Abstract
Disabled-1 (Dab1) plays a key role in reelin-mediated neuronal migration during brain development. Tyrosine phosphorylation of Dab1 at two YQXI and two YXVP motifs recruits multiple SH2 domains, resulting in activation of a wide range of signaling cascades. However, the molecular mechanisms underlying the coordinated regulation of Dab1 downstream effectors remain poorly understood. Here, we show that alternative splicing results in inclusion of different combinations of YQXI and YXVP motifs in Dab1 isoforms during development. Dab1 variants with partial or complete loss of YQXI motifs are preferentially expressed at early developmental stages, whereas the commonly studied Dab1 is predominantly expressed at late developmental stages. Expression of Dab1 variants in 293T and Neuro2a cells reveals reduced levels or absence of tyrosine phosphorylation in variants that have lost one or both YQXI motifs. We further demonstrate that Dab1 variants differ in their abilities to activate Src and recruit distinct SH2 domains involved in specific downstream signaling pathways. We propose that coordinated expression of specific Dab1 isoforms in different populations of cells in the developing brain contributes to precise neuronal migration by modulating the activity of subsets of Dab1 downstream effectors.
INTRODUCTION
Brain development requires coordinated migration of neurons that results in the formation of distinct layers in laminated brain structures. The reelin–disabled-1 (Dab1) signaling pathway plays a key role in regulating neuronal cell migration (19, 38). Binding of reelin to its receptors, apolipoprotein E receptor 2 (ApoER2) and very-low-density lipoprotein receptors (VLDLR), activates Src family kinases (SFK) and induces tyrosine phosphorylation of the intracellular protein Dab1 (2, 6, 21, 45). Tyrosine-phosphorylated Dab1 transmits the signal to downstream molecular targets, which regulate neuronal migration. Inactivation of Dab1 in mice (scrambler, yotari, and Dab1−/− mutants) results in extensive migration defects with layer inversion in the cerebral cortex and lamination disruption in the cerebellum and hippocampus (11, 22, 35, 40, 45). These phenotypes are indistinguishable from those observed in mice deficient for reelin (reeler) and reelin receptors (Vldlr−/−; Apoer2−/− mice) (11, 22, 35, 40, 45), highlighting the pivotal importance of Dab1 in reelin signaling.
Dab1 contains an N-terminal protein interaction/phosphotyrosine binding (PI/PTB) domain that associates with the NPXY motifs in ApoER2 and VLDLR (23, 44). The PI/PTB domain is followed by a tyrosine-rich region, which contains five conserved tyrosine residues (Y185, Y198, Y200, Y220, and Y232). These residues correspond to two consensus SFK recognition sites (YQXI, at Y185 and Y198) and two consensus Abl family kinase recognition sites (YXVP, at Y220 and Y232) (42). Tyrosine phosphorylation of all four sites is required for maximal Dab1 activity (14, 27). Mice expressing Dab1 with nonphosphorylatable phenylalanine substitutions at these five tyrosine residues exhibit a phenotype similar to that of reelin-deficient (reeler) and Dab1−/− mice, suggesting that tyrosine phosphorylation is essential for reelin signaling (22).
Tyrosine-phosphorylated Dab1 transmits the reelin signal by recruiting different Src homology 2 (SH2) domain-containing proteins, including Src, the p85 regulatory subunit of phosphatidylinositide-3-kinase (PI3K), cellular adaptors CrkL, Crk, Nckβ, and Socs (suppressor of cytokine signaling) (1, 7, 13–14, 37). Recruitment of the PI3K p85 subunit to Dab1 activates the PI3K-Akt pathway (7), while association of CrkL and Crk with Dab1 links reelin signaling to the C3G-Rap1 pathway (1). Downstream signaling of Dab1-Nckβ interaction is likely involved in actin remodeling (37). Interestingly, Dab1-Socs association terminates reelin signaling by degrading phosphorylated Dab1 through the ubiquitin proteasome system (13). Therefore, Dab1-SH2 domain interactions maintain the balance between activation and termination of downstream signaling cascades. Different tyrosine phosphorylation motifs in Dab1 appear to recruit specific SH2 domains: the Dab1 YXVP motifs (Y220 and Y232) are required for the recruitment of SH2 domains from Crk, CrkL, and Nckβ adaptor proteins (1, 37), whereas Dab1 YQXI motifs (Y185 and Y198) are necessary for recruitment of Src, PI3K, and Socs SH2 domains (13, 14). How these motifs coordinate complex downstream events to regulate neuronal migration is not well understood.
It is noteworthy that nucleotide sequences encoding the four consensus Dab1 tyrosine phosphorylation motifs are located within four adjacent exons (exons 6 to 9). When alternative splicing occurs at these exons, it generates isoforms with modified tyrosine phosphorylation sites. For example, alternative splicing of exons 7 and 8, which removes the Y198Q and Y220Q residues and converts the Y185QXI motif into a Y185QVP motif, has been documented in chick retina, human retina and pig liver (28–29, 31). Skipping of exons 8 and 9, which removes both Y220QVP and Y232QVP, has been observed in zebrafish brain (10). In mouse, a Dab1 isoform called Dab1555* or Dab1.7bc is specifically expressed in nonneuronal cells (3, 46). Based on sequences from GenBank (accession numbers NC_000070 and NM_177259) and ENSEMBL (ENSMUSG00000028519), this Dab1 isoform includes duplicated exons 9b and 9c, which were previously referred to as 7b and 7c (46). To date, Dab1 variants excluding tyrosine-encoding exons have not been reported in mouse.
In this study, we have identified 11 alternatively spliced Dab1 variants in the developing mouse brain that involve two tyrosine-encoding exons (7 and 8) and exons 9b/9c. Importantly, exclusion of exons 7 and/or 8 reconstitutes tyrosine phosphorylation motifs, generating isoforms with different combinations of consensus SFK and Abl kinase recognition sites. We demonstrate that differentially phosphorylated Dab1 isoforms recruit distinct SH2 domains involved in different downstream pathways. Combined with the specific distribution of exons 7/8 versus exons 9b/9c-containing transcripts in the developing brain, our study supports a critical role for alternative splicing in regulating Dab1-mediated downstream signaling.
MATERIALS AND METHODS
Antibodies and plasmids.
Anti-Dab1-exon 9b (Dab1-ex9b) and anti-Dab1-exon 8 (Dab1-ex8) antisera were generated by immunizing rabbits with keyhole limpet hemocyanin (KLH)-conjugated peptides PLEDFDSRFAAATP and GHEPIRDPETEENI, respectively. The antisera were affinity purified using the immunizing peptides (Genscript). Rabbit anti-Dab1 antibody (Rockland), raised against the C terminus (amino acids [aa] 440 to 555) of mouse Dab1, has been described previously (17, 27). Rabbit anti-Dab1 B3 antibody was provided by Jonathan Cooper (Fred Hutchison Cancer Centre). Mouse anti-Src antibody was provided by Joan Brugge (Harvard Medical School). The other antibodies used were mouse antiactin (Sigma), mouse anti-phosphotyrosine 4G10 (anti-pY 4G10) (Millipore), mouse anti-Abl (Santa Cruz), mouse anti-TUJ1 (Covance), mouse anti-CrkL antibody (Millipore), and phospho-Src (pTyr418) (Affinity Bioreagents). The pCI-c-Src plasmid was a gift from Don Fujita (University of Calgary, Canada).
Expression and purification of His-tagged SH2 domains.
pET28 SacB alkaline phosphatase (AP) expression constructs containing the SH2 domains of human Src, the p85 subunit of PI3K (N- or C-terminal SH2 domain), Nckβ, Crk, CrkL, and Socs2 (9) were transformed into BL21 Escherichia coli cells. Expression of His-tagged SH2 domains was induced in the presence of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C for 4 h, except for Socs2 and CrkL SH2s, which were induced at 18°C overnight. His-tagged SH2s were purified using Ni-nitrilotriacetic acid (Ni-NTA) spin columns (Qiagen) according to the manufacturer's protocol.
Mice.
The strains of mice used in this study were FVB and C57BL/6. The Reln allele was Relnrl/J from Jackson Laboratory. The reeler and Dab1−/− mice were maintained and genotyped by PCR as previously described (11, 20). The morning when the vaginal plug was found is referred to as day 0.5 of gestation. Mouse care, husbandry, and handling were performed in compliance with federal and institutional regulations and policies.
Reverse transcription-PCR (RT-PCR) analysis and cloning of mouse Dab1 cDNAs.
Total RNA was extracted from FVB and C57BL/6 mouse brain and retinal tissues at different developmental stages using an UltraClean tissue RNA isolation kit (MO BIO Laboratories) according to the manufacturer's instructions. Five micrograms of RNA was reverse transcribed using oligo(dT) primer and Superscript II reverse transcriptase (Invitrogen). For analysis of alternatively spliced Dab1 isoforms, cDNAs were PCR amplified using the following primer sets: (i) primers flanking exons 7 and 8 of Dab1 (P1/P2), (ii) primers flanking exon 7 (P1/P15), (iii) primers flanking exons 9b and 9c (P3/P4), and (iv) primers flanking exons 6 to 10 (P5/P4). PCR products were electrophoresed in 8% polyacrylamide gels. For sequencing, DNA fragments were cloned into the pCRII-TOPO vector (Invitrogen) and sequenced using T7 and SP6 primers.
The entire coding region of Dab1 cDNA was generated by PCR amplification of embryonic day 16.5 (E16.5) and postnatal day 1 (P1) retina cDNAs using primers P7/P8 flanking the entire Dab1 open reading frame (ORF). Amplified DNA fragments were cloned into pCRII-TOPO for sequence analysis. Inserts of interest were then transferred into the pCIG2 mammalian expression vector (gift from Franck Polleux, University of North Carolina) at the EcoRI site or into pEGFP-c1 at the Bgl11/EcoRI site, followed by sequence verification. All the primer sequences are shown in Table 1.
Table 1.
PCR primers
| Primer | Sequencea |
|---|---|
| Dab1-P1 | CGGCTGAACCTGTTATCCTG |
| Dab1-P2 | CTTCCTTCTTTTGGCTGGTG |
| Dab1-P3 | TGTGCCAAAAAGTCAACCTG |
| Dab1-P4 | CAGCAGTGCCGAAAGACATA |
| Dab1-P5 | GCGAAGGACATCACAGATCA |
| Dab1-P7 | AGCTGGATCCACCATGTCAACTGAGACAGAAC |
| Dab1-P8 | AGCTCTCGAGCTAGCTACCGTCTTGTGGAC |
| Dab1-P9 | AGCTGGATCCTTCATGCCCACACAAACTGT |
| Dab1-P10 | AGCTCTCGAGATGGATCACTGGTGGAGGAG |
| Dab1-P11 | AGCTGGATCCGTCAACCTAATAGCCAGCCG |
| Dab1-P12 | AGCTCTCGAGCACAGCACTCACCTTGGTTG |
| Dab1-P13 | AGCTGGATCCCCAGACCATTTTGGAAGAGG |
| Dab1-P14 | AGCTCTCGAGTGGTGGGAACCTGGTAAATG |
| Dab1-P15 | CTCATGTCCAGCCTCAAACA |
Underlined sequences indicate restriction enzyme sites.
Cell culture and DNA transfections.
P19 mouse embryonic carcinoma cells (105 cells/ml) were cultured in petri dishes in Dulbecco's modified Eagle's medium (DMEM) supplemented with 7.5% bovine serum and 2.5% fetal calf serum. Neural differentiation of P19 cells was carried out by treating aggregated cells with all-trans-retinoic acid (RA) (Sigma R2625) as previously described (25). Cells were treated with 0.5 μM RA on day 0 and day 2. At day 4, cell aggregates were harvested, trypsinized, and then triturated into single-cell suspensions using a Pasteur pipette. Dissociated cells were replated on tissue culture plates or coverslips coated with 100 μg/ml poly-d-lysine and cultured in DMEM supplemented with 10% fetal calf serum. Two days after replating (day 6), 5 μg/ml ara-C (Sigma C1768) was added to kill the proliferating cells. Two days later (day 8), cells were fixed in 4% paraformaldehyde for immunofluorescence analysis. Cells were harvested at different stages (as indicated) and used for either RNA isolation using the TRIzol reagent (Invitrogen) or protein extraction in radioimmunoprecipitation (RIPA) buffer.
HEK293T cells were cultured in DMEM supplemented with 10% fetal calf serum and antibiotics. Transfections were carried out using Extreme Gene 9 transfection reagent (Roche Diagnostics). Cells in six-well plates were transfected with 0.15 μg of Dab1 DNA, with or without 0.05 μg of Src DNA.
Neuro2a cells were cultured in DMEM supplemented with 10% fetal calf serum and antibiotics. Cells cultured on coverslips were transfected with polyethylenimine (PEI) (5 μl of reagent to 1 μg of DNA per ml) (Polysciences Inc.). Neuro2a cells cultured on plates were transfected by calcium phosphate-DNA precipitation.
Western blot and far-Western blot analysis.
For Western blotting, mouse brain tissue from different developmental stages was lysed in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM Na3VO4, tyrosine phosphatase cocktail [Sigma], and 1× Complete protease inhibitor cocktail [Roche]). For transfected 293T cells, cells were lysed in RIPA buffer at 24 to 48 h posttransfection. Proteins were resolved by 8% SDS-PAGE and transferred to nitrocellulose or polyvinylidene difluoride (PVDF) membranes, followed by immunodetection using anti-Dab1 (1:5,000; Rockland), anti-pY 4G10 (1:5,000; Millipore), anti-Dab1-exon 9b (1:8,000), or anti-Dab1-exon 8 (1:500) antibody.
Far-Western blot analysis was carried out as previously described with minor modifications (12, 32, 36). 293T cells were lysed in KLB buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 10 mM NaF, 1 mM PMSF, 1 mM Na3VO4, tyrosine phosphatase cocktail [Sigma], and 1× Complete protease inhibitor cocktail [Roche]) 24 to 48 h posttransfection. The concentration of each SH2 domain to be used for far-Western blotting was determined by examining the binding of SH2 domains to phosphatase- and pervanadate-treated lysates, which served as negative and positive controls, respectively. Ten micrograms of cell lysates was electrophoresed in an 8% SDS-polyacrylamide gel and transferred to PVDF membranes in CAPS [3-(cyclohexylamino)-1 propane sulfonic] transfer buffer (10 mM CAPS, pH 11.0). Membranes were blocked in 10% nonfat milk in Tris-buffered saline (TBS) containing 1 mM Na3VO4 at room temperature (RT) for 1 h and incubated with 30 to 100 nM purified His-tagged SH2 domains (diluted in 10% milk) at 4°C overnight. The membranes were then washed in TBS–0.1% Tween 20 (TBS-T) five times for 10 min, and incubated with 1:3,000 to 1:4,000 mouse antipolyhistidine antibody (diluted in 5% nonfat milk in TBS) (H1029; Sigma). After five washes in TBS-T, membranes were incubated in horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody, and the bound SH2 domains were detected by chemiluminescence using Immobilon reagent (Millipore). The membranes were stripped in acidic buffer (100 mM glycine, pH 2.0) at room temperature for 30 min, followed by blocking and reprobing with Dab1 antibody.
Immunofluorescence analysis.
RA-treated P19 cells (at day 8) and Neuro2a cells were fixed in 4% phosphate-buffered saline (PBS)-buffered paraformaldehyde for 10 min and permeabilized in PBS–0.1% Triton X-100 (PBS-T) for 5 min at room temperature. Cells were blocked in 10% donkey serum containing 1% bovine serum albumin (BSA) for 1 h and incubated in primary antibody (1:2,000 mouse anti-β III tubulin [TUJ1; Covance]) or rabbit anti-pSrc (1:500) at 4°C overnight, followed by incubation with Alexa Fluor 488- or Alexa Fluor 555-conjugated secondary antibody for 2 h at room temperature. Cells were mounted using Prolong Gold mounting medium (Invitrogen) containing 4′,6′-diamidino-2-phenylindole (DAPI) to stain the nuclei.
In situ hybridization.
Dab1 riboprobes were generated for detection of all Dab1 isoforms (514 bp, prepared with P9/P10 primers), exons 9b/9c (118 bp, prepared with P11/P12 primers), and exons 7/8 (119 bp, prepared with P13/P14 primers). Primer sequences are shown in Table 1. PCR was carried out using pCIG2-Dab1 as a template, and PCR fragments were cloned into pGEM-T Easy and transferred to pBluescript. pBluescript constructs were linearized with either BamHI or XhoI.
Digoxigenin (DIG)-labeled antisense riboprobes were generated by in vitro transcription of linearized pBluescript plasmids using T7 RNA polymerase (Roche Diagnostics) according to the manufacturer's protocol. E11.5 mouse brain tissues were fixed in 4% PBS-buffered paraformaldehyde, cryoprotected in sucrose, and embedded in OCT compound (Tissue-Tek). E14.5 mouse brain tissues were fixed in formalin and embedded in paraffin. Consecutive sections (6 to 8 μm) were prehybridized at 55°C in 40% formamide, 10% dextran sulfate, 1× Denhardt's solution, 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10 mM dithiothreitol (DTT), 1 mg/ml Saccharomyces cerevisiae tRNA, and 1 mg/ml heat-denatured herring testis sperm DNA. Tissue sections were hybridized to heat-denatured riboprobes overnight at 55°C, followed by washing and incubation with alkaline phosphatase (AP)-conjugated anti-DIG antibody. The signal was detected with 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NBT) after polyvinyl alcohol enhancement. Micrographs from similar regions of consecutive sections were captured with a Zeiss Axioskop 2 Plus microscope using a 10× lens.
Immunohistochemical analysis of mouse brain tissues.
Cryostat sections (E11.5) were used for immunohistochemical analysis. For antigen retrieval, tissue sections were microwaved in a pressure cooker in 10 mM citrate (pH 6.0) for 10 min. Consecutive sections were stained with rabbit anti-Dab1 C-terminal antibody (1:500) or Dab1-B3 antibody (1:500). The signal was detected using a DakoCytomation EnVision+ secondary system. Tissues were counterstained with hematoxylin for nuclear staining. Micrographs from similar regions of consecutive sections were captured with a Zeiss Axioskop 2 Plus microscope using a 10× lens.
RESULTS
Alternative splicing generates multiple Dab1 isoforms.
We have previously identified two alternatively spliced Dab1 isoforms in the chick retina: Dab1-E, expressed in proliferating cells at early developmental stages, and Dab1-L, found at later developmental stages in differentiated neurons. Dab1-L, which represents the canonical form of Dab1, includes exons 7 and 8 but excludes exon 9b, whereas Dab1-E excludes exons 7 and 8 but includes exon 9b (17, 29). To examine whether similar Dab1 isoforms are present in mouse, we carried out RT-PCR analysis of RNA prepared from E16.5 and P1 mouse retina using primers flanking exons 6 to 10 (primers P5/P4), as well as primers flanking the entire ORF of Dab1 (primers P7/P8). The PCR fragments were cloned into pCRII-TOPO vector and sequenced. Of the 35 sequenced cDNAs (15 from E16.5 and 20 from P1 retina), 13 encoded the commonly studied Dab1 (referred to as Dab1 or variant 1), whereas six clones encoded an isoform containing exons 9b and 9c (referred to as variant 5) (46). The remaining clones corresponded to previously unidentified alternatively spliced Dab1 transcripts (Table 2).
Table 2.
Alternatively spliced murine Dab1 isoforms
| Dab1 isoform [alias(es)] | Alternative splicing event(s) | Predicted difference(s) in protein sequence relative to Dab1 | No. of clones |
|---|---|---|---|
| Variant 1 (Dab1, Dab1-L, Dab1555) | 13 | ||
| Variant 2 | Exclusion of exon 7 | Deletion of 13 aa containing Y198 | 1 |
| Variant 3 | Exclusion of exon 8 | Deletion of 22 aa containing Y200 and Y220 | 3 |
| Variant 4 | Exclusion of exons 7 and 8 | Deletion of 35 aa containingY198, Y200, and Y220 | 3 |
| Variant 5 (Dab1555*, Dab1.7bc) | Inclusion of exons 9b and 9c | Insertion of 33 aa | 6 |
| Variant 6 (Dab1-E) | Exclusion of exons 7 and 8, inclusion of exons 9b and 9c | Deletion of 35 aa containingY198, Y200, and Y220; insertion of 33 aa | 2 |
| Variant 7 | Exclusion of exon 8, inclusion of exons 9b and 9c | Deletion of 22 aa containing Y200 and Y220; insertion of 33 aa | 1 |
| Variant 8 | Alternative 5′ splice site of intron 9 | Deletion of 2 aa (valine and serine) | 3 |
| Variant 9 | Alternative 5′ splice site of intron 9, exclusion of exons 7 and 8 | Deletion of 2 aa (valine and serine); deletion of 35 aa containingY198, Y200, and Y220 | 1 |
| Variant 10 | Alternative 5′ splice site of intron 9, inclusion of exons 9b and 9c | Deletion of 2 aa (valine and serine); insertion of 33 aa | 1 |
| Variant 11 | Alternative 5′ splice site of intron 9, exclusion of exons 7 and 8, inclusion of exons 9b and 9c | Deletion of 2 aa (valine and serine); deletion of 35 aa containing Y198, Y200, and Y220; insertion of 33 aa | 1 |
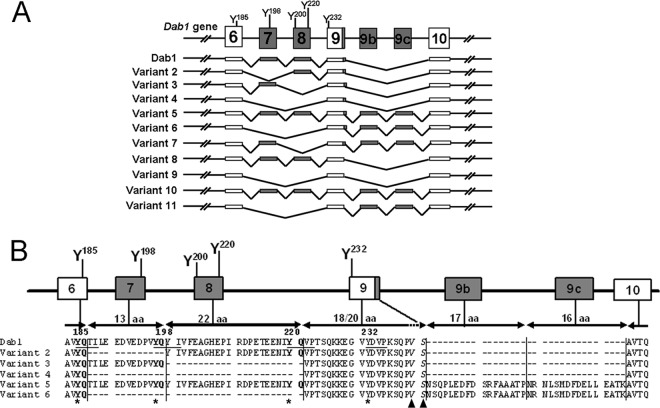
The Dab1 alternatively spliced transcripts identified in this study are generated by a single splicing event such as alternative exclusion of exon 7 or 8, alternative inclusion of exons 9b and 9c, alternative 5′ splice site usage in intron 9, or different combinations of these splicing events (Fig. 1A). Of the 16 possible combinations of these events, we identified 11 alternatively spliced Dab1 transcripts in mouse retina (schematically shown in Fig. 1A). Among these isoforms, variant 6, which excludes exons 7 and 8 but includes exons 9b and 9c, has the same genomic architecture as Dab1-E. The mRNA sequences and predicted protein sequences of six representative isoforms (variants 1 to 6) are aligned and shown in Fig. 1B and 2. Detailed information regarding the excluded and included exons in each RNA variant relative to Dab1 is given in Table 2.
Fig 1.
Identification of multiple alternatively spliced Dab1 isoforms. (A) Schematic of Dab1 gene structure and alternatively spliced variants. Constitutive exons are shown in white, whereas alternative exons are indicated in gray. Tyrosine residues encoded by exons 6 to 9 are shown. Exons and introns are not drawn to scale. (B) Alignment of predicted protein sequences of representative Dab1 isoforms. The exon-intron structure (exons 6 to 10) of the mouse Dab1 gene is depicted. Amino acid sequences encoded by these exons are indicated. Note that the last two residues encoded by exons 6 to 8 are the same (Y185Q, Y198Q, and Y220Q, in bold). The four consensus tyrosine phosphorylation sites (Y185QTI, Y198QYI, Y220QVP, and Y232DVP) are underlined in Dab1, and tyrosines are indicated with asterisks. The two amino acids affected by the alternative 5′ splice site in intron 9 are italicized and indicated with triangles. Amino acid numbering is based on the Dab1 protein sequence (accession number NP_796233).
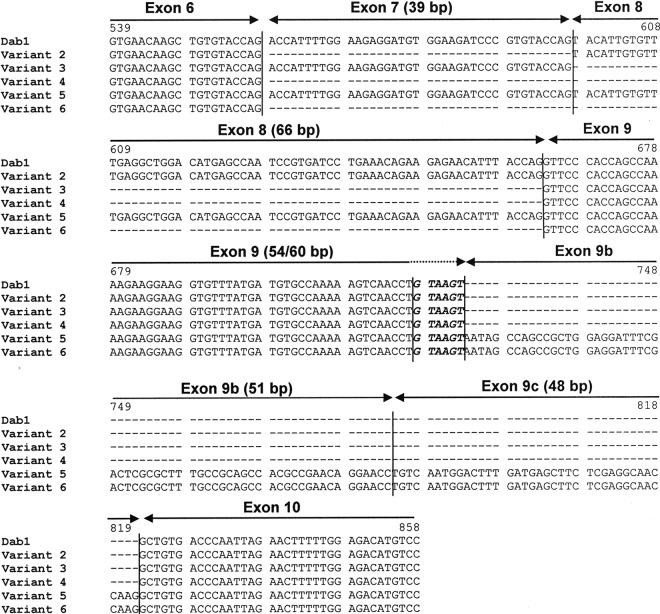
Fig 2.
mRNA sequence alignment of representative Dab1 variants. Exons 6 to 10 are indicated by arrows. The italicized and bold nucleotides at the 3′ end of exon 9 represent the 5′ alternative splice site. Nucleotide numbering is based on Dab1 mRNA (accession number NC_000070.5) and starts from the ATG codon.
Alternative splicing not only removes but also rearranges the tyrosine phosphorylation motifs in Dab1 variants.
None of the alternative splicing events identified here result in frameshift changes. For example, exclusion of exons 7 and/or 8 leads to in-frame deletion of residues in the tyrosine-enriched region, whereas the alternative 5′ splice site selection of intron 9 results in microdeletion of valine 240 (V240) and serine 241 (S241) (Fig. 1B). Inclusion of exons 9b and 9c results in an in-frame insertion of a 33-aa fragment immediately after V240 S241. Therefore, all the Dab1 isoforms retain an intact PI/PTB domain and C terminus but show variations in the central (tyrosine-enriched or adjacent) region.
The four Dab1 consensus tyrosine phosphorylation sites are encoded by four adjacent exons (exons 6, 7, 8, and 9). Of note, the last two codons (TACCAG), encoding tyrosine (Y) and glutamine (Q), are identical in exons 6 to 8 (Fig. 2). In Dab1, the encoded amino acids correspond to Y185Q (exon 6), Y198Q (exon 7), and Y220Q (exon 8) (Fig. 1B). As the second codons in exons 7 and 8 both encode isoleucine (I), Y185Q and Y198Q form two YQXI motifs (Fig. 1B and 3). These tyrosine phosphorylation motifs are preferentially recognized by SFK SH2 domains (42). In contrast, the second codon in exon 9 encodes a proline (P). Thus, Y220Q corresponds to a YXVP motif, an optimal binding site for Crk, Nckβ, and Abl SH2 domains (42).
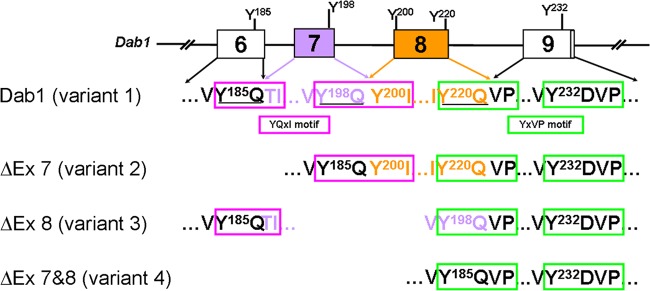
Fig 3.
Reconstitution of consensus tyrosine phosphorylation sites in alternatively spliced Dab1 isoforms. The exon-intron structure (exons 6 to 9) of the mouse Dab1 gene is illustrated, with constitutive exons shown in white and alternative exons in purple or gold. Amino acid sequences constituting the four consensus tyrosine phosphorylation sites are shown in colors corresponding to the coding exons. Dab1 contains two YQXI motifs (Y185QTI and Y198QYI, boxed in magenta) and two YXVP motifs (Y220QVP and Y232DVP, boxed in green). Note that nucleotides encoding amino acids comprising Y185QXI, Y198QXI, and Y220QVP) are located at the 5′ or 3′ end of neighboring exons. Exclusion of exon 7 (ΔEx 7) removes Y198Q residues but connects Y185Q to Y199I200, thus reconstituting a Y185QYI motif identical to the original Y198QYI. Exclusion of exon 8 (ΔEx 8) removes Y220Q but rejoins Y198Q with V222P223, thus converting Y198QXI into a Y198QVP site that mimics the original Y220QVP motif. Exclusion of exons 7 and 8 removes both Y198Q and Y220Q and rejoins Y185Q to V222P223, resulting in loss of both YQXI motifs but retention of two YXVP motifs.
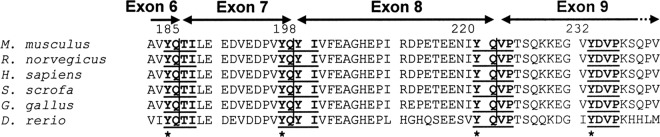
Skipping of exon 7 and/or exon 8 in Dab1 variants not only removes the coding tyrosine residues but also changes the contextual residues within the motif. For example, exclusion of exon 7 removes Y198Q in Dab1 variant 2; however, rejoining exons 6 and 8 in this variant links Y185Q to Y200I201 (Fig. 3), thereby reconstituting a Y185QYI motif identical to the original Y198QYI motif. In contrast, skipping of exon 8 in variant 3 does not affect the Y198Q199 residues but removes the Y220Q221 residues, resulting in conversion of the original Y198QYI motif to a Y198QVP motif (Y198Q199 to V222P223) (Fig. 3). Likewise, when both exons 7 and 8 are excluded (variants 4, 6, 9, and 10), the Y185 Q186 residues are directly connected to the V222P223 residues, thereby producing an isoform that retains two YXVP motifs with no YQXI motifs. As the organization of Dab1 tyrosine coding exons is highly conserved in vertebrates (Fig. 4), similar alternative splicing-induced rearrangements of Dab1 tyrosine sites may be widespread throughout vertebrate species.
Fig 4.
Alignment of Dab1 amino acid sequences. Dab1 sequences from the following species were aligned: mouse (Mus musculus; accession number NP_796233.2), rat (Rattus norvegicus; NP_705885.1), human (Homo sapiens; NP_066566.3), pig (Sus scrofa; NP_001090911.1), chicken (Gallus gallus; NP_989569.1), and zebrafish (Danio rerio; NP_001128514.1) (genus names are abbreviated on the figure). Alignment was performed with CLUSTALW (http://www.genome.jp/tools/clustalw/). The four consensus tyrosine phosphorylation motifs in Dab1 are in bold and underlined. Asterisks indicate the critical tyrosine residues in each motif.
Partial or complete loss of YQXI motifs in Dab1 isoforms results in reduced levels or absence of Dab1 tyrosine phosphorylation and Src activation.
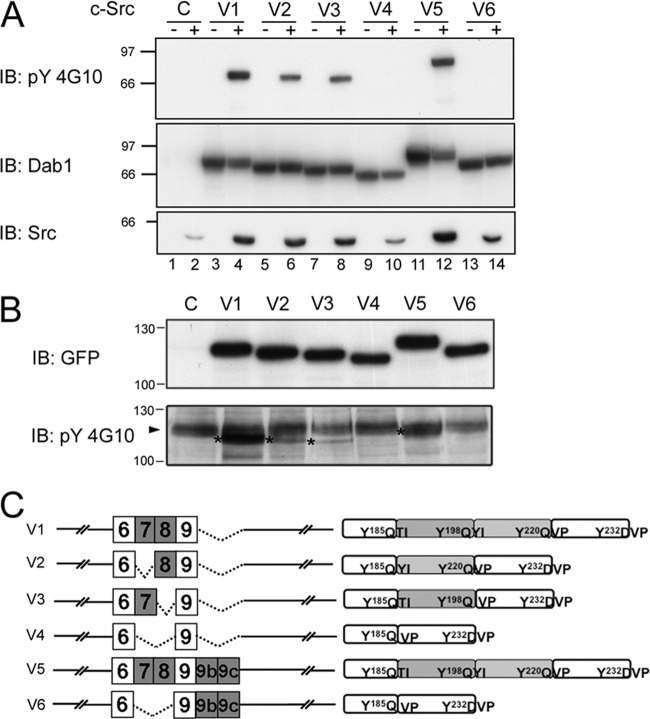
As alternative splicing affects Dab1 consensus SFK tyrosine phosphorylation sites, we reasoned that these isoforms might be differentially phosphorylated at tyrosines. To examine the phosphorylation state of tyrosine residues in mouse Dab1 isoforms, we transfected expression constructs encoding different Dab1 isoforms (variants 1 to 6) into 293T cells along with wild-type c-Src. In the absence of exogenous Src, tyrosine phosphorylation of all the Dab1 variants was undetectable (Fig. 5A). Cotransfection with c-Src robustly increased tyrosine phosphorylation of Dab1 variant 1 and variant 5 (Fig. 5A, lanes 4 and 12), whereas variants 2 and 3 were tyrosine phosphorylated to a lesser extent. In contrast, neither variant 4 nor variant 6 was phosphorylated in the presence of c-Src. These results are in agreement with our previous observations that Dab1-E, the chicken counterpart of variant 6 which lacks exons 7 and 8, is not tyrosine phosphorylated (16, 17).
Fig 5.
Differential tyrosine phosphorylation of Dab1 isoforms. (A) 293T cells were cotransfected with Dab1 variants and c-Src expression constructs. Cell lysates were analyzed by Western blotting with anti-phosphotyrosine 4G10 (top panel), anti-Dab1 (middle panel), and anti-Src antibodies. (B) Neuro2a cells were transfected with GFP-Dab1 variants 1 to 6. Cell lysates from control and transfected cells were electrophoresed in a 10% SDS-polyacrylamide gel, and proteins were transferred to a PVDF membrane. The membrane was immunoblotted with anti-phosphotyrosine 4G10 and anti-GFP antibodies. Asterisks mark the positions of the GFP-Dab1 variants. The arrowhead points to an unidentified protein recognized by the 4G10 antibody. (C) The Dab1 variants transfected into 293T and Neuro2a cells are indicated. Data shown are representative of two independent experiments. IB, immunoblotting.
To further verify the phosphorylation state of Dab1 variants, particularly the novel variants 2 and 3 that have lost one YQXI motif, in a neural context, we introduced green fluorescent protein (GFP)-tagged Dab1 variants into Neuro2a cells, a mouse neural crest-derived cell line that has been extensively used to study neuronal cell differentiation, particularly as related to signaling pathways and axonal growth (43). Neuro2a cells express the key players in Dab1 downstream signaling (Crk, CrkL, Src, Socs2, p85 subunit of PI3K, and Nckβ) (data not shown). Western blot analysis of lysates from transfected cells revealed elevated expression of all GFP-tagged variants in Neuro2a cells (Fig. 5B). Tyrosine phosphorylation of variants 2 and 3 was significantly reduced compared to that of variant 1 (Fig. 5B). Together, our data show that Dab1 isoforms are differentially phosphorylated and that both exons 7 and 8 are required for maximal Dab1 tyrosine phosphorylation.
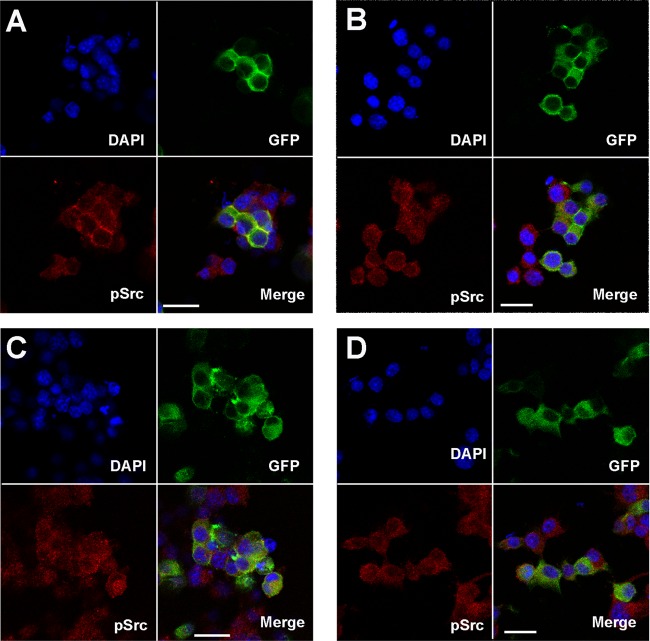
Src, a key player in Dab1 signaling, has previously been shown to be activated (phosphorylated at tyrosine 416/418) by the canonical Dab1 form (variant 1) (2, 6, 27–29). We therefore carried out immunostaining of Neuro2a cells transfected with Dab1 variants 1 to 3, with variant 6 serving as a negative control. Cells were fixed and immunostained with an antibody that specifically recognizes activated Src. As expected, cells expressing the canonical Dab1 form showed induction of Src activation, whereas Src activation was either attenuated or undetectable in cells transfected with variant 3 or with variants 2 and 6, respectively (Fig. 6).
Fig 6.
Src activation by GFP-tagged Dab1 variants in Neuro2a cells. Neuro2a cells were transfected with GFP-Dab1 variant 1 (A), variant 2 (B), variant 3 (C), and variant 6 (D). Cells were immunostained with anti-pSrc418 antibody. GFP was detected by epifluorescence.
Dab1 isoforms recruit distinct sets of SH2 domains.
Tyrosine-phosphorylated Dab1 acts as a hub to recruit SH2 domain-containing proteins implicated in the transmission and regulation of downstream reelin signaling. SH2 domains bind to phosphotyrosine ligands in a context-dependent manner, based on the 3 to 6 residues that are C-terminal to phosphotyrosine (30, 42). As alternative splicing of Dab1 removes tyrosines and changes the sequences of contextual residues, we hypothesized that the ability of Dab1 to recruit SH2 domains may vary in different isoforms.
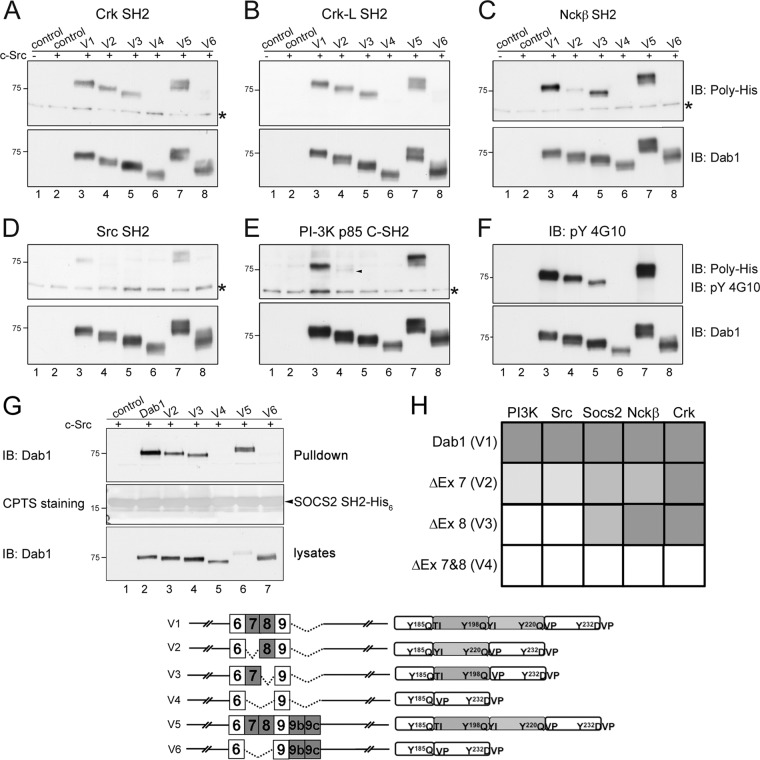
To assess the interactions between phosphorylated Dab1 and SH2 domains, we carried out far-Western blot analysis in 293T cells cotransfected with Src and Dab1. In total, seven SH2 domains were tested: SH2s from Crk, CrkL, Nckβ, Socs2, and Src and N- and C-terminal SH2s from the p85 subunit of PI3K. These SH2-containing proteins have been previously shown to bind phosphorylated Dab1 (1, 6–8, 13, 37). Of the SH2 domains tested, SH2s from Crk, CrkL, Nckβ, and Src and the C-terminal (but not the N-terminal) SH2 from p85 demonstrated strong binding to Dab1 (Fig. 7A to E, lanes 3). Interaction between Dab1 and Socs2 was not detected. As previous studies have shown interaction between Dab1 and Socs proteins (13), we used the pulldown assay to detect possible interaction between Dab1 and Socs2 under native conditions. Strong interaction with Socs2 was observed using this assay (Fig. 7G, lane 2).
Fig 7.
Dab1 variants interact with different SH2 domains. (A to E) Far-Western blot analysis of interactions between Dab1 variants and SH2 domains. Ten micrograms of cell lysate prepared from 293T cells cotransfected with Dab1 variants and c-Src was resolved by 8% SDS-PAGE and transferred to PVDF membranes. His-tagged SH2 probes were applied to the membranes, and bound SH2 domains were detected using antipolyhistidine antibody, followed by secondary antibody and visualization by chemiluminescence. Membranes were stripped and reprobed with anti-Dab1 antibody to ensure the presence of Dab1 variants and match the location of bound SH2 domains. Asterisks indicate nonspecific bands recognized by the antipolyhistidine antibody. The arrowhead points to a weakly stained band in panel E. (F) Western blot analysis of the same cell lysates using anti-phosphotyrosine 4G10 and anti-Dab1 antibodies. (G) Twenty microliters (∼10 μg of protein) of Socs2 SH2-coupled Ni-NTA magnetic beads was incubated with 50 μg of lysates prepared from 293T cells cotransfected with Dab1 variants and c-Src. The bead resuspensions (top panel) and lysates (bottom panel) were resolved by 10% SDS-PAGE, followed by immunoblotting using anti-Dab1 antibody. The membrane was stained with 3,4′,4″,4‴-copper phthalocyanine tetrasulfonic acid, tetrasodium salt (CPTS) to visualize the Socs2 SH2 peptide (middle panel). (H) Schematic representation of Dab1-SH2 interactions. Dark gray, strong association; medium gray, reduced association; light gray, substantially reduced association; white, no association. The six Dab1 variants used for these analyses are diagrammatically represented. Data shown are representative of two independent experiments.
Comparison of the relative ability of Dab1 isoforms to recruit different SH2 domains revealed a number of patterns. First, Dab1 variant 5, in which all four tyrosine phosphorylation sites are present, along with exons 9b and 9c, recruited all the Dab1-interacting SH2 domains to the same extent as Dab1 variant 1 (Fig. 7A to E, lanes 7). Second, the non-tyrosine-phosphorylated Dab1 variants 4 and 6 showed no associations with SH2 domains (Fig. 7A to E, lanes 6 and 8), establishing the specificity of phosphotyrosine-SH2 associations using this approach. Third, Dab1 variants 1 (containing exons 7 and 8), 2 (minus exon 7), and 3 (minus exon 8) interacted strongly with Crk and CrkL SH2 domains (Fig. 7A and B). Fourth, similar binding was observed between Dab1 variants 1 and 3 and the Nckβ SH2 domain; however, variant 2 showed reduced interaction with the Nckβ SH2 domain (Fig. 7C). Fifth, both Dab1 variants 2 and 3 demonstrated significantly reduced binding to the PI3K p85 C-terminal SH2 domain, with the most dramatic reduction observed with variant 3 (Fig. 7E). The predominant difference between variants 2 and 3, both of which contain a single YQXI site and two YXVP sites, is that exon 7 exclusion in variant 2 rearranges the Y185QXI motif, whereas exon 8 exclusion in variant 3 reconstitutes the Y198QVP site (Fig. 3).
Both Dab1 variants 1 and 5 were effectively pulled down by the Socs2 SH2 domain (Fig. 7G, top panel, lanes 2 and 6), suggesting that the Socs2 SH2 domain interacts with Dab1 either directly in its native state or as part of a complex. As expected, neither variant 4 nor variant 6 showed binding to the Socs2 SH2 domain. Both variants 2 and 3 were bound equally well by Socs2 SH2. The associations between different Dab1 isoforms and SH2 are schematically summarized in Fig. 7H.
Dab1 alternative splicing parallels mouse brain development and neural differentiation in P19 cells.
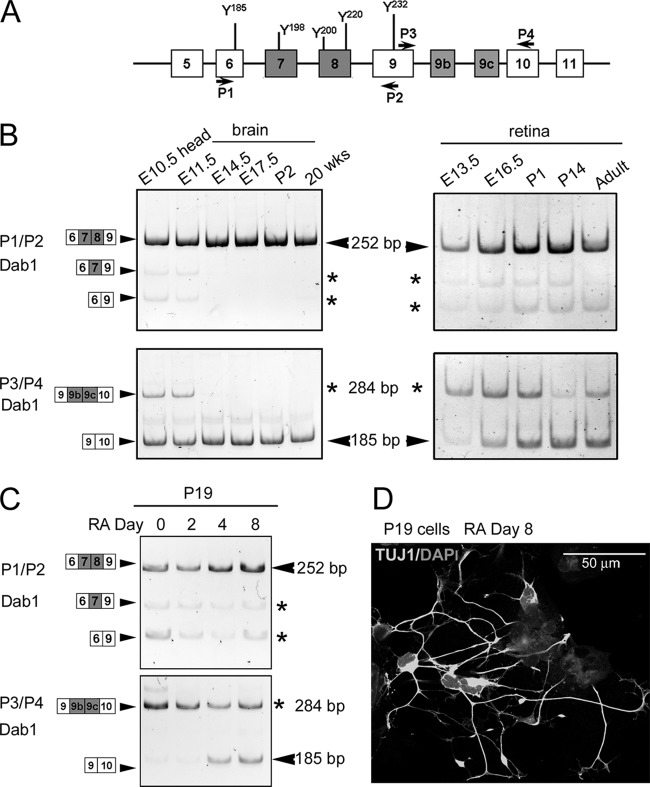
Dab1 plays an important role in regulating neuronal migration during brain development. To examine Dab1 alternative splicing as a function of development, we carried out RT-PCR analysis of mouse brain and retina at different developmental stages using primers that flanked either the deletion region (exons 7 and 8) (primers P1/P2) or the insertion region (exons 9b and 9c) (primers P3/P4) (schematically diagrammed in Fig. 8A). As shown in Fig. 8B (top left panel), RT-PCR analysis of the deletion region in E10.5 head revealed three bands of 252 bp, 186 bp, and 147 bp. The two bottom bands disappeared at later stages of development. Sequence analysis confirmed that the top band contains exons 7 and 8 (variant 1), whereas the bottom two bands are missing either exon 8 (186-bp band) (variant 3) or exons 7 and 8 (147-bp band) (variants 4 and/or 6). Sequencing of cDNA clones containing the entire ORF of Dab1 (P7/P8 primer pair) revealed similar variants. As variant 2 was not detected by either direct DNA sequencing or by RT-PCR, we carried out RT-PCR with primers flanking exon 7 (primer P1 in exon 6 and primer P15 in exon 8). A faint band was detected from E10.5 (head) to E17.5 (brain) (Fig. 9), indicating that variant 2 is expressed at very low levels in developing brain. The presence of variant 2 in the faint band was confirmed by DNA sequencing.
Fig 8.
Differential expression of Dab1 isoforms during development and P19 cell differentiation. (A) Locations of primers used for analysis of inclusion and exclusion regions of Dab1 are indicated. (B) RT-PCR analysis of Dab1 in the developing mouse brain and retina. cDNAs from mouse brain and retina at different developmental stages (as indicated) were amplified using Dab1 primer set P1/P2 or P3/P4. Sizes of amplified bands and exons included are indicated at the sides of the panels. (C) RT-PCR analysis of Dab1 in P19 cells. cDNAs from P19 cells treated with 0.5 μM retinoic acid (RA) for 0, 2, 4, and 8 days were amplified as described for panel B. (D) Immunofluorescence analysis of P19 cells at day 8 post-RA treatment using a mouse anti-TUJ1 antibody that specifically labels neurons. Nuclei are labeled with DAPI. Micrographs were collected with a Zeiss LSM 710 confocal microscope equipped with a 40× lens.
Fig 9.
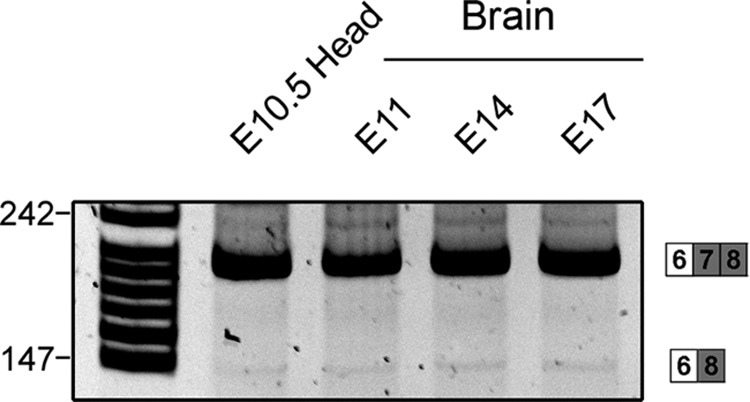
Detection of variant 2 in developing mouse brain. RT-PCR analysis of Dab1 variant 2 in the developing mouse brain. cDNA from mouse brain at different developmental stages (as indicated) was amplified using primers P1 (exon 6) and P15 (exon 8).
RT-PCR analysis of the insertion region revealed two bands of 284 bp and 185 bp (Fig. 8B, bottom left panel). Both the 284-bp and 185-bp bands were prominent in E10.5 mouse head and E11.5 brain. However, the 284-bp band was no longer detected by E17.5 in brain, in contrast to the 185-bp band which remained abundant throughout development as well as in adult brain. The presence and absence of exons 9b/9c in the 284-bp and 185-bp bands, respectively, were verified by sequence analysis. While the same-sized bands and similar expression trends were observed in retina, differentiation of this tissue was not accompanied by the complete loss of exons 9b/9c observed in the differentiating brain (Fig. 8B, right panels). These results suggest that Dab1 alternative splicing is differentially regulated in brain and retina.
To directly address Dab1 alternative splicing as a function of neuronal cell differentiation, we used the P19 teratocarcinoma model of neural cell differentiation. Exponentially growing P19 cells were aggregated in the presence of 0.5 μM RA to induce differentiation along the neuronal cell lineage (25). Based on immunostaining with the neuronal-specific marker TUJ1, most of the P19 cells had differentiated into neurons post-RA treatment (Fig. 8D). An increase in Dab1 transcripts that excluded exons 9b and 9c was observed upon P19 differentiation (Fig. 8C, bottom panel), in agreement with a previous report (3). In addition, we observed a concomitant increase in exons 7 and/or 8 inclusion during the course of P19 cell differentiation (Fig. 8C, top panel), consistent with concurrent inclusion of exons 7 and 8 and exclusion of exons 9b and 9c observed in mouse brain (Fig. 8B, left panels).
Temporal and spatial expression of Dab1 variants in mouse brain.
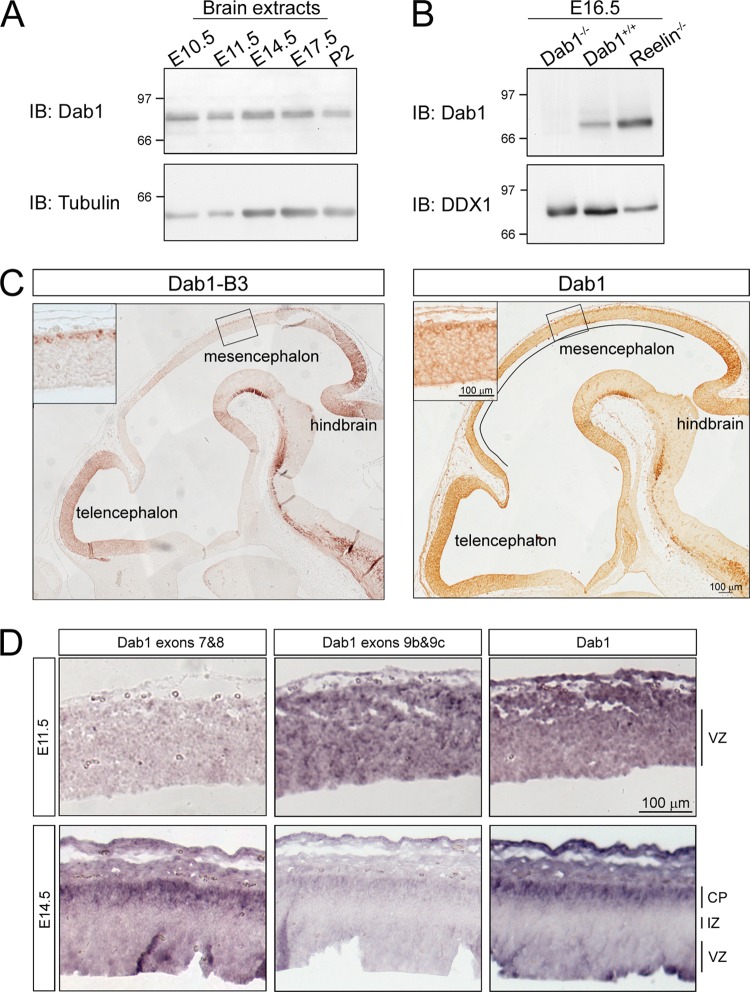
Expression of endogenous Dab1 protein in brain was examined by Western blot analysis of mouse brain lysates using the anti-Dab1 antibody raised against the C terminus of Dab1, which recognizes all Dab1 isoforms. A diffuse band of ∼80 kDa (likely representing multiple isoforms) was detected at all stages tested (Fig. 10A). The specificity of this antibody was verified using Dab1−/− (complete absence of bands on a Western blot) and Reln−/− mice (increased signal at ∼80 kDa) (Fig. 10B).
Fig 10.
Expression of Dab1 isoforms in the developing mouse brain. (A) Western blot analysis of brain lysates prepared at different developmental stages (as indicated) using anti-Dab1 (1:2,000) antibody. Tubulin was used a loading control. (B) Western blot analysis of E16.5 brain lysates from wild-type, Dab1−/−, and Reeler mice demonstrates the specificity of the Dab1 antibody. DDX1 was used as a loading control. (C) Immunohistochemical analysis of Dab1 in E11.5 mouse brain tissue using rabbit anti-Dab1 (1:500 dilution) or Dab1-B3 (1:500) antibody. To visualize the entire brain, partially overlapped photos were taken from different regions and automatically merged in Photoshop. Insets show higher magnification of Dab1 expression in the mesencephalon. The black line marks the region of the brain (mesencephalon) that is differentially stained by Dab1 and Dab1-B3 antibodies. (D) In situ hybridization analysis of Dab1 isoforms in the developing mouse brain. Mouse brain tissue sections at E11.5 and E14.5 were hybridized with DIG-labeled Dab1 exons 7/8 and exons 9b/9c or Dab1 3′ end antisense RNA probes. The signal was detected by alkaline phosphatase-coupled antibody, which produces a purple image. All photographs were captured with a 10× lens using a Zeiss Axioskop 2 Plus microscope. CP, cortical plate; IZ, intermediate zone; VZ; ventricular zone.
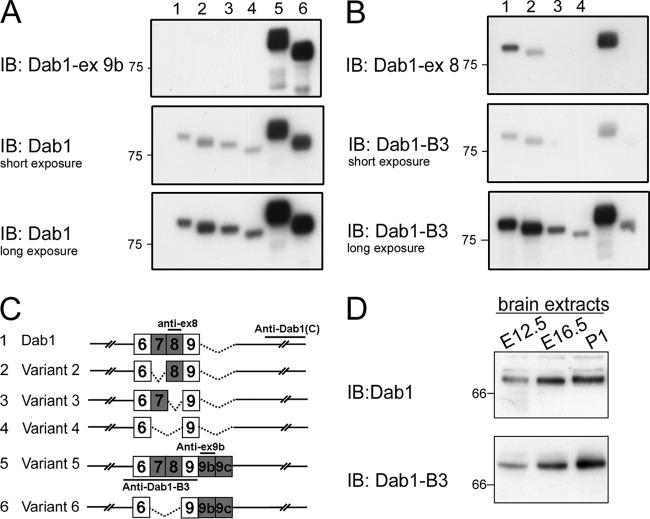
To distinguish isoforms containing exons 7/8 from those containing exons 9b/9c, we attempted to generate antibodies specific to exon 9b (Dab1-ex9b) or exon 8 (Dab1-ex8) (schematically shown in Fig. 11). While these antibodies specifically recognized exogenously expressed Dab1 isoforms (Fig. 11A and B), recognition of nonspecific proteins by both antibodies precluded analysis of endogenous Dab1 isoforms (data not shown). Temporal and spatial analysis of Dab1 in the developing mouse brain was therefore carried out with two Dab1 antibodies shown to specifically recognize endogenous Dab1 proteins: Dab1 C terminus antibody and B3 antibody, which preferentially recognizes exon 8-containing isoforms (Fig. 11D) (17). Dab1 isoforms were found throughout the developing brain at E11.5, including telencephalon, mesencephalon, and hindbrain (Fig. 10C). Exon 8-containing Dab1 isoforms were primarily found in telencephalon and hindbrain (Fig. 10C).
Fig 11.
Specificity of Dab1 isoform antibodies. (A and B) Western blot analysis of cell lysates prepared from 293T cells expressing different Dab1 isoforms using the following Dab1 antibodies, as indicated on the figure: anti-mouse Dab1-ex 9b, anti-Dab1, anti-mouse Dab1-ex 8, and anti-Dab1-B3. (C) Schematic representation of the immunogens used to generate Dab1 antibodies. (D) Western blot analysis of tissue extracts from E12.5, E16.5, and postnatal day 1 (P1) mouse brain using anti-Dab1 and anti-Dab1-B3 antibodies.
To examine the tissue distribution of Dab1 variants 1, 2, and 3 at the transcriptional level, we attempted to generate splice junction-specific probes that span the boundary of exons 6, 7, 8, and 9. However, our attempts at producing exon 7- and exon 8-specific splice junction probes were not successful because the last 6 nucleotides of exons 6, 7, and 8 are identical (Fig. 2). We then carried out in situ hybridization using probes prepared against exons 7/8 and exons 9b/9c. In general agreement with the immunostaining results obtained with the B3 antibody, weak staining was detected in the outermost layer of the E11.5 mesencephalon using the exon 7/8 probe (Fig. 10D). In contrast, intense staining was observed throughout the E11.5 mesencephalon with the exon 9b/9c probe and a 3′ end probe which labels all forms of Dab1 (Fig. 10D).
Radial migration of cortical neurons peaks between E14.5 and E16.5 (18). In situ hybridization of E14.5 cerebral cortex revealed intense staining with the exon 7/8 probe in the outermost layer of the cortical plate. Staining was virtually undetectable using the exon 9b/9c probe (Fig. 10D). The staining pattern obtained with the Dab1 3′ end probe was similar to that observed with the exon 7/8 probe. These results demonstrate mutually exclusive expression of exons 7/8 and exons 9b/9c during brain development, with inclusion of exons 7/8 and exclusion of exons 9b/9c correlating with prominent cortical neuron migration.
DISCUSSION
The role of canonical Dab1 (variant 1), an isoform that contains two YQXI and two YXVP phosphorylation sites, has been well characterized in reelin-mediated neuronal migration (14, 15, 21, 22). Of note, three of these four tyrosine phosphorylation sites are encoded by nucleotides located at the 5′ or 3′ end of neighboring exons (6, 7, and 8) so that each site requires coding sequences from two adjacent exons. Here, we identify 11 alternatively spliced Dab1 variants in the developing mouse brain and retina. The majority of these variants are generated by alternative splicing of exon 7 and/or exon 8, resulting in alterations in the number of YQXI sites, as well as contextual changes within and in the vicinity of YQXI sites. We also demonstrate that Dab1 variants are differentially phosphorylated and recruit distinct subsets of SH2 domains involved in the reelin-Dab1 signal. Our study highlights the importance of alternative splicing in modulating Dab1 function through key tyrosine phosphorylation motifs required for activation of downstream signaling cascades.
Alternative splicing regulates Dab1 activity and downstream interactions.
In comparison to Dab1 variant 1, variants 2 and 3 each contain one YQXI motif and two YXVP motifs. The significant impairment of tyrosine phosphorylation and reduced ability to activate Src in variants 2 and 3 indicate that YQXI motifs play a predominant role in Dab1 tyrosine phosphorylation and that both YQXI motifs are required for full activation of Dab1 (14, 27). These results are consistent with the processive phosphorylation mechanism proposed by Mayer et al. (34), whereby Dab1 YQXI motifs recruit SFK, which subsequently phosphorylate the adjacent YXVP motifs.
SFK-mediated Dab1 tyrosine phosphorylation recruits multiple SH2 domain-containing proteins. We were able to confirm previously documented interactions with Dab1 (variant 1), including Src, Socs2, PI3K (p85 subunit), Crk/CrkL, and Nckβ. It is noteworthy that in contrast to other SH2 domains which were recruited under denaturing conditions (far-Western assay), interaction of Dab1 with Socs2 SH2 domains was observed only under native conditions (pulldown assay).
Importantly, we found that SH2 domains are preferentially recruited depending on which exons are retained in Dab1. For example, exclusion of exon 7 in variant 2 (Y185QYI, Y220QVP, and Y232DVP) significantly reduced its ability to recruit p85 and Src SH2s, whereas exclusion of exon 8 in variant 3 (Y185QTI, Y198QVP, and Y232DVP) prevented PI3K and Src SH2s interactions. In addition, exclusion of either exon 7 or exon 8 impaired Dab1-Socs SH2 interaction. As both variants 2 and 3 contain only one YQXI motif, the significantly decreased binding of these variants to p85, Src, and Socs2 SH2s indicates that both YQXI motifs need to be present for efficient interactions with these domains. The difference between Dab1 variants 2 and 3 in recruiting p85 SH2 is likely due to variations in residues surrounding the YQXI motif as these have been shown to affect SH2 recognition (30). The reduced tyrosine phosphorylation and attenuated Src activation of Dab1 variants 2 and 3 observed in Neuro2a cells lend support to the importance of tyrosine-encoding exons in determining the recruitment of SH2 domains and activation of downstream signaling.
Crk/CrkL and Nckβ have previously been shown to specifically bind to Dab1 YXVP sites (1, 8, 24) and would therefore be predicted to bind with the same affinity to Dab1 variants 2 and 3, both of which contain two YXVP sites. As expected, Crk and CrkL SH2s effectively bound variants 2 and 3. However, Nckβ SH2 showed a clear binding preference to variant 3 compared to variant 2. We speculate that the two YXVP sites may be differentially phosphorylated in variants 2 and 3 due to the presence of different YQXI motifs. As Crk is directly linked to the C3G-Rap1 pathway (1) and as Nckβ is linked to the p130Cas pathway (39), it is likely that variants 2 and 3 connect to different downstream cascades. Although these binding experiments were carried out in 293T cells, our data provide clear evidence that Dab1 variants have different affinities for different SH2 domains and emphasize the importance of splicing-mediated changes at YQXI and YXVP sites and contextual residues in recruiting specific SH2 domains.
Conservation and implication of Dab1 alternative splicing.
The genomic organization of Dab1 is unusually complex and highly conserved in different species (3, 10, 31). The mammalian Dab1 gene spans over 1 Mb with an open reading frame of 1,668 bp (3). Different alternative splicing events, including alternative promoter usage, alternative cassette exons, and mutually exclusive alternative exons, have been documented for Dab1 in multiple species, including zebrafish, lizard, chick, pig, mouse, and human (3, 10, 20, 29, 31, 46).
Other than the canonical Dab1 isoform (variant 1), three additional Dab1 isoforms, Dab1217, Dab1271, and Dab1555* (Dab1.7bc, referred to as variant 5 in this study), have been described in mice (3, 20, 46). All three Dab1 isoforms retain the intact PI/PTB domain at the N terminus but differ in the middle tyrosine-enriched region or in the C terminus. Dab1217 contains an alternative exon (57 bp) inserted after exon 7, whereas Dab1271 contains an alternative exon (93 bp) inserted after exon 9 (20). We did not detect either Dab1217 or Dab1271 in our study; however, it should be noted that both of these isoforms introduce in-frame premature termination codons (PTC), which may promote RNA degradation through nonsense-mediated mRNA decay (NMD) (5). Dab1 variant 5 contains exons 9b/c (3, 20, 46), whose homologues have also been detected in chicken, lizard, and pig (3, 29, 31). Unlike the inclusion of exons 9b/9c in mouse variant 5, 9b/9c inclusion in chicken and pig is accompanied by concurrent exclusion of exons 7 and 8, producing an isoform homologous to the variant 6 identified in the present study. Intriguingly, a number of studies have shown that exon 9b/9c-containing Dab1 isoforms are expressed in nonneuronal cells, either in proliferative neural tissue or adult nonneuronal tissue such as liver (17, 29). As the canonical Dab1 isoform has been proposed to act as a scaffold as well as an adaptor to direct neuronal migration (14), it will be important to investigate whether different Dab1 isoforms may be involved in reelin-independent signaling pathways in nonneuronal cells.
Despite the identification of multiple Dab1 isoforms, the vast majority of studies carried out to date have focused on the characterization of Dab1 variant 1. Recent studies suggest that alternatively spliced Dab1 isoforms also contribute to overall Dab1 signaling. For example, we have described a role for chicken Dab1 variant 6 (Dab1-E) in the maintenance of retinal progenitor cells (17). Furthermore, a recent study by Yano et al. (46) has demonstrated that Nova2 specifically excludes exons 9b/9c in neurons. Ablation of Nova2 in mice increases the levels of exon 9b/9c-containing Dab1 transcripts and results in aberrant neuronal migration of late-born, but not early-born, neurons (46). Introduction of wild-type Dab1 into Nova2−/− mice rescues the migration defects, in support of an antagonistic role for Dab1 and exon 9b/9c-containing Dab1 isoforms (46). However, it is not clear how these different Dab1 isoforms counteract each other since both wild-type and exon 9b/9c-containing forms are tyrosine phosphorylated and recruit SH2 domains.
Coordination of reelin signaling by Dab1 alternative splicing—a model.
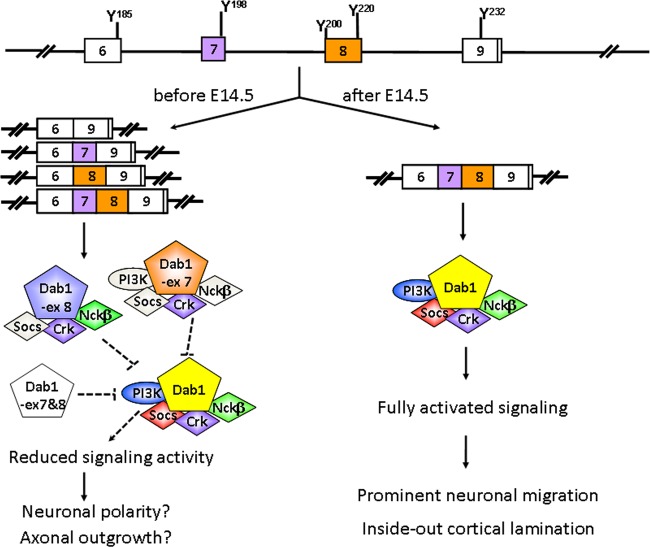
In a recent study, reelin has been shown to regulate the migration of both early- and late-born neurons, with individual neurons responding differently to the reelin signal in terms of migratory mode, speed, and polarity (41). However, it is still unclear how different neuronal cell populations coordinately migrate to their correct destination in response to reelin. The presence of multiple Dab1 isoforms expressed in distinct cell populations at different developmental stages, along with differential tyrosine phosphorylation and isoform-specific differences in recruiting downstream SH2 domains, suggests that Dab1 alternative splicing may be key to controlling the intricate processes underlying neuronal migration.
Based on our combined data, we propose a model whereby different Dab1 isoforms, generated through alternative splicing, coordinately regulate reelin-mediated neuronal migration in a space- and time-dependent manner (schematically shown in Fig. 12). Early in development (prior to E14.5), both Dab1 variant 1 and Dab1 isoforms that exclude exon 7 and/or exon 8 are present, with the latter found primarily in proliferating cells. Isoforms that exclude exon 7 and/or exon 8 cannot assemble the complete set of Dab1 downstream signaling complexes and may not be able to fully transmit the reelin signal. Thus, we propose that the presence of these truncated isoforms may either mitigate the reelin signal or uncouple the reelin signal from subsets of downstream signaling cascade effectors. As a consequence, migration of cells expressing Dab1 isoforms that exclude exons 7 and/or 8 may be attenuated, while allowing early events associated with neuronal migration such as polarization and axonal outgrowth to take place (4, 26, 33). As development proceeds (∼E14.5), Dab1 variant 1 becomes the predominant form expressed in neuronal cells. Dab1 responds fully to the reelin signal and activates all the downstream pathways required for neuronal migration, leading to the inside-out lamination of the cerebral cortex. Therefore, the presence of multiple Dab1 isoforms allows fine-tuning of reelin signaling in a spatiotemporal manner, which in turn directly contributes to accurate neuronal positioning during brain development. Further studies using in utero electroporation or knock-in approaches are needed to characterize the physiological functions of the different Dab1 isoforms.
Fig 12.
Model of proposed roles for Dab1 alternative splicing in regulating reelin signaling and neuronal migration during development. Early in development (E10.5 to E12.5), alternative splicing excludes Dab1 exons 7 and/or 8 in proliferating precursor cells and early-born neurons. Depending on the variant, reelin-Dab1 downstream effectors are either not activated or partially activated, events compatible with neuronal polarization and early-born neuronal migration. As development progresses (E14.5), splicing results in inclusion of both exons 7 and 8 in late-born neurons. Cells expressing this Dab1 form are fully responsive to reelin and migrate to form the inside-out lamination in the cerebral cortex. Red, blue, green, and purple indicate strong association with Dab1, whereas gray indicates reduced interaction with Dab1.
Taken together, our data illustrate the importance of alternative splicing in regulating Dab1 function during brain development. In particular, the role of Dab1 appears to be governed through inclusion and exclusion of exons that result in Dab1 isoforms with different combinations of YQXI and YXVP phosphorylation sites. The ability of the different Dab1 isoforms to recruit distinct sets of SH2 domains suggests a fine-tuning role for Dab1 splicing in the intricate cascade of events that underlie neuronal migration and their proper positioning within the laminated structures of the brain. Splicing-controlled switching and reconstitution of tyrosine phosphorylation sites also reveal a novel mechanism to regulate the specificity of signal transduction in complex pathways.
ACKNOWLEDGMENTS
We are grateful to Jonathan Cooper, Joan Brugge, Don Fujita, and Franck Polleux for antibodies and plasmids. We thank the Cell Imaging Facility, Xuejun Sun, and Gerry Barron for their help with micrograph collection and Devon Germain for reading the manuscript and helpful comments.
This work was supported by the Canadian Institutes of Health Research.
Footnotes
Published ahead of print 14 May 2012
REFERENCES
- 1. Ballif BA, et al. 2004. Activation of a Dab1/CrkL/C3G/Rap1 pathway in Reelin-stimulated neurons. Curr. Biol. 14:606–610 [DOI] [PubMed] [Google Scholar]
- 2. Ballif BA, Arnaud L, Cooper JA. 2003. Tyrosine phosphorylation of Disabled-1 is essential for Reelin-stimulated activation of Akt and Src family kinases. Brain Res. Mol. Brain Res. 117:152–159 [DOI] [PubMed] [Google Scholar]
- 3. Bar I, Tissir F, Lambert de Rouvroit C, De Backer O, Goffinet AM. 2003. The gene encoding disabled-1 (DAB1), the intracellular adaptor of the Reelin pathway, reveals unusual complexity in human and mouse. J. Biol. Chem. 278:5802–5812 [DOI] [PubMed] [Google Scholar]
- 4. Barnes AP, et al. 2007. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell 129:549–563 [DOI] [PubMed] [Google Scholar]
- 5. Blencowe BJ. 2006. Alternative splicing: new insights from global analyses. Cell 126:37–47 [DOI] [PubMed] [Google Scholar]
- 6. Bock HH, Herz J. 2003. Reelin activates SRC family tyrosine kinases in neurons. Curr. Biol. 13:18–26 [DOI] [PubMed] [Google Scholar]
- 7. Bock HH, et al. 2003. Phosphatidylinositol 3-kinase interacts with the adaptor protein Dab1 in response to Reelin signaling and is required for normal cortical lamination. J. Biol. Chem. 278:38772–38779 [DOI] [PubMed] [Google Scholar]
- 8. Chen K, et al. 2004. Interaction between Dab1 and CrkII is promoted by Reelin signaling. J. Cell Sci. 117:4527–4536 [DOI] [PubMed] [Google Scholar]
- 9. Colwill K, et al. 2011. A roadmap to generate renewable protein binders to the human proteome. Nat. Methods 8:551–558 [DOI] [PubMed] [Google Scholar]
- 10. Costagli A, Felice B, Guffanti A, Wilson SW, Mione M. 2006. Identification of alternatively spliced Dab1 isoforms in zebrafish. Dev. Genes Evol. 216:291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. D'Arcangelo G, et al. 1995. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374:719–723 [DOI] [PubMed] [Google Scholar]
- 12. Dierck K, Machida K, Mayer BJ, Nollau P. 2009. Profiling the tyrosine phosphorylation state using SH2 domains. Methods Mol. Biol. 527:131–155, ix [DOI] [PubMed] [Google Scholar]
- 13. Feng L, Allen NS, Simo S, Cooper JA. 2007. Cullin 5 regulates Dab1 protein levels and neuron positioning during cortical development. Genes Dev. 21:2717–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng L, Cooper JA. 2009. Dual functions of Dab1 during brain development. Mol. Cell. Biol. 29:324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. 2011. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron 69:482–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao Z, Godbout R. 2011. Serine phosphorylation regulates disabled-1 early isoform turnover independently of Reelin. Cell Signal. 23:555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao Z, Monckton EA, Glubrecht DD, Logan C, Godbout R. 2010. The early isoform of disabled-1 functions independently of Reelin-mediated tyrosine phosphorylation in chick retina. Mol. Cell. Biol. 30:4339–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta A, Tsai LH, Wynshaw-Boris A. 2002. Life is a journey: a genetic look at neocortical development. Nat. Rev. Genet. 3:342–355 [DOI] [PubMed] [Google Scholar]
- 19. Herz J, Chen Y. 2006. Reelin, lipoprotein receptors and synaptic plasticity. Nat. Rev. Neurosci. 7:850–859 [DOI] [PubMed] [Google Scholar]
- 20. Howell BW, Gertler FB, Cooper JA. 1997. Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 16:121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howell BW, Herrick TM, Cooper JA. 1999. Reelin-induced tyrosine phosphorylation of disabled 1 during neuronal positioning. Genes Dev. 13:643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Howell BW, Herrick TM, Hildebrand JD, Zhang Y, Cooper JA. 2000. Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr. Biol. 10:877–885 [DOI] [PubMed] [Google Scholar]
- 23. Howell BW, Lanier LM, Frank R, Gertler FB, Cooper JA. 1999. The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol. Cell. Biol. 19:5179–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang Y, et al. 2004. Tyrosine phosphorylated Disabled 1 recruits Crk family adapter proteins. Biochem. Biophys. Res. Commun. 318:204–212 [DOI] [PubMed] [Google Scholar]
- 25. Jones-Villeneuve EM, Rudnicki MA, Harris JF, McBurney MW. 1983. Retinoic acid-induced neural differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 3:2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jossin Y, Cooper JA. 2011. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat. Neurosci. 14:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katyal S, Gao Z, Monckton E, Glubrecht D, Godbout R. 2007. Hierarchical disabled-1 tyrosine phosphorylation in Src family kinase activation and neurite formation. J. Mol. Biol. 368:349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katyal S, Glubrecht DD, Li L, Gao Z, Godbout R. 2011. Disabled-1 alternative splicing in human fetal retina and neural tumors. PLoS One 6:e28579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katyal S, Godbout R. 2004. Alternative splicing modulates Disabled-1 (Dab1) function in the developing chick retina. EMBO J. 23:1878–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu BA, et al. 2010. SH2 domains recognize contextual peptide sequence information to determine selectivity. Mol. Cell. Proteomics 9:2391–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Long H, et al. 2011. Identification of alternatively spliced Dab1 and Fyn isoforms in pig. BMC Neurosci. 12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Machida K, Mayer BJ. 2009. Detection of protein-protein interactions by far-western blotting. Methods Mol. Biol. 536:313–329 [DOI] [PubMed] [Google Scholar]
- 33. Matsuki T, et al. 2010. Reelin and stk25 have opposing roles in neuronal polarization and dendritic Golgi deployment. Cell 143:826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mayer BJ, Hirai H, Sakai R. 1995. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 5:296–305 [DOI] [PubMed] [Google Scholar]
- 35. Niu S, Renfro A, Quattrocchi CC, Sheldon M, D'Arcangelo G. 2004. Reelin promotes hippocampal dendrite development through the VLDLR/ApoER2-Dab1 pathway. Neuron 41:71–84 [DOI] [PubMed] [Google Scholar]
- 36. Nollau P, Mayer BJ. 2001. Profiling the global tyrosine phosphorylation state by Src homology 2 domain binding. Proc. Natl. Acad. Sci. U. S. A. 98:13531–13536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pramatarova A, et al. 2003. Nck beta interacts with tyrosine-phosphorylated disabled 1 and redistributes in Reelin-stimulated neurons. Mol. Cell. Biol. 23:7210–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rice DS, Curran T. 2001. Role of the reelin signaling pathway in central nervous system development. Annu. Rev. Neurosci. 24:1005–1039 [DOI] [PubMed] [Google Scholar]
- 39. Rivera GM, et al. 2006. Requirement of Nck adaptors for actin dynamics and cell migration stimulated by platelet-derived growth factor B. Proc. Natl. Acad. Sci. U. S. A. 103:9536–9541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheldon M, et al. 1997. scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature 389:730–733 [DOI] [PubMed] [Google Scholar]
- 41. Simo S, Jossin Y, Cooper JA. 2010. Cullin 5 regulates cortical layering by modulating the speed and duration of Dab1-dependent neuronal migration. J. Neurosci. 30:5668–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Songyang Z, et al. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell 72:767–778 [DOI] [PubMed] [Google Scholar]
- 43. Tremblay RG, et al. 2010. Differentiation of mouse Neuro 2A cells into dopamine neurons. J. Neurosci. Methods 186:60–67 [DOI] [PubMed] [Google Scholar]
- 44. Trommsdorff M, Borg JP, Margolis B, Herz J. 1998. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J. Biol. Chem. 273:33556–33560 [DOI] [PubMed] [Google Scholar]
- 45. Trommsdorff M, et al. 1999. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97:689–701 [DOI] [PubMed] [Google Scholar]
- 46. Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. 2010. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron 66:848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]