Abstract
The yeast high-osmolarity glycerol response (HOG) mitogen-activated protein (MAP) kinase pathway is activated in response to hyperosmotic stress via two independent osmosensing branches, the Sln1 branch and the Sho1 branch. While the mechanism by which the osmosensing machinery activates the downstream MAP kinase cascade has been well studied, the mechanism by which the machinery senses and responds to hyperosmotic stress remains to be clarified. Here we report that inhibition of the de novo sphingolipid synthesis pathway results in activation of the HOG pathway via both branches. Inhibition of ergosterol biosynthesis also induces activation of the HOG pathway. Sphingolipids and sterols are known to be tightly packed together in cell membranes to form partitioned domains called rafts. Raft-enriched detergent-resistant membranes (DRMs) contain both Sln1 and Sho1, and sphingolipid depletion and hyperosmotic stress have similar effects on the osmosensing machinery of the HOG pathway: dissociation of an Sln1-containing protein complex and elevated association of Sho1 with DRMs. These observations reveal the sphingolipid-mediated regulation of the osmosensing machinery of the HOG pathway.
INTRODUCTION
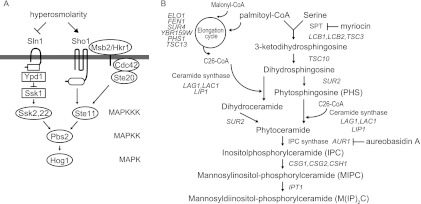
The osmolarity response is a fundamental stress response that is observed in all cells. The high-osmolarity glycerol response (HOG) mitogen-activated protein kinase (MAPK) pathway in Saccharomyces cerevisiae plays a central role in survival and adaptation in high-osmolarity environments (Fig. 1A) (25). In S. cerevisiae, high-osmolarity stress induces rapid activation of the MAPK Hog1 through phosphorylation by the MAPK kinase (MAPKK) Pbs2. The HOG pathway has two osmosensing branches, the Sln1 branch and the Sho1 branch, and these two branches independently activate Pbs2. In the Sln1 branch, the transmembrane histidine kinase Sln1, which is a homolog of bacterial two-component signal transducers, serves as the osmosensor. Under normal-osmolarity conditions, Sln1 forms a dimer and autophosphorylates a histidine residue in its own histidine kinase domain. This phosphate group is sequentially transferred to an aspartic acid residue in the Sln1 receiver domain, then to a histidine residue in the histidine phosphotransfer protein Ypd1, and finally to an aspartic acid residue in the response regulator Ssk1. High osmolarity inactivates Sln1 and thereby increases unphosphorylated Ssk1, which, in turn, binds to and activates the redundant MAPKK kinases (MAPKKKs) Ssk2 and Ssk22. Activated Ssk2 and Ssk22 then phosphorylate and activate Pbs2 (18, 27, 28, 35). In the Sho1 branch, the recently identified mucin-like proteins Msb2 and Hkr1 interact with, and transmit the signal to, the tetraspanning protein Sho1. Sho1 then serves as a facilitator of a signaling module assembly that includes Cdc42, Ste20, MAPKKK Ste11, Ste50, and Pbs2 (36, 37). This assembly leads to Ste11 activation by Cdc42-activated Ste20 and to subsequent Pbs2 activation by thus-activated Ste11. Although Sln1 is proposed to monitor changes in turgor pressure induced by osmotic stress (29), the exact mechanism by which the osmosensing machinery, especially that of the Sho1 branch, senses and responds to osmotic stress, and by which it is kept inactive in normal osmotic environments, remains to be clarified. To address these issues, we performed a genetic screen to isolate mutants that exhibit constitutive activation of the HOG pathway in normal osmotic environments. We found that a mutation in LCB2, which encodes a component of serine palmitoyltransferase (SPT) (19), results in activation of the HOG pathway. Yeast SPT is comprised of Lcb1, Lcb2, and a minor subunit, Tsc3, and catalyzes the first step of sphingolipid biosynthesis, condensation of serine with palmitoyl coenzyme A (palmitoyl-CoA) (Fig. 1B). SPT is essential for yeast growth and is conserved among all organisms that make sphingolipids.
Fig 1.
(A) Schematic representation of the HOG pathway. (B) Outline of the de novo sphingolipid synthesis pathway in S. cerevisiae. The target processes of two inhibitors (myriocin and aureobasidin A) are also shown.
Sphingolipids and sterols are tightly packed together with specific proteins in cell membranes, resulting in the formation of partitioned membrane domains called rafts or membrane microdomains (15). Rafts have been suggested to have roles in cell signaling and membrane trafficking through directly changing local membrane structure or by allowing protein-protein interactions to occur with higher affinity or specificity, thereby functioning as platforms for up- or downregulation of these processes (6, 8). Recent studies have shown that certain proteins are transiently associated with rafts, by either entering or leaving these compartments in response to various stimuli such as ligand binding, heat, or oxidative stress (3, 26, 34). These observations suggest that rafts dynamically control physiological events, including stress responses, via regulation of the localization of those proteins. While it has been suggested that heat stress affects raft structure through hyperfluidization of the membrane (20), whether and how other stresses, including osmotic stress, act on rafts and lead to cellular responses remain unknown.
In this study, we found that inhibition of sphingolipids or ergosterol biosynthesis induces activation of the HOG pathway. We also show that both Sln1 and Sho1 are distributed in raft-enriched detergent-resistant membranes (DRMs). Furthermore, sphingolipid depletion and osmotic stress similarly lead to dissociation of an Sln1-containing protein complex and elevated association of Sho1 with DRMs. Based on these observations, we suggest that rafts are involved in the regulation of the osmosensing mechanisms of the HOG pathway.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The yeast strains used are listed in Table 1. All strains used were in the S288C background except where noted otherwise. SHO1, SSK2, and SSK22 genes were disrupted as previously described (17). The STE11 gene in MH273 and MH322 was disrupted by transformation with pNC276 (31), and that in MH276 was disrupted by replacement with a loxP-kanMX-loxP cassette that was amplified by PCR as described previously (9). MH293, MH296, MH305, MH302, and MH314 were constructed by replacing ELO1, CSG1, or SUR2 of TM100, CSH1 of TM101, and FEN1 of TM141, respectively, with corresponding kanMX4 cassettes that were amplified with PCR using genomic DNA of the knockout collection strains as templates. MH417 and MH425 were constructed by replacing ERG3 of TM225 and ERG2 of TM141, respectively, with His3MX6 cassettes that were amplified by PCR as previously described (16). MH309, MH311, and MH421 were constructed by crossing MH296 with MH302, TM221 with 5281, and MH280 with MH417, respectively. Epitope tags and enhanced green fluorescent protein (EGFP) were attached to the C termini of Sho1 and Sln1 using a standard PCR-based gene modification (12, 16). Chromosomal LCB2 was replaced with lcb2-M1 using a standard two-step gene replacement method (33). MH335 and MH337 were constructed by crossing MH329 with MH331. For all experiments except for those shown in Fig. 2A and B and 6B, cells were grown to early log phase in yeast extract-peptone-dextrose (YPD) medium at 30°C. For Fig. 2A and B, the cells were cultured in synthetic complete media lacking Trp (SC-Trp) and SC-Trp Ura, respectively, supplemented with 180 μg/ml Leu medium overnight, and were spotted onto plates. For Fig. 6B, the cells were cultured in SC-Ura medium to mid-log phase.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| TM415 | MATa leu2Δ1 trp1Δ63 his3Δ200 ade2 URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3 | L40-derived lab stock |
| TM415α | Same as TM415 except for MATα | Lab stock |
| MH266 | Same as TM415α except for lcb2-M1 | This study |
| TM100 | MATa ura3-52 leu2Δ1 trp1Δ63 | 17 |
| TM101 | MATα ura3-52 leu2Δ1 his3Δ200 | 18 |
| TM141 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 | 17 |
| TM221 | MATα ade8 | Lab stock |
| TM225 | MATα ura3-52 leu2Δ1 his3Δ200 lys2Δ202 | 11 |
| TM252 | MATa ura3-52 leu2Δ1 trp1Δ63 ssk2::LEU2 ssk22::LEU2 | 18 |
| TM287 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 sho1::HIS3 | Lab stock |
| TM370 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 ssk2::LEU2 ssk22::LEU2 sho1::TRP1 | Lab stock |
| TM537 | MATa ura3-52 leu2Δ1 trp1Δ63 erg3::His3MX6 | Lab stock |
| TM540 | MATa ura3-52 leu2Δ1 trp1Δ63 erg6::His3MX6 | Lab stock |
| 4007 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 ipt1::kanMX4 | Open Biosystems |
| 5281 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 sur4::kanMX4 | Open Biosystems |
| MH273 | MATα ura3-52 leu2Δ1 his3Δ200 ste11::URA3 | This study |
| MH276 | MATa ura3-52 leu2Δ1 trp1Δ63 ssk2::LEU2 ssk22::LEU2 ste11::loxP-kanMX-loxP | This study |
| MH280 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lcb2-M1 | This study |
| MH293 | MATa ura3-52 leu2Δ1 trp1Δ63 elo1::kanMX4 | This study |
| MH296 | MATa ura3-52 leu2Δ1 trp1Δ63 csg1::kanMX4 | This study |
| MH302 | MATα ura3-52 leu2Δ1 his3Δ200 csh1::kanMX4 | This study |
| MH305 | MATa ura3-52 leu2Δ1 trp1Δ63 sur2::kanMX4 | This study |
| MH309 | MATα ura3-52 leu2Δ1 trp1Δ63 csg1::kanMX4 csh1::kanMX4 | This study |
| MH311 | MATa leu2Δ0 his3Δ1 met15Δ0 sur4::kanMX4 | This study |
| MH314 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 fen1::kanMX4 | This study |
| MH317 | Same as MH280 except for ssk2::LEU2 ssk22::LEU2 | This study |
| MH319 | Same as MH280 except for sho1::TRP1 | This study |
| MH321 | Same as TM370 except for lcb2-M1 | This study |
| MH322 | Same as MH280 except for ste11::URA3 | This study |
| MH324 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 SLN1-EGFP::His3MX6 | This study |
| MH329 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 SHO1-GFP::His3MX6 lcb2-M1 | This study |
| MH331 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 SLN1-9myc::His3MX6 | This study |
| MH335 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 SLN1-9myc::His3MX6 SHO1-GFP::His3MX6 | This study |
| MH337 | Same as MH335 except for lcb2-M1 | This study |
| MH417 | MATα ura3-52 leu2Δ1 his3Δ200 lys2Δ202 erg3::His3MX6 | This study |
| MH421 | MATa ura3-52 leu2Δ1 his3Δ200 lys2Δ202 lcb2-M1 erg3::His3MX6 | This study |
| MH425 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 erg2::His3MX6 | This study |
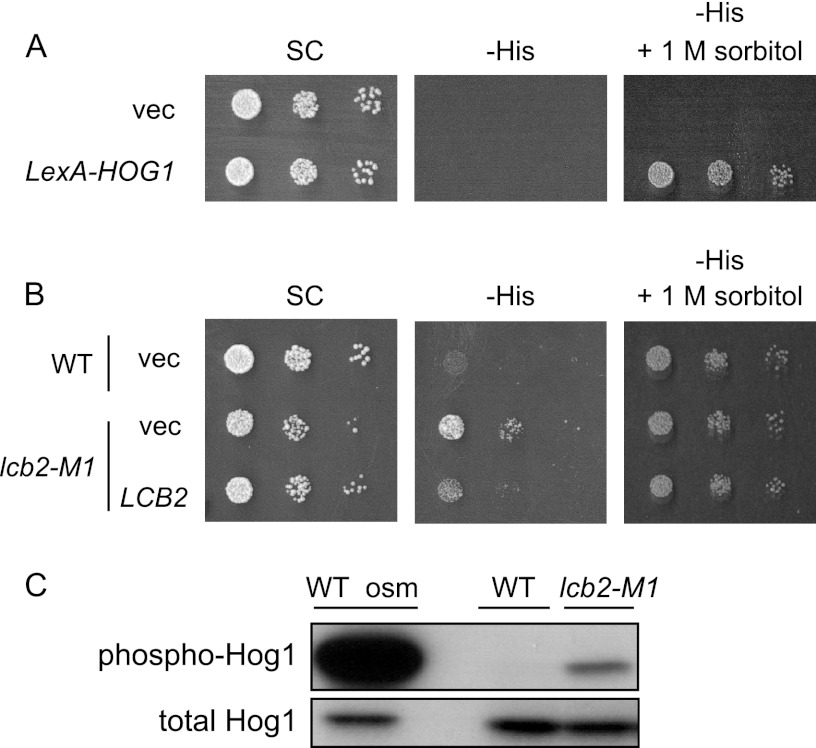
Fig 2.
Screen for mutants exhibiting constitutive activation of the HOG pathway. (A) The parental strain, which has a LexAop-HIS3 reporter gene and a LexA-Hog1 expressing plasmid, grows on His-depleted medium in an osmotic stress-dependent manner. TM415, harboring the vector (p414-LexA) or the LexA-HOG1 coding plasmid (pMT304), was spotted onto SC-Trp (SC), SC-Trp His (-His), and SC-Trp His supplemented with 1 M sorbitol (-His + 1 M sorbitol) plates and incubated for 2 days at 30°C. (B) The lcb2-M1 mutant cells grow on the -His plate in the absence of osmotic stress. Wild-type (TM415, WT) and lcb2-M1 (MH266) cells, harboring pMT304 and a vector (pRS416) or an LCB2 plasmid (pMH29), were spotted onto SC-Trp Ura (SC) and SC-Trp His Ura (-His) plates and incubated for 2 days at 30°C. (C) lcb2 mutant cells exhibit elevated Hog1 phosphorylation. Phosphorylated Hog1 in wild-type (TM141) and lcb2-M1 (MH280) cells was detected by Western blotting using an anti-phospho-p38 specific antibody. Wild-type cells treated with 0.4 M KCl for 3 min (WT osm) were used as a positive control.
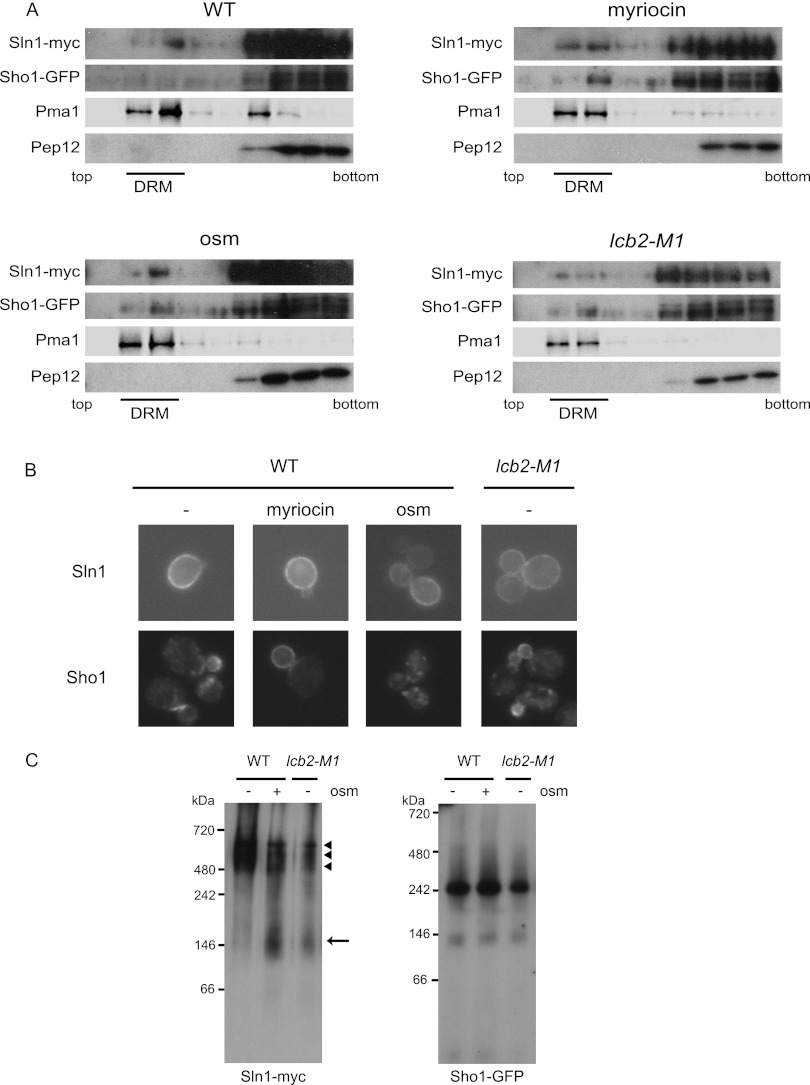
Fig 6.
DRM localization and complex formation of Sln1 and Sho1. (A) Sln1 and Sho1 localize in DRMs. SLN1-9myc SHO1-GFP (MH335) cells that were treated with 0.4 M KCl for 5 min (osm) or with 2 μg/ml myriocin for 2.5 h (myriocin) and untreated wild-type (MH335, WT) or lcb2-M1 SLN1-9myc SHO1-GFP (MH337, lcb2-M1) cells were disrupted, and their membrane fractions were extracted with 20 mM CHAPS and subjected to Optiprep density gradient centrifugation. Nine fractions were collected from the top and were analyzed by Western blotting using anti-myc, -GFP, -Pma1, and -Pep12 antibodies. Pma1 was used as a marker for DRM-associated proteins and Pep12 as a marker for non-DRM-associated membrane proteins. Fractions 2 and 3 contain DRMs. (B) Cellular localization of Sln1 and Sho1 in sphingolipid synthesis-deficient cells. Wild-type (TM141, WT) and lcb2-M1 (MH280) cells harboring an SLN1-GFP- or an SHO1-GFP-encoding plasmid were grown in SC-Ura. Wild-type cells were treated or not treated (−) with 2 μg/ml myriocin for 2.5 h (myriocin) or 0.4 M KCl for 5 min (osm). (C) Detection of Sln1 and Sho1 complexes by BN-PAGE. The membranes of SLN1-9myc SHO1-GFP (MH335, WT) cells and lcb2-M1 SLN1-9myc SHO1-GFP (MH337, lcb2-M1) cells that had been treated (osm +) or not treated (osm −) with 0.4 M KCl for 5 min were isolated, solubilized with 1% digitonin, and then subjected to BN-PAGE. After electrophoresis, Western blotting was performed using anti-myc and anti-GFP antibodies. An arrow and arrowheads indicate the fast- and the slow-migrating species of Sln1, respectively.
Plasmids.
pMT304 was created as follows. BamHI sites were introduced both upstream and downstream of the HOG1 open reading frame (ORF) by PCR, and the fragment was cloned into the BamHI site of pBTM116 (40) to create pNS493. pNS493 was digested with SphI, and the fragment containing LexA-HOG1, together with the ADH1 promoter and the ADH1 terminator, was subcloned into the SphI site of YCplac111 to create pMT301. Finally, the 2.7-kb XbaI-PvuII fragment of pMT301 was cloned into the SpeI-HincII sites of pRS414. The NaeI-ScaI fragment containing lacZ driven by eight tandem repeats of the ENA1-derived CRE sequence (8×CRE-lacZ) of pKT760 (gift from Haruo Saito, the University of Tokyo) was introduced into the NaeI-ScaI site of pRS415 to create pMH41. To create pMH29, a genomic EcoRI-ApaLI (blunted) fragment containing LCB2 was cloned into the EcoRI-SmaI sites of pRS416. pFP15 (pRS416-SHO1-EGFP) was a gift from Haruo Saito. An SLN1-EGFP fragment containing the SLN1 promoter region was obtained from MH324 by a gap repair method using pPD2146 (18) as a recipient and was cloned into pRS426 to create pMH54.
Selection for Hog1-activating mutants.
TM415 carrying pMT304 was cultivated in SC-Trp supplemented with 180 μg/ml Leu medium overnight, mutagenized with ethyl methanesulfonate (EMS) as described previously (11) (viability, 55%), diluted, and plated onto SC-Trp His plates supplemented with 180 μg/ml Leu. After 3 days of incubation at 30°C, candidate His+ mutants were selected and retested on the same plates. The 467 His+ mutants thus selected from 2.5 × 105 mutagenized cells were then subjected to Western blotting in order to identify mutants that exhibit elevated Hog1 phosphorylation compared with wild-type cells, and 24 mutants were selected. Among these, mutants that showed temperature-sensitive growth as well as relatively high Hog1 phosphorylation were crossed with TM415α and were subjected to tetrad analysis to test their monogenic segregation patterns, as well as cosegregation of two phenotypes: Hog1 phosphorylation and temperature-sensitive growth. Only one mutant met these conditions, and a healthy tetrad product of the backcross (MH266) was used for subsequent experiments.
Treatment of cells with sphingolipids, inhibitors, or filipin.
Five or 10 μM phytosphingosine (PHS) (Sigma, St. Louis, MO) or 10 μM C2-ceramide (Sigma) was added to the culture of TM141 (wild-type) or MH280 (lcb2-M1) cells, and the cells were incubated for 1 to 4 h at 30°C. Two micrograms/milliliter myriocin (Sigma) and 500 ng/ml aureobasidin A (TaKaRa Bio, Shiga, Japan) were added to the culture of TM141 or MH335 (SLN1-9myc SHO1-GFP) cells, which were then incubated for various times at 30°C. Five micrograms/milliliter filipin complex (Sigma) was added to the logarithmically growing YPD culture of TM141, TM252 (ssk2Δ ssk22Δ), or TM287 (sho1Δ) cells.
Detection of phosphorylated Hog1, Fus3, and Kss1.
Cells were harvested by centrifugation at 1,200 × g for 30 s, suspended in Laemmli sample buffer, and immediately boiled for 4 min. Samples were then centrifuged at 9,000 × g for 3 min, and the supernatant was subjected to SDS-PAGE. After electrophoretic transfer onto polyvinylidene difluoride (PVDF) membranes, the membranes were immunoblotted with the polyclonal anti-phospho-p38 antibody D3F9 (Cell Signaling Technology, Danvers, MA), the polyclonal anti-Hog1 antibody yC-20 (Santa Cruz Biotechnology, Santa Cruz, CA), the polyclonal anti-phospho-p44/p42 MAPK antibody 9101 (Cell Signaling Technology), or the polyclonal anti-Kss1 antibody y-50 (Santa Cruz Biotechnology). Membranes were then treated with the appropriate secondary antibody conjugated with horseradish peroxidase, anti-rabbit IgG (NA934; GE Healthcare, Little Chalfont, United Kingdom), anti-goat IgG (sc-2056; Santa Cruz Biotechnology), or anti-mouse IgG (NA931; GE Healthcare), and the signal was developed using ECL Plus (GE Healthcare).
Quantification of sphingolipid levels.
Yeast cells were precultured in 2 ml YPD medium containing 10 μCi of [9,10-3H]palmitic acid (60 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO), which had been complexed with 4 mg/ml (final concentration of 0.4 mg/ml) fatty acid-free bovine serum albumin (BSA; Sigma) for 18 h at 30°C. Cells were then diluted into 0.5 ml YPD medium containing 2.5 μCi [9,10-3H]palmitic acid complexed with BSA as described above to an absorbance at 600 nm of 0.15 and grown for 8 h at 30°C. After labeling, lipid extraction and alkaline treatment were performed as described previously (13). Lipids were separated by thin-layer chromatography (TLC) on Silica Gel 60 high-performance TLC plates (Merck, Whitestation, NJ) with chloroform-methanol–4.2 N ammonia (9:7:2, vol/vol) as the solvent system and visualized by autoradiography. Densitometry and calibration of the autoradiogram were performed using the NIH ImageJ software (http://rsbweb.nih.gov/ij/).
Northern blot analysis.
Total RNA was isolated from logarithmically growing cells that were treated or not treated with 0.4 M KCl for 1 h or with 2 μg/ml of myriocin for 4 h. A total of 12 μg of RNA per lane were separated in formaldehyde gels and transferred to a positively charged nylon membrane (Pall-Life Sciences, Ann Arbor, MI). The GRE2 and HSP12 probes were amplified from genomic DNA by PCR, labeled with digoxigenin using DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, Mannheim, Germany), and hybridized with the membrane according to the manufacturer's instructions.
Fluorescence microscopy.
Logarithmically growing cells carrying pMH54 (pRS426-SLN1-EGFP) or pFP15 (pRS416-SHO1-EGFP) were microscopically observed. Images were captured with an Olympus BX51 microscope (Olympus Optical, Tokyo, Japan) using a U-MNIBA2 filter (Olympus). Image acquisition was performed using a 1394-based digital camera (Hamamatsu Photonics, Tokyo, Japan) and the AQUA-Lite software program (Hamamatsu Photonics).
Isolation of DRMs and Western blotting.
DRMs were isolated essentially as described by Umebayashi and Nakano (38), with the following modifications. All procedures were carried out on a scale three times larger than that of the protocol of Umebayashi and Nakano. After 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) extraction, the lysates were subjected to Optiprep density gradient flotation by centrifugation for 14 h at 170,000 × g in an SW40 Ti rotor (Beckman Instruments, Fullerton, CA) at 4°C. After centrifugation, nine fractions of equal volume were collected starting from the top. Each fraction was mixed with Laemmli sample buffer, incubated at 37°C for 10 min, and resolved by SDS-PAGE. Western blotting was performed as described above using the following primary antibodies: anti-GFP (Roche, Indianapolis, IN), anti-myc (9E10, ascites, gift from Yutaka Hoshikawa, the Cancer Institute of Japanese Foundation for Cancer Research), anti-Pep12 (2C3; Invitrogen, Carlsbad, CA), and anti-Pma1 (40B7; Abcam, Cambridge, United Kingdom).
Detection of membrane protein complexes by blue native PAGE.
Logarithmically growing cells cultured in YPD medium were collected, suspended in breaking buffer (50 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 40 μg/ml aprotinin, 10 μg/ml pepstatin A, 20 μg/ml leupeptin), and then disrupted by vortexing with 425- to 600-μm-diameter glass beads (Sigma). After dilution with an equal amount of breaking buffer, the sediment of glass beads was removed and the supernatant was centrifuged at 1,200 × g for 5 min. After the supernatant was centrifuged at 64,300 × g for 1 h, the pellets were washed once with breaking buffer and were then suspended in solubilization buffer (50 mM imidazole [pH 7.0], 50 mM NaCl, 2 mM 6-aminohexanoic acid, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 40 μg/ml aprotinin, 10 μg/ml pepstatin A, 20 μg/ml leupeptin). Membrane protein complexes were solubilized by addition of digitonin (final concentration, 1%), and the sample was centrifuged at 20,000 × g for 20 min. A 1/40 volume of 5% CBB dye stock (5% CBB G-250, 500 mM 6-aminohexanoic acid) was added to the supernatant, and blue native PAGE (BN-PAGE) was carried out as described previously (41) using a 3 to 12% acrylamide gradient gel. After electrophoresis, the gel was washed with the Laemmli running buffer for 10 min, and then Western blotting was performed as described above.
RESULTS
Screen for mutants exhibiting constitutive activation of the HOG pathway.
To identify novel regulatory components of the HOG pathway, we performed a genetic screen aimed at isolating mutants that exhibit constitutive activation of the HOG pathway in normal osmotic environments. We utilized the LexA-Hog1 reporter system for this screen (1). It has been shown that phosphorylated Hog1 is recruited to specific osmostress-responsive promoters in the nucleus and induces their expression by facilitating recruitment of the transcription initiation complex (1). Thus, the LexA-Hog1 fusion protein can induce transcription of the LexA operator-fused HIS3 reporter when Hog1 is phosphorylated upon osmotic stress, thereby supporting growth of the reporter strain on a histidine-depleted medium (Fig. 2A) (1). The reporter strain was mutagenized using EMS, and mutants were selected that grew on a histidine-depleted medium without osmotic stress. We then identified mutants in which Hog1 was activated by directly examining the phosphorylation status of Hog1. One of these isolated mutants (MH266) showed temperature-sensitive growth (data not shown), and LCB2 was the gene responsible for this phenotype, as well as for the histidine prototrophy (Fig. 2B). MH266 has a mutation in LCB2 (lcb2-M1) in which Ala377 is replaced by Val. Lcb2 is a component of SPT, the enzyme that is responsible for the first committed step in sphingolipid de novo synthesis (Fig. 1B), and is essential for cell viability. To confirm that the observed phenotype was indeed caused by this mutation, and that the phenotype was not specific to the parental strain used for the screen, we introduced the lcb2-M1 mutation into an S288C-derived wild-type strain. The resultant lcb2-M1 cells (MH280) also exhibited elevated phosphorylation of Hog1 (Fig. 2C) as well as temperature-sensitive growth (Fig. 3C). These results indicate that a single lcb2-M1 mutation is sufficient to induce activation of the HOG pathway.
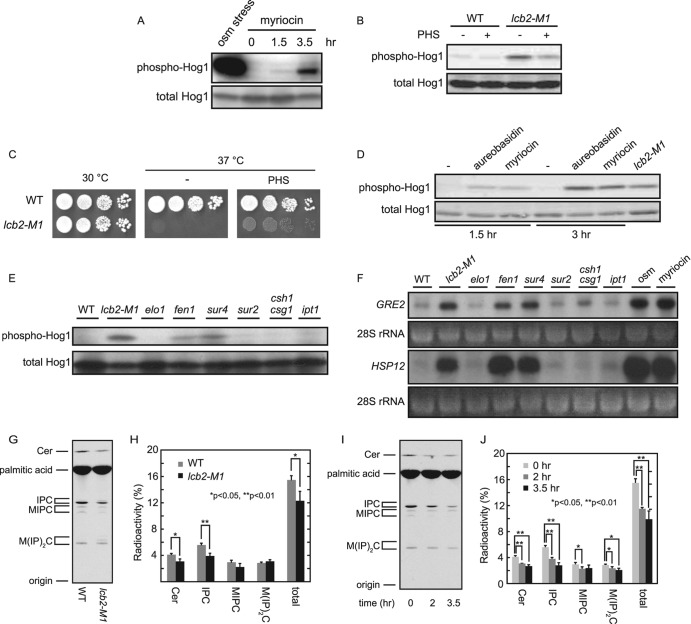
Fig 3.
Inhibition of the sphingolipid biosynthetic pathway induces activation of the HOG pathway. (A, B, D, and E) Hog1 phosphorylation was assayed by Western blotting for phospho-Hog1 and total Hog1. (A) Myriocin treatment induces phosphorylation of Hog1. Wild-type (TM141) cells were treated or not treated with 2 μg/ml of myriocin for the indicated times or with 0.4 M KCl for 3 min (osm stress), followed by Western blotting. (B) Exogenous addition of PHS attenuates elevated Hog1 phosphorylation in lcb2-M1 cells. Wild-type (TM141) and lcb2-M1 (MH280) cells were cultivated with YPD in the absence or presence of 5 μM PHS for 4 h and were then subjected to Western blotting. (C) Temperature-sensitive growth of lcb2-M1 cells is partially rescued by exogenous addition of PHS. Wild-type (TM141) and lcb2-M1 (MH280) cells were spotted onto YPD plates with or without supplementation with 5 μM PHS and were incubated at the indicated temperatures for 3 days. (D) Inhibition of IPC synthase by aureobasidin A treatment induces Hog1 phosphorylation. Wild-type (TM141) cells were treated with 500 ng/ml aureobasidin A or 2 μg/ml myriocin for the indicated times and were then subjected to Western blotting. lcb2-M1 cells were used as a positive control. (E and F) Mutant cells in which complex sphingolipid synthesis is inhibited show activation of the HOG pathway. Wild-type (TM141), lcb2-M1 (MH280), elo1Δ (MH293), fen1Δ (MH314), sur4Δ (MH311), sur2Δ (MH305), csg1Δ csh1Δ (MH309), and ipt1Δ (4007) cells were tested. For panel E, Hog1 phosphorylation of the indicated strains was examined by Western blotting. For panel F, transcript levels of GRE2 and HSP12 in the indicated strains were monitored by Northern analysis. The ethidium bromide-stained rRNA was used as a loading control. Wild-type cells were treated or not treated with 0.4 M KCl for 1 h (osm) or with 2 μg/ml myriocin for 4 h (myriocin) before the cells were collected. (G) Decreased sphingolipid levels in lcb2-M1 mutant cells. Wild-type (TM141) and lcb2-M1 (MH280) cells were cultured in YPD containing [3H]palmitic acid for 26 h at 30°C. Labeled lipids were extracted, treated with alkaline to hydrolyze ester linkages in glycerolipids, separated by TLC, and detected by autoradiography. (H) Quantification of sphingolipid levels that were labeled as in panel G. Radioactivity associated with sphingolipids was quantified from the autoradiogram. Values represent the means ± standard deviations from three independent experiments. (I) Myriocin treatment results in decreased sphingolipids levels. Wild-type (TM141) cells were cultured in YPD containing [3H]palmitic acid for 26 h at 30°C. Myriocin (5 μM) was added 0, 2, or 3.5 h prior to termination of the labeling. Labeled lipids were detected as in panel G. (J) Quantification of sphingolipid levels that were labeled as in panel I. The data were presented as in panel H.
Inhibition of complex sphingolipid synthesis results in activation of the HOG pathway.
To confirm that inhibition of SPT induces activation of the HOG pathway, the effect of a specific pharmacological inhibitor of SPT, myriocin, was assayed (Fig. 1B). Like lcb2-M1 cells, wild-type cells treated with myriocin exhibited elevated phosphorylation of Hog1 (Fig. 3A). We next tested if activation of the HOG pathway in lcb2-M1 cells could be reversed by rescuing sphingolipid metabolism. It was reported that exogenous addition of some sphingolipid synthesis intermediates, including phytosphingosine (PHS), enables lcb1Δ or lcb2Δ cells to grow (4, 19). We therefore examined whether PHS addition can also suppress the activation of the HOG pathway in lcb2-M1 cells. As expected, addition of PHS attenuated the level of Hog1 phosphorylation in lcb2-M1 cells (Fig. 3B), as well as partially rescued their temperature-sensitive growth (Fig. 3C). These results indicate that depletion of sphingolipids results in activation of the HOG pathway.
Ceramide, a metabolite of sphingolipids, activates protein phosphatases termed ceramide-activated protein phosphatases (CAPPs) in yeast and mammals. We therefore tested if CAPPs are involved in regulation of the HOG pathway. CDC55 encodes a regulatory B subunit of protein phosphatase type 2A (PP2A) and is essential for yeast CAPP activity (22). CDC55-disrupted cells exhibited neither elevated Hog1 phosphorylation nor expression of 8×CRE-lacZ, a HOG pathway reporter gene (data not shown). Furthermore, addition of C2-ceramide, which activates yeast CAPP in vitro (22), to wild-type cells did not suppress myriocin-induced activation of the HOG pathway (data not shown). These results suggest that Hog1 phosphorylation induced by depletion of sphingolipids is not mediated by failure to activate known CAPPs.
Next, in order to identify which sphingolipid metabolites are necessary for maintaining the HOG pathway in an inactive state in normal osmotic environments, we inhibited various steps of the de novo sphingolipid synthesis pathway by using a specific inhibitor or by disrupting genes involved in this pathway (see Fig. 1B). Wild-type cells treated with aureobasidin A, an inositolphosphorylceramide (IPC) synthase inhibitor, as well as fen1Δ and sur4Δ cells exhibited elevated Hog1 phosphorylation (Fig. 3D and E). Consistently, fen1Δ and sur4Δ cells induced transcription of Hog1 target genes, GRE2 and HSP12 (Fig. 3F) (5, 39). All these conditions under which the HOG pathway is activated show impaired synthesis of complex sphingolipids, including IPC, mannosylinositolphosphorylceramide (MIPC), and mannosyldiinositolphosphorylceramide [M(IP)2C]. In addition, elevated Hog1 phosphorylation and induced transcription of Hog1 target genes were hardly observed in csg1Δ csh1Δ and ipt1Δ cells, suggesting that MIPC and M(IP)2C are not required for maintaining the HOG pathway in an inactive state. Therefore, IPC appears to be the critical sphingolipid for proper regulation of the HOG pathway. It also should be noted that sur2Δ cells exhibited neither elevated Hog1 phosphorylation nor induction of GRE2 and HSP12 genes (Fig. 3E and F). Sur2 hydroxylates dihydrosphingosine to form phytosphingosine (Fig. 1B). Hence, sur2Δ cells lack mature forms of IPC but contain immature forms of IPC that lack a hydroxyl group at the C-4 position of the long-chain base (10). This suggests that IPC is required, but mature forms of IPC are not required, for proper regulation of the HOG pathway.
It is unlikely for the following reasons that disturbance of each biosynthetic step results in abnormal accumulation of intermediate metabolites, which, in turn, leads to activation of the HOG pathway. First of all, inhibition of SPT, which reduces the overall synthesis of sphingolipids, causes activation of the HOG pathway. Second, although fen1Δ and sur4Δ cells accumulate PHS (24), wild-type cells treated with 10 μM PHS, which is a toxic concentration for yeast, did not alter the Hog1 phosphorylation level (data not shown). The combined data suggest that it is the abundance of complex sphingolipids, rather than the proper balance of their intermediate metabolites, that appears to be necessary for proper regulation of the HOG pathway.
The lcb2-M1 mutation and myriocin treatment caused only moderate Hog1 activation compared with osmotic stress, estimated at about 1/10 to 1/20 (Fig. 2C and 3A and data not shown). This can be due to modest effects of these conditions on sphingolipid levels or modest effects of sphingolipid depletion on Hog1 activation. To test these possibilities, we quantified the sphingolipid levels of cells under these conditions. Total sphingolipid level in lcb2-M1 cells was 79% that of wild-type cells, where the decrease of IPC was most obvious (70%) among complex sphingolipids (Fig. 3G and H). Total sphingolipid levels in myriocin-treated (2 μg/ml) wild-type cells were 74% (2 h) and 64% (3.5 h) those of untreated cells (Fig. 3I and J). These results suggest that the modest Hog1 activation under these conditions resulted from their moderate effects on sphingolipid levels.
SPT inhibition induces activation of Hog1 via both the Sln1 and the Sho1 branches.
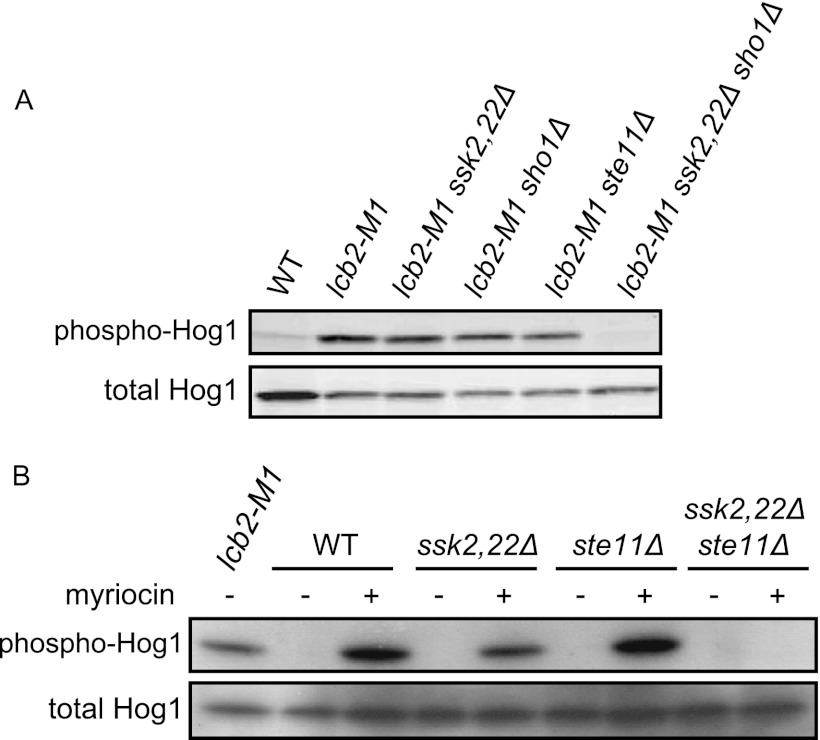
The HOG pathway has two osmosensing branches, the Sln1 and the Sho1 branches (Fig. 1A). We next investigated which of these branches is activated by impaired biosynthesis of sphingolipids. Both lcb2-M1 cells with ssk2Δ ssk22Δ mutations and those with an sho1Δ or an ste11Δ mutation still exhibited Hog1 phosphorylation, while lcb2-M1 ssk2Δ ssk22Δ sho1Δ cells did not (Fig. 4A). These mutants are defective in the Sln1 branch alone, the Sho1 branch alone, and both branches, respectively. In addition, myriocin treatment induced activation of Hog1 in either ssk2Δ ssk22Δ or ste11Δ cells, but not in ssk2Δ ssk22Δ ste11Δ cells (Fig. 4B). These results indicate that SPT inhibition causes activation of both the Sln1 and the Sho1 branches.
Fig 4.
SPT inhibition induces activation of Hog1 via both the Sln1 and the Sho1 branches. ssk2Δ ssk22Δ is abbreviated as ssk2,22Δ. (A) Wild-type (TM141, WT), lcb2-M1 (MH280), lcb2-M1 ssk2,22Δ (MH317), lcb2-M1 sho1Δ (MH319), lcb2-M1 ste11Δ (MH322), and lcb2-M1 ssk2,22Δ sho1Δ (MH321) cells were subjected to Western blotting for phospho-Hog1 and total Hog1. (B) Wild-type (TM141, WT), ssk2,22Δ (TM252), ste11Δ (MH273), and ssk2,22Δ ste11Δ (MH276) cells were treated with 2 μg/ml myriocin for 3 h and then subjected to Western blotting as in panel A. lcb2-M1 (MH280) cells were used as a positive control.
Inhibition of ergosterol synthesis results in activation of the HOG pathway.
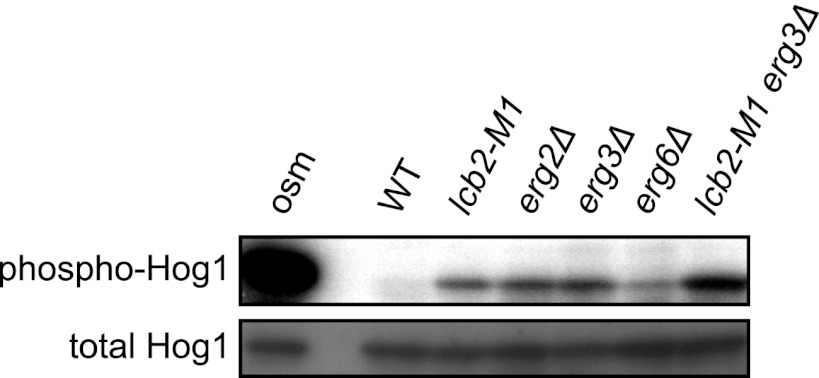
The above observations indicate that depletion of sphingolipids activates the osmosensing machinery of the HOG pathway. Sphingolipids, together with ergosterol, a major sterol in yeast, form detergent-insoluble membrane domains called rafts. We therefore hypothesized that depletion of sphingolipids perturbs raft structure and thereby leads to activation of the osmosensors. If this is the case, then depletion of ergosterol may also activate the HOG pathway. To test this possibility, we examined mutants of the ergosterol synthesis pathway. This study showed that erg2Δ, erg3Δ, and erg6Δ cells indeed exhibited elevated Hog1 phosphorylation (Fig. 5), supporting the possibility that perturbation of raft structure activates the HOG pathway. In addition, lcb2-M1 erg3Δ double mutant cells exhibited even higher Hog1 phosphorylation than cells of each single mutant (Fig. 5). This additive effect of two mutations may result from severer defects they may cause in raft structure than the single mutants.
Fig 5.
Inhibition of the ergosterol biosynthetic pathway induces activation of the HOG pathway. Wild-type (TM141), lcb2-M1 (MH280), erg2Δ (MH425), erg3Δ (TM537), erg6Δ (TM540), and lcb2-M1 erg3Δ (MH421) cells were subjected to Western blotting for phospho-Hog1 and total Hog1. Wild-type cells treated with 0.4 M KCl for 3 min (osm) were used as a positive control.
Sln1 and Sho1 localize in DRMs.
We next tested whether Sln1 and Sho1 localize in rafts. To this end, raft-enriched DRMs were separated using Optiprep density gradient centrifugation, and fully functional (29, 30) (data not shown) epitope-tagged Sln1 and Sho1 were probed for by Western blotting. Sln1 and Sho1 were indeed detected in the DRM fraction (Fig. 6A). The distribution of Sln1 into DRMs was not significantly affected by myriocin treatment or by lcb2-M1 mutation, although this result may be due to incomplete disruption of the raft structure under these conditions, as evidenced by the similar distribution of the Pma1, a typical raft marker (2), under these conditions to that in nonstressed wild-type cells (Fig. 6A). The distribution of Sln1 was not affected by osmotic stress either. In contrast, the amount of Sho1 distributed in DRMs was increased both in myriocin-treated cells and in lcb2-M1 cells (Fig. 6A). The correlation between the increased distribution of Sho1 in DRMs and activation of the Sho1 branch suggested that translocation of Sho1 to rafts may underlie the activation. We then tested if osmotic stress induces a similar change in Sho1 distribution. The amount of Sho1 in DRMs was moderately, but reproducibly, increased in cells exposed to osmotic stress (Fig. 6A). These results suggest that translocation of Sho1 to rafts may be an essential step in osmosensing by the Sho1 branch.
Next, we microscopically observed localization of GFP-tagged Sln1 and Sho1 in living cells (Fig. 6B). These proteins were shown to be functional in previous reports (29, 30). Sln1-GFP localized at the plasma membrane, and Sho1-GFP localized mainly at the bud tip or bud neck in a dot-like pattern, as previously reported (29, 30, 36). Neither lcb2-M1 mutation nor myriocin treatment apparently altered the localization of Sln1-GFP. On the other hand, myriocin treatment altered the localization of Sho1-GFP from a punctate pattern to plasma membrane uniformly. This mislocalization of Sho1 seemed to be irrelevant for activation of the Sho1 branch, because lcb2-M1 cells maintained intact localization of Sho1-GFP, despite the fact that the Sho1 branch is similarly activated in both lcb2-M1 cells and myriocin-treated cells. The different localization of Sho1-GFP between myriocin-treated and lcb2-M1 mutant cells might be dependent on the degree of SPT inhibition: sphingolipid synthesis was more strongly inhibited by myriocin treatment than by the lcb2-M1 mutation (Fig. 3G to J). Since sphingolipids are known to be necessary for endocytosis, myriocin might cause the delocalization of Sho1 through impaired endocytosis of Sho1 from the plasma membrane.
We then tested if sphingolipid depletion alters protein-protein interactions or complex formation of the Sln1 or the Sho1 branch. Cell membranes were solubilized with digitonin and separated by blue native PAGE (Fig. 6C). The lcb2-M1 mutation, as well as osmotic stress, increased the fast-migrating species, and decreased the slow-migrating species, of Sln1. This result suggested that both sphingolipid depletion and osmotic stress induce dissociation of the Sln1 complex. On the other hand, the migration pattern of Sho1 was not affected by lcb2-M1 mutation or by osmotic stress. The apparent lack of change in Sho1 may reflect the transient nature of the interaction of Sho1 with upstream or downstream components.
Filipin modulates the activation of the HOG pathway.
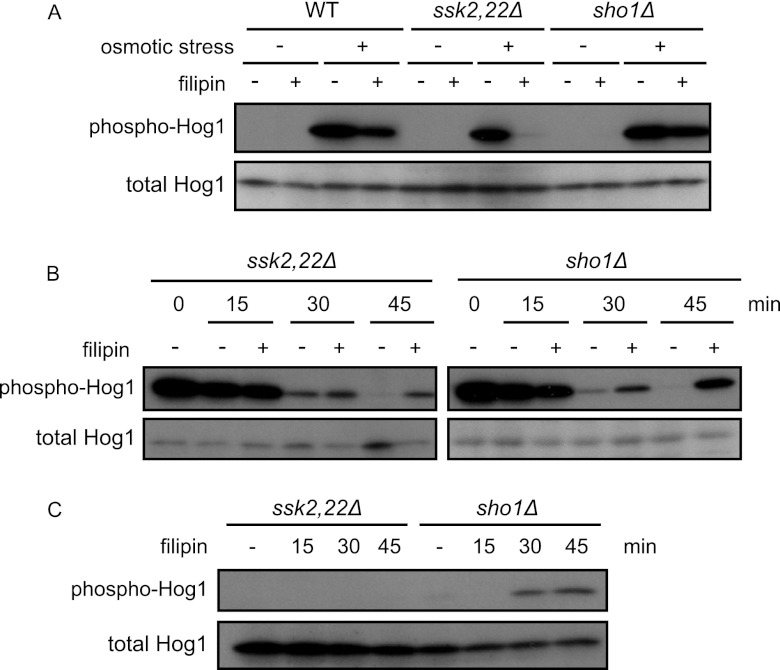
To further examine the possible relevance of rafts to the regulation of the HOG pathway, we tested the effects of filipin. Filipin is an antifungal polyene macrolide that stoichiometrically interacts and thereby forms stable complex with sterol (7, 23). When yeast cells are treated with filipin, the sterol-rich membrane compartment of Can1 (MCC), which appears to represent a specific type of rafts, is immobilized and lateral redistribution of the MCC-resident transporter Can1 is inhibited (7). We treated cells with filipin and then monitored activation of the HOG pathway upon osmotic stress. Filipin pretreatment hindered activation of the Sho1 branch (shown in ssk2Δ cells), while it hardly affected the Sln1 branch (shown in ssk22Δ sho1Δ cells) (Fig. 7A). On the other hand, when cells were treated in the opposite order, first with osmotic stress and then with filipin, phosphorylation of Hog1 was sustained longer than in untreated cells both in ssk2Δ ssk22Δ and sho1Δ cells (Fig. 7B). The sustained phosphorylation of Hog1 in filipin-treated ssk2Δ ssk22Δ cells implies that filipin treatment prolongs activation of the Sho1 branch induced by osmotic stress. In contrast, slow activation of the Sln1 branch induced by filipin treatment may account for the sustained phosphorylation of Hog1 in filipin-treated sho1Δ cells, as filipin treatment alone caused phosphorylation of Hog1 in sho1Δ cells, but not in ssk2Δ ssk22Δ cells, at later time points (Fig. 7C). These results further suggest that raft integrity affects the activation of the HOG pathway.
Fig 7.
Filipin treatment affects activation of the HOG pathway. (A) Filipin treatment impedes activation of the Sho1 branch. Wild-type (TM141, WT), ssk2,22Δ (TM252), and sho1Δ (TM287) cells were treated or not treated with 5 μg/ml filipin for 5 min and then treated with 0.4 M KCl. After 5 min of incubation, cells were collected and subjected to Western blotting. (B) Filipin treatment after exposure to osmotic stress sustains activation of the HOG pathway. ssk2,22Δ (TM252) and sho1Δ (TM287) cells were exposed to 0.4 M KCl and incubated for 5 min (0 min). Then, cells were treated with 5 μg/ml filipin, incubated for the indicated times, and collected and subjected to Western blotting. (C) Filipin treatment induces activation of the Sln1 branch. ssk2,22Δ (TM252) and sho1Δ (TM287) cells were treated with 5 μg/ml filipin for the indicated times and subjected to Western blotting.
The lcb2-M1 mutation causes activation of Fus3 and Kss1 in a Sho1-independent but Ste11-dependent manner.
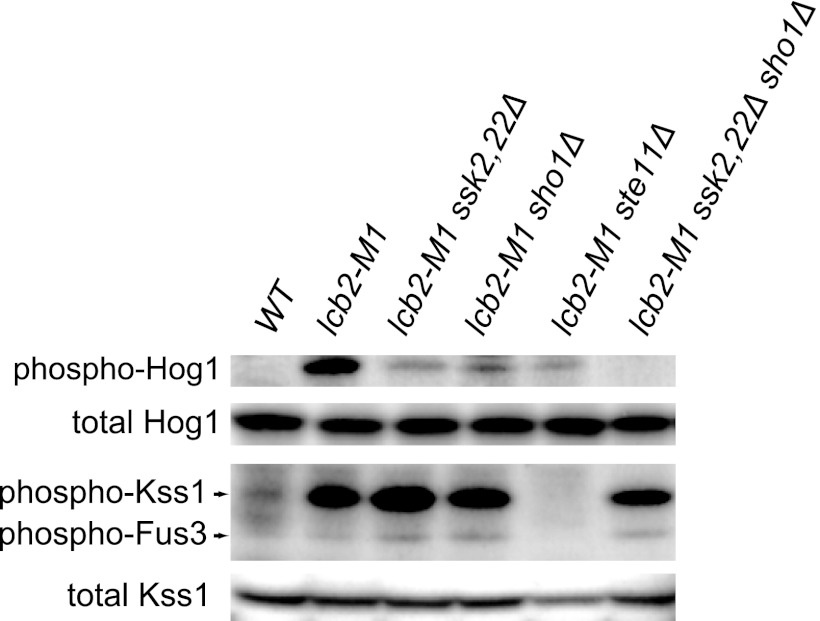
Finally, to examine whether sphingolipid depletion affects other MAPK pathways, phosphorylation of Fus3 and Kss1 was monitored in lcb2-M1 cells. Fus3 and Kss1 were indeed activated in lcb2-M1 cells in a Sho1-independent but Ste11-dependent manner (Fig. 8). These results suggest that sphingolipid depletion also causes activation of the mating pathway but not the filamentous growth pathway, as activation of the latter is strictly dependent on Sho1.
Fig 8.
The lcb2-M1 mutation causes activation of Kss1 and Fus3 in a Sho1-independent but Ste11-dependent manner. Wild-type (TM141, WT), lcb2-M1 (MH280), lcb2-M1 ssk2,22Δ (MH317), lcb2-M1 sho1Δ (MH319), lcb2-M1 ste11Δ (MH322), and lcb2-M1 ssk2,22Δ sho1Δ (MH321) cells were subjected to Western blotting using anti-phospho-p38 (which detects phospho-Hog1), anti-Hog1, anti-phospho-p44/p42 MAPK (which detects phospho-Kss1 and phospho-Fus3), and anti-Kss1 antibodies.
DISCUSSION
In this report, we show that inhibition of sphingolipid biosynthesis results in activation of the HOG pathway. lcb2-M1, fen1Δ, and sur4Δ mutations, as well as myriocin and aureobasidin A treatment, cause activation of the HOG pathway (Fig. 2 and 3). All of these cells show impaired synthesis of complex sphingolipids. On the other hand, csg1Δ csh1Δ and ipt1Δ mutations hardly activate the HOG pathway, suggesting that IPC is the critical sphingolipid for proper regulation of the HOG pathway (Fig. 3). We conclude that it is depletion of complex sphingolipids, rather than accumulation of minor sphingolipid biosynthetic intermediates, that causes activation of the HOG pathway. It is unlikely that the HOG pathway activation is caused by gross general defects in membrane structure, because growth of the lcb2-M1 strains is apparently normal at 30°C.
If this is the case, then how does depletion of sphingolipids result in activation of the HOG pathways? One possible explanation is that intact rafts organized with sphingolipids are required for proper regulation of the osmosensors or of other regulatory molecules. Our observations that inhibition of ergosterol biosynthesis also results in activation of the HOG pathway (Fig. 5), and that both Sln1 and Sho1 localize in DRMs (Fig. 6A), support this possibility, although we should keep in mind that association with DRMs is not necessarily an accurate criterion for raft residency, because detergent treatment could cause artificial aggregation of lipids and proteins (14).
Sphingolipid depletion, like osmotic stress, activates the HOG pathway via both the Sln1 and the Sho1 branches (Fig. 4). However, sphingolipid depletion affects each branch in a distinctive manner. In the case of the Sho1 branch, we observed that both sphingolipid depletion and osmotic stress promote association of Sho1 with DRMs. If this redistribution indeed reflects translocation of Sho1 to rafts, then it should be elucidated whether this translocation has a regulatory role in activation as well as in the osmosensing mechanism of the Sho1 branch. We also found that treatment of cells with filipin, which forms stable complex with sterols and therefore is suggested to immobilize raft structure, impedes osmotic stress-induced activation of the Sho1 branch (Fig. 7A). This is consistent with the idea that redistribution of Sho1 to rafts is a key step for activation of the Sho1 branch. Furthermore, filipin treatment after exposure to osmotic stress prolonged duration of the Sho1 branch activation (Fig. 7B). This might also be due to the fact that filipin treatment impedes Sho1 from leaving rafts and thereby sustains the active state of the Sho1 branch. These indirect pieces of evidence support the idea that redistribution of Sho1 to rafts has an important regulatory role in the activation of the Sho1 branch, although potential toxic or artifactual effects of filipin have to be kept in mind.
If translocation of Sho1 to rafts is a key step in the activation of the Sho1 branch, how can the translocation be regulated? The structural change induced by osmotic stress in Sho1 itself may change Sho1's affinity to rafts versus nonraft compartments. Alternatively, it is possible that other molecules that are regulated by rafts recruit Sho1 to rafts in response to stress, as Sho1 is known to interact with many components of the Sho1 branch. Cdc42 is a possible target of raft-mediated regulation. Recently, sphingolipids were reported to be necessary for activation of Fpk1 and -2 (32). Since Fpk1 and -2 are activators of the aminophospholipid flippase Dnf1 and -2 (21), which transport phosphatidylethanolamine (PtdEth) and phosphatidylserine (PtdSer) from the external leaflet to the cytosolic leaflet of a bilayer membrane, sphingolipid depletion may cause depletion of aminophospholipids from the cytosolic leaflet through downregulation of Dnf1 and -2. As it was also reported that aminophospholipids stimulate the GTPase-activating protein (GAP) activity of Rga1 and -2 toward Cdc42 in vitro (21), sphingolipid depletion may result in elevated activation of Cdc42 through failure in Rga1 and -2 activation. Our observation that sphingolipid depletion also causes activation of Fus3 and Kss1, which are MAPKs of another Cdc42-controlled MAPK pathway, the mating pathway, in a Sho1-independent but Ste11-dependent manner supports this idea (Fig. 8). As for the Sln1 branch, we have demonstrated that depletion of sphingolipids, as well as osmotic stress, caused partial dissociation of the Sln1 complex, although the DRM distribution of Sln1 and the localization of Sln1-GFP were apparently not affected (Fig. 6). It has been suggested that Sln1 is functional as a homodimer (18, 28, 35). We therefore propose the model that Sln1 has to be tightly packed in rafts, in which it forms a dimeric complex, in order to efficiently conduct phosphotransfer relay under nonstressed environments, and that osmotic stress or sphingolipid depletion perturbs the raft-mediated packing of the Sln1 complex, resulting in inhibition of phosphotransfer relay and thereby activation of the Sln1 branch.
In this study, we have demonstrated a possible link between osmotic stress-sensing and membrane sphingolipids. Sphingolipid depletion and osmotic stress induce similar effects on the osmosensing machinery of the HOG pathway, even though specific effects on the Sln1 and Sho1 branches are distinct. These observations may imply that yeast cells sense osmotic stress as changes in the structural and/or physical properties of rafts. Further study is necessary to solve the precise mechanism by which sphingolipids regulate the osmosensing machinery.
ACKNOWLEDGMENTS
We thank Takahito Wada for excellent technical assistance in the initial screening. We also thank Paul J. Cullen and Takehiko Yoko-o for valuable technical comments, Haruo Saito and Beverly Errede for plasmids, Yutaka Hoshikawa for an antibody, and all members of the Maeda laboratory for valuable support and discussions.
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas (KAKENHI 21025008) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), a Grant-in-Aid for Challenging Exploratory Research (KAKENHI 23651233) from the Japan Society for the Promotion of Science (JSPS), grants from the Noda Institute for Scientific Research and the Salt Science Research Foundation (no. 11D2) (all to T.M.), and a Grant-in-Aid for JSPS Fellows (to T.T.). T.T. was the recipient of the Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
Published ahead of print 14 May 2012
REFERENCES
- 1. Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F. 2003. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 22:2433–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagnat M, Kernen S, Shevchenko A, Shevchenko A, Simons K. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. U. S. A. 97:3254–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Broquet AH, Thomas G, Masliah J, Trugnan G, Bachelet M. 2003. Expression of the molecular chaperone Hsp70 in detergent-resistant microdomains correlates with its membrane delivery and release. J. Biol. Chem. 278:21601–21606 [DOI] [PubMed] [Google Scholar]
- 4. Buede R, Rinker-Schaffer C, Pinto WJ, Lester RL, Dickson RC. 1991. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J. Bacteriol. 173:4325–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garay-Arroyo A, Covarrubias AA. 1999. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast 15:879–892 [DOI] [PubMed] [Google Scholar]
- 6. Golub T, Wacha S, Caroni P. 2004. Spatial and temporal control of signaling through lipid rafts. Curr. Opin. Neurobiol. 14:542–550 [DOI] [PubMed] [Google Scholar]
- 7. Grossmann G, Opekarová M, Malinsky J, Weig-Meckl I, Tanner W. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grzybek M, Kozubek A, Dubielecka P, Sikorski AF. 2005. Rafts—the current picture. Folia Histochem. Cytobiol. 43:3–10 [PubMed] [Google Scholar]
- 9. Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haak D, Gable K, Beeler T, Dunn T. 1997. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272:29704–29710 [DOI] [PubMed] [Google Scholar]
- 11. Hayashi M, Fukuzawa T, Sorimachi H, Maeda T. 2005. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol. Cell. Biol. 25:9478–9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janke C, et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947–962 [DOI] [PubMed] [Google Scholar]
- 13. Kihara A, Sakuraba H, Ikeda M, Denpoh A, Igarashi Y. 2008. Membrane topology and essential amino acid residues of Phs1, a 3-hydroxyacyl-CoA dehydratase involved in very long-chain fatty acid elongation. J. Biol. Chem. 283:11199–11209 [DOI] [PubMed] [Google Scholar]
- 14. Lingwood D, Simons K. 2007. Detergent resistance as a tool in membrane research. Nat. Protoc. 2:2159–2165 [DOI] [PubMed] [Google Scholar]
- 15. Lingwood D, Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science 327:46–50 [DOI] [PubMed] [Google Scholar]
- 16. Longtine MS, et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 17. Maeda T, Takekawa M, Saito H. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554–558 [DOI] [PubMed] [Google Scholar]
- 18. Maeda T, Wurgler-Murphy SM, Saito H. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242–245 [DOI] [PubMed] [Google Scholar]
- 19. Nagiec MM, Baltisberger JA, Wells GB, Lester RL, Dickson RC. 1994. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc. Natl. Acad. Sci. U. S. A. 91:7899–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagy E, et al. 2007. Hyperfluidization-coupled membrane microdomain reorganization is linked to activation of the heat shock response in a murine melanoma cell line. Proc. Natl. Acad. Sci. U. S. A. 104:7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakano K, Yamamoto T, Kishimoto T, Noji T, Tanaka K. 2008. Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol. Biol. Cell 19:1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nickels JT, Broach JR. 1996. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 10:382–394 [DOI] [PubMed] [Google Scholar]
- 23. Norman AW, Demel RA, de Kruyff B, van Deenen LL. 1972. Studies on the biological properties of polyene antibiotics. Evidence for the direct interaction of filipin with cholesterol. J. Biol. Chem. 247:1918–1929 [PubMed] [Google Scholar]
- 24. Oh CS, Toke DA, Mandala S, Martin CE. 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272:17376–17384 [DOI] [PubMed] [Google Scholar]
- 25. O'Rourke SM, Herskowitz I, O'Shea EK. 2002. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18:405–412 [DOI] [PubMed] [Google Scholar]
- 26. Park SJ, et al. 2009. Oxidative stress induces lipid-raft-mediated activation of Src homology 2 domain-containing protein-tyrosine phosphatase 2 in astrocytes. Free Radic. Biol. Med. 46:1694–1702 [DOI] [PubMed] [Google Scholar]
- 27. Posas F, Saito H. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17:1385–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Posas F, et al. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865–875 [DOI] [PubMed] [Google Scholar]
- 29. Reiser V, Raitt DC, Saito H. 2003. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J. Cell Biol. 161:1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reiser V, Salah SM, Ammerer G. 2000. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2:620–627 [DOI] [PubMed] [Google Scholar]
- 31. Rhodes N, Connell L, Errede B. 1990. STE11 is a protein kinase required for cell-type-specific transcription and signal transduction in yeast. Genes Dev. 4:1862–1874 [DOI] [PubMed] [Google Scholar]
- 32. Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. 2010. A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. U. S. A. 107:34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rothstein R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281–301 [DOI] [PubMed] [Google Scholar]
- 34. Suzuki KG, et al. 2007. GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177:717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tao W, Malone CL, Ault AD, Deschenes RJ, Fassler JS. 2002. A cytoplasmic coiled-coil domain is required for histidine kinase activity of the yeast osmosensor, SLN1. Mol. Microbiol. 43:459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tatebayashi K, et al. 2007. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 26:3521–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tatebayashi K, et al. 2006. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 25:3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Umebayashi K, Nakano A. 2003. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 161:1117–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Varela JC, Praekelt UM, Meacock PA, Planta RJ, Mager WH. 1995. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol. Cell. Biol. 15:6232–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vojtek AB, Hollenberg SM, Cooper JA. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205–214 [DOI] [PubMed] [Google Scholar]
- 41. Wittig I, Braun HP, Schägger H. 2006. Blue native PAGE. Nat. Protoc. 1:418–428 [DOI] [PubMed] [Google Scholar]