between NS5A and NS5B (18). Recently, a cell culture-adapted J6/JFH1 variant containing novel mutations in C (K78E), NS2 (W879R), and NS4B (V1761L) was found to have longer infectious stability than wild-type virus, suggesting a different mechanism for enhancing HCV morphogenesis (29). Distinct from these earlier studies, our studies demonstrated for the first time that adaptive mutations could promote physical interactions among HCV structural and NS proteins. The increased protein-protein interactions by adaptive mutations most likely contributed to the enhancement of HCV assembly rather than virion egression, as suggested by marked increases of intracellular HCV titers (Fig. 3C and data not shown). The enhancement of physical interactions by adaptive mutations was exemplified by increased protein-protein interactions between C, E1, NS2, NS3, and NS5A containing adaptive mutations compared to those between corresponding wild-type proteins, as demonstrated by both mammalian two-hybrid assays and co-IP (Fig. 6 and Fig. 7). These findings are in line with those recently reported by others that NS2 plays a central role in HCV assembly through physical interactions with other viral structural (E1/E2) and NS proteins (16, 35). In addition to NS2, our findings suggest that complex protein-protein interactions among HCV structural and NS proteins are involved in virion assembly. Future studies are needed to determine whether NS proteins have any structural functions in HCV morphogenesis. It should be noted that the enhancement of infectious HCV titers by adaptive mutation does not perfectly match the increases in protein-protein interactions (Fig. 3 and Fig. 6), suggesting other mechanism likely involved in HCV morphogenesis. For instance, adaptive mutations could alter the association of core with lipid droplets (LDs), which was previously demonstrated to determine HCV morphogenesis and/or egression (33). The J6 core, which remarkably enhanced HCV production by 100-fold, was found to only have partial colocalization with LDs, unlike the lower-HCV-producing JFH1 core, which perfectly colocalized with LDs in the perinuclear region (33). Similar to these findings, we found that the double mutations Q1012R (NS2) and V2440L (NS5A) resulted in partial colocalization between core and LDs, in contrast to the core of wild-type HCV, which was predominantly colocalized with LDs in the perinuclear region. However, the colocalization between core and NS5A was not affected by any of the adaptive mutations (data not shown). These observations suggest that adaptive mutations in NS protein can alter the association of core with LDs and therefore facilitate HCV assembly and/or egression. The question of how enhanced protein-protein interactions by adaptive mutations alter the colocalization between core and LDs and consequently promote HCV production/egression remains enigmatic. Future investigations are warranted to further determine the underlying mechanism of HCV NS proteins in virion morphogenesis.
ACKNOWLEDGMENTS
We thank Charles M. Rice (Rockefeller University) for the Huh-7.5 cell line and the NS2 and NS5A monoclonal antibodies, Takaji Wakita (Japan NIAID) for the JFH1 replicon cDNA, and Jake Liang (NIDDK) for core and E1 monoclonal antibodies.
This work was supported by NIH/NIAID grants AI091953, AI097318, and AI092074 and in part by a grant from the Kentucky Science and Engineering Foundation (KSEF-148-502-10-272).
Footnotes
Published ahead of print 6 June 2012
REFERENCES
- 1. Appel N, et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035 doi:10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cai Z, et al. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963–13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang KS, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81:13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choo QL, et al. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362 [DOI] [PubMed] [Google Scholar]
- 5. Cun W, Jiang J, Luo G. 2010. The C-terminal alpha-helix domain of apolipoprotein E is required for interaction with nonstructural protein 5A and assembly of hepatitis C virus. J. Virol. 84:11532–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Delgrange D, et al. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J. Gen. Virol. 88:2495–2503 [DOI] [PubMed] [Google Scholar]
- 7. El-Hage N, Luo G. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761–2769 [DOI] [PubMed] [Google Scholar]
- 8. Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goh PY, et al. 2001. The hepatitis C virus core protein interacts with NS5A and activates its caspase-mediated proteolytic cleavage. Virology 290:224–236 [DOI] [PubMed] [Google Scholar]
- 10. Gottwein JM, et al. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 133:1614–1626 [DOI] [PubMed] [Google Scholar]
- 11. Gottwein JM, et al. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364–377 [DOI] [PubMed] [Google Scholar]
- 12. Goueslain L, et al. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J. Virol. 84:773–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han Q, et al. 2009. Compensatory mutations in NS3 and NS5A proteins enhance the virus production capability of hepatitis C reporter virus. Virus Res. 145:63–73 [DOI] [PubMed] [Google Scholar]
- 14. Jiang J, Luo G. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83:12680–12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jirasko V, et al. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546–28562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jirasko V, et al. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 6:e1001233 doi:10.1371/journal.ppat.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaul A, Woerz I, Meuleman P, Leroux-Roels G, Bartenschlager R. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J. Virol. 81:13168–13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leung JY, et al. 2008. The role of nonstructural protein Ns2a in flavivirus assembly. J. Virol. 82:4731–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lindenbach BD, Rice CM. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933–938 [DOI] [PubMed] [Google Scholar]
- 21. Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication, 5th ed, vol 1 Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 22. Liu WJ, Chen HB, Wang XJ, Huang H, Khromykh AA. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 78:12225–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo G, Xin S, Cai Z. 2003. Role of the 5′-proximal stem-loop structure of the 5′ untranslated region in replication and translation of hepatitis C virus RNA. J. Virol. 77:3312–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masaki T, et al. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moradpour D, et al. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 60:103–109 [DOI] [PubMed] [Google Scholar]
- 26. Murray CL, Jones CT, Rice CM. 2008. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 6:699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murray CL, Jones CT, Tassello J, Rice CM. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patkar CG, Kuhn RJ. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 82:3342–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pokrovskii MV, et al. 2011. Novel mutations in a tissue culture-adapted hepatitis C virus strain improve infectious-virus stability and markedly enhance infection kinetics. J. Virol. 85:3978–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robertson B, et al. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch. Virol. 143:2493–2503 [DOI] [PubMed] [Google Scholar]
- 31. Russell RS, et al. 2008. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 105:4370–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seeff LB, Hoofnagle JH. 2002. National Institutes of Health Consensus Development Conference: management of hepatitis C: 2002. Hepatology 36:S1–S2 [DOI] [PubMed] [Google Scholar]
- 33. Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169 [DOI] [PubMed] [Google Scholar]
- 34. Shi ST, et al. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198–210 [DOI] [PubMed] [Google Scholar]
- 35. Stapleford KA, Lindenbach BD. 2011. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 85:1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steinmann E, et al. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103 doi:10.1371/journal.ppat.0030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032 doi:10.1371/journal.ppat.1000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. WHO 1998. WHO concerns on hepatitis C. Lancet 351:1415 [Google Scholar]
- 40. Yi M, Ma Y, Yates J, Lemon SM. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhong J, et al. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]