Abstract
The ability of HIV-1 to establish a latent infection presents a barrier to curing HIV. The best-studied reservoir of latent virus in vivo is resting memory CD4+ T cells, but it has recently been shown that CD34+ hematopoietic progenitor cells (HPCs) can also become latently infected by HIV-1 in vitro and in vivo. CD34+ cells are not homogenous, however, and it is not yet known which types of CD34+ cells support a latent infection. Furthermore, the mechanisms through which latency is established in this cell type are not yet known. Here we report the development of a primary cell model for latent HIV-1 infection in HPCs. We demonstrate that in this model, latent infection can be established in all subsets of HPCs examined, including HPCs with cell surface markers consistent with immature hematopoietic stem and progenitor cells. We further show that the establishment of latent infection in these cells can be reversed by tumor necrosis factor alpha (TNF-α) through an NF-κB-dependent mechanism. In contrast, we do not find evidence for a role of positive transcription elongation factor b (P-TEFb) in the establishment of latent infection in HPCs. Finally, we demonstrate that prostratin and suberoylanilide hydroxamic acid (SAHA), but not hexamethylene bisacetamide (HMBA) or 5-aza-2′-deoxycytidine (Aza-CdR), reactivate latent HIV-1 in HPCs. These findings illuminate the mechanisms through which latent infection can be established in HPCs and suggest common pathways through which latent virus could be reactivated in both HPCs and resting memory T cells to eliminate latent reservoirs of HIV-1.
INTRODUCTION
Although highly active antiretroviral therapy (HAART) for HIV-1 suppresses viral replication and extends the life spans of infected individuals, it cannot eliminate the virus (25, 30, 31, 84). A major barrier to viral eradication is the ability of HIV-1 to establish a latent infection (25, 84). Latent infection occurs when a replication-competent HIV-1 provirus integrates into a host cell's genomic DNA, but viral genes are not transcribed (reviewed in reference 29). Because no viral proteins are produced, the immune system cannot eliminate latently infected cells. At any point, however, cellular changes can reactivate the latent virus, leading to the production of new viral particles and, in the absence of antiretroviral therapy, the infection of additional cells (reviewed in reference 69). Thus, reactivation of latent virus can lead to a resurgence of viremia in patients who have discontinued antiretroviral therapy.
Although reactivation of latent virus can lead to increased viremia, reactivating latent virus in a patient who is receiving HAART therapy could clear latent reservoirs while preventing new infection events. Cells harboring reactivated virus would be eliminated because they would produce viral proteins, leading to cell death through the action of the immune system or viral cytotoxic effects. Reactivation of virus from latently infected cells has thus been proposed as a strategy to cure HIV-1. Unfortunately, efforts to use this strategy to eliminate HIV-1 infection have thus far been unsuccessful (reviewed in reference 29). Thus, there remains an urgent need for research to determine both the cell types that harbor latent virus and the mechanisms underlying the establishment and reactivation of latent infection in all latently infected cell types.
Although resting memory T cells are the best-studied and likely the largest reservoir of latent HIV-1, several studies have found that not all viral genomes in the plasma can be matched to sequences in these cells (3, 9, 61). These findings suggest that additional reservoirs of latent virus exist and contribute to residual plasma viremia. Recently, we proposed that CD34+ hematopoietic progenitor cells (HPCs) in the bone marrow serve as a reservoir of HIV-1, and we demonstrated that active and latent infection of these cells occurs in vitro and in vivo (13). These findings are supported by additional studies demonstrating that HPCs are infected by HIV-1 in vivo (54, 67). However, there are also conflicting studies reporting that HIV-1 genomes could not be detected in CD34+ cells in the bone marrow of patients with undetectable viral loads on HAART (21, 36).
CD34+ HPCs are rare cells, and only an extremely low rate of latent infection would be expected in most HAART-treated patients. This is particularly true for patients with predominantly CCR5-tropic virus, since only CXCR4-utilizing HIV is able to infect immature HPCs (12). In addition, it is difficult to rule out the possibility that contaminating CD4+ T cells in HPC samples contribute to the detection of HIV genomes in these cells. Thus, it may be difficult to show definitively whether latent infection of HPCs occurs in a majority of individuals: indeed, disagreement on this point has persisted through more than 2 decades of study (reviewed in reference 45). However, latent infection can be readily established in HPCs in vitro (13), and thus, in vitro systems can be used to assess which subtypes of HPCs become latently infected. Such studies can illuminate the potential of HPCs to serve as a reservoir in vivo by demonstrating whether long-lived HPCs can be latently infected. In addition, they can identify the HPC types most likely to harbor latent infection, suggesting cell types to be selectively purified in future efforts to identify latent reservoirs in vivo. Finally, in vitro studies can assess whether the mechanisms that promote the establishment and reactivation of latent infection in HPCs are comparable to those at work in CD4+ T cells and thus whether both reservoirs might be targeted and eliminated with similar reactivation strategies. For these reasons, an in vitro examination of latent infection of CD34+ HPCs provides valuable information to aid in both the search for latent reservoirs and the development of strategies to reactivate and eliminate latent virus in vivo.
CD34+ HPCs are not homogenous, and it is not clear which subsets of CD34+ cells support latent HIV-1 infection. The most immature HPCs are hematopoietic stem cells (HSCs), which can differentiate into all blood cell lineages and have unlimited self-renewal capacity. HSCs differentiate into multipotent progenitor cells (MPPs), which also differentiate into all blood cell lineages but have a reduced self-renewal capacity (20). MPPs give rise to common myeloid progenitors (CMPs), which give rise to all myeloid lineages, and multilymphoid progenitors (MLPs), which generate the lymphoid lineages as well as monocytes and dendritic cells (20). These progenitors give rise to committed progenitor cells that eventually differentiate into mature blood cells. While we recently showed that HIV-1 is capable of infecting multipotent HPCs, including hematopoietic stem cells, the ability of HIV-1 to establish a latent as opposed to an active infection in these cells is unknown (12).
The mechanisms that promote latency in HPCs have not been investigated, but models of latent infection in T cells have identified mechanisms that promote latent infection in this cell type (8, 41, 62, 70, 79). In T cells, epigenetic modifications result in a heterochromatic structure at the HIV-1 long terminal repeat (LTR) (37, 70). Histone methylation and histone deacetylase (HDAC) recruitment contribute to the formation of this restricted transcriptional state (70). DNA methylation has also been associated with latent infection in both Jurkat and primary T cell models of latency (6, 37). Furthermore, resting memory T cells have restricted levels of nuclear factor κB (NF-κB) in the nucleus (70). NF-κB, a key transcriptional regulator in many hematopoietic cells, is sequestered in the cytoplasm of resting cells by the inhibitor of κB (IκB) (reviewed in reference 71). Upon T cell activation, phosphorylation of IκB by IκB kinase (IKK) triggers the release of NF-κB from IκB and its translocation to the nucleus (42; reviewed in reference 71). There, NF-κB can bind to the HIV-1 LTR, displacing inhibitory factors and recruiting histone acetyltransferases to relieve heterochromatic repression (47, 73).
NF-κB activation alone is sufficient to reactivate latent virus in Jurkat cells (22, 70), but in primary resting memory T cells, activation of positive transcription elongation factor b (P-TEFb) is also required (70). P-TEFb, a complex made up of the cyclin-dependent kinase 9 (CDK9) and cyclin T1 (CycT1), is present only at low levels in resting CD4+ T cells, and much of the complex is sequestered by the 7SK small nuclear RNA and hexamethylene bisacetamide (HMBA)-induced protein (HEXIM1) (48, 80, 81). T cell activation results in increased P-TEFb mRNA transcription and protein translation (33) as well as in the rapid phosphorylation of preexisting CDK9 at Thr186 (53) and the dissociation of P-TEFb from 7SK RNA (38). HIV-1 Tat assists in this process by competitively displacing HEXIM1 to free P-TEFb from the inhibitory complex (4, 64). Activated P-TEFb is then recruited to nascent HIV-1 transcripts, where it facilitates transcript elongation and thus reactivation of latent virus (38, 63, 70, 76).
Compounds that counteract the factors promoting latency in T cells have been shown to reactivate latent virus in primary T cells or cell line models. Among these compounds are prostratin, which activates NF-κB through protein kinase C (40, 77); HDAC inhibitors, including suberoylanilide hydroxamic acid (SAHA) and valproic acid (VPA) (19, 24, 82); the DNA methylation inhibitor 5-aza-2′-deoxycytidine (Aza-CdR) (37); and HMBA, a compound that appears to facilitate the release of P-TEFb from its inhibitory complex with 7SK and HEXIM1 (18). Several of these compounds, notably SAHA, have been proposed as therapies to reactivate latent virus in vivo (19, 24, 82). However, if resting memory T cells are not the sole reservoir for latent virus, these compounds will be effective therapies only if they can reactivate virus in all additional HIV reservoirs as well.
In this paper, we develop an in vitro model system of latent HIV-1 infection in HPCs that permits detailed study of the factors promoting latency in these cells. We use this model to show that HIV-1 is able to establish a latent infection in all subsets of HPCs examined, including cells with surface markers consistent with HSCs and MPPs. We further show that CD34+ HPCs have low levels of NF-κB in the nucleus and that NF-κB activation can reactivate latent virus in these cells. Meanwhile, P-TEFb is readily detectable in the nuclei of unstimulated HPCs, and its levels are not increased under conditions that reactivate latent virus. Finally, we assess the ability of compounds that reactivate latent virus in T cell systems to perform a similar function in HPCs. We find that while prostratin and SAHA can reactivate latent infection in HPCs, HMBA and Aza-CdR cannot. These findings enhance our understanding of the cellular factors required to establish a latent HIV-1 infection in HPCs and suggest common pathways in HPCs and T cells that could be targeted to purge latent reservoirs.
MATERIALS AND METHODS
Cell isolation and culture.
Whole umbilical cord blood (CB) was obtained from the New York Blood Center, and whole bone marrow (BM) was obtained commercially (AllCells Ltd.); mononuclear cells were purified by Ficoll-Hypaque centrifugation and were either frozen or used fresh. Cells were adherence depleted for 1 to 2 h at 37°C in StemSpan medium (STEMCELL Technologies), and then CD133+ cells were isolated by magnetic separation (Miltenyi Biotec). Isolated cells were cultured in STIF medium (StemSpan medium supplemented with 100 ng/ml stem cell factor [SCF], 100 ng/ml thrombopoietin [TPO], 100 ng/ml Flt3 ligand [Flt3L] [all from STEMCELL Technologies], and 100 ng/ml insulin-like growth factor binding protein 2 [IGFBP-2] [R&D Systems]). Presorted CD133+ BM or CB cells were obtained commercially (AllCells Ltd.) and were cultured as described above.
Resting memory CD4+ T cells were purified from buffy coats obtained from the New York Blood Center. Mononuclear cells were purified as described above, and then memory CD4+ T cells were isolated by magnetic separation using the Memory CD4+ T cell isolation kit (Miltenyi Biotec). The resulting cells were incubated with biotinylated antibodies against the activation markers HLA-DR, CD69, and CD25 (eBioscience). Resting memory cells were then isolated by negative selection using an anti-biotin magnetic separation kit (Miltenyi Biotec). Resting memory CD4+ T cells were either used immediately or cultured overnight with anti-CD3/anti-CD28 beads (Life Technologies).
U1 cells (26) and J-Lat clones 6.3, 8.4, and 9.2 (35) were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS) and 2 mM penicillin, streptomycin, and glutamine (PSG). 293T cells were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS and 2 mM PSG.
HIV-1 preparation and transductions.
NL4-3-ΔGPE-GFP was generated by removing a SwaI-SwaI fragment from NL4-3-ΔE-GFP (83) and religating.
Infectious supernatants were prepared by transfection of proviral plasmids into 293T cells using polyethylenimine. HXB-ePLAP (15) or NL4-3-ΔGPE-GFP was cotransfected with a plasmid encoding either the vesicular stomatitis virus glycoprotein (VSV-G) or the HXB envelope; the helper plasmid pCMV-HIV-1 (28) was additionally cotransfected to permit infectious particles to be formed with NL4-3-ΔGPE-GFP. Viruses pseudotyped with the HXB envelope were concentrated using high-molecular-weight polyethylene glycol precipitation as described previously (39). Pellets were resuspended in 1/25 the original volume and were used immediately. Cells were infected by spin inoculation at 1,048.6 × g for 2 h at room temperature.
Sorting and stimulation of latently infected cells.
Actively infected cells from HPCs infected with HXB-ePLAP were removed by magnetic sorting using a biotin-conjugated antibody to placental alkaline phosphatase (PLAP) and anti-biotin beads (Miltenyi Biotec). Actively infected cells were removed from samples infected with NL4-3-ΔGPE-GFP by flow cytometric sorting using a MoFlo XDP (Beckman Coulter), MoFlo Astrios (Beckman Coulter), or FACSAria (BD Biosciences) flow cytometer. After sorting, cells were incubated in STIF or StemSpan medium with 8 μM raltegravir (Selleck Chemicals) alone or with one or more of the following: granulocyte-macrophage colony-stimulating factor (GM-CSF; R&D Systems), tumor necrosis factor alpha (TNF-α; Biolegend), prostratin (Sigma), SAHA (Cayman Chemical), HMBA (Sigma), Aza-CdR (Sigma), IKK2 VI (Calbiochem), or functional-grade anti-TNF-α (eBioscience).
Time course of stimulation in uninfected cells.
Uninfected HPCs were cultured for 5 days and were then incubated in StemSpan medium with the solvent dimethyl sulfoxide (DMSO) as a control or with TNF-α, prostratin, or SAHA for 3 days. Images of cells under these conditions were acquired using a QICAM digital camera and Q-Capture Pro 7 software (QImaging). The brightness and contrast of images were minimally adjusted using Canvas 8 (ACD Systems).
Flow cytometry and antibodies.
Antibodies to the following proteins were used for flow cytometry: CD133 (phycoerythrin [PE] conjugated; Miltenyi), CD3 (conjugated with allophycocyanin [APC] or Alexa Fluor 647; eBioscience), CD34 (conjugated with fluorescein isothiocyanate [FITC], APC, Alexa Fluor 647, or biotin; Miltenyi, eBioscience, and Invitrogen), CD45RA (PE conjugated; eBioscience), CD38 (PE-Cy7 conjugated; eBioscience), PLAP (Serotec) as provided or conjugated to biotin using the EZ-Link Sulfo-NHS biotinylation kit (Pierce), and HIV-1 Gag (clone KC57, FITC conjugated; Coulter). The secondary reagents used were streptavidin (conjugated with Alexa Fluor 488, PE, or Alexa Fluor 647; Life Technologies) and an antibody to mouse IgG1 (Alexa Fluor 647 conjugated; Invitrogen). Cells were stained with 7-aminoactinomycin D (7-AAD) to exclude dead cells. Analysis was performed using a BD FacsCanto cytometer or a BD FACScan system with Cytek 6-color upgrade.
For staining of cell surface proteins, cells were first incubated in fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline [PBS] with 2% FBS, 1% human serum, 2 mM HEPES, and 0.025% NaN3) on ice for 10 min with directly conjugated antibodies; then they were washed and fixed in PBS with 2% paraformaldehyde. For stains utilizing antibodies against PLAP, cells were incubated first in 10% Fc receptor block (Accurate Chemical) in FACS buffer for 30 min and then with antibodies against PLAP in FACS buffer with 10% Fc receptor block for 15 min on ice. Cells were then washed and stained with a secondary antibody for an additional 5 (streptavidin) or 15 (anti-mouse IgG1) min on ice. For stains using antibodies against HIV Gag, cells were first fixed and then permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature. Cells were then washed and incubated with an antibody against Gag for 30 min on ice.
Transcription factor ELISA.
Nuclear and cytoplasmic extracts were prepared using a commercially available kit (Active Motif). Levels of activated nuclear NF-κB and AP-1 family members were determined using commercially available transcription factor enzyme-linked immunosorbent assays (ELISAs) (Active Motif). The protein contents of nuclear and cytoplasmic extracts were quantified, and an equal amount of protein was plated for each condition. Commercially obtained Raji cell nuclear extracts were used as a positive control for NF-κB activation, and commercially obtained nuclear extracts from K-562 cells stimulated with 12-O-tetradecanoylphorbol-13-acetate (TPA) were used as a positive control for AP-1 activation (Active Motif). Absorbance readings from blank wells were subtracted from all sample and positive-control absorbance readings. Readings were then normalized to that for the positive control on each plate to control for differences in color development between plates. The normalized absorbance in cytoplasmic extracts was subtracted from the absorbance in the nuclear extracts from each sample to control for nonspecific binding. These normalized differences were expressed in arbitrary units (AU) and were used to compare samples. In all cases, the signal from the positive-control extract was set to 1 AU.
Western blotting.
Nuclear extracts were collected as described above. Antibodies to the following proteins were used for Western blot analysis: cyclin T1 (Santa Cruz catalog no. sc-8127), pCDK9 (Cell Signaling catalog no. 2549), and histone H1.4 (Sigma catalog no. H7665). Secondary reagents used were rabbit antibodies directed against goat IgG and conjugated to horseradish peroxidase (HRP) (Zymed) and goat antibodies directed against rabbit IgG and conjugated to HRP (Invitrogen).
RESULTS
Development of an in vitro model of HIV-1 latency in HPCs.
Although latent infection of HPCs has been studied previously in vitro (13), the methods used required relatively long culture periods. Because HPCs differentiate rapidly in culture, these methods did not permit the examination of latent infection in immature HPC subsets such as HSCs and MPPs. To better assess latent infection in immature HPCs, we developed a short-term primary cell model of latency in CD133+ HPCs (Fig. 1A). This short-term model allowed us to examine latent infection in immature HPCs, which is an important benefit since these cells are the longest-lived and thus the most likely to serve as a long-term reservoir of virus in vivo. However, the short culturing times required to study these cells in an undifferentiated state meant that while we could examine factors important for the establishment of latency, we could not assess whether additional factors contribute to the maintenance of proviral silencing at later time points.
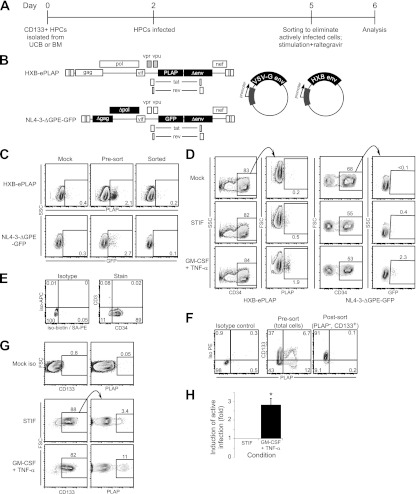
Fig 1.
(A to D) GM-CSF and TNF-α reactivate latent virus in a primary cell model for HIV-1 infection of CD34+ HPCs. (A) Time course for latency experiments. (B) Schematic diagrams of the HIV-1 reporter viruses used for these studies. Viruses were pseudotyped with the VSV-G envelope (center) or the CXCR4-tropic HIV-1 HXB envelope (right). The shading of vpr and vpu in HXB-ePLAP indicates that these genes are defective in full-length HXB. Filled rectangles represent genes that have been added to or deleted from the wild-type viral clone. (C) Flow cytometric analysis of cells sorted for reactivation protocols. (Top) Cord blood-derived HPCs infected with HXB-ePLAP/VSV-G and PLAP− cells isolated by magnetic sorting; (bottom) cells infected with NL4-3-ΔGPE-GFP/VSV-G and GFP− cells isolated by flow sorting. The percentages of live cells that are PLAP+ or GFP+ are given in each panel. Live cells were gated using forward scatter (FSC) and side scatter (SSC) parameters. (D) Flow cytometric analysis of cells for which results are shown in panel C after overnight incubation with STIF medium or GM-CSF (100 ng/ml) and TNF-α (2.5 ng/ml). CD34+ cells are gated on at the left; then PLAP or GFP expression in uninfected, STIF-treated, or GM-CSF- and TNF-α-treated cells is shown on the right. Numbers indicate the percentages of cells within the labeled gate. Live cells were gated based on FSC, SSC, and 7-AAD. (E) CD3+ T cells are not present in the CD34+ HPC population. Cord blood-derived HPCs were sorted and infected with NL4-3-ΔGPE-GFP/VSV-G as outlined in panel A. CD3 and CD34 staining was assessed on day 6 following flow sorting and overnight incubation in STIF medium. Live cells were gated using FSC, SSC, and 7-AAD. Data are representative of two independent experiments, one each using cord blood- or bone marrow-derived HPCs. (F to H) Latent infection that can be reactivated with GM-CSF and TNF-α occurs in immature CD133+ HPCs. (F) Flow cytometric analysis of CD133+ cells treated with HXB-ePLAP/VSV-G and sorted to remove actively infected (PLAP+) cells. The percentages of live cells are given in each quadrant. Live cells were gated using forward scatter and side scatter. (G) Flow cytometric analysis of cells sorted as in panel F and then stimulated overnight with STIF medium or GM-CSF and TNF-α. Live cells were defined by FSC, SSC, and 7-AAD. Numbers indicate the percentages of cells falling within the indicated gate. (H) Quantitation of reactivation in CD133+ cells (cells sorted for CD133 prior to stimulation) relative to that in cells incubated in STIF medium. Means and standard errors for 4 independent experiments are shown. The asterisk indicates a significant difference (P = 0.01) from the expected fold increase of 1 by a 1-sample t test.
In our model, primary HPCs were isolated on day zero from umbilical cord blood (CB) or bone marrow (BM) (Fig. 1A). Two days later, HPCs were infected with replication-defective HIV-1 reporter viruses, either HXB-ePLAP or NL4-3-ΔGPE-GFP (Fig. 1B). Both of these viruses have a deletion in the env gene, while NL4-3-ΔGPE-GFP has additional deletions in gag and pol, resulting in viruses that are capable of a single round of infection but not of continuing spread. This feature of the viruses simplified our analysis by allowing us to distinguish reactivation of latent infection from stimulation of new infection events. HXB-ePLAP encodes placental alkaline phosphatase (PLAP) as a reporter protein, which is expressed on the cell surface; NL4-3-ΔGPE-GFP expresses enhanced green fluorescent protein (EGFP; shortened to GFP throughout). These viruses were pseudotyped with either the VSV-G envelope or the HIV-1 HXB envelope (Fig. 1B). Most experiments were conducted using the VSV-G envelope to maximize the infection rate and increase the sensitivity of analysis, but major findings were confirmed using the HXB envelope. The CXCR4-tropic HXB envelope was selected for these experiments because we have shown previously that only HIV-1 envelopes that can use CXCR4 as a coreceptor for entry can infect immature HPCs (12).
Three days after infection, actively infected cells expressing the reporter protein were removed by magnetic or flow cytometric sorting (Fig. 1C); the remaining cells, a mixture of uninfected and latently infected cells, were resuspended in media containing 8 μM raltegravir (Fig. 1A). The integrase inhibitor raltegravir was routinely included in these experiments to ensure that increases in reporter gene expression would stem solely from activation of integrated virus and not from new integration events. Then cells were either incubated in STIF medium, which contains a cocktail of cytokines that minimizes differentiation in culture, or stimulated with GM-CSF and TNF-α or other compounds overnight. Finally, samples were analyzed by flow cytometry the next day (Fig. 1A). In all cases, cells that remained CD34+ or CD133+ at this time point were gated on so that only HPCs would be included in our analysis of latent infection.
We tested our model by stimulating latently infected cells with GM-CSF and TNF-α, which we have previously shown to be capable of reactivating latent virus in HPCs in vitro and ex vivo (13). In agreement with our previous work, we found that overnight (13-h) stimulation with GM-CSF and TNF-α resulted in a 2- to 6-fold increase in reporter protein expression over that for unstimulated controls (Fig. 1D). The rate of latent infection that could be reactivated with GM-CSF and TNF-α was comparable to the rate of active infection observed prior to sorting and stimulation (compare Fig. 1C and D). These results cannot be attributed to contamination of the culture with CD4+ T cells, since CD3+ cells are largely absent from our HPC cultures on the day of reactivation and are readily excluded by gating on CD34+ cells (Fig. 1E).
Latent infection occurs in multiple HPC subsets.
Although we have reported that HIV-1 can infect multipotent HPCs, the ability of the virus to establish a latent infection in this cell subset was not established. We therefore used our model to examine latent infection in HPCs expressing CD133, a cell surface marker found primarily on immature HPCs and associated with colony-forming capacity (12). HPCs infected with HXB-ePLAP were magnetically sorted to isolate PLAP− CD133+ cells (Fig. 1F); then these cells were incubated overnight in STIF medium or GM-CSF and TNF-α. Compared with the rate of infection in cells immediately after sorting (0.1%) (Fig. 1F), we observed spontaneous induction of active infection in a subset of cells (Fig. 1G), which may result from spontaneous differentiation that occurs even in STIF medium. Additionally, we observed a 2- to 4-fold increase in PLAP expression in CD133+ cells stimulated with GM-CSF and TNF-α over that in the STIF medium-treated control (Fig. 1G and H), demonstrating that HIV-1 can establish a latent infection in CD133+ HPCs that can be reactivated by GM-CSF and TNF-α treatment.
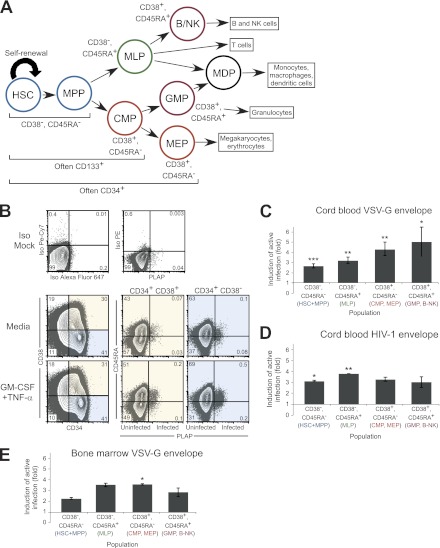
To further define HPC populations in which latent HIV-1 infection occurs, we stained HPCs with antibodies against CD38 and CD45RA, cell surface markers for which expression on various HPC populations has recently been defined (20) (Fig. 2A). HSCs and MPPs stain CD34+ CD38− CD45RA−; multilymphoid progenitors (MLPs) stain CD34+ CD38− CD45RA+; common myeloid progenitors (CMPs) and megakaryocyte/erythroid progenitors (MEPs) stain CD34+ CD38+ CD45RA−; and finally, committed granulocyte/monocyte progenitors (GMPs) and B/NK progenitors (B/NK) stain CD34+ CD38+ CD45RA+. We infected and sorted the cells as described for Fig. 1; then we analyzed the reactivation of latent virus in each of the populations defined by these cell surface antigens. We found that we could reactivate latent virus in each population, including the CD34+ CD38− CD45RA− subset, which includes the most immature, longest-lived HPCs (Fig. 2B). Similar results were obtained whether cells had been infected using the VSV-G (Fig. 2C and E) or the HXB (Fig. 2B and D) envelope and whether HPCs were derived from cord blood (Fig. 2B to D) or bone marrow (Fig. 2E). These results demonstrate that latent infection does not occur in a specific type of HPCs but rather in all cell subsets identified, including long-lived HSCs and MPPs with the potential to serve as a long-term reservoir for latent virus in vivo.
Fig 2.
Latent infection occurs in all subsets of CD34+ HPCs defined by CD38 and CD45RA expression. (A) Diagram of hematopoiesis showing CD34, CD133, CD38, and CD45RA expression on each cell subset. HSC, hematopoietic stem cell; MPP, multipotent progenitor; MLP, multilymphoid progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte/erythrocyte progenitor; GMP, granulocyte/monocyte progenitor; B-NK, B and NK cell progenitor. (B) Flow cytometric analysis of CD133+ cells treated with HXB-ePLAP/HXB Env, sorted to remove actively infected (PLAP+) cells, and stimulated overnight with STIF medium or GM-CSF and TNF-α. Live cells were defined using forward scatter (FSC), side scatter (SSC), and 7-AAD; then they were analyzed for CD34, CD38, CD45RA, and PLAP expression. The percentage of total cells is given in each quadrant. (C) Summary of experiments similar to those shown in panel B but using the HXB-ePLAP/VSV-G envelope to infect cells. Shown is the fold induction of active infection in cells treated with GM-CSF and TNF-α or with TNF-α alone relative to cells incubated in medium. Labels are colored to match cell types in panel A. Means and standard errors for 6 independent experiments are given. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) by a 1-sample t test from the expected fold increase of 1,. Differences in the fold induction of active infection between cell subsets are not significant (P, 0.26 by 1-way analysis of variance). (D) Summary of experiments for which results are shown in panel B. Shown is the fold induction of active infection in cells treated with GM-CSF and TNF-α or with TNF-α alone relative to unstimulated cells. Means and standard errors for 2 independent experiments are shown. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) by a 1-sample t test from the expected fold increase of 1. Differences in the fold induction of active infection between cell subsets are not significant (P, 0.53 by 1-way analysis of variance). (E) Summary of experiments similar to those shown in panel B but using the NL4-3-ΔGPE-GFP/VSV-G envelope to infect bone marrow cells. Shown is the fold induction of active infection in cells treated with TNF-α relative to unstimulated cells. Means and standard errors for 2 independent experiments are given. Asterisk indicates a significant difference (*, P < 0.05) by a 1-sample t test from the expected fold increase of 1. Differences in the fold induction of active infection between cell subsets are not significant (P, 0.11 by 1-way analysis of variance).
Conditions that reactivate latent virus in HPCs are associated with the activation of NF-κB but not with that of AP-1.
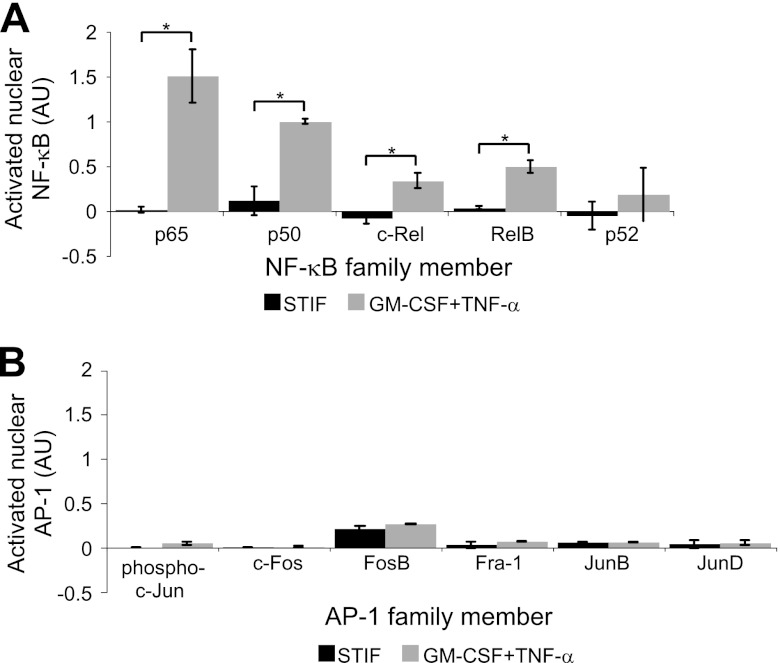
We next assessed the cellular changes that occur when CD34+ HPCs are stimulated with GM-CSF and TNF-α in order to elucidate the mechanism by which these cytokines reactivate latent HIV-1. We first examined whether nuclear levels of the transcription factor NF-κB or AP-1 are increased under these conditions. We examined these factors because both have binding sites on the HIV-1 LTR, have been shown to promote HIV-1 transcription (11, 47, 55, 57, 58, 72; reviewed in reference 51), and can be activated by TNF-α (reviewed in references 2 and 16).
Uninfected, cord blood-derived CD133+ HPCs were expanded in STIF medium for 5 days and were then split into STIF versus GM-CSF-plus-TNF-α cultures for overnight incubation. We isolated cytoplasmic and nuclear extracts from these cells and assessed levels of activated nuclear NF-κB and AP-1 transcription factor proteins by a transcription factor enzyme-linked immunosorbent assay (ELISA). We found that levels of activated nuclear NF-κB family members p65, p50, c-Rel, and RelB were all stimulated by overnight incubation with GM-CSF and TNF-α (Fig. 3A), suggesting that these cytokines activate NF-κB through the classical pathway (reviewed in reference 71). In contrast, we found that nuclear NF-κB p52, which is activated mainly through the nonclassical NF-κB pathway (reviewed in reference 71), was not upregulated by GM-CSF and TNF-α stimulation (Fig. 3A). Additionally, no AP-1 family member was upregulated by GM-CSF and TNF-α (Fig. 3B). Of the proteins studied, only AP-1 FosB and JunB could be consistently detected in the nuclei of HPCs incubated in STIF medium (Fig. 3B). The finding that NF-κB is usually undetectable in the nuclei of unstimulated HPCs suggests that NF-κB restriction promotes latent infection in this cell type.
Fig 3.
GM-CSF- and TNF-α-treated HPCs have higher nuclear NF-κB DNA binding activity than STIF medium-treated cells. (A) Quantitation of the results of a transcription factor ELISA measuring NF-κB DNA-binding activity in nuclear lysates from HPCs incubated with STIF medium or with GM-CSF plus TNF-α. Absorbance was normalized to that of the positive control on each plate; then nonspecific activity (defined by the activity obtained using cytoplasmic extracts) was subtracted from the nuclear absorbance, and the difference was graphed as arbitrary units (AU). The value for the positive control, a Raji nuclear cell extract, was set to 1 AU. Means and standard errors for 3 independent experiments are shown. Asterisk indicates a significant difference (*, P < 0.05) by a paired t test. (B) Quantitation of the results of a transcription factor ELISA measuring AP-1 DNA-binding activity in nuclear lysates from HPCs incubated with STIF medium or with GM-CSF plus TNF-α overnight. Data were analyzed as described for panel A except that nuclear extracts of K-562 cells stimulated with 12-O-tetradecanoylphorbol-13-acetate were used as the positive control. Means and standard errors for 2 independent experiments are shown. AP-1 nuclear activity was not significantly elevated by GM-CSF and TNF-α treatment (P, >0.2 for all subunits by a paired t test); however, JunB expression in STIF medium-treated cells (P < 0.05) and FosB (P < 0.01) and Fra-1 (P < 0.05) expression in GM-CSF- and TNF-α-treated cells were significantly different from zero by a 1-sample t test.
TNF-α reactivates latent virus in HPCs through an NF-κB-dependent mechanism.
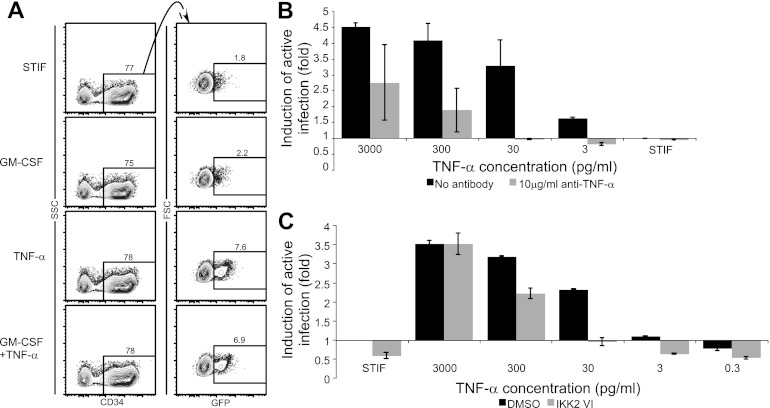
In other cell types, TNF-α has been reported to activate NF-κB through the classical pathway, which requires activation of the β subunit of IKK (42). We therefore asked whether TNF-α alone could reactivate latent virus in HPCs and whether this reactivation required IKKβ activity. To test whether TNF-α alone was sufficient to reactivate latent virus, we incubated latently infected HPCs with STIF medium, GM-CSF alone, TNF-α alone, or GM-CSF plus TNF-α. We observed that while GM-CSF alone had no significant effect on viral gene expression, TNF-α was able to induce viral gene expression at the same level as the combination of the cytokines (Fig. 4A). We confirmed that this effect was specifically due to TNF-α by stimulating cells with TNF-α in the presence and absence of antibodies to TNF-α. Induction of viral gene expression was substantially reduced in the presence of antibodies to TNF-α (Fig. 4B).
Fig 4.
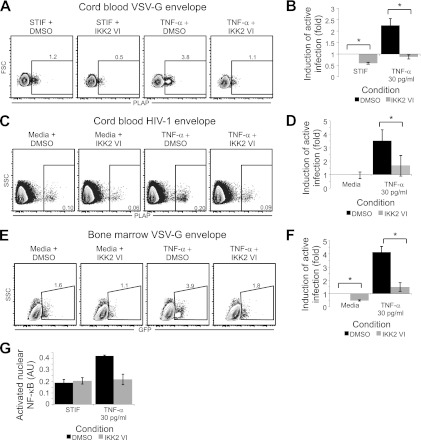
TNF-α reactivates latent infection in HPCs via an NF-κB-dependent pathway. (A) TNF-α is sufficient to reactivate latent virus. CD133+ cells treated with NL4-3-ΔGPE-GFP/VSV-G, sorted to remove actively infected cells, and incubated overnight with the indicated cytokines were analyzed by flow cytometry. Numbers show the percentages of cells falling within the indicated gates. Live cells were gated using forward scatter (FSC), side scatter (SSC), and 7-AAD. (B) Quantitative analysis of reactivation in cells treated as described for panel A and incubated with 10-fold dilutions of TNF-α in the presence or absence of an antibody to TNF-α. The mean fold increase in the percentage of cells expressing GFP over that for cells incubated in STIF medium alone is shown. Error bars represent standard errors for two independent experiments. (C) Quantitation of reactivation in cells treated as described for panel B except that cells were treated with HXB-ePLAP/VSV-G and were reactivated with or without the classical NF-κB pathway inhibitor IKK2 VI (10 μM). The mean fold increase in the percentage of cells expressing PLAP over that for cells incubated in STIF medium plus DMSO is shown. Error bars represent standard errors for two independent experiments.
To assess the dependence of TNF-α-induced viral reactivation on activation of the classical NF-κB pathway, we stimulated latently infected HPCs with varying concentrations of TNF-α with and without an inhibitor of IKKβ activity, IKK2 VI (5). While high doses of TNF-α overwhelmed the effect of IKK2 VI, at moderate TNF-α doses IKK2 VI had a substantial inhibitory effect (Fig. 4C). IKK2 VI significantly counteracted TNF-α-induced reactivation of latent virus when cells were infected with HIV pseudotyped with either the VSV-G (Fig. 5A and B) or the HIV (HXB) (Fig. 5C and D) envelope and when either cord blood-derived (Fig. 5A to D) or bone marrow-derived (Fig. 5E and F) HPCs were used. Using a transcription factor ELISA to assess nuclear NF-κB p50 levels in these samples, we observed that in samples where IKK2 VI inhibited the ability of TNF-α to reactivate latent virus, IKK2 VI also blocked TNF-α-induced NF-κB activation (Fig. 5G).
Fig 5.
The NF-κB inhibitor IKK2 VI counteracts TNF-α-induced reactivation of latent virus in HPCs. (A, C, and E) Flow cytometric analysis of CD133+ cells infected using the indicated envelope, sorted to remove actively infected cells, and stimulated overnight as indicated. Live cells were gated using forward scatter (FSC), side scatter (SSC), and 7-AAD; numbers indicate the percentage of cells within each gate. Results are representative of 4 (A) or 2 (C and E) independent experiments. (B, D, and F) Summary of reactivation for panels A, C, and E, respectively. The fold increase in the percentage of cells expressing PLAP over that for cells incubated in STIF medium plus DMSO was calculated. Means and standard errors for 4 (B) or 2 (D or F) independent experiments are shown. Asterisk indicates a significant difference (P, <0.01 [B], <0.02 [D], or <0.03 [F]) by a paired t test. (G) Quantitation of nuclear NF-κB p50 DNA binding activity for cells treated as for panel A and analyzed as described for Fig 4A. Means and standard errors for two experiments are shown.
In a majority of experiments, overnight incubation with IKK2 VI also reduced the level of active infection observed in cells cultured without TNF-α (Fig. 4C and 5B, C, and F), indicating that a portion of the low-level spontaneous reactivation observed under these conditions was also due to activation of NF-κB. This low-level spontaneous NF-κB activation may be a component of the spontaneous differentiation that occurs in HPCs under these culture conditions. Such NF-κB activation has been reported as a component of the differentiation of more mature hematopoietic progenitor cells (10, 85).
Conditions that reactivate latent virus in HPCs do not alter P-TEFb expression.
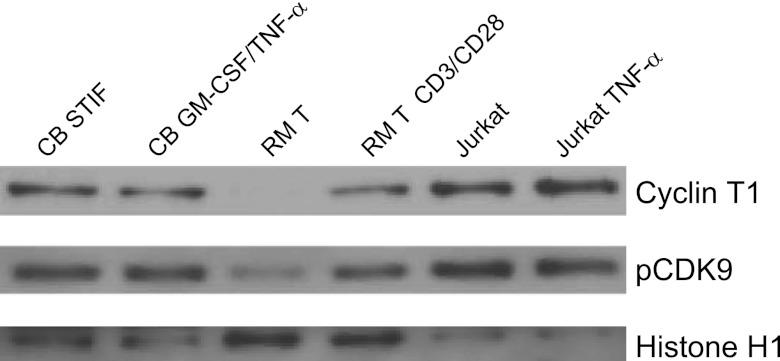
We next asked whether restricted levels of P-TEFb contribute to the establishment of latency in HPCs. To address this question, we analyzed nuclear CycT1 and phosphorylated CDK9 (pCDK9) in cord blood-derived HPCs incubated in STIF medium or stimulated overnight with GM-CSF and TNF-α. We compared CycT1 and pCDK9 levels in these cells to those observed in resting memory T cells or resting memory T cells that had been stimulated overnight with anti-CD3/anti-CD28 beads. As a positive control for CycT1 and pCDK9 expression, we examined the levels of both proteins in Jurkat cells with or without TNF-α stimulation. In agreement with previous findings (33, 38, 53, 70), Jurkat cells had high endogenous levels of CycT1 and pCDK9 that were not affected by TNF-α stimulation, while resting memory T cells had low levels of nuclear CycT1 and pCDK9 that increased dramatically following overnight incubation with anti-CD3/anti-CD28 beads (Fig. 6). Finally, we observed that both CycT1 and pCDK9 were readily detectable in unstimulated HPCs and that levels of these proteins were not affected by incubation with GM-CSF and TNF-α (Fig. 6). These results demonstrate that the ability of these cytokines to induce HIV-1 gene expression in HPCs is not related to alterations in P-TEFb expression.
Fig 6.
Nuclear P-TEFb in HPCs is not increased by stimulation with GM-CSF and TNF-α. Western blot analysis was carried out for cyclin T1 and pCDK9 in nuclear extracts from CD133+ HPCs (CB) incubated for 5 days in STIF medium and then treated with STIF medium or GM-CSF (100 ng/ml GM-CSF) plus TNF-α (2.5 ng/ml). Controls were nuclear extracts from resting memory T (RM T) cells isolated from peripheral blood by magnetic selection for CD4+ CD45RO+ HLA-DR− CD25− CD69− cells and lysed immediately or after stimulation with anti-CD3/anti-CD28 beads overnight, as well as nuclear extracts from Jurkat cells incubated overnight in medium or 3 ng/ml TNF-α. Results are representative of two independent experiments using cells from separate donors (T cells) or separate donor pools (cord blood).
Prostratin and SAHA, but not HMBA, can reactivate latent HIV-1 in HPCs.
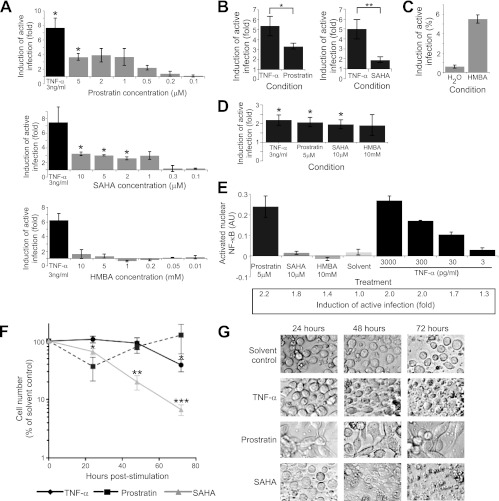
We next asked whether compounds that reactivate latent virus in T cell systems had similar activities in our model of latent infection in HPCs. We first tested prostratin, SAHA, and HMBA, and we found that while prostratin and SAHA were able to induce viral gene expression, HMBA was not (Fig. 7A). Titration of prostratin demonstrated that prostratin effectively reactivated latent virus in HPCs only at high doses (1 to 5 μM) (Fig. 7A). Although the comparative efficacies of prostratin and TNF-α differed between experiments, prostratin was on average significantly less effective than TNF-α at reactivating latent virus from HPCs at the 13-h time point (Fig. 7B). SAHA also reactivated latent virus only at high doses (2 to 10 μM) (Fig. 7A) and also induced reactivation to a significantly lesser extent than TNF-α at 13 h poststimulation (Fig. 7B). Finally, although HMBA could induce HIV-1 Gag expression in the latently infected U1 cell line (Fig. 7C), we did not detect an effect of HMBA on viral gene expression in HPCs at any concentration assessed (Fig. 7A).
Fig 7.
Prostratin and SAHA reactivate latent virus in CD34+ HPCs. (A) Quantitation of viral reactivation in CD133+ cells treated with prostratin, SAHA, or HMBA for 13 h. Reactivation was measured using the latency reactivation assay described for Fig 1 after infection with NL4-3-ΔGPE-GFP/VSV-G or HXB-ePLAP/VSV-G. The mean fold increase in the percentage of live CD34+ cells expressing PLAP or GFP over that with the solvent control is shown. Error bars represent standard errors for 3 (prostratin, SAHA) or 2 (HMBA) independent experiments. Asterisk indicates a significant difference (*, P < 0.05) by a 1-sample t test from the expected fold increase of 1. (B) Quantitation of reactivation by cells treated with prostratin (5 μM) or SAHA (10 μM) for 13 h by using the reactivation assay described for Fig 1. Means and standard errors for 7 independent experiments in cord blood (6 experiments) or bone marrow (1 experiment) are shown. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) by a paired t test. (C) Quantitation of reactivation in U1 cells stimulated for 48 h with 5 mM HMBA or a solvent control (H2O) and analyzed by flow cytometry to assess induction of HIV-1 Gag expression. Means and standard errors for three replicates are shown. (D) Quantitation of reactivation by cells treated with TNF-α, prostratin, SAHA, or HMBA for 24 h using the reactivation assay described for Fig. 1. Means and standard errors for 6 (TNF-α), 5 (HMBA), or 4 (prostratin or SAHA) independent experiments are shown. Asterisk indicates a significant difference (*, P < 0.03) by a 1-sample t test from the expected fold increase of 1. Induction of active infection with prostratin, SAHA, and HMBA is not significantly different from induction with TNF-α stimulation. (E) Quantitation of the results of a transcription factor ELISA measuring NF-κB DNA binding activity in nuclear extracts from HPCs infected with NL4-3-ΔGPE-GFP/VSV-G and stimulated for 24 h with the indicated compounds. ELISA data were analyzed as described for Fig 4A. Means and standard deviations for two replicates are shown. The fold increase in the percentage of cells expressing GFP over that with the solvent control is given under the bar graph. Results are representative of two independent experiments using cells from separate donor pools. (F) Time course of survival of HPCs cultured with TNF-α (3 ng/ml), prostratin (5 μM), or SAHA (10 μM). CD133+ HPCs were isolated from cord blood and were cultured in STIF medium for 5 days; then they were split into a solvent control or subjected to the conditions shown for an additional 3 days. The number of treated cells remaining is displayed as a percentage of the number of cells remaining in the solvent control (taken as 100%), as a function of time. Means and standard errors for 3 independent experiments are shown. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) by a 1-sample t test from the expected cell count of 100% of the cell count in the solvent. (G) Images, obtained with light microscopy at ×400 magnification, of the cells analyzed in panel F. Images were collected at the indicated times poststimulation. Data are representative of 3 independent experiments.
Although SAHA and prostratin were less effective than TNF-α at 13 h poststimulation, different results were obtained at 24 h. At this time point, we again found that SAHA, prostratin, and TNF-α were all able to induce statistically significant reactivation of latent virus in HPCs (Fig. 7D); however, there was no significant difference between the induction of active infection caused by TNF-α and that caused by prostratin or SAHA (Fig. 7D). HMBA still failed to induce significant reactivation at 24 h (Fig. 7D). Together, these results demonstrate that SAHA and prostratin, but not HMBA, are able to reactivate latent virus in HPCs; however, SAHA and prostratin reactivate virus with slower kinetics than does TNF-α.
Consistent with the established ability of prostratin to activate NF-κB through protein kinase C (77), we found that a 24-h stimulation with prostratin resulted in an increase in nuclear NF-κB p50 activity to levels comparable to those observed with a high dose of TNF-α (Fig. 7E). In contrast, stimulation with HMBA or the histone deacetylase inhibitor SAHA had no effect on nuclear NF-κB activity, even though SAHA reactivated latent HIV-1 to the same extent as a 30-pg/ml dose of TNF-α that did cause detectable NF-κB upregulation (Fig. 7E).
Although SAHA and prostratin reactivated latent virus in our model, both compounds had additional effects on HPCs. We observed that incubating HPCs in a medium containing SAHA caused massive cell death after 48 to 72 h (Fig. 7F and G). Similar cytotoxicity was observed at SAHA concentrations as low as 800 nM (data not shown). Although prostratin was not cytotoxic to HPCs (Fig. 7F), it caused rapid differentiation, resulting in adherent cells after as little as 24 h of exposure (Fig. 7G). In contrast, TNF-α did not induce cellular differentiation, and although somewhat cytotoxic at 72 h, it was substantially less so than SAHA (Fig. 7F and G).
Aza-CdR does not reactivate latent infection in HPCs.
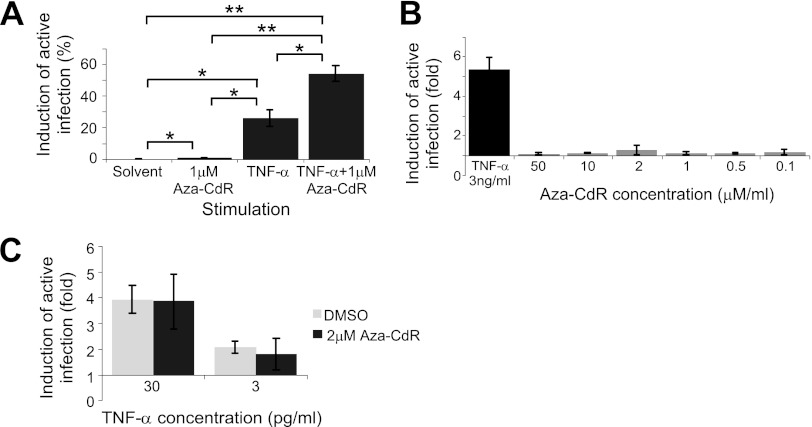
Finally, we examined whether the methylation inhibitor Aza-CdR could induce the reactivation of latent virus in HPCs. We first verified the activity of Aza-CdR in J-Lat cells, clones of Jurkat cells that are latently infected with a GFP-expressing HIV-1 reporter virus (35). In agreement with previous findings (37), we observed that while Aza-CdR alone had minimal ability to induce viral gene expression in three J-Lat clones, it had a synergistic effect on reactivation when combined with TNF-α (Fig. 8A). However, we were unable to detect any effect of Aza-CdR on reactivation of latent virus in HPCs, either alone (Fig. 8B) or in combination with TNF-α (Fig. 8C).
Fig 8.
Aza-CdR does not reactivate latent virus in CD34+ HPCs. (A) Quantitation of reactivation in J-Lat cells treated with the indicated compounds and assayed for GFP expression by flow cytometry. J-Lat cells treated with 30 ng/ml TNF-α and/or 1 μM Aza-CdR for 24 h were assayed for GFP expression (reporter for HIV-1 LTR activity) by flow cytometry after an additional 48 h. Means and standard errors of results from three J-Lat clones (J-Lat 8.4, J-Lat 9.2, and J-Lat 6.3) are shown. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) by a paired t test. (B) Quantitation of reactivation of viral gene expression in CD133+ HPCs using the reactivation assay shown in Fig 1 following treatment with the indicated concentrations of Aza-CdR. The mean fold increase in the percentage of cells expressing GFP over that with the solvent control is shown. Error bars represent standard errors for 2 independent experiments. (C) Quantitation of reactivation of viral gene expression in CD133+ HPCs using the reactivation assay shown in Fig 1 following treatment with TNF-α and either Aza-CdR or the DMSO solvent control. The fold increase in the percentage of cells expressing GFP over that with the solvent control was calculated. Means and standard errors for 2 independent experiments are shown.
DISCUSSION
In this study, we developed an in vitro model to study latency in immature HPCs, allowing us to investigate the types of HPCs that support latent infection and to study the factors that promote the establishment of latency. We found that HIV-1 could establish a latent infection in all of the subsets of HPCs that we examined, including an immature population that includes hematopoietic stem cells and multipotent progenitors. Because hematopoietic stem cells and multipotent progenitors are long-lived cell types, this result has important implications for the ability of HPCs to serve as a latent reservoir in vivo. Hematopoietic stem cells in particular are capable of indefinite self-renewal, and thus, latent infection in this cell type could result in a reservoir with an infinite half-life. Such a reservoir has previously been proposed based on the decay kinetics of the virus in plasma (50).
While our results suggest that many classes of progenitor cells might serve as short-term reservoirs of latent virus, immature progenitors are more likely to serve as latent reservoirs in patients treated with antiretroviral therapy. The reason for this is that these cells have the greatest self-renewal capacity, as demonstrated by the fact that only HSCs are capable of sustaining long-term, multilineage engraftment in mice (49). Similarly, these immature cells are the most likely to persist in humans after months or years of treatment. Thus, efforts to assess the extent of the latent reservoir in HPCs in patients on long-term antiretroviral therapy might be most successful if they focus on the immature CD34+ CD38− CD45RA− population of HPCs to maximize the chance of finding latently infected cells.
Our analysis revealed that TNF-α-induced activation of NF-κB in the absence of P-TEFb induction reactivates latent virus in HPCs. This result differs from recent findings in resting memory T cells, where TNF-α is not sufficient to reactivate latent virus, and induction of both NF-κB and P-TEFb activity appears to be required (41, 70, 79). Instead, our result is reminiscent of Jurkat cell line models of latency (22, 35, 38). Thus, our analysis suggests that HPCs may have fewer factors reinforcing latent infection than do resting memory T cells. In vivo, this might result in a rate of latent infection in HPCs that is lower than that observed in resting memory CD4+ T cells, where about 1 in 1 million cells harbors replication-competent provirus in treated patients (25).
In addition to TNF-α, which reactivated latent virus to the maximum level that we were able to achieve, we found that prostratin and SAHA can also reactivate latent virus in HPCs but with slower kinetics. Prostratin, like TNF-α, efficiently induces NF-κB activation; however, the two compounds activate NF-κB through distinct pathways (23, 27, 32, 34, 43, 44, 46, 52, 59, 60, 68, 75, 78). Thus, our finding that prostratin reactivates virus with slower kinetics than does TNF-α suggests that the pathway through which prostratin acts is less efficient in at least a subset of HPCs. Alternatively, it is possible that in addition to NF-κB, TNF-α activates another factor that promotes reactivation in HPCs at early time points. However, since we found that TNF-α treatment of HPCs does not result in AP-1 activation, the identity of such an additional factor is not clear. Since TNF-α reactivates latent virus from HPCs more rapidly than prostratin does and without inducing the rapid cellular differentiation observed during prostratin stimulation, our results suggest that NF-κB activation may be a successful strategy for reactivating latent virus in HPCs but that prostratin may not be an ideal compound with which to initiate NF-κB activation in these cells.
Consistent with the function of SAHA as a histone deacetylase inhibitor (24), we found that this compound reactivates latent virus in HPCs in the absence of NF-κB activation. Reactivation occurred using SAHA concentrations comparable to those that are effective at reactivating latent virus in T cells (19, 24). The relatively low efficacy of SAHA at 13 h poststimulation compared to that of the NF-κB activators TNF-α and prostratin suggests that although histone acetylation enhances HIV-1 transcription in HPCs, low NF-κB levels may still restrict rapid reactivation from latency. In addition, although SAHA has been shown to be nontoxic to T cells even over long culture periods (65), we found that doses of SAHA that could reactivate latent virus were extremely toxic in HPCs. This finding suggests that SAHA may not be effective at clearing latent virus from HPCs without significant detrimental effects on hematopoiesis.
Finally, we found that neither HMBA nor Aza-CdR could reactivate latent HIV-1 in our system. These findings are not surprising given the mechanisms of action of these compounds. HMBA is believed to activate P-TEFb (18); thus, the inability of this compound to reactivate virus in HPCs confirms our finding that P-TEFb levels are not restricted in this cell type. Finally, methylation has been shown to be a late event in the silencing of an HIV-1 provirus (6, 22, 37); since our model is designed to examine only early events in the establishment of latency in HPCs, it is not surprising that the methylation inhibitor Aza-CdR would have no effect in this system. Unfortunately, the difficulty of maintaining HPCs in culture in an undifferentiated state precludes examination of whether methylation might similarly occur as a late event in proviral silencing in HPCs.
To maximize the number, viability, and immaturity of HPCs in our experiments, we cultured HPCs in STIF medium, which contains the cytokines stem cell factor, thrombopoietin, IGFBP-2, and Flt3L. In addition to maintaining HPCs in an immature state, this medium also promotes the expansion of immature HPCs. Of note, we found that latency could be established in HPCs in spite of the fact that these cells proliferated during the course of our assay with a mean doubling time of less than 48 h (data not shown). This observation parallels findings from resting memory T cells, where homeostatic proliferation does not reactivate latent virus and instead helps to maintain the latent reservoir (7, 17). Similarly, our observations suggest that self-renewal of HPCs in vivo would not reactivate latent virus but instead would help to maintain this latent reservoir. However, additional study is needed to better define the effects of cell cycling on latently infected HPCs.
While further research to understand the extent of the latent reservoir in HPCs in patients on antiretroviral therapy will be informative, the very low rate of infection anticipated in these cells may make it difficult to determine definitively the size of this reservoir in vivo. Technical issues in culturing HPCs ex vivo may also contribute to the variable results that have been obtained when these cells are examined for infection; for instance, since active HIV infection is cytotoxic to HPCs (13), the reactivation of latent virus under differentiating culture conditions may result in cell death and false-negative results. Therefore, protocols that include a culturing step prior to cell purification may result in a reduced yield of provirus-containing cells (21). In addition, cell preparations that have been depleted of CD4+ CD34+ cells may yield false-negative cells, as CD4 is required for the infection of HPCs (12, 36). Conversely, false-positive results could be obtained if HPC samples are contaminated with CD4+ T cells. Together, these factors may make it difficult to define the role of HPCs in viral persistence.
Further complicating the detection of infected HPCs is the fact that only CXCR4-utilizing viruses can infect immature HPCs (12), which may result in widely differing infection rates in these cells in patients with different proportions of CXCR4- or dual-tropic viruses. Early in the course of infection, most HIV isolates utilize CCR5 (56, 74, 86); however, several recent reports have demonstrated that CXCR4-utilizing viral subpopulations are found in as many as 10 to 50% of recently infected individuals (1, 14). In addition, CXCR4-utilizing viruses emerge in many patients later in the course of infection (55, 66). A substantial fraction of HIV-infected patients would thus be expected to harbor virus that is capable of infecting HPCs; however, patients for whom CXCR4-utilizing virus is a minority population would be expected to have only a very low rate of infection in these cells. Additional studies are needed to more fully understand these variables.
Nonetheless, an eradication strategy for HIV must reactivate latent virus from all latently infected cells to eliminate all reservoirs of virus in the body. Thus, our observations on the mechanisms underlying the establishment and reactivation of latency in HPCs are of great utility in designing and evaluating eradication strategies, because they allow us to assess whether strategies that are effective in T cells will be equally effective in the proposed reservoir for latent virus in HPCs. Our findings demonstrate that latent infection can occur in all HPC subsets studied, including cells with surface markers consistent with long-lived, multipotent hematopoietic stem and progenitor cells. We found that latent infection could be reversed by NF-κB activation but that nuclear P-TEFb levels are not restricted in HPCs. Finally, we observed that TNF-α, prostratin, and SAHA are all capable of reactivating latent infection in HPCs but that prostratin and SAHA also induce differentiation or death, respectively, in the HPC population. Together, these data show that immature HPCs have the potential to serve as a reservoir of latent virus and suggest that strategies that activate NF-κB can reactivate latent virus from these cells. Proposed reactivation strategies can be assessed in this model of latent infection in HPCs as well as in models of latently infected resting memory T cells in order to assess the ability of different strategies to reactivate latent virus in both cell types and thus to maximize the chance of eliminating all latent reservoirs in vivo.
ACKNOWLEDGMENTS
This work was supported by the Burroughs Wellcome Foundation, U.S. National Institutes of Health grants RO1 A1096962 and R21 A1086599, a U.S. National Science Foundation Graduate Student Research Fellowship (grant DGE 0718128, to L.A.M.), and a University of Michigan Bernard Maas Fellowship (to L.A.M.). Flow sorting at the University of Michigan flow core was supported by Cancer Center Support grant 5 P30 CA46592.
We thank the members of the Collins lab for insightful discussions of the data in this paper, and in particular we thank F. Taschuk, A. Onafuwa-Nuga, and N. Sebastian for careful reading of the manuscript. We thank the University of Michigan flow core for assistance with flow sorting. For assistance with transcription factor ELISAs, we thank S. Rains-Lynema, J. Dalton, A. East Obi, N. Sutton, Y. Kanthi, N. Hariadi, C. Ebens, L. Nicholls, M. Vander Lugt, K. Scott, R. Jan, Y. Rezk, C. Marsh, T. Elarini, and K. Sugg. The following reagents were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, U.S. National Institutes of Health: J-Lat clones 8.4, 9.2, and 6.3 from E. Verdin (Gladstone Institute of Virology and Immunology, University of California, San Francisco), U1 cells from T. Folks (Texas Biomedical Research Institute), and NL4-3-ΔE-GFP from H. Zhang, Y. Zhou, and R. Siliciano (Johns Hopkins University). pCMV-HIV-1 was a gift from S.-J.-K. Yee (City of Hope National Medical Center).
Footnotes
Published ahead of print 20 June 2012
REFERENCES
- 1. Abbate I, et al. 2011. Detection of quasispecies variants predicted to use CXCR4 by ultra-deep pyrosequencing during early HIV infection. AIDS 25:611–617 [DOI] [PubMed] [Google Scholar]
- 2. Aggarwal BB. 2000. Tumour necrosis factors receptor associated signalling molecules and their role in activation of apoptosis, JNK and NF-κB. Ann. Rheum. Dis. 59(Suppl. 1):i6–i16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey JR, et al. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80:6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barboric M, et al. 2007. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35:2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baxter A, et al. 2004. Hit-to-lead studies: the discovery of potent, orally active, thiophenecarboxamide IKK-2 inhibitors. Bioorg. Med. Chem. Lett. 14:2817–2822 [DOI] [PubMed] [Google Scholar]
- 6. Blazkova J, et al. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554 doi:10.1371/journal.ppat.1000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V. 2011. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 7:e1002288 doi:10.1371/journal.ppat.1002288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bosque A, Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brennan TP, et al. 2009. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J. Virol. 83:8470–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burkly L, et al. 1995. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature 373:531–536 [DOI] [PubMed] [Google Scholar]
- 11. Canonne-Hergaux F, Aunis D, Schaeffer E. 1995. Interactions of the transcription factor AP-1 with the long terminal repeat of different human immunodeficiency virus type 1 strains in Jurkat, glial, and neuronal cells. J. Virol. 69:6634–6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carter CC, et al. 2011. HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host Microbe 9:223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carter CC, et al. 2010. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat. Med. 16:446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chalmet K, et al. 2012. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J. Infect. Dis. 205:174–184 [DOI] [PubMed] [Google Scholar]
- 15. Chen BK, Gandhi RT, Baltimore D. 1996. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J. Virol. 70:6044–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen G, Goeddel DV. 2002. TNF-R1 signaling: a beautiful pathway. Science 296:1634–1635 [DOI] [PubMed] [Google Scholar]
- 17. Chomont N, et al. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Contreras X, Barboric M, Lenasi T, Peterlin BM. 2007. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3:e146 doi:10.1371/journal.ppat.0030146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Contreras X, et al. 2009. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 284:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doulatov S, Notta F, Eppert K, Nguyen L. 2010. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11:585–593 [DOI] [PubMed] [Google Scholar]
- 21. Durand CM, et al. 2012. HIV-1 DNA is detected in bone marrow populations containing CD4+ T cells but is not found. in purified CD34+ hematopoietic progenitor cells in most patients on antiretroviral therapy. J. Infect. Dis. 205:1014–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duverger A, et al. 2009. Determinants of the establishment of human immunodeficiency virus type 1 latency. J. Virol. 83:3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ea C-K, Deng L, Xia Z-P, Pineda G, Chen ZJ. 2006. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22:245–257 [DOI] [PubMed] [Google Scholar]
- 24. Edelstein LC, Micheva-Viteva S, Phelan BD, Dougherty JP. 2009. Activation of latent HIV type 1 gene expression by suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor approved for use to treat cutaneous T cell lymphoma. AIDS Res. Hum. Retroviruses 25:883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Finzi D, et al. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517 [DOI] [PubMed] [Google Scholar]
- 26. Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. 1987. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238:800–802 [DOI] [PubMed] [Google Scholar]
- 27. Gaide O, et al. 2002. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-κB activation. Nat. Immunol. 3:836–843 [DOI] [PubMed] [Google Scholar]
- 28. Gasmi M, et al. 1999. Requirements for efficient production and transduction of human immunodeficiency virus type 1-based vectors. J. Virol. 73:1828–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geeraert L, Kraus G, Pomerantz RJ. 2008. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu. Rev. Med. 59:487–501 [DOI] [PubMed] [Google Scholar]
- 30. Gulick RM, et al. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734–739 [DOI] [PubMed] [Google Scholar]
- 31. Hammer SM, et al. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 337:725–733 [DOI] [PubMed] [Google Scholar]
- 32. Hara H, et al. 2003. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity 18:763–775 [DOI] [PubMed] [Google Scholar]
- 33. Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72:9881–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsu H, Shu HB, Pan MG, Goeddel DV. 1996. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84:299–308 [DOI] [PubMed] [Google Scholar]
- 35. Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Josefsson L, et al. 2012. Hematopoietic precursor cells isolated from patients on long term suppressive HIV therapy did not contain HIV-1 DNA. J. Infect. Dis. 206:28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kauder S, Boque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495 doi:10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim YK, Mbonye U, Hokello J, Karn J. 2011. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 410:896–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kohno T, et al. 2002. A new improved method for the concentration of HIV-1 infective particles. J. Virol. Methods 106:167–173 [DOI] [PubMed] [Google Scholar]
- 40. Korin Y, Brooks D, Brown S, Korotzer A, Zack JA. 2002. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J. Virol. 76:8118–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lassen KG, Hebbeler AM, Bhattacharyya D, Lobritz MA, Greene WC. 2012. A flexible model of HIV-1 latency permitting evaluation of many primary CD4 T-cell reservoirs. PLoS One 7:e30176 doi:10.1371/journal.pone.0030176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li SW, et al. 1999. The IKKβ subunit of IκB kinase (IKK) is essential for nuclear factor κB activation and prevention of apoptosis. J. Exp. Med. 189:1839–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lucas PC, et al. 2001. Bcl10 and MALT1, independent targets of chromosomal translocation in MALT lymphoma, cooperate in a novel NF-κB signaling pathway. J. Biol. Chem. 276:19012–19019 [DOI] [PubMed] [Google Scholar]
- 44. McAllister-Lucas LM, et al. 2001. Bimp1, a MAGUK family member linking protein kinase C activation to Bcl10-mediated-NF-κB induction. J. Biol. Chem. 276:30589–30597 [DOI] [PubMed] [Google Scholar]
- 45. McNamara LA, Collins KL. 2011. Hematopoietic stem/precursor cells as HIV reservoirs. Curr. Opin. HIV AIDS 6:43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Micheau O, Tschopp J. 2003. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114:181–190 [DOI] [PubMed] [Google Scholar]
- 47. Nabel G, Baltimore D. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713 [DOI] [PubMed] [Google Scholar]
- 48. Nguyen VT, Kiss T, Michels AA, Bensaude O. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322–325 [DOI] [PubMed] [Google Scholar]
- 49. Notta F, et al. 2011. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333:218–221 [DOI] [PubMed] [Google Scholar]
- 50. Palmer S, et al. 2008. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 105:3879–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. 2000. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 28:663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pobezinskaya YL, et al. 2008. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat. Immunol. 9:1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramakrishnan R, Dow EC, Rice AP. 2009. Characterization of Cdk9 T-loop phosphorylation in resting and activated CD4+ T lymphocytes. J. Leukoc. Biol. 86:1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Redd AD, Avalos A, Essex M. 2007. Infection of hematopoietic progenitor cells by HIV-1 subtype C, and its association with anemia in southern Africa. Blood 110:3143–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Richman DD, Bozzette SA. 1994. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 169:968–974 [DOI] [PubMed] [Google Scholar]
- 56. Rieder P, et al. 2011. Characterization of human immunodeficiency virus type 1 (HIV-1) diversity and tropism in 145 patients with primary HIV-1 infection. Clin. Infect. Dis. 53:1271–1279 [DOI] [PubMed] [Google Scholar]
- 57. Roebuck KA, Brenner DA, Kagnoff MF. 1993. Identification of c-fos-responsive elements downstream of TAR in the long terminal repeat of human immunodeficiency virus type-1. J. Clin. Invest. 92:1336–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Roebuck KA, Gu DS, Kagnoff MF. 1996. Activating protein-1 cooperates with phorbol ester activation signals to increase HIV-1 expression. AIDS 10:819–826 [DOI] [PubMed] [Google Scholar]
- 59. Ruefli-Brasse AA, French DM, Dixit VM. 2003. Regulation of NF-κB-dependent lymphocyte activation and development by paracaspase. Science 302:1581–1584 [DOI] [PubMed] [Google Scholar]
- 60. Ruland J, et al. 2001. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-κB and neural tube closure. Cell 104:33–42 [DOI] [PubMed] [Google Scholar]
- 61. Sahu GK, et al. 2009. Low-level plasma HIVs in patients on prolonged suppressive highly active antiretroviral therapy are produced mostly by cells other than CD4 T-cells. J. Med. Virol. 81:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Saleh S, et al. 2007. CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: a novel model of HIV-1 latency. Blood 110:4161–4164 [DOI] [PubMed] [Google Scholar]
- 63. Salerno D, et al. 2007. Direct inhibition of CDK9 blocks HIV-1 replication without preventing T-cell activation in primary human peripheral blood lymphocytes. Gene 405:65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sedore SC, et al. 2007. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 35:4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shan L, et al. 2012. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36:491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shankarappa F, et al. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stanley SK, et al. 1992. CD34+ bone marrow cells are infected with HIV in a subset of seropositive individuals. J. Immunol. 149:689–697 [PubMed] [Google Scholar]
- 68. Ting AT, Pimentel-Muiños FX, Seed B. 1996. RIP mediates tumor necrosis factor receptor 1 activation of NF-κB but not Fas/APO-1-initiated apoptosis. EMBO J. 15:6189–6196 [PMC free article] [PubMed] [Google Scholar]
- 69. Trono D, et al. 2010. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 329:174–180 [DOI] [PubMed] [Google Scholar]
- 70. Tyagi M, Pearson RJ, Karn J. 2010. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vallabhapurapu S, Karin M. 2009. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733 [DOI] [PubMed] [Google Scholar]
- 72. Van Lint C, et al. 1997. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J. Virol. 71:6113–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Van Lint C, Emiliani S, Ott M, Verdin E. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112–1120 [PMC free article] [PubMed] [Google Scholar]
- 74. van't Wout AB, et al. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Invest. 94:2060–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang D, et al. 2002. A requirement for CARMA1 in TCR-induced NF-κB activation. Nat. Immunol. 3:830–835 [DOI] [PubMed] [Google Scholar]
- 76. Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462 [DOI] [PubMed] [Google Scholar]
- 77. Williams SA, et al. 2004. Prostratin antagonizes HIV latency by activating NF-κB. J. Biol. Chem. 279:42008–42017 [DOI] [PubMed] [Google Scholar]
- 78. Wu C-J, Conze DB, Li T, Srinivasula SM, Ashwell JD. 2006. NEMO is a sensor of Lys 63-linked polyubiquitination and functions in NF-κB activation. Nat. Cell Biol. 8:398–406 [DOI] [PubMed] [Google Scholar]
- 79. Yang H-C, et al. 2009. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Invest. 119:3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang Z, Zhu Q, Luo K, Zhou Q. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317–322 [DOI] [PubMed] [Google Scholar]
- 81. Yik JHN, et al. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971–982 [DOI] [PubMed] [Google Scholar]
- 82. Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18:1101–1108 [DOI] [PubMed] [Google Scholar]
- 83. Zhang H, et al. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang L, et al. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605–1613 [DOI] [PubMed] [Google Scholar]
- 85. Zhang M-Y, Sun S-C, Bell L, Miller BA. 1998. NF-κB transcription factors are involved in normal erythropoiesis. Blood 91:4136–4144 [PubMed] [Google Scholar]
- 86. Zhu T, et al. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179–1181 [DOI] [PubMed] [Google Scholar]