Abstract
Members of the Alphavirus genus are arboviruses that alternate replication in mosquitoes and vertebrate hosts. In vertebrate cells, the alphavirus resists the activation of antiviral RNA-activated protein kinase (PKR) by the presence of a prominent RNA structure (downstream loop [DLP]) located in viral 26S transcripts, which allows an eIF2-independent translation initiation of these mRNAs. This article shows that DLP structure is essential for replication of Sindbis virus (SINV) in vertebrate cell lines and animals but is dispensable for replication in insect cells, where no ortholog of the vertebrate PKR gene has been found. Sequence comparisons and structural RNA analysis revealed the evolutionary conservation of DLP in SINV and predicted the existence of equivalent DLP structures in many members of the Alphavirus genus. A mutant SINV lacking the DLP structure evolved in murine cells to recover a wild-type phenotype by creating an alternative structure in the RNA that restored the translational independence for eIF2. Genetic, phylogenetic, and biochemical data presented here support an evolutionary scenario for the natural history of alphaviruses, in which the acquisition of DLP structure in their mRNAs probably allowed the colonization of vertebrate host and the consequent geographic expansion of some of these viruses worldwide.
INTRODUCTION
The natural cycle of many viruses involves replication in more than one host species. Arthropod-borne viruses (arboviruses) are examples of broad-host-range viruses that can alternate replication in insects (the primary vector) and many vertebrate hosts, including humans. This versatility reflects the notable adaptation capability of these viruses for replicating in evolutionarily distant hosts with remarkable genetic differences (7, 13, 47, 57). Assuming that arthropods were the primitive hosts of the arboviruses, the further colonization of vertebrates hosts had to involve an enormous challenge to these viruses by the presence of innate and adaptive (immune) antiviral responses. Although insects are endowed with a small interfering RNA (siRNA) interference-based defense mechanism that can attenuate virus replication (22, 31, 45), protective responses against viruses generally require a more complex coordinated action of both innate and adaptive systems that are present only in vertebrates (4, 9, 25, 36). Thus, the secretion of interferons (IFNs) and other proinflammatory cytokines greatly influences the outcome of virus replication in animals, by limiting the virus tropism to specific tissues or by thwarting interspecies transmission in some cases (29, 55). The double-stranded RNA (dsRNA)-activated protein kinase (PKR) (coded for by the EIK2AK2 or PKR gene) induced by interferon is an important component of this innate response against viruses and other pathogens (30, 36, 44). As soon as dsRNA molecules accumulate in the cell as a consequence of virus replication, PKR is activated and phosphorylates translation initiation factor 2 (eIF2) in an attempt to block translation of viral mRNAs. Phosphorylation of eIF2 at S51 of its alpha subunit (eIF2α) prevents the recycling of this factor, which is necessary for ongoing translation, causing a rapid halt in viral protein synthesis and virus multiplication (12, 23). However, many viruses express proteins (or RNAs) that prevent PKR activation by allosteric inhibition or by acting as pseudosubstrates of the kinase (reviewed in reference 23). This intimate host-parasite coevolution has been recently illustrated by the fast evolution rate (and positive selection) experienced by the PKR gene during the last 50 million years of evolutionary divergence of primates, most probably driven by intense episodes of antagonist evolution with some viruses encoding proteins that act as inhibitors of the kinase (16, 17, 41, 60).
Among arboviruses, the Alphavirus group shows one of the broadest host ranges known. Alphavirus is composed of seven antigenic groups, and members of this genus have been isolated from mosquitoes, birds, rodents, fish, pigs, bats, sheep, marsupials, horses, monkeys, and humans (39, 47, 57). Moreover, the prototype alphavirus, Sindbis virus (SINV), is widely distributed in four of the six continents. Alphaviruses are maintained in natural cycles involving transmission by an arthropod vector (mainly mosquitoes from the Culex and Aedes genera) among susceptible vertebrate hosts. Replication of alphavirus in insect vectors is generally persistent, whereas in mammalian hosts, these viruses replicate producing an acute (and generally short duration) infection that is often associated with rash or encephalitis, depending on the virus species (34, 40, 47). Contrary to what has been observed for most viruses, infection of murine cells with SINV or Semliki Forest virus (SFV) resulted in a strong activation of PKR, which leads a complete phosphorylation (and inactivation) of eIF2α (14, 54). Viral 26S mRNAs that encode the structural proteins of SINV are efficiently translated under these conditions by means of a prominent cis-acting secondary structure in this mRNA that allows location of 40S ribosome on initiator AUG in the absence of eIF2 (50, 54). This structure, called the downstream loop (DLP), is located 28 nucleotides (nt) downstream from the AUG in SINV 26S mRNA and in other members of the Alphavirus group (see below). Elimination of DLP resulted in a SINV mutant unable to replicate in normal murine fibroblasts but which replicated to wild-type (WT) levels in cells lacking the PKR gene (54). An evolutionary interpretation of this result is that the DLP structure may have been acquired by some alphavirus in the past, during adaptation to vertebrate hosts. This article shows molecular evidence supporting this idea.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with the recommendations in Directive 86/609/EEC of the European Union on the protection of animals used for experimental and other scientific purposes (17a), and which was implemented by the Spanish Government under approval no. 1201/2005. The protocol was approved by the Committee on the Ethics of Animal Experiments of Universidad Autónoma de Madrid (permit no. CEI 20-419). All surgery was performed under isoflurane anesthesia, and all efforts were made to minimize suffering.
Cells, viruses, and animals.
Mouse embryonic fibroblast (MEF) cells derived from wild-type and PKRo/o animals (61) were grown as described previously. Only low-passage-number stocks of MEFs and NIH 3T3 cells were used, given the tendency of these cells to undergo spontaneous transformation that leads to the loss of PKR response to infection. MEFs were prepared from 129sv mouse embryos at the E13 stage, whereas chicken embryo fibroblasts (CEFs) were prepared from 10- to 12-day-old chicken eggs according to standard procedures. All vertebrate cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal or normal calf serum. H5 cells (from Trichoplusia ni eggs; Invitrogen) were grown in TC100 medium supplemented with 10% of fetal calf serum and gentamicin. C6/36 cells were grown in M3 medium supplemented with 10% fetal calf serum, gentamicin, antibiotics, and antimycotic. SINV (AR339 strain) and SFV were amplified in BHK21 cells and purified through a sucrose cushion as described previously (50). The Aura virus (AURAV; strain BeAR 10315) was purchased from ATCC (catalog no. VR-368), amplified in BHK21 cells, and purified by polyethylene glycol 6000 (PEG 6000) precipitation (26). The SINV ΔDLP mutant was obtained after electroporation of BHK21 cells with viral RNA derived from the pT7-Toto1101 infectious clone as described previously (54). The SINV expressing the enhanced green fluorescent protein (EGFP) mRNA from a duplicated subgenomic promoter was described previously (54). Wild-type and Pkr knockout mice of strain 129Sv (2) were propagated according to standard methods.
Virus infections.
Cultured cells growing in 24-well plates were inoculated with purified viruses at the indicated multiplicity of infection (MOI) in 0.3 ml of DMEM lacking serum. After 1 h of adsorption, the inoculum was replaced by a growth medium, and samples were analyzed at the indicated times for Western blotting or metabolic labeling with [35S]Met/Cys as described previously (54). For replication studies, cells were infected with a low multiplicity of infection (0.1 to 1) and viral yields produced at 48 h postinfection were quantified by plaque assay on BHK21 cells as described previously (54). For mouse infections, 107 PFU of virus was used to inoculate animals by the intranasal route under isoflurane anesthesia. At the times indicated, animals were sacrificed and brains were extracted for virus yield determination or immunofluorescence analysis using the anti-SINV capsid antisera as described previously (50).
RNA structure analysis by SHAPE.
In vitro-synthesized RNAs encompassing nt 35 to 250 of 26S mRNA from WT and ΔDLP viruses and nt 35 to 273 from revertant virus were probed with N-methylisatoic anhydride (NMIA) as described previously (10, 59). About 2 pmol of RNA was mixed with 2 pmol of VIC-labeled reverse primer (54) in 0.5× Tris-EDTA (TE) buffer, heated at 95°C for 5 min, and then quenched on ice. Refolding buffer was added (100 mM Tris-HCl [pH 8.0], 100 mM KCl, 6 mM MgCl2), and RNAs were incubated first at 37°C and then at room temperature (both for 30 min). NMIA was added to a final concentration of 13 mM and allowed to react for 45 min, and then RNAs were precipitated with isopropanol and reversed transcribed with Superscript III (Invitrogen) at 52°C. Products were analyzed by capillary electrophoresis, and data were processed using ShapeFinder (51). Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) data were used to constrain folding predictions with the RNA Vienna package and MC-Fold/MC-Sym pipeline (33).
Bioinformatic analysis.
All bioinformatic analyses were carried out using web servers with open access programs. The BLAST data from the NCBI and the Flybase page (http://flybase.org/blast/) were used for searching PKR homologs. For secondary RNA structure prediction, we routinely used the Vienna RNA webServer (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) and MC-Fold/MC-Sym pipeline (http://www.major.iric.ca/MC-Fold/). Three criteria were used to detect stable DLP structures in viral subgenomic mRNAs. (i) The first criterion was the presence of low free energy values predicted by folding programs (<−20 kcal mol−1), which are indicative of stability. (ii) The G+C content of DLP should not be <60%. (iii) The DLP structure should represent an independent folding domain when placed into a heterologous context.
To analyze genetic variability within the first 400 nucleotides of 26S mRNA SINVs, we first selected 16 sequences from GenBank that are representative of the main 5 genotypes (or geographic subspecies) of SINV with available sequence data (28). The following sequences were used, with accession numbers given in parentheses: EgAR338 (AF061206.1), EgAR339 (AF061205.1), Babanki (AF339477.1), BH40503 (AF06121 9.1), Girdwood (U38304.1), Ockelbo (M69205.1), Kyzylagach (AF339478.1), MK6962 (AF061209.1), MM840 (AF061216.1), MRE16 (AF061210.1), RRD764 (AF061234.1), S.A.AR86 (U38305.1), SW6562 (AF06123 6.2), XJ-160 (AF103728.1), YN87448 (A F103734.1), MRM39 (AF061208.1), and Whataroa (AF339479). To avoid bias of data toward the overrepresented Australian isolates annotated in GenBank, these were first grouped into five subtypes with 1 or 2 representative members for each group: A (MRM39), B (MK6962 and MM840), C (MRE16 and BH40503), D (RRD764), and E (SW6562) (46). Sequences were aligned with MUSCLE and T-Coffee, gaps were removed, and variability was calculated by the Shannon entropy method (http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy.html). For capsid variability analysis, the following viral sequences were used, with the GenBank accession numbers shown: SINV, NC_001547.1; Aura virus (AURAV), AF126284; Western equine encephalitis virus (WEEV), NC_0039 08; Nduma virus (NDUV), AF339487; Venezuelan equine encephalitis virus (VEEV), L01442; Chikungunya virus (CHIKV), NC_004162; O'nyong-nyong virus (ONNV), NC_0015 12; Barmah Forest virus (BFV), NC_001786; Eastern equine encephalitis virus (EEEV), South America, AF159559; EEEV, North America, U01558; Mayaro virus (MAYV), DQ 001069; SFV, NC_003215; Ross River virus (RRV), DQ226993; RRV, Sagiyama virus, AB032553; Getah virus (GETV), NC_006558; Middelburg virus (MIDV), EF536323; Unah virus (UNAV), AF33948; Bebaru virus (BEBV), AF339480; Southern elephant seal virus (SESV), HM147990; sleeping disease virus (SDV), AJ316246; and salmon pancreas disease virus (SPDV), AJ316244.
For phylogenetic analysis, the first 150 nt of 26S mRNA coding sequences that include DLP structures were aligned with the T-Coffee program by the MAGNOLIA webserver (18), which incorporates the coding frame information of mRNA to guide sequence alignments. GTR+I+Γ, HKY+I+Γ, and TN93+I+Γ substitution models were used according to MODELTEST prediction (37). I also used amino acid sequences of capsid or from the entire structural polyprotein that were aligned with T-Coffee (32). In this case, the JTT+I+Γ substitution model was used for evolutionary estimates. Phylogenetic analysis were performed with MEGA 5.0 (48) and Phylogeny.fr (11).
Antibodies, immunoblotting, and immunofluorescence.
The following antibodies were used: anti-PKR (Santa Cruz Biotech.), anti-eIF2α (Santa Cruz Biotech.), anti-phospho eIF2α (Invitrogen and Cell Signaling), and anti-SINV capsid (rabbit antisera). For immunoblotting, crude cell extracts made in sample buffer were separated by 12% acrylamide SDS-PAGE and transferred to nitrocellulose using a semidry apparatus (Bio-Rad). Membranes were incubated with primary and secondary antibodies conjugated to horseradish peroxidase (HRP) in phosphate-buffered saline (PBS) with 5% dry milk, and revealed by enhanced chemiluminescence (ECL) as described previously (54). For immunofluorescence analysis, brains fixed in 4% paraformaldehyde were cryoprotected with 10% sucrose overnight (O/N) and frozen. Fifteen-micrometer slides were made in a cryostat, incubated with primary antibodies O/N at 4°C in a wet chamber, washed, incubated with secondary antibodies conjugated to Alexa Fluor 488, and mounted. Preparations were examined and photographed in a Leica confocal microscope.
Virus evolution.
To force the reversion of the ΔDLP mutant virus, we first prepared a ΔDLP virus stock free of wild-type contamination. For this, the pT7-SVΔDLP plasmid (54) was recloned three times by consecutive rounds of transformation in Escherichia coli, followed by colony isolation and plasmid purification. Then, SINV ΔDLP RNA was synthesized in vitro and electroporated in BHK21 cells, which allowed replication of the mutant virus to a level comparable to that of the wild type. Two flasks of low-passage-number NIH 3T3 cells (about 2 × 107 cells per flask) were infected with virus at an MOI of 0.1, and 5 days later, both intracellular and extracellular viruses were recovered, amplified in BHK21 cells, and titrated. This preparation was used to infect new flasks of NIH 3T3 cells as described above. Eleven passages were done. Viruses with large lysis plaques that arose from the 5th to 6th passage were isolated and amplified. Genomic RNAs from these biological clones were isolated and retrotranscribed with Superscript II reverse transcriptase (RT) (Invitrogene) using the primers SINV-6863 (GGTACTGGAGACGGATATCGC) and SINV-8101 (GATAGCACAGGGTGGTCGATGG). Amplified cDNA was sequenced, and the fragment encompassing the HpaI (position 6916) and PlmI (position 8066) sites was subcloned into the pT7-SINVΔDLP plasmid. The resulting reconstituted revertant virus from this molecular clone was obtained as described above.
Proteomic analysis.
For N-terminal sequence determination of capsid proteins, about 108 cells were infected with revertant SINV at an MOI of 5 PFU/cell. After 8 h postinfection (hpi), the growth medium was washed out, and cells were incubated in medium lacking serum for 14 h. The medium then was recovered and clarified by centrifugation at 8,000 × g for 35 min, and viral particles were precipitated with 9% PEG 6000 (Sigma) at 4°C O/N. After centrifugation, the sediment was resuspended in TNE buffer (50 mM Tris [pH 7.5], 140 mM NaCl, 5 mM EDTA) and spun in a 15 to 40% sucrose gradient (made in TNE) for 4 h at 35,000 rpm in an SW40 rotor (Beckman). Fractions 10 and 11 containing the viral particles were pooled and separated by SDS-PAGE, and gel slices containing capsid proteins were digested with trypsin and subjected to fingerprint analysis by matrix-assisted laser desorption ionization–tandem time of flight mass spectrometry (MALDI-TOF/TOF MS) as described previously (54).
RESULTS
Differential responses of insect and vertebrate cells to SINV infection.
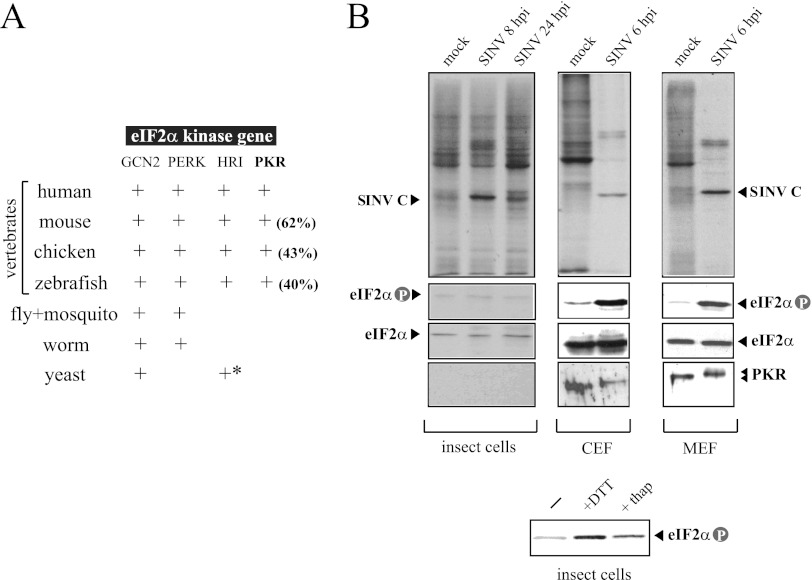
Among eIF2α kinases, PKR is only present in vertebrates, whereas other members of this family, such as GCN2 and PERK, have been found in all metazoans with available genomic sequence information (Fig. 1A) (12). BLAST searching for homologs of the human PKR gene in Drosophila melanogaster and three mosquitoes species databases (Aedes aegypti, Culex pipiens, and Aedes gambiae) confirmed the absence of a PKR ortholog in insects. Since alphavirus can infect both invertebrate and vertebrate hosts, I compared the replication of SINV in insect cells (H5) and avian and murine fibroblasts (CEFs and MEFs, respectively) and the degree of eIF2 phosphorylation in response to infection in these cells. SINV efficiently replicated in invertebrate and vertebrate cells, although the types of infection established in these cells were dramatically different. Infection was highly cytopathic in CEF and NIH 3T3 cells, giving complete cell lysis at 48 hpi. At shorter times of infection, the synthesis of SINV structural proteins was apparent over a profound shutoff of host protein synthesis (Fig. 1B). This correlated with a strong activation of PKR (mobility shift) and the subsequent eIF2α phosphorylation in vertebrate cells (Fig. 1B). In contrast, infection of H5 cells was largely noncytopathic and the virus easily established a persistent infection in these cells, with continuous production of virus as described earlier for arthropod cell lines (34, 40). An equivalent result was obtained in the mosquito cell line C6/36 (data not shown). Neither inhibition of host translation nor eIF2α phosphorylation was observed in H5 cells upon infection with SINV, despite the massive synthesis of viral proteins detected in these cells (Fig. 1B). This shows that insect cells lack of PKR-like activity, but they express other eIF2α kinases such as PERK that responded to endoplasmic reticulum (ER) stress as occurred in mammalian cells (Fig. 1B) (1).
Fig 1.
Differential responses of insect and vertebrate cells to SINV infection. (A) The PKR gene is only present in vertebrates. The result of a BLAST searching for orthologs of eIF2α kinases in different species is summarized in this table. Amino acid sequence identity with human PKR is shown in parentheses. An asterisk denotes the presence of HRI-related genes only in Schizosaccharomyces pombe. (B) Effect of SINV infection on translation, PKR activation and eIF2α phosphorylation in insect (H5), avian (CEF), and murine (MEF) cells. Cells were infected at an MOI of 25 PFU/cell and at the indicated times pulsed with [35S]Met/Cys for 30 min, and labeled proteins were analyzed by autoradiography as described in Materials and Methods (upper panels). Extracts were also analyzed by immunoblotting against phospho-eIF2α, total eIF2α, and PKR. Note that the insect eIF2α band migrated more slowly than the vertebrate eIF2α protein. In parallel experiments, H5 cells were treated for 2 h with 1 mM DTT and 10 μM thapsigargin, two stressors that induced eIF2α phosphorylation by activating the endoplasmic reticulum-associated eIF2α kinase (PERK) (lower panel).
DLP structure of SINV 26S mRNA is essential for virus replication in vertebrate hosts.
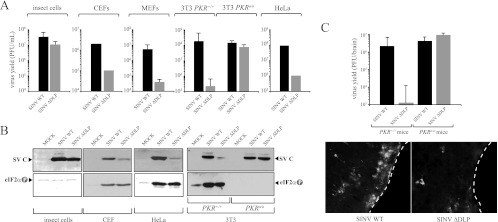
In previous reports, we found that a stable DLP structure of RNA located downstream from the initiation codon was necessary and sufficient to support eIF2-independent translation initiation of subgenomic (26S) mRNAs in SINV-infected 3T3 cells (50, 54). To test the importance of DLP structure in virus replication, I used a SINV mutant in which the structure of DLP has been destroyed by 7 point mutations as described previously (54). Replication of this mutant virus was drastically impaired in human cells (HeLa), mouse fibroblasts (MEFs and NIH 3T3 cells), and chicken fibroblasts (CEF), although it replicated at levels similar to those of wild-type virus in insect cells and in murine cells derived from Pkr knockout mice (PKRo/o) (Fig. 2A and B). This result shows that DLP structure was only required for virus replication in cells that expressed the PKR gene and that responded to infection by phosphorylating eIF2α. The replication capabilities of WT and ΔDLP viruses in animals were also compared. SINV shows a remarkable neurotropism in mice, infecting groups of neurons in the cortex and the hippocampus as well motoneurons of the spinal cord (4, 8, 27). Histological analysis revealed that whereas the WT virus replicated in large foci of neurons, replication of ΔDLP virus was restricted to very few foci of individual neurons (Fig. 2C). This result revealed a defect of the ΔDLP mutant in tissue spreading.
Fig 2.
DLP structure in SINV 26S mRNA is essential for virus replication in vertebrate hosts. (A) Viral yields of WT and ΔDLP mutant viruses in insect cells (H5), chicken cells (CEF), murine cells (MEF and 3T3 derived from WT and PKR knockout mice), and human cells (HeLa). For CEF and HeLa cells, the data are the means of two independent experiments. For the rest, the means ± standard deviations (SD) of four independent experiments are shown. (B) Synthesis of capsid protein and eIF2α phosphorylation in the different cell lines infected with WT and ΔDLP mutant viruses. (C) DLP structure is essential for replication of SINV in mice. Wild-type and PKRo/o mice were infected with the indicated virus, and 4 days later, animals were sacrificed and virus yields in whole brains were titrated on BHK21 cells as described in Materials and Methods. This graph is similar to that published previously (14), but now more animals per group (n = 7) are included. Data are expressed as means ± SD. Brains of some infected animals were cryosectioned and analyzed by immunofluorescence (IF) using an anti-SINV capsid antiserum as described in Materials and Methods. White dashed lines show the external border of cerebral cortex.
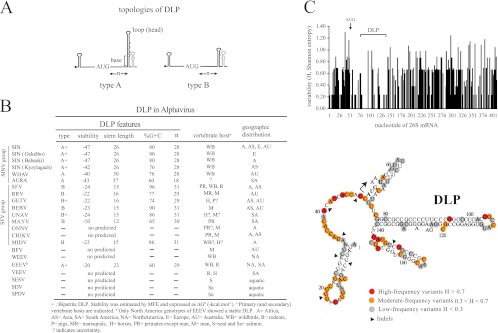
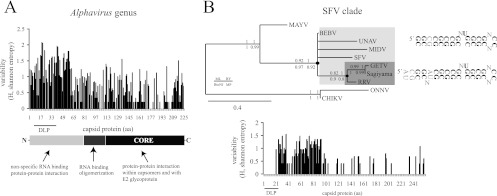
To better understand the importance of DLP structure in alphavirus replication, I searched for equivalent structures in other members of the Alphavirus genus as described in Materials and Methods. All members of the SINV group except the Aura virus showed a very stable DLP structure of similar topology (Fig. 3B). I also found stable DLP structures in SFV, Ross River (RRV), Getah virus (GETV), Middelburg virus (MIDV), Una virus (UNAV), and Bebaru virus (BEBV). Other members of the SFV clade, such as Mayaro virus (MAYV), showed a predictable DLP but of lower stability. However, I was unable to detect a stable DLP structure for O′nyong-nyong virus (ONNV), Chikungunya virus (CHIKV), Barmah Forest virus (BFV), Western equine encephalitis virus (WEEV), or Venezuelan equine encephalitis virus (VEEV) and for aquatic alphaviruses (SDV, SESV, and SPDV). Alphaviral DLPs can be grouped into two topological categories with no obvious sequence homology among them. Type A is composed of a large spiral with some unpaired nucleotides (bulges or small internal loops; e.g., SINV), whereas for type B, the spiral is short and compact (e.g., SFV). In some cases, the first stem-loop (A or B) seems to be propped up by a second stem-loop located immediately downstream of the first one (e.g., SINV) (Fig. 3B and 5D). The distance (n) from the base of DLP to the initiator codon was also conserved, ranging from 28 to 31 nt. We have reported before that this distance allows the appropriate stalling of the 40S ribosome on the initiator AUG codon during the scanning process, thus obviating the participation of eIF2 (20, 21, 50, 54). The only exception found was Aura virus, the most distant member of the SINV group, which shows a lower stable DLP located 18 nt downstream of the initiator codon (see below).
Fig 3.
Structural features of DLPs in Alphavirus. (A) Two main topologies were found; type A, a large spiral with some unpaired nucleotides, and type B, a compact spiral. In some cases, a second stem-loop is localized just downstream from the first (bipartite structure). The distance (n) from the initiation codon (AUG) to the base of DLP is shown. (B) Features of DLP among representative members of Alphavirus genus, including topology, stability, G+C composition, and n distances. The vertebrate hosts that act as primary or secondary virus reservoir, and the geographic distribution for each virus is shown. (Data were taken from the Arbovirus Division of CDC [DVBID] and from the International Committee of Taxonomy of Viruses [ICTVdB].) No vertebrate host for AURAV has been described to date (−). MFE, minimum free energy. (C) DLP sequence and structure are conserved among subspecies, genotypes, and isolates of SINV worldwide (see Materials and Methods). Variability along the first 400 nt of 26S mRNA was calculated by the Shannon entropy (H) method. The position of the initiation codon (AUG) is shown as well as the region that encompasses the DLP structure. The lower panel shows the distribution of variants along the first 168 nt of SINV 26S mRNA. The topology of SINV DLP has been confirmed before by enzymatic probing (54). Data were scored as follows: highly variable positions (red) with H > 0.7 affected for >30% of genotypes, moderately variable positions (yellow) with 0.3 < H < 0.7 affected for 10 to 30% of genotypes, and low-variability positions (gray) with H < 0.3 affected for less than 10% of genotypes. Arrowheads show positions where insertion or deletion of few nucleotides in some variants was found (indels). The initiation codon is marked with an arrow.
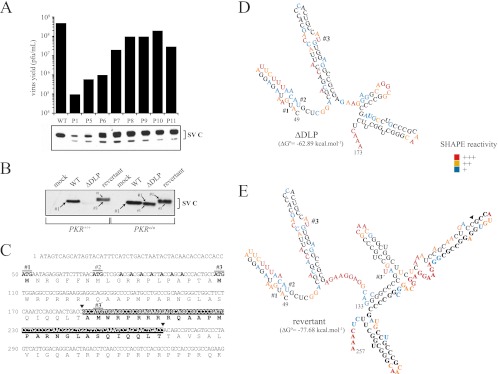
Fig 5.
ΔDLP mutant virus evolved in murine cells to restore DLP activity. (A) Reversion from the 5th to 6th passages allowed fitness gain and rapid enrichment of cultures with revertant viruses. Viral yields for each passage were titrated. Capsid protein synthesis in NIH 3T3 cells infected with virus preparation from each passage is shown. (B) Revertant virus restored translation of 26S mRNA in NIH 3T3 cells. WT and PKRo/o cells were infected with the reconstituted revertant virus, and the synthesis of capsid protein was compared with that of the WT and parental ΔDLP mutant. Capsid bands were numbered according to the AUG codon from which they are translated. The sizes of the arrows are proportional to the translational efficiency from each AUG. (C) Genotype of revertant virus. The first 350 nt of 26S mRNA are shown. Arrowheads mark the insertion of the 84-nt fragment that resulted from the tandem duplication of the RNA fragment (see the text for details). AUG 1 (#1) is the natural initiation codon AUG50, AUG 2 (#2) is AUG72, and AUG 3 (#3) is AUG107. Note that the revertant has a duplicated no. 3 AUG (termed 3′), which was used as an initiation codon at an extremely low rate. The seven A's labeled in boldface are the mutations introduced in the parental WT virus to destroy the DLP structure. (D) RNA secondary structure of ΔDLP virus. MFE corresponds to nt 49 to 257 of 26S mRNA, although only the structure for the fragment from nt 49 to 173 is shown. Shape reactivity was scored as follows: +++, 50 to 100% of maximal; ++, 25 to 50% of maximal; +, <25% of maximal. Noncolored bases show no reactivity. (E) RNA secondary structure of 49 to 257 nt of revertant virus 26S mRNA. Note the new stem-loop formed at 26 nt downstream of AUG 3. Nucleotides that resulted from the tandem duplication are in bold, and the arrowhead indicates the site of insertion. Shape reactivity was scored as indicated above.
To get evidence for DLP selection during the evolutionary history of the SINV group of viruses, I analyzed the changes found in the first 400 nt of 26S mRNA among isolates, subtypes, and subspecies grouped as SINV and SINV-like viruses (except for Aura virus). A prominent degree of sequence conservation was found in the base-paired stretches of DLP, whereas substitutions tended to accumulate in bulges and in the loop (head) of DLP. Outside of DLP, substitutions were much more frequent, especially in the unstructured region of the 5′ untranscribed region (5′ UTR) located immediately before the AUG (Fig. 3C). Clearly, DLP has remained virtually unchanged during the diversification of the SINV group of viruses, suggesting that it has been submitted to a purifying selection. Similar results were found among the closest related members of the SFV complex (see below).
Evolutionary patterns of DLP in Alphavirus.
It has been suggested that evolution in arboviruses is constrained by host alternation among arthropods and vertebrates that impose fitness trade-offs (7, 52). However, this scenario of apparent stability at present might have been different during the speciation, host range expansion, and geographic spreading experienced by the Alphavirus genus (39). Furthermore, as the DLP structure is in the coding sequence of the capsid protein, changes in the RNA sequence that led to DLP appearance could have been constrained by the structural requirements of the capsid protein for virion stability and infectivity (Fig. 4A). To find clues for such evolutionary events, an analysis of sequence variability in the capsid protein among alphaviruses was carried out. Maximal sequence variability among representative alphavirus capsid proteins was concentrated in the first 100 aa, whereas the C-terminal core involved in capsomer assembly (aa 114 to 264) showed a much higher degree of sequence conservation, as reported before (5, 47, 62). The variability found in the N-terminal region of capsid protein also agrees with previous reports showing the tolerability of this region to insertions, deletions, or even partial replacements by heterologous sequences (35, 49). Thus, the appearance of DLP structures in alphaviral 26S mRNAs was probably favored by an intrinsic flexibility of the N-terminal portion of the capsid protein to changes in the coding sequence that allowed RNA to fold in DLP structures without compromising the stability and the infectivity of virions (Fig. 4A). However, once a DLP structure arose in the ancestor of a given branch of highly related alphavirus (e.g., an internal node in the phylogenetic tree), DLP and the corresponding amino acid sequence in the capsid protein should be further selected and conserved. This was found in the closest members of the SFV complex (Fig. 4B). SFV, RRV, GETV, Sagiyama virus, BEBV, MIDV, and UNAV are related viruses in which the DLP is well conserved in sequence and structure so that the few changes found were concentrated in the loop (head) and in internal bulges of the DLP, similar to that found for SINV variants (Fig. 4B). Notably, the region of capsid protein corresponding to the DLP structure also showed an absolute conservation at the amino acid level that contrasted with the variability that accumulated at the immediately adjacent regions, suggesting that once DLP was acquired, it was evolutionarily selected (Fig. 4B). Interestingly, when the first 150 nt of capsid-coding sequences that include the DLP structures were used for phylogenetic analysis, MIDV, MAYV, UNAV, CHIKV, and ONNV were placed with greater confidence than when the entire capsid protein or partial E1 sequences (38, 39) were used (data not shown), suggesting that DLP structures could be used to reconstruct the phylogenetic relationships among alphaviruses.
Fig 4.
Some evolutionary patterns of DLPs in Alphavirus. (A) Flexibility of the N-terminal region of the capsid protein to changes allowed the appearance of DLP structures in the alphavirus. The variability in capsid protein sequences among representative alphaviruses was calculated by the Shannon entropy method as described in Materials and Methods. The domain organization of the SINV prototype of the capsid protein is shown. DLP structures are localized in the coding regions of 26S mRNA corresponding to amino acids (aa) 7 to 40 of the capsid protein. (B) Conservation of DLP structure and the corresponding amino acid sequence among species of the SFV clade (SFV, BEBV, UNAV, MIDV, GETV, Sagiyama virus, and RRV). Shown is a phylogenetic tree of the SFV clade based on the first 150 nt of capsid coding sequences that include the DLP structures. Phylogenetic analysis was carried out by the maximum likelihood method using the GTR+I+Γ substitution model. Neighbor-joining (BioNJ), Bayesian (MrBayes), and maximum parsimony (MP) analysis resulted in an almost equivalent tree topology. The numbers shown are the branch support by 1,000 bootstrap resamplings for BioNJ, PhyML, and MP and the posterior probabilities for MrBayes, as indicated in the legend. Branches with <60% support value were collapsed. According to this model, a stable DLP may have emerged at the base of SFV/BEBV/UNAV/MIDV/GETV/Sagiyama virus/RRV clade that further evolved in the RRV/GETV/Sagiyama virus branch (denoted by black dots). The conserved nucleotide framework of DLPs is in bold, whereas the variable positions among viral species of the same clade are denoted by “N.” The variability in capsid protein sequences (Shannon entropy) among members of the SFV/BEBV/UNAV/MIDV/GETV/Sagiyama virus/RRV complex is shown in the lower panel.
Rapid evolution of SINV ΔDLP in mouse cells that restored DLP activity.
In an attempt to simulate in vitro the evolutionary acquisition of DLP, I forced mutant ΔDLP virus to revert after serial passages in NIH 3T3 cells (15). Some viruses with large lysis plaques arose at around the fifth passage, representing 1 to 5% of the total virus at this stage (Fig. 5A). Further passages progressively increased viral yields and allowed the enrichment of preparations with large-lysis-plaque variants. First, I analyzed the synthesis of structural proteins in NIH 3T3 cells infected with the crude preparation of the virus from each passage. Interestingly, the synthesis of viral proteins increased from passages 6 to 7, although the capsid bands showed an altered electrophoretic mobility in SDS-PAGE (Fig. 5B). Biological clones showing a large-plaque phenotype from passages 5, 7, and 11 were isolated, amplified, and sequenced. All revertants conserved the initial mutations of the parental ΔDLP virus, but showed an in-frame insertion of 84 nt at position 7785 of genomic RNA (nt 188 of 26S mRNA) that resulted from a tandem duplication of the fragment encompassing nt 7701 to 7785 (Fig. 5C). All biological clones from the revertants analyzed displayed an identical genotype, showing that they derived from a unique recombination event. To ensure that the 84-nt insertion alone was responsible for reversion of the parental ΔDLP virus, I subcloned the fragment from positions 6916 to 8067 of revertants P5, P7, and P11 into the cDNA clone of the ΔDLP virus. The resulting viruses showed identical phenotypic properties to parental revertant viruses (Fig. 5B). The 84-nt insertion restored translation up to <60% of wild-type levels in NIH 3T3 cells and increased the size of the capsid band by 2 to 3 kDa after SDS-PAGE analysis. In fact, two capsid protein bands were visible in short exposure of the blots, the lower band being the most abundant in wild-type NIH 3T3 cells, whereas in PKRo/o cells, I found the opposite (Fig. 5B). Proteomic analysis unambiguously showed that the upper and lower bands resulted from initiation at AUG 1 (i.e., the natural initiation codon) and AUG 3, respectively (data not shown). To understand better this change of codon usage in revertant virus, I probed and compared the RNA structures of the first 250 nt of 26S mRNA in revertant, ΔDLP, and WT viruses by means of SHAPE (59). Tandem duplication in revertant virus increased the stability of RNA structure mainly by the formation of a new Y-shape stem-loop located downstream of AUG 3 (Fig. 5D and E). The base of this structure showed a low reactivity in SHAPE experiments, suggesting a base-paired conformation of significant stability similar to that found in WT virus (Fig. 3C). Interestingly, the base of this stem-loop is located ∼26 nt from AUG 3, a distance that is compatible with a DLP activity on AUG 3 that explained the substantial level of translation of revertant virus in wild-type 3T3 cells. Moreover, alteration in initiation codon usage by disruption of DLP in SINV genome had been described previously (20, 21).
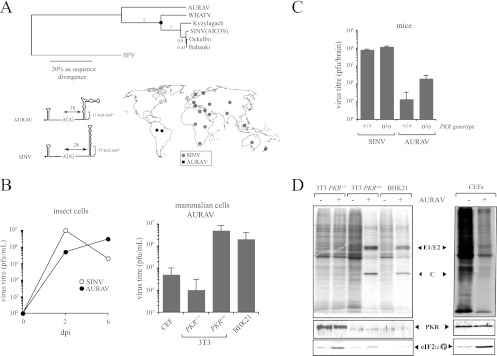
Aura virus represents an isolated member of the SINV group with a suboptimal DLP structure.
Aura virus (AURAV) is the only South American member of the SINV group and showed a DLP of lower stability than the rest of the SINV genotypes or subspecies (Fig. 3B). RNA folding programs predicted the existence of a structure with three remarkable differences with respect to SINV DLP. (i) The stability of DLP was lower than in SINV, especially in the base of the spiral. (ii) The percentage of G+C was only 60%. (iii) The distance from the AUG was 18 nt, which is substantially shorter than in SINV (28 nt). To test the activity of predicted DLP structure, I first obtained a preparation of AURAV by growing the BeAR 10315 strain in BHK21 cells as described earlier (42, 43). Although AURAV grew in BHK21 cells, both the size of lysis plaques and the yields obtained were lower than those for SINV and SFV. Notably, no viral proteins were detected in NIH 3T3 cells or in CEFs infected with AURAV despite the fact that the virus induced a detectable shutoff of host protein synthesis in these cells (Fig. 6D). In PKRo/o cells, however, prominent bands of AURAV structural proteins were detected (C and E1/E2) after [35S]Met/Cys labeling. I also found that our antisera raised against the SINV capsid cross-reacted with AURAV capsid in the Western blot. Interestingly, the absence of AURAV protein synthesis in wild-type 3T3 cells and CEFs correlated with a substantial phosphorylation of eIF2α detected in these cells. No eIF2α phosphorylation was detected in PKRo/o cells or in BHK21 cells infected with AURAV, showing that 26S mRNA of this virus was unable to translate when eIF2 is inactivated by phosphorylation. For BHK21 cells, the result obtained agrees well with a recent finding of our group showing that some tumoral cell lines do not express enough levels of PKR for high-level eIF2 phosphorylation in response to virus infection (53). As predicted from labeling experiments, replication of AURAV was very low in NIH 3T3 and CEF cells (<105 PFU/ml), whereas viral titers of 107 to 108 PFU/ml were obtained in PKRo/o or BHK21 cells. In insect cells, AURAV replicated at levels similar to SINV (Fig. 6B). The replication of AURAV in wild-type and PKRo/o mice was also analyzed (Fig. 6C). Very low viral yields were detected in wild-type mice infected with AURAV compared with SINV, whereas elimination of the PKR gene allowed a partial increase in replication (<1.5 log). Taken together, these results clearly show that AURAV is not adapted to replicate in mammals.
Fig 6.
AURAV is an isolated member of the SINV group with a suboptimal DLP structure. (A) Phylogenetic tree showing the evolutionary relationships among species of the SINV group (upper). AURAV is the only New World representative member of this group and the most divergent. Amino acid sequences of entire structural region were used for maximum likelihood (PhyML) analysis with JTT+I+Γ substitution model and SFV sequence as an outgroup. Bootstrap confidence is shown. BioNJ, Bayesian analysis, and MP resulted in identical topology. A black dot denotes the emergence of the functional DLP in the SINV clade. The topology and stability of AURAV DLP compared to those of SINV are shown (lower). The geographic distribution of AURAV and SINV genotypes is also shown (Division of Vector-Borne Diseases, Centers for Disease Control and Prevention). (B) Replication of AURAV in insect and vertebrate cell lines. Cells were infected at an MOI of 5 PFU/cell, and virus yields were determined at 2 and 6 days postinfection (dpi) for insect cells (H5) and at 2 dpi for avian and rodent cells. (C) Mice of the indicated genotype were infected with 107 PFU of SINV and AURAV by the intranasal route, and virus yields in brains at 4 dpi were estimated by plaque assay on BHK21 cells as described in Materials and Methods. No virus replication was detected in other main organs, such as the lung and liver. Data are means ± SD from four mice per group. (D) Protein synthesis and eIF2α phosphorylation in cell lines infected with AURAV at a high multiplicity of infection (25 PFU/cell). The upper panel shows autoradiography of [35S]Met-labeled proteins at 6 hpi. Viral capsid and glycoprotein (E1/E2) bands are indicated. The lower panels show anti-PKR and anti-phospho eIF2α blots of corresponding samples.
DISCUSSION
This article proposes a scenario for the evolutionary history of Alphavirus, showing how the adaptation to new vertebrate hosts probably involved the acquisition of a new structure in viral mRNA that conferred resistance to PKR activation, by allowing translation of viral 26S mRNA to be independent of eIF2 in vertebrate cells. Given the limited coding capacity of these RNA viruses, DLP structures in alphavirus probably arose as a simple solution (in genetic terms) to overcome PKR activation in vertebrate hosts. This idea was experimentally confirmed here by the observation that a SINV mutant lacking the DLP structure (ΔDLP) rapidly evolved, through recombination events, toward a variant that restored the activity of DLP in murine cells. In other families of viruses, however, a more sophisticated solution of higher genetic cost arose through the appearance of new products (RNA or proteins) or new domains in preexisting proteins that bind PKR in an inhibitory manner or that promote its degradation (23).
The evolutionary model proposed here relies on two assumptions (Fig. 7). First, insects were the primitive hosts of these viruses. Second, 26S mRNA of early alphavirus ancestors did not contain any DLP structure. Overall, the data presented here together with previous studies support this scenario (39, 47, 58). Most phylogenetic evidence supports a New World origin of Alphavirus from an insect-borne plant virus ancestor that further diversified into three main lineages: the EEE virus/VEE virus/WEE virus, SFV/Middelburg virus/Barmah Forest virus/Ndumu virus, and SDV/SPDV complexes. Other possible scenarios, including a marine origin of Alphavirus, have recently been suggested using the entire viral genomes as the sources of the phylogenetic signal (19). In any case, colonization of new hosts such as small mammals and birds probably allowed further evolution and transcontinental expansion and speciation of the SINV and SFV clades (39, 47, 56, 57). Based on sequence, topology, and distance from the initiation AUG, DLP structures in the Alphavirus genus probably arose from at least three independent events in the SINV, SFV, and North America EEEV clades. Given that the most stable and conserved DLP structures were found in SINV and SFV, which are the alphaviruses with the widest geographic distribution, an exciting possibility is that DLP acquisition in these viruses allowed host switching events that promoted geographic dispersion of these viruses worldwide. This idea is supported by the finding that AURAV, the most distant member of the SINV group which is confined to discrete regions of South America, was unable to replicate in murine, avian, or human cells that express the PKR gene. The suboptimal DLP structure predicted for AURAV did not allow 26S mRNA to be translated in an eIF2-independent manner, restricting the replication of this virus to insect cells. In fact, AURAV was no longer detected in other organisms apart from mosquitoes (42; the Arbovirus database from CDC), so that its role as a human pathogen is controversial.
Fig 7.
An evolutionary scenario for the natural history of Alphavirus is proposed. Assuming a New World origin from an insect-borne ancestor, the acquisition of DLP structures in 26S mRNA allowed the colonization of a new vertebrate host (e.g., migratory birds) that may have spread SINV and SFV to the Old World. Cycles involving mosquitoes and vertebrate hosts at present are schematically drawn. In insects, alphaviral 26S mRNA is translated by the canonical mechanism of initiation imposed by eIF2, whereas in vertebrate hosts, it occurs by an eIF2-independent mechanism that requires the DLP structure.
Although the results presented here support a New World origin for the Alphavirus genus and suggest that DLP structures can be used for tracking the evolutionary history of these viruses, the correlation among the presence of DLP structures, the replication in vertebrates, and geographic spreading was not perfect. RNA folding programs failed in predicting stable DLP structure in 26S mRNA of O′nyong-nyong virus (ONNV), Chikungunya virus (CHIKV), WEEV, and VEEE virus, all of which cause medically important outbreaks in humans or horses (56, 57). Collectively, these discrepancies could have two possible explanations. (i) Rather than adapt their translation to eIF2 phosphorylation, ONNV, CHIKV, WEEV, and VEEE virus might have acquired an alternative mechanism to prevent PKR activation in infected cells. (ii) DLP in these viruses could involve two nonadjacent (distant) RNA sequences that folding programs were unable to detect. Supporting the first is our recent finding that replication of the ΔDLP virus can be rescued by coexpression of the vaccinia virus E3 protein (14), a known inhibitor of PKR. Thus, although the appearance of DLP structures in some alphaviruses might have been an easy solution to overcome PKR activation in vertebrate hosts, it is theoretically possible that other mechanisms might have arisen in other alphaviruses with quasiequivalent results. On the other hand, the possibility that two nonadjacent RNA sequences folded to form a functional DLP in these viruses should not be rule out, given some previous reports showing the existence of bipartite regions in SINV RNA that activated GCN2 in infected cells (3).
The recombination event that restored the function of DLP during the evolution of ΔDLP virus in NIH 3T3 cells showed the flexibility of these viral genomes to acquire cis-acting RNA structures with important biological functions. Recombinations (sequence exchange and rearrangements) in RNA molecules are relatively frequent and biologically important events for evolution and diversity of RNA viruses (13, 24). Thus, most alphaviruses, including SINV, show repeated sequence elements 40 to 60 nt in length in the 3′ nontranslated region (3′ NTR) of genomic RNAs (47), so that in terms of probability, a recombination event created by a template switch of RNA replicase seems to be more likely than seven point changes that restored the original sequence of DLP. The effect of fragment duplication on local rearrangement of RNA structure, favoring the formation of a new hairpin loop that promotes translation initiation from a downstream in-frame AUG, emphasized the importance of RNA structure for eIF2-independent translation of alphavirus 26S mRNA and expanded at the same time the conformational possibilities to generate functional DLP structures in these and other viruses.
Finally, the results and the evolutionary scenario presented here could help to understand better the phylogenetic and epidemiological aspects of Alphavirus and other arboviruses, such as West Nile virus, Japanese encephalitis virus, or Dengue virus, where folding programs also predicted the existence of DLP-like structures (6).
ACKNOWLEDGMENTS
I am indebted to J. M. Almendral for having allowed me to work at his laboratory since 2005 and René Toribio for help with virus titrations and Western blot analysis. I am grateful to J. J. Berlanga for providing us with mice and to J. C. Bell (University of Ottawa, Canada) and B. R. Williams (Cleveland Clinic) for Pkr knockout mice and MEFs, respectively. I thank E. Domingo for critical reading of the manuscript and B. Perdiguero and C. Risco for providing us with CEFs and C6736 cells, respectively.
This project was supported in part by the Fundación Mutua Madrileña (FMM 2008), the VIRUS-HOST Interaction Program (Comunidad de Madrid), and by a grant from the Ministerio de Ciencia e Innovación (BFU2010-17411). Institutional support from the Fundación Ramón Areces is also acknowledged. Completion of this project took about 3 years, and the estimated cost was 3,500 €, excluding salaries.
Footnotes
Published ahead of print 3 July 2012
REFERENCES
- 1. Aarti I, Rajesh K, Ramaiah KV. 2010. Phosphorylation of eIF2 alpha in Sf9 cells: a stress, survival and suicidal signal. Apoptosis 15:679–692 [DOI] [PubMed] [Google Scholar]
- 2. Abraham N, et al. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 274:5953–5962 [DOI] [PubMed] [Google Scholar]
- 3. Berlanga JJ, et al. 2006. Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses. EMBO J. 25:1730–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Binder GK, Griffin DE. 2001. Interferon-gamma-mediated site-specific clearance of alphavirus from CNS neurons. Science 293:303–306 [DOI] [PubMed] [Google Scholar]
- 5. Choi HK, et al. 1991. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature 354:37–43 [DOI] [PubMed] [Google Scholar]
- 6. Clyde K, Harris E. 2006. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J. Virol. 80:2170–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coffey LL, et al. 2008. Arbovirus evolution in vivo is constrained by host alternation. Proc. Natl. Acad. Sci. U. S. A. 105:6970–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cook SH, Griffin DE. 2003. Luciferase imaging of a neurotropic viral infection in intact animals. J. Virol. 77:5333–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cullen BR. 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Immunol. 7:563–567 [DOI] [PubMed] [Google Scholar]
- 10. Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U. S. A. 106:97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dever TE. 2002. Gene-specific regulation by general translation factors. Cell 108:545–556 [DOI] [PubMed] [Google Scholar]
- 13. Domingo E, Parrish CR, Holland JJ. 2008. Origin and evolution of viruses, 2nd ed Academic Press, Waltham, MA [Google Scholar]
- 14. Domingo-Gil E, Toribio R, Najera JL, Esteban M, Ventoso I. 2011. Diversity in viral anti-PKR mechanisms: a remarkable case of evolutionary convergence. PLoS One 6:e16711 doi:10.1371/journal.pone.0016711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ebert D. 1998. Experimental evolution of parasites. Science 282:1432–1435 [DOI] [PubMed] [Google Scholar]
- 16. Elde NC, Child SJ, Geballe AP, Malik HS. 2009. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457:485–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elde NC, Malik HS. 2009. The evolutionary conundrum of pathogen mimicry. Nat. Rev. Microbiol. 7:787–797 [DOI] [PubMed] [Google Scholar]
- 17a. European Commission 1986. Directive 86/609/EEC of the European Union on the protection of animals used for experimental and other scientific purposes. European Commission, Brussels, Belgium [Google Scholar]
- 18. Fontaine A, de Monte A, Touzet H. 2008. MAGNOLIA: multiple alignment of protein-coding and structural RNA sequences. Nucleic Acids Res. 36:W14–W18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forrester NL, et al. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J. Virol. 86:2729–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frolov I, Schlesinger S. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70:1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frolov I, Schlesinger S. 1994. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol. 68:8111–8117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 7:590–597 [DOI] [PubMed] [Google Scholar]
- 23. Garcia MA, et al. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hahn CS, Lustig S, Strauss EG, Strauss JH. 1988. Western equine encephalitis virus is a recombinant virus. Proc. Natl. Acad. Sci. U. S. A. 85:5997–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hedrick SM. 2004. The acquired immune system: a vantage from beneath. Immunity 21:607–615 [DOI] [PubMed] [Google Scholar]
- 26. Hernandez R, Sinodis C, Brown DT. 2005. Sindbis virus: propagation, quantification, and storage. Curr. Protoc. Microbiol. 16:15B.1.1-15B.1.41. 1 doi:10.1002/9780471729259.mc15b01s16 [DOI] [PubMed] [Google Scholar]
- 27. Jackson AC, Moench TR, Trapp BD, Griffin DE. 1988. Basis of neurovirulence in Sindbis virus encephalomyelitis of mice. Lab. Invest. 58:503–509 [PubMed] [Google Scholar]
- 28. Lundstrom JO, Pfeffer M. 2010. Phylogeographic structure and evolutionary history of Sindbis virus. Vector Borne Zoonotic Dis. 10:889–907 [DOI] [PubMed] [Google Scholar]
- 29. McFadden G, Mohamed MR, Rahman MM, Bartee E. 2009. Cytokine determinants of viral tropism. Nat. Rev. Immunol. 9:645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meurs E, et al. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379–390 [DOI] [PubMed] [Google Scholar]
- 31. Myles KM, Wiley MR, Morazzani EM, Adelman ZN. 2008. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 105:19938–19943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217 [DOI] [PubMed] [Google Scholar]
- 33. Parisien M, Major F. 2008. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452:51–55 [DOI] [PubMed] [Google Scholar]
- 34. Peleg J. 1969. Behaviour of infectious RNA from four different viruses in continuously subcultured Aedes aegypti mosquito embryo cells. Nature 221:193–194 [DOI] [PubMed] [Google Scholar]
- 35. Perera R, Navaratnarajah C, Kuhn RJ. 2003. A heterologous coiled coil can substitute for helix I of the Sindbis virus capsid protein. J. Virol. 77:8345–8353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pichlmair A, Reis e Sousa C. 2007. Innate recognition of viruses. Immunity 27:370–383 [DOI] [PubMed] [Google Scholar]
- 37. Posada D. 2006. ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 34:W700–W703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Powers AM, et al. 2006. Genetic relationships among Mayaro and Una viruses suggest distinct patterns of transmission. Am. J. Trop. Med. Hyg. 75:461–469 [PubMed] [Google Scholar]
- 39. Powers AM, et al. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75:10118–10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Riedel B, Brown DT. 1977. Role of extracellular virus on the maintenance of the persistent infection induced in Aedes albopictus (mosquito) cells by Sindbis virus. J. Virol. 23:554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. 2009. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat. Struct. Mol. Biol. 16:63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rumenapf T, Strauss EG, Strauss JH. 1995. Aura virus is a New World representative of Sindbis-like viruses. Virology 208:621–633 [DOI] [PubMed] [Google Scholar]
- 43. Rumenapf T, Strauss EG, Strauss JH. 1994. Subgenomic mRNA of Aura alphavirus is packaged into virions. J. Virol. 68:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saleh MC, et al. 2009. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458:346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sammels LM, Lindsay MD, Poidinger M, Coelen RJ, Mackenzie JS. 1999. Geographic distribution and evolution of Sindbis virus in Australia. J. Gen. Virol. 80:739–748 [DOI] [PubMed] [Google Scholar]
- 47. Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 49. Tellinghuisen TL, Perera R, Kuhn RJ. 2001. In vitro assembly of Sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J. Virol. 75:2810–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Toribio R, Ventoso I. 2010. Inhibition of host translation by virus infection in vivo. Proc. Natl. Acad. Sci. U. S. A. 107:9837–9842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. 2008. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14:1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vasilakis N, et al. 2009. Mosquitoes put the brake on arbovirus evolution: experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog. 5:e1000467 doi:10.1371/journal.ppat.1000467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ventoso I, Berlanga JJ, Almendral JM. 2010. Translation control by protein kinase R restricts minute virus of mice infection: role in parvovirus oncolysis. J. Virol. 84:5043–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ventoso I, et al. 2006. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 20:87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang F, et al. 2004. Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 5:1266–1274 [DOI] [PubMed] [Google Scholar]
- 56. Weaver SC. 2006. Evolutionary influences in arboviral disease. Curr. Top. Microbiol. Immunol. 299:285–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weaver SC, Barrett AD. 2004. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2:789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weaver SC, et al. 1997. Recombinational history and molecular evolution of Western equine encephalomyelitis complex alphaviruses. J. Virol. 71:613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610–1616 [DOI] [PubMed] [Google Scholar]
- 60. Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32:569–577 [DOI] [PubMed] [Google Scholar]
- 61. Yang YL, et al. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang W, et al. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645–11658 [DOI] [PMC free article] [PubMed] [Google Scholar]