Abstract
The forkhead-associated (FHA) domain recognizes phosphothreonine (pT) with high specificity and functional diversity. TIFA (TRAF-interacting protein with an FHA domain) is the smallest FHA-containing human protein. Its overexpression was previously suggested to provoke NF-κB activation, yet its exact roles in this signaling pathway and the underlying molecular mechanism remain unclear. Here we identify a novel threonine phosphorylation site on TIFA and show that this phosphorylated threonine (pT) binds with the FHA domain of TIFA, leading to TIFA oligomerization and TIFA-mediated NF-κB activation. Detailed analysis indicated that unphosphorylated TIFA exists as an intrinsic dimer and that the FHA-pT9 binding occurs between different dimers of TIFA. In addition, silencing of endogenous TIFA resulted in attenuation of tumor necrosis factor alpha (TNF-α)-mediated downstream signaling. We therefore propose that the TIFA FHA-pT9 binding provides a previously unidentified link between TNF-α stimulation and NF-κB activation. The intermolecular FHA-pT9 binding between dimers also represents a new mechanism for the FHA domain.
INTRODUCTION
The forkhead-associated (FHA) domain, discovered in 1995 (10) and first suggested to bind phosphoproteins in 1998 (24), is known to specifically recognize phosphothreonine (pT) to exert its function (5, 20). Although the sequence homology among different FHA-containing proteins is relatively low, the structural architecture of FHA domains is highly conserved. It contains a six-stranded β-sheet and another five-stranded β-sheet, forming a β-sandwich. The FHA-pT binding has been shown to regulate diverse biological functions, ranging from DNA damage repair to cell cycle checkpoints to signal transduction (18). Furthermore, the mechanism of FHA-phosphoprotein binding varies greatly among different FHA-containing proteins. The structure, specificity, mechanism, and biological functions of FHA domains have been summarized in recent reviews (16, 18).
TRAF-interacting protein with an FHA domain (TIFA) was first identified as a tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) binding protein. Consisting of 184 amino acids, TIFA is the smallest FHA domain-containing protein in humans (Fig. 1A). In the absence of TNF-α stimulation, TIFA overexpression in HEK 293T cells can activate NF-κB and AP-1 (14), suggesting a direct involvement of TIFA in TNF-mediated immune responses. This involvement of TIFA was further attributed to the binding of TRAF2, which requires the TRAF domain of TRAF2 and almost the entire TIFA protein (residues 1 to 162) (14). TIFA was also reported to bind to TNF-associated factor 6 (TRAF6) (25). The consensus binding site of TIFA for TRAF6 was mapped to be glutamic acid 178 (E178) (11, 25), indicating different binding mechanisms in TIFA-TRAF2 and TIFA-TARF6 interactions. In addition, TIFA overexpression, even in the absence of interleukin-1 (IL-1), was shown to activate NF-κB and c-Jun amino-terminal kinase (JNK), possibly through its enhancement of TRAF6 binding to IL-1 receptor-associated kinase 1 (IRAK-1). On the other hand, mutation of E178 abolished the binding of TIFA to TRAF6 and the ensuing activation of NF-κB (25). In a follow-up report, TIFA was shown to promote oligomerization and ubiquitination of TRAF6, leading to activation of IκB kinase (IKK), based on in vitro studies (6).
Fig 1.
Enhancement of TIFA phosphorylation by TNF-α stimulation. (A) Schematic overview of TIFA domain structure (top) and N-terminal sequences of TIFA orthologues from different species (bottom). T9 is highlighted in dark gray. (B) MS/MS spectrum of the peptide containing pT9 in TIFA. The precursor ion 827.28162+ is from ADPMTSFEDAD(pT)EE. (C) NanoPro immunoassay demonstrates the phosphorylation status of overexpressed Myc-tagged WT and T9A mutant TIFA with or without TNF-α and phosphatase treatment. The peaks at pI 4.75 and pI 4.63 are assigned to unphosphorylated TIFA and singly phosphorylated TIFA, respectively. The bar graph in the lower panel represents the ratios of peak areas of singly phosphorylated TIFA to total TIFA. The results represent means ± standard deviations (SD) from at least 3 independent experiments. (D) NanoPro immunoassay of endogenous TIFA. The conditions were the same as those for panel C. The pI values differ from the peaks in panel C due to lack of the Myc tag. (E) Recombinant TIFA or the T9A mutant was incubated with [γ-32P]ATP and lysates from cells treated with or without TNF-α for 30 min. During incubation, alkaline phosphatase (PPtase) was added as indicated. The reaction mixtures were separated by SDS-PAGE, and the bands were revealed by autoradiography. (F) The experimental conditions were the same as those for panel E except for the inclusion of caffeine or inhibitor cocktails for AKT, IRAK, TAK, or PKC (Go 6976 and Go 6983).
Although the studies of Takatsuna et al. (25) and Ea et al. (6) have previously established the key function of TIFA in its interaction with TRAF6, several issues still remain inconclusive. For example, TIFA has been suggested to be phosphorylated, and the integrity of the FHA domain of TIFA is essential for its function (6, 25), but little information has been unveiled about the molecular basis of TIFA phosphorylation and its functional consequences.
In this work, we report that threonine 9 (T9) is a newly identified phosphorylation site of TIFA and that the phosphorylation level of T9 increases upon TNF-α treatment. Based on data collected here, we concluded that TIFA-FHA binds to this pT9 site. Such a TIFA-FHA/pT9 binding directs TIFA self-association and promotes NF-κB activation through the oligomerization process. We also observed in vivo speckle formation of oligomerized TIFA which colocalizes with TRAF6. Further biophysical analyses indicate that TIFA-FHA/pT9 binding occurs between dimers of TIFA, leading to oligomerization. Moreover, our studies suggest that the TNF-α-mediated signaling is attenuated when endogenous TIFA is knocked down. Thus, we propose that FHA/pT9 binding of TIFA is a critical link between TNF-α stimulation and NF-κB activation. Our findings provide not only a new molecular insight into the TNF-α-mediated signaling pathway but also a new functional mechanism of FHA-containing proteins.
MATERIALS AND METHODS
Cell culture.
The human embryonic kidney (HEK) 293T cell line and human osteosarcoma cell line U2OS were cultured in Dulbecco's modified Eagle medium (DMEM) (Gibco). The human acute monocytic leukemia cell line THP-1 was cultured in Roswell Park Memorial Institute medium (RPMI 1640) (Gibco). All media were supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco), 200 mM l-glutamine (Gibco), 100 U/ml penicillin (Gibco), 100 μg/ml streptomycin (Gibco), and 1% sodium pyruvate (Gibco). Cells were kept in a 37°C incubator with 5% CO2. Cells were starved by replacing the complete medium with serum-free DMEM 8 to 10 h prior to the addition of 50 ng/ml of TNF-α at the indicated time points.
TIFA plasmids and recombinant protein.
The His-tagged human TIFA wild-type (WT) protein and various mutants were expressed in Escherichia coli BL21 Codon Plus and affinity purified with nickel resin (Millipore). Approximately 30 mg of purified recombinant His-TIFA was applied to a HiLoad 16/60 Superdex 75-pg column or a Superdex 75 10/300 GL column (GE Healthcare). The separated fractions were analyzed by 15% SDS-PAGE, and proteins were visualized by Coomassie blue staining. Other plasmids involved in this study, including Flag-tagged pCMV-Tag2a (Stratagene), pcDNA3.1-Myc (EQKLISEEDL) (Invitrogen), pNF-κB-Luc (Clontech), His-tagged pET43.1a (Novagen), and Gal-tagged pBIND (Promega), were obtained from commercial sources.
RNA interference.
Two hundred picomoles of double-stranded small interfering RNA (siRNA) oligomers (Invitrogen) corresponding to the sequence of TIFA (UCAGGACAAACAGGUUUCCCGAGUU) or scramble control oligomers (Invitrogen) was transfected into HEK 293T or THP-1 cells. Cells were collected after 72 h of incubation.
Antibodies.
The anti-TIFA monoclonal antibody (MAb) was produced by the core facility at IBC, Academia Sinica. Other MAbs used in the current study are anti-Flag (Sigma), anti-Myc (Millipore), anti-Gal (Abcam), anti-β-actin (GeneTex), anti-His (Signal Chem), anti-pIKKα (pT23) (Abcam), anti-pIκB (pS32/pS36) (Santa Cruz), and anti-NF-κB p65 (Millipore). The polyclonal antibodies used in this work are anti-NF-κB p65 (Santa Cruz), horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Millipore), fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Millipore), and rhodamine-conjugated anti-rabbit IgG (Millipore).
NanoPro immunoassay.
The NanoPro immunoassay method involves separation of cell lysates in the capillary isoelectric focusing (IEF) step, followed by analysis of specific protein isoforms (i.e., different phosphorylation forms of TIFA) using conventional immune detection (19). Cells were collected and centrifuged at 1.0 krpm for 5 min. The cell pellet was then resuspended in 250 μl Bicine-CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} lysis buffer plus 1× dimethyl sulfoxide (DMSO) inhibitor mix and 1× aqueous inhibitor mix. The cell lysates were cleared by centrifugation at 13.2 krpm for 1 h at 4°C. When indicated, 5 mg total cell lysate was incubated with alkaline phosphatase (Fermentas) together with 10× reaction buffer at 37°C for 1 h. Final supernatants were transferred to a fresh tube and snap-frozen with liquid nitrogen. The nanofluidic proteomic immunoassay was performed with the NanoPro-1000 system. Cell lysate was diluted to 2× the final protein concentration in Bicine-CHAPS buffer in the presence of DMSO inhibitor mix and 20 mM dithiothreitol (DTT). Diluted cell lysate was combined with an equal volume of IEF buffer solution (50% [vol/vol] pH 3 to 10 premix solution, 50% [vol/vol] Servalyte, pH 3 to 6), 1 μM pI standard 4.4, and 1 μM pI standard 5.5. The charge-based separation was performed in a capillary at 45,000 μW for 40 min, and immobilization was by 80 s of irradiation with UV light. After separation and immobilization, the sample was incubated with primary antibody for 120 min. Each overexpressed sample was then incubated with HRP-conjugated goat anti-mouse IgG for 1 h. Endogenous sample was detected by biotin-labeled goat anti-rabbit IgG for 1 h followed by 10 min of incubation with streptavidin (SA)-HRP-conjugated antibody. Chemiluminescence signals for the target proteins were detected by adding luminol and peroxide XDR detection reagents and analyzed with the Compass software.
In vitro kinase assay.
After washing with phosphate-buffered saline (PBS), TNF-α-stimulated or nonstimulated 293T cells were lysed with CHAPS lysis buffer. The cell lysates were precleaned with sheep anti-mouse IgG M-280 Dynabeads (Invitrogen) at 25°C for 30 min and then incubated with the recombinant His-tagged wild-type or T9 mutant TIFA in a buffer containing 40 mM HEPES (pH 7.5), 20 mM MgCl2, 100 μM ATP, and 1 mCi/ml [γ-32P]ATP (Perkin-Elmer) at 37°C for 30 min. His-TIFA was pulled down with M-280 Dynabeads coated with anti-His MAb. The reaction was terminated by addition of SDS sample buffer and heating at 95°C for 10 min prior to SDS-PAGE.
Coimmunoprecipitation analysis.
The protein-coding sequences of TIFA cDNA were subcloned into the expression vector pCMV-Flag or pcDNA3.1-Myc. HEK 293T cells (∼2 × 107) were cotransfected with 3 μg of the expression vectors containing Flag-TIFA and Myc-TIFA using JetPEI (Polyplus transfection). After 36 h, cells were lysed with TNE buffer, containing 10 mM Tris-HCl (pH 7.8), 1% NP-40, 0.15 M NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× protease inhibitor cocktail. An amount of 6.5 μg of anti-Flag MAb was precoated with M-280 Dynabeads overnight at 4°C. The cell lysates were incubated with anti-Flag–Dynabeads at 4°C overnight. The protein-bead complex was then washed with TNE buffer and 1× Tris-buffered saline–Tween 20 (TBST) buffer. The immunoprecipitates were then eluted using 50 μg of the 3× Flag peptide (Sigma) and subjected to Western blot analysis.
Western blotting.
For Western blotting, cell lysates were separated by 15% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Perkin-Elmer). The membranes were then blotted with primary antibody at 4°C overnight followed by HRP-conjugated anti-mouse IgG. The blotted protein bands were revealed by the ECL system (Millipore).
Native PAGE experiment.
For the native PAGE experiment, cells were lysed using the NativePAGE (Invitrogen) sample buffer in the presence of 10% n-dodecyl β-d-maltoside (DDM) and 5% digitonin. After centrifugation at 4°C for 1 h, the whole-cell lysates were separated by gradient PAGE (4 to 16%). A buffer containing 0.5 M Tris base (pH 9.2) and 0.5 M glycine was used for protein transfer. After transfer, the PVDF membrane was incubated in 20 ml of 7.5% acetic acid for 15 min at room temperature to fix the proteins. The membrane was subsequently rinsed with methanol and deionized water to remove the residual Coomassie blue G-250 dye. Finally, TIFA proteins were detected by Western blot analysis using mouse anti-Flag antibody or mouse anti-Myc antibody.
NF-κB activation assay.
293T cells in 6-well plates were transiently transfected with 1.5 μg of the TIFA WT or mutant expression plasmids together with 1.5 μg of the pNF-κB-Luc reporter. After 36 h, cells were lysed with 1× passive lysis buffer (Promega) and 20 μl of cell lysates was dispensed into a 96-well plate, followed by addition of 100 μl of lyophilized luciferase assay substrate (LARII) (Promega) and 100 μl of Stop & Glo reagent (Promega). The luciferase activity was measured as relative luminescence units (RLU). When indicated, Western blot analysis for the total cell lysate was carried out in parallel in order to confirm the expression levels of each sample. The firefly luciferase readings were then normalized to the internal Renilla luciferase readings as well as the corresponding band intensities of the samples.
Immunostaining.
An amount of 7 × 105 U2OS cells seeded on coverslips was transfected with 3 μg of TIFA or mutant vectors. At 36 h posttransfection, cells were washed with cold PBS and fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 30 min. Cells were permeabilized with 0.2% Triton X-100, blocked with PBS containing 10% bovine serum albumin (BSA), and then incubated with primary antibodies, fluorescence-labeled secondary antibodies, and DAPI (4′,6′-diamidino-2-phenylindole) sequentially. Fluorescence image sections were taken by Zeiss LSM 510 confocal microscopy.
Mass spectrometry (MS) analysis.
The phosphorylation sites were analyzed using high-resolution and high-mass-accuracy nanoflow liquid chromatography-tandem MS (LC-MS/MS) on a linear quadrupole ion trap-Fourier transform (LTQ-FT) ion cyclotron resonance mass spectrometer (Thermo Fisher Scientific) with procedures described previously (8).
ITC analysis.
Isothermal titration calorimetry (ITC) experiments were performed using a MicroCal iTC200 instrument (Northampton, MA). Two TIFA N-terminal peptides, i.e., MTSFEDADTEETVT and MTSFEDAD(pT)EETVT (2 mM), were used to titrate TIFA protein (100 μM) at 25°C in the calorimeter cell (0.2044 ml) with automatic injections of 1.6 μl each time. With the use of software provided by the manufacturer, each peak corresponding to the injection was integrated and corrected with baseline. The titration heat had been calculated to eliminate the effect of heat generated from diluting the ligand into buffer. Thermal data were fitted to a two-independent-site binding model to yield the value of the equilibrium dissociation constant (Kd).
Analytical ultracentrifugation (AUC) analysis.
Sample and buffer were loaded into a 12-mm standard double-sector Epon charcoal-filled centerpiece and mounted in an An-60 or An-50 Ti rotor of a Beckman Coulter XL-I analytical ultracentrifuge (Fullerton, CA). The rotor speed was 40,000 rpm at 20°C. The signal was monitored at 280 nm. The partial specific volume of TIFA protein is 0.724. The raw experimental data were analyzed by Sedfit (http://www.analyticalultracentrifugation.com/default.htm), and the plots of c(s, fr) and molecular mass versus the s value were generated by MATLAB (MathWork, Inc.).
RESULTS
TIFA phosphorylation at Thr9.
Given the presence of an FHA domain in the middle of TIFA and five Thr residues at its N terminus (T2, T9, T12, T14, and T19) (Fig. 1A), we investigated whether one of these Thr residues could be phosphorylated and thereby recognized by the FHA domain. The exogenously expressed Flag-TIFA was immunoprecipitated from HEK 293T cells and then subjected to mass spectrometry (MS) analysis. With 80% sequence coverage, the MS data revealed that Flag-TIFA was phosphorylated at T9 (Fig. 1B).
Because both TNF-α stimulation and TIFA overexpression can activate NF-κB (14, 25), it is likely that T9 phosphorylation is correlated with TNF-α-elicited signaling. We thus used the recently developed NanoPro immunoassay (7, 19) to examine the effect of TNF-α treatment on T9 phosphorylation. The method involves separation of cell lysates in the capillary isoelectric focusing step, followed by analysis of specific protein isoforms (i.e., different phosphorylation forms of TIFA) using conventional immune detection (19). As shown in Fig. 1C, trace 1, the exogenously expressed Myc-TIFA, revealed by the anti-Myc antibody, exhibited a major peak at pI 4.75. This was close to the theoretical pI value of 4.92 deduced by the Scansite database (http://scansite.mit.edu/calc_mw_pi.html). Among the several smaller peaks detected, the one with pI 4.63 was consistent with singularly phosphorylated TIFA, since addition of one phosphate group is expected to decrease the pI by 0.12. Treatment with TNF-α also led to an increase of the peak with pI 4.63 (traces 2), suggesting phosphorylation at one single amino acid residue. However, TNF-α did not have such an effect on the T9A mutant (traces 3 and 4). When traces 1 to 4 were repeated in the presence of alkaline phosphatase (traces 5 to 8, respectively), only one single peak at pI 4.75 was observed. These results support that the exogenously expressed TIFA was phosphorylated (at T9 based on the MS result) and that the phosphorylation increased upon TNF-α treatment. To examine whether TNF-α induces T9 phosphorylation of the endogenous TIFA, we raised a monoclonal antibody (MAb) against bacterially expressed full-length TIFA. The endogenous TIFA from 293T cells detected by NanoPro immunoassay showed the same results as seen in Fig. 1C (comparing traces 1 to 4 in Fig. 1D with traces 1, 2, 5, and 6 in Fig. 1C). Of note, since the endogenous TIFA was not tagged, the pI values differed slightly from those in Fig. 1C.
An in vitro kinase assay was then used to further confirm that TIFA T9 phosphorylation is TNF-α dependent. We incubated recombinant TIFA with TNF-α-stimulated cell extracts in the presence of [γ-32P]ATP. Revealed by autoradiography, the band intensity of 32P-labeled His-TIFA increased when TNF-α-stimulated cell extract was used, relative to that from control cells (Fig. 1E, lanes 3 and 4). On the other hand, there was no TNF-α-dependent increase in the band intensity for the T9A mutant (Fig. 1E, lanes 6 and 7), indicating that TNF-α-increased TIFA phosphorylation most likely occurs at T9. In the control lanes 1, 5, 8, and 9, no 32P-labeled TIFA band was observed when the sample was treated with phosphatase or when TIFA protein or cell lysate was omitted.
To explore the kinase that mediates the TNF-α-dependent T9 phosphorylation, cells were treated with caffeine, a general inhibitor of the phosphatidylinositol 3-kinase (PI3K) pathway, or with kinase inhibitor cocktails for AKT, IRAK, TAK, and protein kinase C (PKC) before TNF-α treatment. Autoradiography (Fig. 1F) showed that caffeine and AKT and PKC inhibitors, but not IRAK or TAK inhibitors, decreased the level of TNF-α-induced phosphorylation of wild-type (WT) His-TIFA, while none of these inhibitors affected the basal phosphorylation level of T9A. These results suggest the involvement of Ser/Thr kinases in the PI3K-AKT signaling pathway in T9 phosphorylation.
pT9 and FHA domain are important for TIFA self-association.
Because TIFA molecules can associate with each other to form homo-oligomers (25), we examined whether the phosphorylated T9 binding to TIFA-FHA is the basis of TIFA-TIFA self-association. Flag-tagged WT TIFA and Myc-tagged TIFA (WT or mutants) were overexpressed in HEK 293T cells. The Myc-tagged WT TIFA was detected in the anti-Flag pulldown (Fig. 2A, lane 1). Incubation of anti-Flag pulled-down samples with phosphatase reduced the interaction between Myc- and Flag-tagged WT TIFA (lane 2), while this reduction was rescued by the inclusion of EDTA to block the phosphatase activity (lane 3). In contrast, the T9A mutant was marginally detected under the same conditions, with or without phosphatase (lanes 4 to 6), supporting that T9 phosphorylation is critical for the TIFA self-association.
Fig 2.
pT9 and the FHA domain are required for TIFA self-association. (A) Flag-tagged WT and Myc-tagged WT and T9A TIFA were coexpressed in HEK 293T cells. Anti-Flag-coated Dynabeads was used in immunoprecipitation (IP). The coprecipitated proteins were detected by anti-Myc antibody in immunoblotting. The whole-cell extracts (WCE) used as IP inputs are shown in the right panel. PPtase or EDTA was included as indicated. (B) Sequence alignment of the conserved pT-contacting residues within the FHA domains of different FHA domain-containing proteins. Conserved residues among homologues are in light gray. The residues chosen for mutation in this work are in dark gray. RAD53*, RAD53 FHA1. (C) Effect of FHA domain mutations on the self-association of TIFA. The experiments were performed as described for panel A except that the coprecipitated TIFA and its mutants were detected by anti-Gal antibody. RKN, R51A K88A N89A mutant. (D) ITC analysis of the in vitro binding of WT TIFA and its R51A and R51A K88A (RK) mutants with the T9-containing peptide (1MTSFEDADTEETVT14) or pT9-containing peptide [1MTSFEDAD(pT)EETVT14]. Binding was observed only when the WT TIFA was incubated with the pT9-containing peptide.
We next tested whether FHA mutants impair the TIFA-TIFA association. On the basis of sequence alignment of different FHA domains shown in Fig. 2B, we replaced two of the highly conserved residues, Arg51 and Asn89, and a nonconserved but potentially important (due to its charge neighboring to the conserved Asn89) residue, Lys88, with Ala. As shown in Fig. 2C, Gal-tagged mutant TIFA could no longer be detected in the anti-Flag immunoprecipitates, indicating that each of the three residues is essential for the TIFA-TIFA interaction. A similar result was observed in previous study using a G50E S66A double mutant (25), although the role of pT9 was not then known.
By using isothermal titration calorimetry (ITC), we evaluated further the binding between the FHA domain and pT9 residue of TIFA. The expressed His-tagged WT TIFA and two FHA mutants were purified using a nickel column and then analyzed for association with a synthetic pT9-containing peptide spanning residues 1 to 14 of TIFA. As shown in Fig. 2D (panel II), ITC showed that the pT9-containing peptide binds to the unphosphorylated recombinant wild-type TIFA with a Kd of 50 ± 1.3 μM (stoichiometry, N = 1.04). In contrast, ITC could not detect binding for the corresponding unphosphorylated peptide with wild-type TIFA (panel I) or for the pT9-containing peptide with mutants R51A and R51A K88A (RK) (panels III to IV). These results reinforce the notion that the TIFA self-association is mediated through binding of pT9 to the FHA domain of TIFA.
The TIFA intrinsic dimer is the basic unit for self-association and oligomerization.
As a hallmark of the TRAF family, oligomer formation appears to be critical for TNF-α signaling (2, 12, 21). Since TIFA is able to promote oligomerization of TRAF6 (6), the self-associated TIFA may serve as building blocks for oligomerization. This in turn raises the question of whether the quaternary state of TIFA may exist in high-order protein architecture. The recombinant TIFA has been reported to exist in trimers (25), even though the vast majority of FHA domains are known to exist as monomers (18). Unexpectedly, the results from our experiments using fast protein liquid chromatography (FPLC) (Fig. 3A) or AUC (Fig. 3B) indicated that unphosphorylated WT TIFA existed as a 46-kDa intrinsic dimer which was stable between pH 7.5 and 8.5 in a range of protein concentrations (Fig. 3C). This finding suggests that the TIFA self-association observed in Fig. 2 represents oligomer formation (dimer associates with dimer) through FHA-pT9 interaction rather than dimer formation (monomer associates with monomer). To further investigate the dimerization mechanism of TIFA, we performed FPLC and AUC again to analyze three mutants, the T9A, R51A, and RK mutants. Since these mutants showed reduced self-association (Fig. 2A and C), they should not exist in stable dimers if the self-association reflects the formation of dimers. As the mutants still exist as dimers as shown in Fig. 3A and B, the results provide further support that TIFA exists as intrinsic dimers. In addition, the fact that the peptide-protein complex in Fig. 3D (panel III) remained as a dimer suggests that the dimeric interface does not involve the pT-FHA binding sites so that the dimer is not disrupted by the pT9-containing peptide. Finally, we found that WT TIFA did not dissociate into monomers under nonreduced conditions (Fig. 3E), excluding involvement of disulfide bonds in the intrinsic dimer.
Fig 3.
TIFA exists as an intrinsic dimer in solution. (A and B) Size exclusion chromatography (HiLoad 16/60 Superdex 75 pg column) (A) and AUC analysis (B) show that recombinant TIFA and mutants exist as dimers in solution. (C) Recombinant WT TIFA appeared as a dimer from low (0.5 mg/ml) to high (2 mg/ml) protein concentrations in size exclusion chromatography (Superdex 75 10/300 GL column). (D) AUC analysis of WT recombinant TIFA alone (I) or with the T9-containing peptide (II) or the pT9-containing peptide (III). The concentration of WT TIFA used was 10 mM, and the peptides were presented at a molar ratio of 1:10 (protein/peptide). (E) TIFA remained in the monomeric form on SDS-PAGE under a nonreduced condition, demonstrating that TIFA did not dimerize through disulfide bonds.
FHA-pT9 binding promotes TIFA oligomerization that colocalizes with TRAF6.
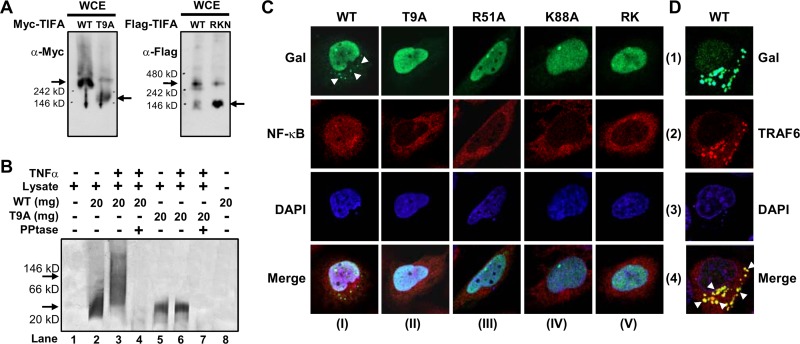
To provide further support that pT9-FHA binding does promote TIFA oligomerization, we evaluated the molecular weights of native TIFA proteins using nondenaturing gels. We examined overexpressed Myc-tagged or Flag-tagged TIFA and mutants from mammalian HEK 293T cells and found that native T9A and R51A K88A N89A (RKN) mutants of TIFA protein migrated faster than the WT protein during electrophoresis, suggesting that the mutations repressed the formation of oligomeric TIFA (Fig. 4A). We also performed an in vitro kinase assay and analyzed the native samples. As a result, the recombinant His-TIFA WT protein that had been phosphorylated by TNF-α-treated cell lysates upshifted to a higher molecular weight, whereas the PBS-treated control WT and both T9A samples remained at a lower molecular weight (Fig. 4B). Taken together, our results suggest that unphosphorylated TIFA forms intrinsic dimers in solution and that the binding between FHA and pT9 of TIFA in cells likely occurs through intermolecular interaction between dimers, presumably leading to oligomerization. This represents a new mechanism for the FHA domain function.
Fig 4.
TIFA pT9-FHA interaction is required for TIFA oligomerization. (A) The exogenously expressed Myc-TIFA, Flag-TIFA, and their T9A and RKN mutants were separated by native gel and detected by Western blotting. Arrows indicate the major bands. (B) Autoradiography of the native gel shows that the phosphorylated WT His-TIFA in TNF-α-treated cell lysate was upshifted to a higher molecular mass (indicated by arrows). No signal was detected when samples were treated with alkaline phosphatase. (C) U2OS cells ectopically expressing Gal-tagged TIFA mutants were fixed and incubated with anti-Gal and anti-NF-κB antibodies followed by FITC-conjugated anti-mouse IgG and rhodamine-conjugated anti-rabbit IgG. The nuclei were counterstained with DAPI. The fluorescence images were obtained with a Zeiss LSM 510 confocal microscope. Arrowheads indicate the aggregations/speckles of WT Gal-TIFA. (D) Colocalization of WT Gal-TIFA and Flag-tagged TRAF6 is indicated by arrowheads in the superimposed images. The experimental conditions were the same as for panel C except that Flag-TRAF6 was recognized by anti-TRAF6 antibody.
Next, we employed immunostaining to study the role of FHA-pT9 binding in TIFA oligomerization in cells. U2OS cells were transfected with Gal-tagged WT TIFA or its various mutants. As shown in the first panel of Fig. 4C (column I, row 1), a number of discrete punctate spots recognized by anti-Gal (indicated by arrowheads) were found in the perinuclear region of the cytoplasm of U2OS cells (representing possible protein aggregates or oligomers) transfected with WT Gal-TIFA. In parallel experiments in which cells were transfected with T9A or FHA domain mutants (columns II to V, row 1), no aggregates of Gal-TIFA were observed.
Moreover, since this newly found aggregation/oligomerization of exogenously expressed TIFA behaved similarly to what has been observed for members involved in the same inflammatory signaling pathway, including TRAF6, IRAK, P62, sequestosome, etc. (9, 22, 23, 26, 27, 30), we extrapolated that the speckles observed in cells exogenously expressing TIFA could be part of this huge signalosome protein complex. Indeed, the confocal section (Fig. 4D) shows that when WT and Flag-tagged TRAF6 were coexpressed, most of the punctuate spots of TIFA (row 1) colocalized with that of TRAF6 (row 2) in the cytoplasm of the cells (indicated by arrowheads) (row 4). These results suggest that the pT9-FHA binding is crucial for TIFA/TRAF6 oligomerization.
FHA-pT9 binding is required for TNF-α-mediated activation of NF-κB.
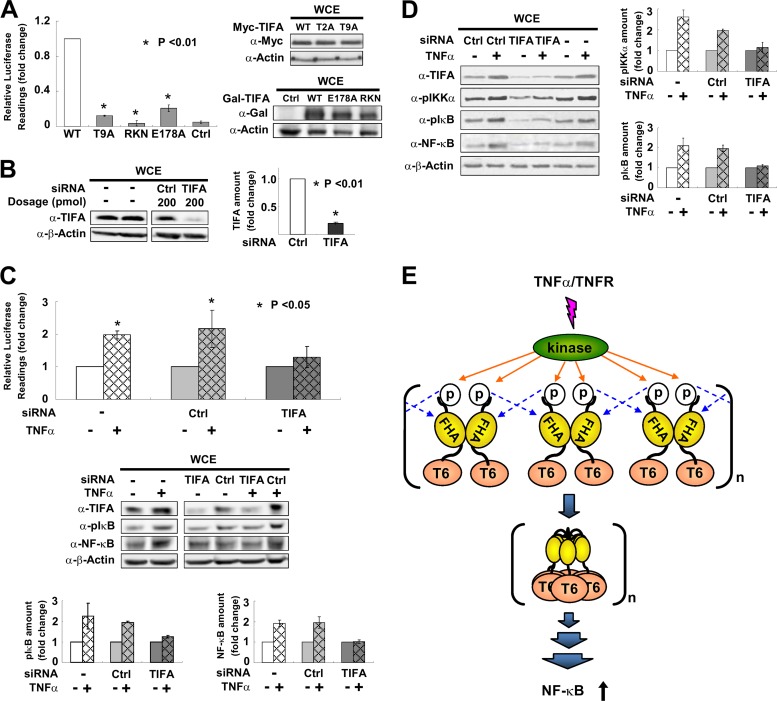
Previous reports showed that oligomeric forms of TIFA can activate IKK (6) and that interaction of TIFA with TRAF6 is indispensable for NF-κB activation (25). Given that TIFA-TIFA interaction occurs via pT9-FHA binding between TIFA dimers leading to oligomerization, we tested whether the pT9-FHA interaction is necessary for NF-κB activation. HEK 293T cells were transfected with a NF-κB luciferase reporter together with expressing plasmids encoding WT TIFA or its various mutants. As shown in Fig. 5A, the NF-κB-driven luciferase activity was much lower in cells cotransfected with the unphosphorylatable T9A or RKN FHA domain mutant than in those cotransfected with WT TIFA. In line with previous observations (25), the NF-κB activity mediated by E178A mutant was also drastically decreased, which supports that TRAF6 binds to TIFA via E178. We next monitored the subcellular localization of endogenous NF-κB affected by exogenously expressed TIFA. In agreement with the reporter assay, Gal-tagged wild-type TIFA in U2OS cells did promote the nuclear translocation of NF-κB (Fig. 4C, column I, row 2). Such a nuclear translocation of NF-κB was not observed when the T9A, R51A, K88A, or RK mutant was transfected (Fig. 4C, columns II to V, row 2).
Fig 5.
pT9-FHA binding is involved in TNF-α-mediated activation of NF-κB. (A) HEK 293T cells were transfected with an NF-κB luciferase reporter together with WT TIFA or its various mutants. NF-κB activation was evaluated by luciferase activity assays. The bar graph represents fold changes of normalized relative luminescence units (RLU). The results are mean ± SD from at least 3 independent experiments. Western blotting on the right shows the expression profiles of mutants used in the experiments. (B) HEK 293T cells were transfected with TIFA siRNA or scramble RNA (Ctrl). The efficiency of RNA interference was verified by Western blotting in at least 3 independent experiments and plotted in the bar graph shown on the right. (C) HEK 293T cells receiving TIFA siRNA or scramble RNA (Ctrl) were cotransfected with an NF-κB luciferase reporter and then stimulated with TNF-α as indicated. NF-κB activation was evaluated by luciferase activity assays. The values for the untreated groups were set as 1. The cell lysates were also analyzed by Western blotting. The bar graphs at the bottom represent the fold changes of phosphorylated IκB (pS32/pS36) and total NF-κB (p65) proteins after TNF-α treatment. (D) The experimental conditions for siRNA transfection were the same as those described for panel C except that the cell line used was THP-1 (a human acute monocytic leukemia cell line). The bar graphs represent the fold changes of phosphorylated IKKα (pT23) and phosphorylated IκB (pS32/pS36) after TNF-α stimulation. (E) Proposed model for TIFA oligomerization, which first takes place via intermolecular FHA-pT9 binding between TIFA dimers upon TNF-α stimulation and then leads to TRAF6 (labeled T6) oligomerization and subsequent NF-κB activation.
In a complementary experiment, small interfering RNA (siRNA) was used to silence the endogenous TIFA in HEK 293T cells, which was down to approximately 20% of the original level (Fig. 5B). Reporter assay and Western blot analysis were performed again to examine NF-κB activation while endogenous TIFA was silenced in HEK 293T cells. Compared with those in positive controls using nontransfected cells or cells transfected with scramble RNA, the TNF-α-induced activation of NF-κB (Fig. 5C, upper bar graph) and elevation level of phosphorylated IκB and total NF-κB proteins (Fig. 5C, lower bar graphs) were reduced to levels similar to those for the PBS control for the TIFA-silenced group. Similar attenuation of TNF-α-mediated signaling was also observed when endogenous TIFA was silenced in the human acute monocytic leukemia cell line, THP-1 (Fig. 5D). These findings together demonstrate that TIFA is necessary, and the FHA-pT9-induced TIFA oligomerization is important, for TNF-α-mediated NF-κB activation.
DISCUSSION
The principal finding of the current study is that TIFA oligomerizes through intermolecular FHA-pT9 binding between TIFA dimers upon TNF-α stimulation, which leads to TRAF6 oligomerization and subsequent NF-κB activation. Such a mechanism indicates that TIFA is an imperative molecule linking inflammatory cytokines, such as TNF-α, to the NF-κB-driven gene expression. Based on the deduced mechanism, a model is proposed for the function of TIFA, as shown in Fig. 5E. Upon TNF-α stimulation, an activated serine/threonine kinase involved in the PI3K-AKT pathway phosphorylates Thr9 of TIFA. This phosphorylation event then triggers the pT9-FHA interaction between different dimers of TIFA, leading to their oligomerization. Since TRAF6 constitutively binds to TIFA E178 located at the C terminus of TIFA, the oligomerization of TIFA then induces TRAF6 oligomerization, which in turn enhances the E3 ligase activity of the RING (“really interesting new gene”) domain of TRAF6 and activates the downstream signaling (6). Thus, this newly defined mechanism has important translational implications, particularly for inflammatory responses in mammalian cells.
Using MS, NanoPro immunoassay, and in vitro kinase assay, we confirmed the T9 phosphorylation and semiquantified its changes responding to TNF-α stimulation. These assays also showed that in addition to T9, there are other basal phosphorylations for both exogenous and endogenous TIFA (Fig. 1C and D, traces 1). However, it is not clear whether these basal phosphorylations are specific. The exact kinases responsible for the TNF-α-induced phosphorylation of T9 and the basal phosphorylation of TIFA remain to be established. Furthermore, because the pT9-FHA binding plays a pivotal role not only in mediating TIFA self-association/oligomerization but also in provoking TRAF6 oligomerization/NF-κB activation, antagonizing such a putative kinase may interfere with the TNF-α-related inflammation.
Based on recent reports, the mechanism of FHA domain function appears to be highly diversified (18). The “mechanism” here is defined as how, at the protein level, the FHA domain binds to its biological ligand and confers its biological function. To date, three mechanisms have been characterized: (i) a monomer of the FHA domain binds the pT residue(s) of a different monomer of its biological ligand intermolecularly (3), (ii) a monomer of the FHA domain binds with the pT residue of another monomer of the same FHA protein intermolecularly to enhance homodimerization (4, 15, 29), and (iii) intramolecular binding occurs between an FHA domain and a pT site within the same protein molecule (1). The model for the aggregation of TIFA therefore uncovers a new mechanism for FHA domain functions: the unphosphorylated FHA domain-containing protein exists as an intrinsic dimer that oligomerizes via the intermolecular FHA-pT bindings between dimers. Interestingly, the FHA domains of both human and mouse MDC1 (mediator of DNA damage checkpoint 1) proteins were recently shown to exist as intrinsic dimers in solution and in crystals (13, 17, 28).
Finally, the detailed atomic structure of full-length TIFA dimer complexed with a pT9-containing peptide, when available, will be very valuable for interpretation of molecular mechanisms of the signaling pathway. Most importantly, it will provide us chances to design regulatory compounds against NF-κB activation and properly manage inflammatory responses, autoimmune diseases, viral infection, and cancer development.
Overall, our work here, along with previous reports, has provided a mechanistic insight into TIFA oligomerization and the critical role of TIFA in NF-κB activation. The TIFA pT9-directed oligomerization of molecules involved in TNF-α-mediated inflammatory responses may have a broad impact on understanding of mammalian immunity. Our results have provided a basis for future studies into these significant issues.
ACKNOWLEDGMENTS
We thank Y.-M. Chou and P. K. Tai of Cold Spring Harbor Inc. (Taipei, Taiwan) and members of Cell Biosciences Inc. for their contributions to the NanoPro immunoassay, Y.-Y. Kao for preliminary ITC analysis, Y.-L. Huang for peptide synthesis, L.-P. Ting (National Yang-Ming University) for the pNF-κB-Luc plasmid, and E. S.-W. Chen, M.-I. Su, and R. H. Chen for useful discussions.
This work was supported by National Health Research Institute grant NHRI-EX100-10002NI and the Academia Sinica Investigator Award to M.-D.T.
C.-C.F.H., P.-Y.G.W., J.-H.W.,T.-Y.W.W., D.Q., and M.-D.T. designed the research; C.-C.F.H., J.-H.W., T.-Y.W.W., P.-H.H., Y.-H.C., S.-C.W., C.-C.H., and S.-T.C. performed the research; C.-C.F.H., P.-Y.G.W., J.-H.W., T.-Y.W.W., A.H.-J.W., J.Y.-J.S., and M.-D.T. interpreted the data; and C.-C.F.H., P.-Y.G.W., J.-H.W., J.Y.-J.S., and M.-D.T. wrote the paper.
We declare that we have no competing interests.
Footnotes
Published ahead of print 7 May 2012
REFERENCES
- 1. Barthe P, et al. 2009. Dynamic and structural characterization of a bacterial FHA protein reveals a new autoinhibition mechanism. Structure 17:568–578 [DOI] [PubMed] [Google Scholar]
- 2. Baud V, et al. 1999. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13:1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Byeon IJ, Li H, Song H, Gronenborn AM, Tsai MD. 2005. Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67. Nat. Struct. Mol. Biol. 12:987–993 [DOI] [PubMed] [Google Scholar]
- 4. Cai Z, Chehab NH, Pavletich NP. 2009. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol. Cell 35:818–829 [DOI] [PubMed] [Google Scholar]
- 5. Durocher D, Jackson SP. 2002. The FHA domain. FEBS Lett. 513:58–66 [DOI] [PubMed] [Google Scholar]
- 6. Ea CK, Sun L, Inoue J, Chen ZJ. 2004. TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6. Proc. Natl. Acad. Sci. U. S. A. 101:15318–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan AC, et al. 2009. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat. Med. 15:566–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fang CY, et al. 2010. Global analysis of modifications of the human BK virus structural proteins by LC-MS/MS. Virology 402:164–176 [DOI] [PubMed] [Google Scholar]
- 9. Force WR, et al. 2000. Discrete signaling regions in the lymphotoxin-beta receptor for tumor necrosis factor receptor-associated factor binding, subcellular localization, and activation of cell death and NF-kappaB pathways. J. Biol. Chem. 275:11121–11129 [DOI] [PubMed] [Google Scholar]
- 10. Hofmann K, Bucher P. 1995. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. Sci. 20:347–349 [DOI] [PubMed] [Google Scholar]
- 11. Inoue J, et al. 2005. Identification and characterization of Xenopus laevis homologs of mammalian TRAF6 and its binding protein TIFA. Gene 358:53–59 [DOI] [PubMed] [Google Scholar]
- 12. Ishida T, et al. 1996. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J. Biol. Chem. 271:28745–28748 [DOI] [PubMed] [Google Scholar]
- 13. Jungmichel S, et al. 2012. The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator. Nucleic Acids Res. 40:3913–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanamori M, Suzuki H, Saito R, Muramatsu M, Hayashizaki Y. 2002. T2BP, a novel TRAF2 binding protein, can activate NF-kappaB and AP-1 without TNF stimulation. Biochem. Biophys. Res. Commun. 290:1108–1113 [DOI] [PubMed] [Google Scholar]
- 15. Li J, et al. 2008. Chk2 oligomerization studied by phosphopeptide ligation: implications for regulation and phosphodependent interactions. J. Biol. Chem. 283:36019–36030 [DOI] [PubMed] [Google Scholar]
- 16. Liang X, Van Doren SR. 2008. Mechanistic insights into phosphoprotein-binding FHA domains. Acc. Chem. Res. 41:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J, et al. 2012. Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain. Nucleic Acids Res. 40:3898–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahajan A, et al. 2008. Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 1:re12 doi:10.1126/scisignal.151re12 [DOI] [PubMed] [Google Scholar]
- 19. O'Neill RA, et al. 2006. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc. Natl. Acad. Sci. U. S. A. 103:16153–16158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pennell S, et al. 2010. Structural and functional analysis of phosphothreonine-dependent FHA domain interactions. Structure 18:1587–1595 [DOI] [PubMed] [Google Scholar]
- 21. Pullen SS, et al. 1998. CD40-tumor necrosis factor receptor-associated factor (TRAF) interactions: regulation of CD40 signaling through multiple TRAF binding sites and TRAF hetero-oligomerization. Biochemistry 37:11836–11845 [DOI] [PubMed] [Google Scholar]
- 22. Sanz L, Diaz-Meco MT, Nakano H, Moscat J. 2000. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 19:1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seibenhener ML, et al. 2004. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 24:8055–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun Z, Hsiao J, Fay DS, Stern DF. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272–274 [DOI] [PubMed] [Google Scholar]
- 25. Takatsuna H, et al. 2003. Identification of TIFA as an adapter protein that links tumor necrosis factor receptor-associated factor 6 (TRAF6) to interleukin-1 (IL-1) receptor-associated kinase-1 (IRAK-1) in IL-1 receptor signaling. J. Biol. Chem. 278:12144–12150 [DOI] [PubMed] [Google Scholar]
- 26. Wang KZ, Galson DL, Auron PE. 2010. TRAF6 is autoinhibited by an intramolecular interaction which is counteracted by trans-ubiquitination. J. Cell Biochem. 110:763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang KZ, et al. 2006. TRAF6 activation of PI 3-kinase-dependent cytoskeletal changes is cooperative with Ras and is mediated by an interaction with cytoplasmic Src. J. Cell Sci. 119:1579–1591 [DOI] [PubMed] [Google Scholar]
- 28. Wu HH, Wu PY, Huang KF, Kao YY, Tsai MD. 2012. Structural delineation of MDC1-FHA domain binding with CHK2-pThr68. Biochemistry 51:575–577 [DOI] [PubMed] [Google Scholar]
- 29. Xu X, Tsvetkov LM, Stern DF. 2002. Chk2 activation and phosphorylation-dependent oligomerization. Mol. Cell. Biol. 22:4419–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zapata JM, et al. 2001. A diverse family of proteins containing tumor necrosis factor receptor-associated factor domains. J. Biol. Chem. 276:24242–24252 [DOI] [PubMed] [Google Scholar]