Abstract
The mitochondrial respiratory chain is organized within an array of supercomplexes that function to minimize the generation of reactive oxygen species (ROS) during electron transfer reactions. Structural models of supercomplexes are now known. Another recent advance is the discovery of non-OXPHOS complex proteins that appear to adhere to and seal the individual respiratory complexes to form stable assemblages that prevent electron leakage. This review highlights recent advances in our understanding of the structures of supercomplexes and the factors that mediate their stability.
DESCRIPTION OF RESPIRATION AND THE RESPIRATORY CHAIN
Glycolysis, fatty acid oxidation, and the citric acid cycle generate the reducing molecules NADH and reduced flavin adenine dinucleotide (FADH2), which are subsequently oxidized within the mitochondria. Energy from these oxidation steps is converted into a proton gradient across the inner mitochondrial membrane (IM) that drives ATP synthesis. Electron transfer through a series of respiratory complexes ends with the terminal reduction of molecular oxygen to water. This oxidative phosphorylation process was initially shown to involve a series of heme-containing cytochromes by David Keilin in 1925 (42). By 1940, the respiratory chain was defined as a simple chain: dehydrogenase → cytochrome b → cytochrome c → cytochrome a → cytochrome a3 → oxygen (42). By 1960, David Green's group demonstrated that these components existed within four respiratory complexes, designated complex I (CI, NADH dehydrogenase), complex II (CII, succinate dehydrogenase), complex III (CIII, ubiquinol cytochrome c reductase), and complex IV (CIV, cytochrome c oxidase). Electron transfer between these complexes is dependent on diffusible electron shuttles; the isoprenyl moiety ubiquinone mediates electron transfer from CI and CII to CIII, and the small cytochrome c protein serves as the mediator from CIII to CIV. Electron transfer through complexes I, III, and IV is coupled to proton pumping, creating the electrochemical gradient used by complex V (ATP synthase) to synthesize ATP from ADP and inorganic phosphate. An additional component of the respiratory chain in metazoans is the electron transfer flavoprotein-ubiquinone oxidoreductase (ETF-QO), which is a branch point responsible for transferring electrons from a myriad of flavodehydrogenases to the respiratory chain as ubiquinol (51).
These membrane-embedded respiratory complexes contain multiple redox centers, including quinones, flavins, iron-sulfur clusters, hemes, and copper ions. The oxidation of NADH by CI generates two electrons that are transferred through flavin mononucleotide (FMN) and multiple Fe/S cluster centers to the ubiquinone reduction site (34). Succinate oxidation by CII occurs with the initial reduction of FAD and subsequent electron transfer through 3 distinct Fe/S cluster centers to the ubiquinone reduction site. Ubiquinone reduction by the ETF-QO complex occurs by electron transfer through a FAD and a single Fe/S center. Reduced ubiquinol is oxidized by CIII with a bifurcated electron flow that sends one electron through a 2Fe-2S cluster and a covalently bound c-type cytochrome prior to reduction of the diffusible cytochrome c. The second electron from each ubiquinone passes through two heme b moieties as part of the proton-pumping Q cycle (12). Oxidation of cytochrome c occurs by CIV with electron flux through a binuclear copper center to a heme a moiety and, subsequently, to a heterobimetallic heme a3-copper center where oxygen is reduced.
Structures for the individual mitochondrial respiratory complexes are known. The eukaryotic CI forms an L-shaped structure composed of a membrane arm and a hydrophilic arm protruding into the matrix. The hydrophilic arm projecting into the matrix contains all the redox cofactors necessary for electron transfer. The long, slightly curved α-helical membrane arm of the structure is responsible for proton translocation. The bacterial CI X-ray structure is known, along with a low-resolution CI structure of the fungal C1 (19, 26). In addition, electron microscopy (EM) reconstructions of CI have been reported (10, 23). In CII, a hydrophilic arm consisting of 2 subunits containing the redox cofactors is associated with a membrane anchor (46). CIII is a stable dimeric enzyme, whereas CIV is a monomeric entity, although the X-ray structure of the bovine enzyme consisted of a dimeric complex (27, 31, 48).
ORGANIZATION OF THE RESPIRATORY CHAIN
The respiratory complexes are enriched in segments of the IM that invaginate into tubular entities designated cristae (49). Keilin proposed in 1947 that the respiratory components were “more or less rigidly held together in a framework that ensures their mutual accessibility and a consequent high catalytic activity” (29, 42). However, a vigorous debate ensued on this model of organization. Evidence was provided for both a random distribution and an ordered association of components. In the random distribution model, electron transfer would be limited by diffusion of components, whereas the model of ordered association predicted electron channeling between the complexes. Studies addressing the behavior of ubiquinone and cytochrome c pools failed to support the diffusion model of independent respiratory complexes randomly distributed in the IM (6). Although physical association of bacterial respiratory complexes had been reported, the ordered association of eukaryotic complexes was not verified until the isolation and visualization of supercomplexes first reported by Cruciat et al. and Schägger and Pfeiffer (13, 41). A protocol to isolate membrane protein complexes using a mild, nonionic detergent and negatively charged Coomassie blue dye led to the visualization of the respiratory supercomplex on native gels (blue native polyacrylamide gel electrophoresis, BN-PAGE). Yeast mitochondria were shown to form two high-mass supercomplexes consisting of CIII and CIV with the predicted stoichiometry of III2-IV1-2 (13, 41). CI is not present in the yeast respiratory supercomplexes, since Saccharomyces cerevisiae lacks the multimeric CI complex. In contrast, mammalian mitochondria were shown to contain high-order complexes consisting of CI, CIII, and CIV (41). Since these three respiratory complexes provide the bulk of the proton motive force, these supercomplexes were designated respirasomes (41). The respirasome concept was validated in 2008 with the isolation of CI-CIII-CIV supercomplexes from mouse fibroblast cells and the demonstration that they contained ubiquinone and cytochrome c and were functional in NADH-stimulated oxygen consumption (2).
Supercomplexes are preserved during membrane solubilization with a variety of detergents, including digitonin, Triton X-100, NP-40, and Tween 20 (2, 41). The association of respiratory complexes is therefore not specific for one detergent, so their existence is unlikely to be a detergent artifact. However, supercomplexes are dissociated into individual components with the harsher detergent dodecyl-maltoside. Respiratory supercomplexes have now been reported in a broad range of organisms, from bacteria to plants, fungi, and animals, and can be resolved by either sucrose gradient centrifugation or BN-PAGE (5).
COMPOSITE NATURE OF RESPIRATORY SUPERCOMPLEXES
A myriad of supercomplexes formed from electron transfer complex (ETC) components exist. The composition and abundance of these supercomplexes vary with growth or physiological conditions (22, 41). In yeast, lacking the proton-translocating CI, two main supercomplexes of 650 and 850 kDa are evident, consisting of CIII and CIV (III2-IV1 and III2-IV2) (24, 39). The abundance of the III2-IV2 supercomplex is enhanced in yeast propagating on nonfermentable carbon sources (41). In addition to the two high-mass complexes, less abundant and lower mass supercomplexes exist that likely represent assemblies in which one component is an immature assembly intermediate (33, 36). Although the bulk of CIV is present in supercomplexes in yeast, the absence of either CIII or CIV does not generally attenuate the abundance of the other individual complex (13, 41).
Mammalian and plant respiratory supercomplexes are a composite of ETC complexes. Complexes containing only CI and CIII exist, in addition to the respirasome complexes of CI, CIII, and CIV (20, 39, 41). Within the Arabidopsis genus, the most abundant supercomplex is the CI-CIII assemblage (17); the CIV-containing assemblies are much lower in abundance (20). In contrast, the CI-CIII-CIV supercomplex is higher in abundance than the CI-CIII complex in mammalian cells (35, 39). The bulk of CI is associated with supercomplexes, whereas CIV is present predominantly as a monomeric unit. Nearly half of CIII resides within supercomplexes. The relative ratios of ETC complexes in bovine heart mitochondria were determined to be 1.1 (CI), 1.3 (CII), 3 (CIII), and 6.7 (CIV) (40). The abundances of the various supercomplexes are predicted to be I-III2-IV1, I-III2, I-III2-IV2, and I-III2-IV3-4 in order of decreasing abundance (40).
The mammalian supercomplexes appear to have a stabilizing role in each component respiratory complex. Mutations in a given respiratory complex can present in patients as multicomponent defects. For instance, mutations in the cytochrome b subunit of CIII were shown to result in the instability of CI in human and mouse cells, although CIII was not perturbed in mutations precluding the assembly of CI (1). Likewise, mutations in CIV can also destabilize CI (14, 15, 32), and CI dysfunction can also destabilize CIII (8). Although not all mutations within a given respiratory complex present with multicomplex deficiencies, there are apparent stabilizing interconnections between the respiratory complexes, probably arising from supercomplex formation. Stabilizing interactions by respiratory supercomplexes are also apparent in bacterial CI-CIII-CIV assemblies (44). Paracoccus denitrificans lacking either CIII or CIV shows defects in the assembly of CI.
The citric acid cycle and respiratory enzyme succinate dehydrogenase (CII) forms supercomplexes involving CIII in mouse mitochondria, and purified mouse respirasomes are competent for succinate-induced oxygen consumption (2). Although CII is not typically seen in supercomplexes (13, 35), mutations in the yeast CIII subunit Qcr8 were reported to impair CII activity without compromising bc1 function (7). One potential implication of these results is that CIII has a stabilizing interaction with CII in yeast, although such a stabilizing interaction has not been reported with mammalian CII (1).
STRUCTURE OF SUPERCOMPLEXES
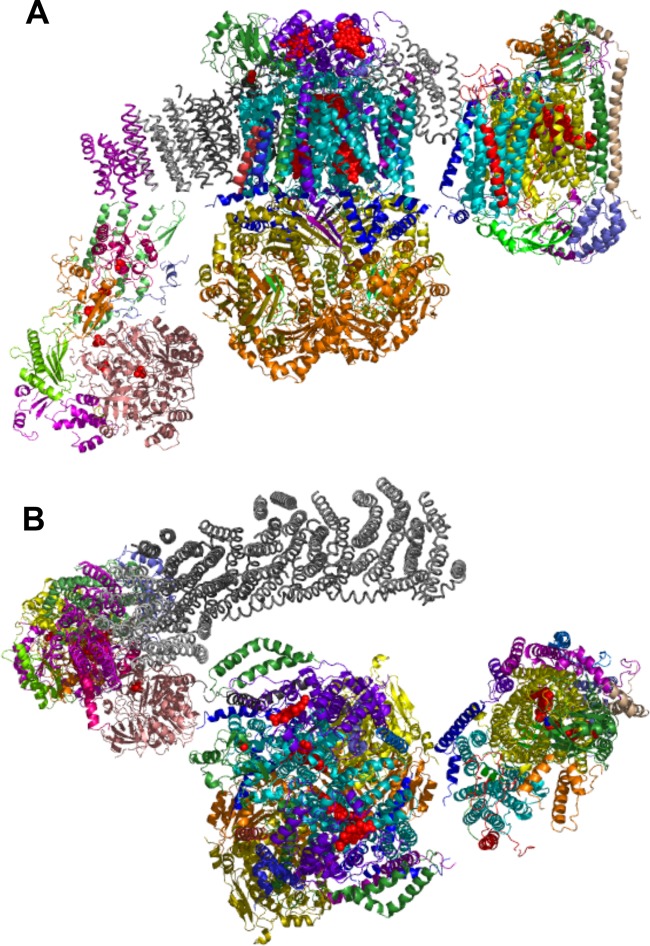
ETC supercomplexes have been visualized by single-particle electron microscopy. Image analysis of both uranyl acetate-stained I-III2-IV1 supercomplex solubilized with detergent and cryo-EM of the amphipol-solubilized supercomplex revealed that CIII is attached to the inner curve of the CI membrane arm, with CIV attached near the tip of the CI membrane arm (3, 38). The docked atomic structures of CI, CIII, and CIV complexes account for approximately 60% of the supercomplex electron density map (3) (Fig. 1). Electron tomography on cryo-EM preparations of the bovine heart I-III2-IV1 supercomplex was recently reported (18). Single-particle reconstruction of these images confirmed the overall orientation of the assembly observed by the earlier single-particle analysis of uranyl acetate-stained complex; however, the predicted interaction interfaces between CIII and CIV differed from those in an earlier model (38). The recent reconstructions predict that CIII contacts CIV at a face distinct from that mediating CIV dimerization in the X-ray structure (48). Modeling of the EM projection maps of the yeast III2-IV1 supercomplex predicts a different interaction interface on CIV, although in both the yeast and bovine models, the CIV dimerization interface is free (24). A second III2-IV2 yeast supercomplex was also apparent, and this showed a monomeric CIV on either side of the dimeric CIII. Supercomplexes are possible with dimeric CIV entities associated with CIII.
Fig 1.
Atomic structures of CI, CIII, and CIV that were docked into the electron density maps of the bovine I-III2-IV1 supercomplex obtained from single-particle cryo-electron tomography (18). The coordinates were kindly provided by Natalya Dudkina. (A) Side view. Subunits in the membrane arm of CI are colored gray to provide contrast with the CIII and CIV subunits. Heme moities in CIII and CIV are shown as red spheres. Iron atoms in FeS clusters are also shown as red spheres. The center complex is CIII, with CIV on the right. Among the key subunits in CIII, the two Cor2 subunits are orange, Cor1 subunits are yellow, Cob is cyan, Cyt1 is purple, and Qcr6 is green. Within CIV, Cox3 (III) is cyan, Cox12 (VIb) is orange, Cox13 (VIa) is red, VIIa is blue, Cox1 is yellow, and Cox2 is dark green. (B) Top view from IMS side.
The predicted interaction interface based on the yeast reconstruction model consists of CIV subunits I, II, III, IV, VIc, VIIa, and VIIc (bovine nomenclature) and CIII subunits cytochrome b (Cob), cytochrome c1 (Cyt1), and Qcr6, Qcr7, Qcr8, and Qcr9 (24). In contrast, the bovine model predicts CIV subunits III, VIa, and VIIa (not IV, VIc, and VIIc) and CIII subunits cytochrome b, Rieske protein, and subunit 11 (18). Additional studies are needed to confirm whether the yeast and bovine supercomplexes have a distinct rotational orientation of each complex at the interface. Yeast mutants lacking candidate interaction subunits do not provide clear insight on the interface. Yeast lacking the corresponding subunit to VIa (Cox13) have no defects in CIII-CIV supercomplexes (36, 39). Likewise, yeast lacking subunits corresponding to bovine subunit 11 (Qcr10) or Qcr6 retain supercomplexes, but cells lacking Qcr9 or the Rieske Rip1 subunits exhibit destabilized, partially assembled CIII-CIV supercomplexes (13, 39).
STABILIZATION OF SUPERCOMPLEXES
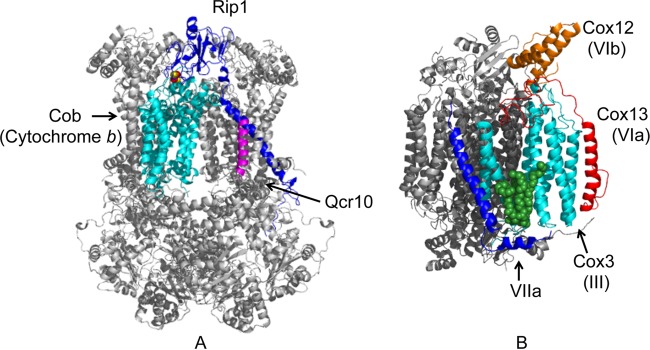
In the reconstructed models of the CI-CIII-CIV supercomplex, only limited apparent interaction sites exist between neighboring complexes. Some of the gap volumes may be lipid filled. Cardiolipin associates with Cox3 at the face predicted in the bovine model to associate with CIV (45) (Fig. 2B). Cardiolipin is also associated with cytochrome b in CIII; however, that face is not predicted to be at the CIV interface. Consistent with the observation that cardiolipin is at the CIII-CIV interface, yeast impaired in cardiolipin synthesis (crd1Δ cells) exhibits destabilized CIII-CIV supercomplexes by BN-PAGE; however, the complexes are apparent by gel filtration or clear native PAGE (36, 53). Coomassie stain may disrupt weak interactions.
Fig 2.
Structures of CIII (A) and CIV (B) showing the subunits at the candidate CIII-CIV interface. Cob is cytochrome b. The 2Fe-2S center in the Rieske protein Rip1 is shown as spheres. The structure shown is the bovine enzyme (1BGY) (27). In the CIV structure, the convex face subunits are shown in color (3ASO structure) (45). Bovine VIIa does not have a yeast equivalent. The bound cardiolipin is shown as green spheres.
Another scenario is that additional, non-ETC proteins may associate with the supercomplexes, providing further stabilization. A series of proteins have been reported that associate with the yeast respiratory supercomplexes. The major isoform of the ADP/ATP exchanger Aac2 is associated with the yeast CIII-CIV supercomplexes (11, 16). The interaction of Aac2 with the respiratory supercomplexes is dependent on the presence of cardiolipin (11). Cells lacking Aac2 exhibited reduced CIV activity and steady-state protein levels (16). In addition, the abundance of the III2-IV2 supercomplex was diminished, while the III2-IV1 supercomplex intensified (11, 16). A significant fraction of CIII was present not in supercomplexes but as the individual dimeric complex in aac2Δ cells. Additionally, Aac2 appears to be associated with CIV rather than CIII within the supercomplex (16). Under solubilization conditions with reduced digitonin levels, the fraction of Aac2 associated with the respiratory supercomplex becomes very prominent, and the TIM23 protein import machinery is also associated under these conditions (16, 37, 52). The association of Aac2 with the supercomplexes may impart stability to the complexes and may also facilitate the ATP/ADP exchange function of the translocator. Alternatively, Aac2 was reported to have an additional function in the import of protoporphyrin IX into mitochondria for heme synthesis (4). Since respiratory complexes are major heme proteins within mitochondria, the Aac2-CIII-CIV supercomplexes may coordinate sites of heme synthesis.
A subset of CIV biogenesis assembly factors are reported to be in association with the Aac2-containing CIII-CIV supercomplexes in yeast (33). Cox14 and ShyI function in the maturation of Cox1 prior to its association with Cox2 and Cox3. A small fraction of each of these assembly factors associates with the III2-IV2 and III2-IV1 supercomplexes, and these factors are also probably associated with assembly intermediates of the supercomplexes.
An evaluation of the physiological significance of these interactions and the significance of respiratory supercomplexes has been complicated by the lack of dedicated supercomplex assembly factors whose existence would enable evaluation of the importance of supercomplexes for mitochondrial respiration (14). Recently, three separate research groups reported the discovery of two proteins, Rcf1 and Rcf2 (where Rcf designates respiratory supercomplex factor), that appear to serve that function (9, 43, 50).
Strogolova et al. reported in this journal that purification of the yeast Aac2-CIII-CIV supercomplexes led to the identification of the two previously unannotated proteins Rcf1 and Rcf2 (43). Vukotic and colleagues identified Rcf1 and Rcf2 from the affinity purification of the CIII complex (50). Chen et al. discovered the link between Rcf1 and respiratory supercomplexes from the affinity purification of Rcf1 in an attempt to functionally define this previously unannotated yeast protein (9).
Rcf1 associates both with individual CIII and CIV complexes but appears to have a stronger association with CIV through its Cox3 subunit (mammalian subunit III) that is predicted to be at the CIII-CIV interface. Rcf1 is also associated with unassembled Cox3, although it does not appear to be a Cox3-stabilizing chaperone. The CIII-CIV supercomplexes isolated by affinity purification of Rcf1 also contain Aac2. With the addition of a chemical cross-linker, a specific Rcf1-Aac2 cross-linked adduct was isolated even from rho0 mitochondria, suggesting a close physical association of Rcf1 and Aac2 in mitochondrial membranes (43). While both Rcf1 and Rcf2 associate with CIII-CIV supercomplexes, Rcf1 and Rcf2 do not copurify (43). The implication is that a heterogeneous population of supercomplexes exists within yeast, some of which have Rcf1 and a distinct population having Rcf2.
Yeast lacking Rcf1 has reduced abundance of the III2-IV2 supercomplex, and CIV enzymatic activity was diminished without any appreciable change in steady-state CIV subunits (9). Although cells lacking Rcf2 are not significantly impaired with respect to supercomplex stability, rcf1Δ rcf2Δ double mutant cells are markedly compromised in supercomplex stability and CIV enzymatic activity (43). The CIV peripheral subunits Cox12 and Cox13 are destabilized in rcf1Δ cells, and this destabilization is augmented in rcf1Δ rcf2Δ cells (43, 50). Thus, the binding of these peripheral subunits in CIV is dependent on Rcf1 and the dissociation of these subunits in rcf1Δ cells probably accounts for the diminished CIV activity (30). Cox12 is located close to the cytochrome c binding site in CIV, so activity impairment may relate to the efficiency of electron transfer by cytochrome c.
One apparent consequence of the impaired formation of the III2-IV2 supercomplex in rcf1Δ cells is the elevation of reactive oxygen species (ROS) (9, 50). The mutant cells have diminished aconitase activity and are sensitive to hydrogen peroxide in growth tests. If the electron transfer function of the CIII-CIV supercomplexes is attenuated, side reactions generating superoxide anion may be enhanced, leading to the oxidative stress phenotype in rcf1Δ cells.
Mammalian cells contain a family of Rcf1-related proteins, one of which is highly induced under hypoxic conditions (28). Thus, the family designation is HIG, for hypoxia-induced gene. Depletion of one HIG protein, HIG2A, in a mouse myoblast cell line led to the destabilization of high-mass CIV supercomplexes (9). Thus, Rcf1 and Rcf2 may have a conserved function in metazoans in regulating supercomplex formation and/or stability.
SUPERCOMPLEX ASSEMBLY
Studies addressing the pathway for supercomplex formation suggest that supercomplexes are assembled in stages (35). CIII and CIV complexes appear to be assembled independent of each other. In contrast, CI is assembled in stages (34) and supercomplex formation occurs prior to the final stage in maturation, namely, the addition of the NADH dehydrogenase N module (35). This supercomplex assembly intermediate lacks respirasome activity. The final addition of the N module appears to occur on this supercomplex assembly intermediate. One proposed function of the respirasome is as a platform for the sequential assembly of CI (35). This observation provides an alternative interpretation of the presentation of multicomplex deficiencies in patients with mutations within a single respiratory complex. The interpretation of the candidate stabilizing interactions' interconnections between the complexes given above under Composite Nature of Respiratory Supercomplexes may not be the major answer. Defects in CIII or CIV may result in CI deficiency through impaired CI assembly.
Another intriguing CI-CIV interconnection based on a human cybrid cell line containing a truncated CIV Cox1 subunit was recently reported (25). The truncation conferred a marked steady-state CIV instability; although the mutant CIV assembled within the usual CI-CIII-CIV supercomplexes, stability was compromised, leading to a relative increase in the CI-CIII supercomplex. The steady-state levels of CI were attenuated in the mutant cybrid cells. Compromising the mAAA quality control protease within the IM was able to partially restore CIV levels.
Supercomplex intermediates can also form, as mentioned previously, in cells stalled in the maturation of CIII or CIV. The final step in CIII biogenesis is the addition of the Rieske Fe/S catalytic subunit, and cells impaired in Rieske maturation form supercomplexes (13, 21). The last step in the maturation of both CI and CIII is the addition of a key catalytic subunit or module. This may ensure that the supercomplex intermediates are not activated until all components are present.
SUMMARY
A heterogeneous mixture of respiratory supercomplexes exists in eukaryotic cells, and these assemblages are sealed together by lipids and accessory proteins. Rcf1 and Rcf2 are novel components of the mitochondrial ETC supercomplexes, and these factors separately associate with supercomplexes. As with Aac2 and the cardiolipin lipid, Rcf1 and Rcf2 are important for the stability of the assemblage. Their roles in supercomplex stability open the possibility that respiration may be modulated through these factors. The induction of one mammalian homolog by hypoxia conditions may result in remodeling of respiratory supercomplexes under varying growth conditions. The observation that CI maturation may occur in the context of the supercomplex suggests that Rcf1 may also modulate CI assembly.
Rcf1 appears to link the CIII and CIV components within the supercomplexes, and its presence stabilizes the binding of Cox12 and Cox13 peripheral subunits. The observation that Rcf1 can readily cross-link with Aac2 suggests that Aac2 may also exist near the CIII-CIV interface. As mentioned above, modeling of EM tomographs predicts that an edge of the convex face of CIV associates with CIII (24). This face, showing the candidate subunits involved in the CIII interaction, is shown in Fig. 2. In the supercomplex model shown in Fig. 1, CIV subunits III (Cox3), VIa (Cox13), and VIIa are prominent at the CIII interface. Since Cox12 and Cox13 are destabilized in rcf1Δ cells, Rcf1, as well as Aac2, may associate with Cox3 either on the convex CIV face near the bound cardiolipin (Fig. 2B) at the CIII-CIV interface or, alternatively, on the concave CIV face. Rcf1 is a small, 18-kDa protein with two candidate transmembrane helices and a long C-terminal tail projecting into the intermembrane space (IMS) (43). This long C-terminal tail is not conserved and is not essential for the interaction with CIV. Higher-resolution structures of the supercomplexes are needed to identify the positions of Rcf1/2 and Aac2 within the complex.
One unresolved question concerns why the loss of Rcf1 (or HIG2A) leads to depletion of the III2-IV2 supercomplex in yeast and the I-III2-IV2 supercomplex in mouse cells. The selective loss of the I-III2-IV2 supercomplex in mouse cells depleted of HIG2A may be accounted for by the asymmetric association of CIII-CIV on the membrane arm of CI. However, the basis for the loss of the yeast III2-IV2 but not the III2-IV1 supercomplex in yeast must arise from a different phenomenon.
The Rcf1-stabilized III2-IV2 supercomplex may allow more efficient electron transfer between CIII and CIV by cytochrome c, thereby restricting ROS generation. One major function of supercomplex formation appears to be this limitation of ROS production. The apparent greater efficiency of electron transfer within respirasomes is probably not due to direct electron channeling between donor/acceptor sites, based on EM models. Supercomplexes may restrict the diffusion space for ubiquinol and cytochrome c (17), yet this conclusion was recently challenged by the observation in yeast showing that cytochrome c is not restricted in its diffusion (47).
Although CII has been observed in supercomplexes, further work is required to verify the composition of such assemblages and its structural position. In addition, no information is available on whether the flavoprotein-ubiquinone oxidoreductase (ETF-QO) forms respiratory supercomplexes with CIII and CIV.
ACKNOWLEDGMENTS
D.R.W. is supported by grants ES03817 and GM083292.
I appreciate discussions with Natalya Dudkina on the cryo-EM projection maps and permission to use the atomic coordinate model from her image reconstruction. I appreciate comments from Rosemary Stuart and my research group.
Footnotes
Published ahead of print 14 May 2012
REFERENCES
- 1. Acin-Perez R, et al. 2004. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13:805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. 2008. Respiratory active mitochondrial supercomplexes. Mol. Cell 32:529–539 [DOI] [PubMed] [Google Scholar]
- 3. Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. 2011. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 30:4652–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Azuma M, et al. 2008. Adenine nucleotide translocator transports haem precursors into mitochondria. PLoS One 3:e3070 doi:10.1371/journal.pone.0003070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boekema EJ, Braun HP. 2007. Supramolecular structure of the mitochondrial oxidative phosphorylation system. J. Biol. Chem. 282:1–4 [DOI] [PubMed] [Google Scholar]
- 6. Boumans H, Grivell LA, Berden JA. 1998. The respiratory chain in yeast behaves as a single functional unit. J. Biol. Chem. 273:4872–4877 [DOI] [PubMed] [Google Scholar]
- 7. Bruel C, Brasseur R, Trumpower BL. 1996. Subunit 8 of the Saccharomyces cerevisiae cytochrome bc1 complex interacts with succinate-ubiquinone reductase complex. J. Bioenerg. Biomembr. 28:59–68 [PubMed] [Google Scholar]
- 8. Budde SM, et al. 2000. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem. Biophys. Res. Commun. 275:63–68 [DOI] [PubMed] [Google Scholar]
- 9. Chen YC, et al. 2012. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 15:348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clason T, et al. 2010. The structure of eukaryotic and prokaryotic complex I. J. Struct. Biol. 169:81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. 2008. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182:937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Covian R, Trumpower BL. 2008. Regulatory interactions in the dimeric cytochrome bc1 complex: the advantages of being a twin. Biochim. Biophys. Acta 1777:1079–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. 2000. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275:18093–18098 [DOI] [PubMed] [Google Scholar]
- 14. D'Aurelio M, Gajewski CD, Lenaz G, Manfredi G. 2006. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Hum. Mol. Genet. 15:2157–2169 [DOI] [PubMed] [Google Scholar]
- 15. Diaz F, Fukui H, Garcia S, Moraes CT. 2006. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 26:4872–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dienhart MK, Stuart RA. 2008. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol. Biol. Cell 19:3934–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. 2005. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. U. S. A. 102:3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. 2011. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 108:15196–15200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Efremov RG, Baradaran R, Sazanov LA. 2010. The architecture of respiratory complex I. Nature 465:441–445 [DOI] [PubMed] [Google Scholar]
- 20. Eubel H, Heinemeyer J, Sunderhaus S, Braun HP. 2004. Respiratory chain supercomplexes in plant mitochondria. Plant Physiol. Biochem. 42:937–942 [DOI] [PubMed] [Google Scholar]
- 21. Fernandez-Vizarra E, et al. 2007. Impaired complex III assembly associated with BCS1L gene mutations in isolated mitochondrial encephalopathy. Hum. Mol. Genet. 16:1241–1252 [DOI] [PubMed] [Google Scholar]
- 22. Gomez LA, Monette JS, Chavez JD, Maier CS, Hagen TM. 2009. Supercomplexes of the mitochondrial electron transport chain decline in the aging rat heart. Arch. Biochem. Biophys. 490:30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guenebaut V, Schlitt A, Weiss H, Leonard K, Friedrich T. 1998. Consistent structure between bacterial and mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 276:105–112 [DOI] [PubMed] [Google Scholar]
- 24. Heinemeyer J, Braun HP, Boekema EJ, Kouril R. 2007. A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 282:12240–12248 [DOI] [PubMed] [Google Scholar]
- 25. Hornig-Do HT, et al. 2012. Nonsense mutations in the COX1 subunit impair the stability of respiratory chain complexes rather than their assembly. EMBO J. 31:1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hunte C, Zickermann V, Brandt U. 2010. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329:448–451 [DOI] [PubMed] [Google Scholar]
- 27. Iwata S, et al. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281:64–71 [DOI] [PubMed] [Google Scholar]
- 28. Kasper LH, Brindle PK. 2006. Mammalian gene expression program resiliency: the roles of multiple coactivator mechanisms in hypoxia-responsive transcription. Cell Cycle 5:142–146 [DOI] [PubMed] [Google Scholar]
- 29. Keilin D, Hartree EF. 1947. Activity of the cytochrome system in heart muscle preparations. Biochem. J. 41:500–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LaMarche AE, Abate MI, Chan SH, Trumpower BL. 1992. Isolation and characterization of COX12, the nuclear gene for a previously unrecognized subunit of Saccharomyces cerevisiae cytochrome c oxidase. J. Biol. Chem. 267:22473–22480 [PubMed] [Google Scholar]
- 31. Lange C, Hunte C. 2002. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc. Natl. Acad. Sci. U. S. A. 99:2800–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y, et al. 2007. An assembled complex IV maintains the stability and activity of complex I in mammalian mitochondria. J. Biol. Chem. 282:17557–17562 [DOI] [PubMed] [Google Scholar]
- 33. Mick DU, et al. 2007. ShyI couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26:4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. 2012. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta. 1817:851–862 [DOI] [PubMed] [Google Scholar]
- 35. Moreno-Lastres D, et al. 2012. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 15:324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pfeiffer K, et al. 2003. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278:52873–52880 [DOI] [PubMed] [Google Scholar]
- 37. Saddar S, Dienhart MK, Stuart RA. 2008. The F1F0-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J. Biol. Chem. 283:6677–6686 [DOI] [PubMed] [Google Scholar]
- 38. Schafer E, et al. 2006. Architecture of active mammalian respiratory chain supercomplexes. J. Biol. Chem. 281:15370–15375 [DOI] [PubMed] [Google Scholar]
- 39. Schagger H. 2001. Respiratory chain supercomplexes. IUBMB Life 52:119–128 [DOI] [PubMed] [Google Scholar]
- 40. Schagger H, Pfeiffer K. 2001. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 276:37861–37867 [DOI] [PubMed] [Google Scholar]
- 41. Schägger H, Pfeiffer K. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19:1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Slater EC. 2003. Keilin, cytochrome, and the respiratory chain. J. Biol. Chem. 278:16455–16461 [DOI] [PubMed] [Google Scholar]
- 43. Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. 2012. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 32:1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stroh A, et al. 2004. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J. Biol. Chem. 279:5000–5007 [DOI] [PubMed] [Google Scholar]
- 45. Suga M, et al. 2011. Distinguishing between Cl- and O22− as the bridging element between Fe3+ and Cu2+ in resting-oxidized cytochrome c oxidase. Acta Crystallogr. D Biol. Crystallogr. 67:742–744 [DOI] [PubMed] [Google Scholar]
- 46. Sun F, et al. 2005. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121:1043–1057 [DOI] [PubMed] [Google Scholar]
- 47. Trouillard M, Meunier B, Rappaport F. 2011. Questioning the functional relevance of mitochondrial supercomplexes by time-resolved analysis of the respiratory chain. Proc. Natl. Acad. Sci. U. S. A. 108:E1027–E1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsukihara T, et al. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272:1136–1144 [DOI] [PubMed] [Google Scholar]
- 49. Vogel F, Bornhovd C, Neupert W, Reichert AS. 2006. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175:237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vukotic M, et al. 2012. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 15:336–347 [DOI] [PubMed] [Google Scholar]
- 51. Watmough NJ, Frerman FE. 2010. The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim. Biophys. Acta 1797:1910–1916 [DOI] [PubMed] [Google Scholar]
- 52. Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. 2007. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179:1115–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang M, Mileeykovskaya E, Dowhan W. 2002. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277:43553–43556 [DOI] [PubMed] [Google Scholar]