Abstract
Epigenetic mechanisms maintain the specific characteristics of differentiated cells by ensuring the inheritance of gene expression patterns through DNA replication and mitosis. We examined the mechanism of epigenetic inheritance of Sir protein-dependent transcriptional silencing in Saccharomyces cerevisiae by examining gene expression and molecular markers of silencing at the silent mating type loci under conditions of limiting Sir3 protein. We observed that silencing at HMR, as previously reported for HML, is epigenetically inherited. This inheritance is accompanied by an increased ability of previously silenced cells to retain or recruit limiting Sir3 protein to cis-acting silencer sequences. We also observed that the low H4-K16 histone acetylation and H3-K79 methylation associated with a silenced HMR locus persist in recently derepressed cells for several generations at levels of Sir3 insufficient to maintain these marks in long-term-derepressed cells. The unique ability of previously silenced cells to retain Sir3 protein, maintain silencing-specific histone modifications, and repress HMR transcription at levels of Sir3 insufficient to mediate these effects in long-term-derepressed cells suggests that a cis-acting, chromatin-based mechanism drives epigenetic inheritance at this locus.
INTRODUCTION
Differentiation into distinct cell types involves establishing unique patterns of gene expression. Once established, epigenetic mechanisms operate to maintain these patterns as cells divide. Chromatin structure plays a fundamental role in dictating gene expression states; chromatin-based models for epigenetic inheritance propose that nucleosomes bearing specific histone modifications are randomly distributed to sister chromatids during DNA replication. In these models specific histone modifications help recruit the enzymes responsible for creating the modification; these enzymes then modify adjacent new nucleosomes and recapitulate the starting chromatin state (for reviews, see references 21 and 32).
The budding yeast Saccharomyces cerevisiae uses a transcriptional silencing mechanism to regulate genes controlling its developmental fate. Silencing depends on the locus-specific action of the Sir proteins (for reviews, see references 10 and 42). Once recruited to cis-acting silencer sequences, the Sir2 protein deacetylates histone H3 and H4 tails on adjacent nucleosomes (48, 49). This deacetylation increases the affinity of the Sir3 and Sir4 proteins for these tails (14, 24). Reiterative binding of the Sir complex and histone deacetylation provide a model for Sir protein spreading across the silenced region. Mutations that weaken silencing, such as deletion of the SIR1 gene (37) or specific mutations in the silencers (26) or RAP1 (46), reveal an epigenetic pattern of silencing at the HML locus, in which previously silenced cells are far more likely to be silenced in subsequent generations than previously unsilenced cells. Silencing at yeast telomeres also exhibits an epigenetic pattern of inheritance (1).
The presence of a positive-feedback loop in Sir-protein-dependent silencing, in which a Sir2-induced histone modification recruits Sir3 and Sir4, which in turn can recruit Sir2, is consistent with the chromatin-based epigenetic model described above. However, basic predictions of these models have not been tested. To examine the possibility that the epigenetic pattern observed for yeast silencing is due to a self-templating mechanism, we examined the stability of silencing at the HML and HMR loci under conditions of steadily decreasing Sir3 protein levels. We found evidence that silencing at HMR, like at HML, is also epigenetically inherited and that this inheritance is accompanied by an increased ability of previously silenced cells to retain or recruit limiting Sir3 protein to silencer sequences. We also observed that the low H4-K16 histone acetylation and H3-K79 methylation associated with a silenced HMR locus persist in recently derepressed cells for several generations at levels of Sir3 insufficient to maintain these marks in long-term-derepressed cells.
MATERIALS AND METHODS
Media.
All the strains were grown in YPraffinose medium (1% Bacto yeast extract, 2% Bacto peptone extract, and 2% raffinose). To induce expression of GAL-SIR3, galactose was added to YPraffinose medium to 2%. For solid medium, Bacto agar was added to 2%.
Strains.
Strains used in this study are listed in Table 1. To create strains YSH831 and YSH832, the SIR3 alleles of BY4735 (3) and YSH811 (28) were tagged at their C terminus with a 9-myc epitope (23). The MAT locus of YSH494 (16) was replaced with the hphMX4 (hygromycin resistance) gene (12) to create YSH829. To make YSH1089, we first excised URA3 from TELVR in YSH832 and then inserted the GFP-lacI gene at the URA3 locus using plasmid pAFS152 (45), creating strain YSH1088. An HMR proximal fragment (chromosome III coordinates 296200 to 297039, SGD) was cloned into plasmid pAFS59.1 (45) at the KpnI-SacI sites to create plasmid pTM2. This plasmid was cut with BglII and then integrated into YSH1088, introducing an ∼10-kb array of 256 lac operators ∼2 kb downstream of HMR, creating YSH1089.
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| YSH556 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ppr1Δ::HIS3 URA3-TELVR |
| YSH829 | MATa ade2 lys1 his5 leu2 can1 sir3Δ::LEU2 ura3::URA3-sir3-8 2μm mataΔ::HYG |
| YSH831 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 SIR3-9MYC KAN |
| YSH832 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 met15Δ0 ura3Δ0 ppr1Δ::HIS3 sir3Δ::NAT1 mataΔ::HYG URA3-TELVR trp1Δ63::GAL10p-SIR3-9MYC-KAN-TRP1 |
| YSH872 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ppr1Δ::HIS3 URA3-TELVR sir3Δ::NAT1 mataΔ::HYG |
| YSH1088 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 met15Δ0 ura3Δ0 ppr1Δ::HIS3 sir3Δ::NAT1 mataΔ::HYG trp1Δ63::GAL10p-SIR3-9MYC-KAN-TRP1 13GFP-LacI-URA3 |
| YSH1089 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 met15Δ0 ura3Δ0 ppr1Δ::HIS3 sir3Δ::NAT1 mataΔ::HYG trp1Δ63::GAL10p-SIR3-9MYC-KAN-TRP1 13GFP-LacI-URA3 HMR-I-tDNA-256xlacop-LEU2 |
To examine repression at HML and HMR under conditions of limiting Sir3 protein, we grew strain YSH832 to early log phase in YPraffinose medium, at which time Sir3 production was induced with galactose for 16 h. During this induction phase and throughout the subsequent time course of this experiment, the culture was diluted in fresh medium approximately every 2 h to ensure that the culture remained in early log phase (optical density at 600 nm [OD600], between 0.1 and 0.5). After galactose induction the cells were washed by filtration and resuspended in YPraffinose medium for the duration of the experiment.
qPCR.
RNA extraction, cDNA synthesis, and quantitative PCR (qPCR) were performed as previously described (28). Primers specific for HMRa1, HMLα1, and ACT1 are listed in Table 2. ACT1 was used as an internal control. An average of three independent experiments is shown in the illustrations, unless otherwise indicated.
Table 2.
Primers used in this study
| Primer no. | Primer sequence | Region amplified and method(s)a |
|---|---|---|
| SP65 | GGCGGAAAACATAAACAGAACTCTG | HMRa1, RT-PCR and ChIP (B) |
| SP66 | CCGTGCTTGGGGTGATATTGATG | |
| SP221 | CCAGATTCCTGTTCCTTCC | HMLα1, RT-PCR and ChIP (D) |
| SP222 | GTCCCATATTCCGTGCTG | |
| SP 236 | CTGAATTAACAATGGATTCTG | ACT1, RT-PCR |
| SP 237 | CATCACCAACGTAGGAGTC | |
| SP638 | ATCGTTATGTCCGGTGGTACC | ACT1, ChIP |
| SP639 | TGGAAGATGGAGCCAAAGC | |
| SP1335 | TGCAAAAACCCATCAACCTTG | HMR-E, ChIP (A) |
| SP1336 | ACCAGGAGTACCTGCGCTTA | |
| SP1337 | GGATGGATCTAGGGTTTTATGCC | HML-E, ChIP (C) |
| SP1338 | TTTGGCCCCCGAAATCG |
Probe (A, B, C, or D) used in ChIP assay is indicated in parentheses.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (29). Immunoprecipitation was carried out using the following antibodies: 1 μl myc-epitope antibody (9B11; Cell Signaling Technology), 2.5 μl anti-acetyl-histone H4 (K16) antibody (07-329; Millipore), and 4 μl anti-dimethyl-histone H3 (K79) antibody (ab3594; Abcam). Primers for ChIP are listed in Table 2. Enrichments relative to an endogenous control were calculated as described previously (29). An average of three independent experiments is shown in the illustrations, unless otherwise indicated.
Western blotting.
Protein was extracted from whole cells using the trichloroacetic acid (TCA) precipitation method as described previously (51). Whole-cell protein extracts were fractionated on 12% polyacrylamide-SDS gels, transferred to nitrocellulose membranes (Amersham), and probed with anti-myc antibody (clone 9E11; Chemicon International; or clone 9B11; Cell Signaling Technology) and with anti-α-tubulin antibody (YOL1/34, sc-53030; Santa Cruz Biotechnology Inc.). Secondary detection was performed using horseradish peroxidase coupled to goat anti-mouse secondary antibody (sc-2005; Santa Cruz Biotechnology Inc.) and goat anti-rat secondary antibody (sc-2006; Santa Cruz Biotechnology Inc.). A chemifluorescent reagent (Amersham ECL plus Western blotting detection reagent RPN 2132 from GE Healthcare) was used for detection of the protein, and the membrane was scanned by using a Storm 840 PhosphorImager (GE Healthcare) or Typhoon 9400 (GE Healthcare). The Sir3p bands were quantified with respect to corresponding α-tubulin bands using Image-Quant software. An average of three independent experiments is shown in the illustrations.
Immunostaining and microscopy.
Semisquash preparations were adapted from published protocols (18, 39) with minor modifications. Cultures were grown to log phase (OD600, 0.3 to 0.5), 5 ml was removed, and formaldehyde was added to a final concentration of 4%. Fixation was carried out at room temperature for 1 h. Cells were then washed with 1 ml of 1% potassium acetate (KAc)–1 M sorbitol solution and collected by centrifugation (2,000 rpm for 4 min). The pellet was resuspended in 500 μl 1% KAc–1 M sorbitol solution, and 10 μl 1 M dithiothreitol and 20 μl of a Zymolyase 100T (Seikagaku Co., Tokyo) stock solution (10 mg/ml) were added to carry out spheroplasting at 37°C. After 30 to 40 min, digestion was halted by the addition of 500 μl stop solution consisting of 0.1 M 2-(N-morpholino)ethane acid (MES), 1 mM EDTA, 0.5 mM MgCl2, and 1 M sorbitol in distilled water. Cells were then collected by centrifugation, and the pellets were washed in 1 ml stop solution. Cell pellets were resuspended in 80 μl cold MES solution (0.1 M MES, 1 mM EDTA, 0.5 mM MgCl2), and 200 μl of fixative (4% paraformaldehyde and 3.4% sucrose) was added. The mixture was evenly spread over the surface of the slide with a coverslip, and the slides were processed for immunostaining.
Immunostaining was performed as described previously (18), using a mouse monoclonal antibody against Nsp1p (ab4641; Abcam) at a 1:100 dilution to mark the nuclear periphery and Alexa Fluor 568–goat anti-mouse IgG (H+L) (A11004; Molecular Probes) at a 1:200 dilution as the secondary antibody.
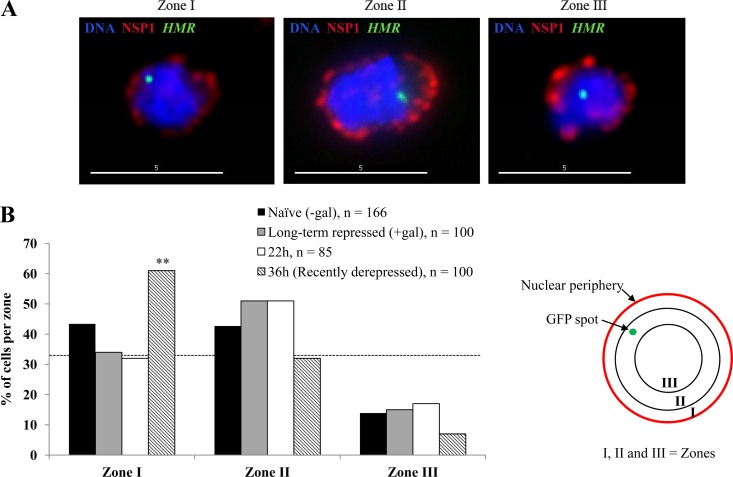
Quantitative analysis of green fluorescent protein (GFP) dot position within the nucleus was performed as described previously (15). Softworx software, in conjunction with the Deltavision RT imaging system (Applied Precision) adapted to an Olympus (IX70) microscope, was used to acquire image stacks at 0.2-μm spacing along the z axis. The three-dimensional position of the HMR-lacI-GFP spot was determined relative to the center of the cell and to the nuclear periphery. In each cell the distance of the GFP dot to the nuclear periphery (x) and the nuclear diameter (y) were determined. Dividing the first value by one-half of the second (i.e., the radius), we get a value that relates the position of HMR relative to the nuclear periphery and normalized to the diameter of the nucleus. We classified each spot as falling into one of the three concentric zones based on this value (see Fig. 7B). The spot is considered to be in zone I if this value is <(0.184 × r), where r is the nuclear radius, in zone II if the value is between 0.184r and 0.422r, and in zone III if the value is >0.422r.
Fig 7.
Peripheral localization of HMR in recently derepressed cells. (A) The position of HMR relative to the nuclear envelope was measured by tracking a GFP-lac repressor fusion bound to the ∼256 lacOP array inserted ∼2 kb from the locus, as described in Materials and Methods. The nuclear envelope was visualized using an α-Nsp1 antibody; DNA was stained with DAPI (4′,6-diamidino-2-phenylindole). Representative images showing HMR localized to the three zones are shown. Scale bars, 5 μm. (B) Distribution of HMR in three zones from naïve, long-term-repressed cells and induced/washed cells at 22 h or 36 h. The dotted line at 33% indicates a random distribution. The number of cells in zone I at 36 h is significantly different from that of naïve cells (two-sided Fisher's exact test; **, P = 0.007). No statistically significant difference was observed between long-term-repressed and naïve cells (P = 0.15) or between results for 22 h cells and naïve cells (P = 0.07).
RESULTS
In limiting amounts of Sir3 protein, silencing at HMR is more stable than at the HML locus.
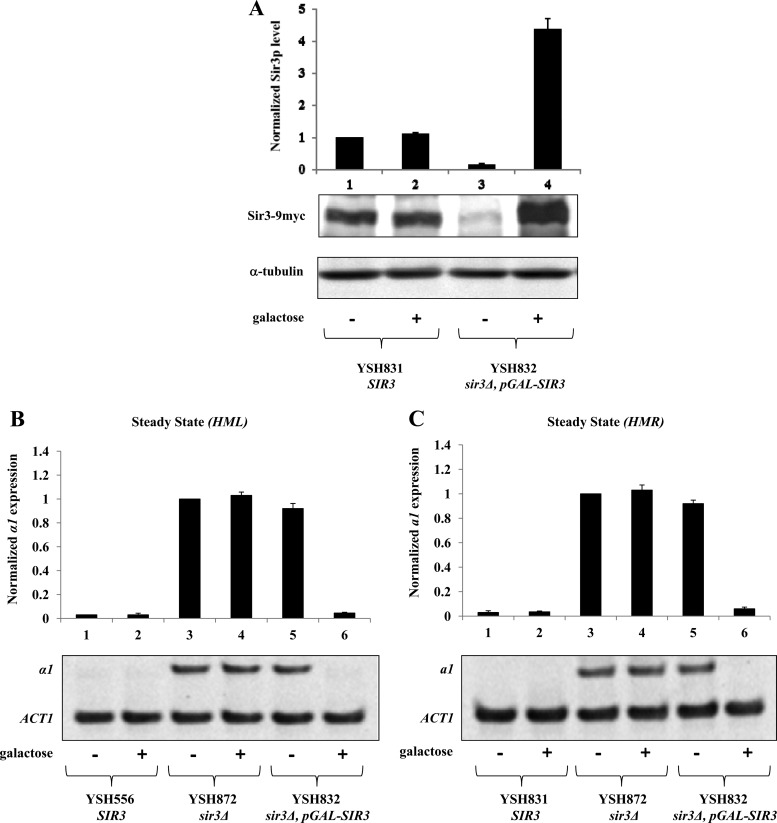
To study the mechanism of epigenetic inheritance of heterochromatin in budding yeast, we monitored the persistence of heterochromatin at the silent mating type loci under conditions of steadily decreasing Sir3 protein levels. We used YSH832, a strain in which the sole source of Sir3 was supplied by an integrated galactose-inducible SIR3 allele (pGAL10-SIR3). Sir3 production and silencing are tightly controlled by the carbon source in these cells; Western blot analysis (Fig. 1A) revealed that Sir3 protein levels in the absence of galactose are at least 10-fold lower than wild-type levels and are increased to approximately 4.5 times the wild-type levels upon galactose induction. Using qPCR to assess mRNA levels, we observed no significant silencing of HML or HMR in cells grown in noninducing YPraffinose medium and wild-type levels of silencing in galactose-induced cells (Fig. 1B and C).
Fig 1.
Steady-state examination of Sir3 protein α1 and a1 mRNA levels. (A) Sir3 protein levels were determined by Western blotting, normalized to an α-tubulin internal control, and expressed relative to a control strain bearing a wild-type SIR3 gene (YSH831, −galactose). (B and C) Galactose-inducible repression of transcription at HML and HMR. Strains YSH556, YSH872, YSH831, and YSH832 were grown to steady state in YPraffinose medium with or without galactose. Levels of α1, a1, and ACT1 mRNA were determined by qPCR analysis (see Materials and Methods) and expressed relative to the uninduced (−galactose) control of the sir3Δ strain. A representative gel scan is shown.
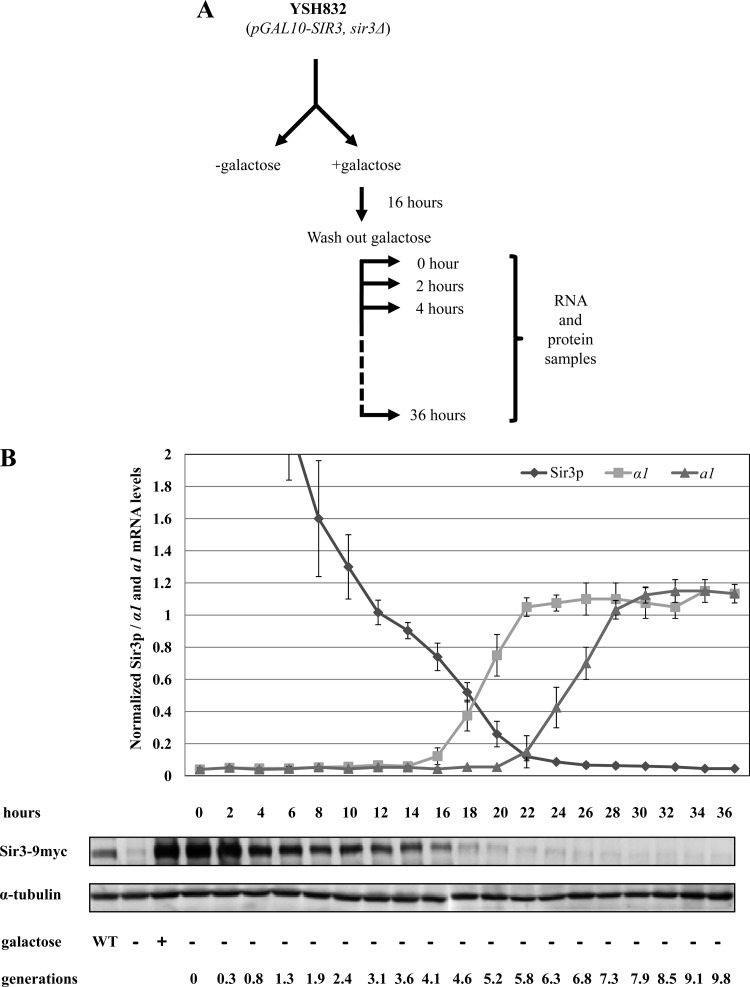
To examine the pattern of silencing in cells experiencing limiting levels of Sir3, we grew YSH832 in YPraffinose medium to early log phase and then carried out Sir3 induction for at least 16 h; a previous study using an inducible Sir3 strain reported that inductions of this duration were sufficient for complete repression at HMR and loss of histone modifications associated with active chromatin (20). Following Sir3 induction, galactose was washed out of the culture and the cells were grown in YPraffinose. Every 2 h we measured levels of α1 and a1 mRNA, transcribed from the HML and HMR genes, respectively, and also determined Sir3 protein levels (Fig. 2A). Sir3 protein returned to wild-type levels approximately 12 h (∼3.1 generations) after removal of galactose and reached the uninduced level by 22 h (∼5.8 generations) (Fig. 2B). The half-life of Sir3 in our experiments is roughly equal to the generation time, indicating that Sir3 protein is relatively stable across the time course of our experiment and that decreases in concentration are primarily due to dilution following cell division. This stability is consistent with published reports (9).
Fig 2.
Stability of silencing at HML and HMR under conditions of steadily decreasing Sir3 protein. (A) Experimental design. Strain YSH832 was grown to early log phase in YPraffinose medium. This culture was divided, and one half was induced with 2% galactose, while the other half was not induced. After 16 h galactose was washed out, and cells were cultured in YPraffinose medium. Every 2 h, samples were collected for analyzing mRNA levels from HML and HMR and to monitor Sir3 protein levels. (B) Stability of silencing at HML and HMR under conditions of steadily decreasing Sir3 protein. α1 and a1 mRNA levels were determined by qPCR analysis (see Materials and Methods) and expressed relative to their uninduced (−galactose) levels. Sir3 protein levels were determined by Western blotting, normalized to an α-tubulin internal control, and expressed relative to a control strain bearing a wild-type SIR3 gene (YSH831, WT lane). A representative Western blot is shown. Lanes “−galactose” and “+galactose” indicate Sir3 levels in long-term-uninduced and induced cultures, respectively. The numbers of hours and cell generations following galactose washout are listed.
Our qPCR analysis revealed that under conditions of limiting Sir3 protein, silencing at HMR is more stable than at HML. The transcriptional repression of α1 message from HML started to decay by 16 h and was completely lost by 22 h (Fig. 2B). In contrast, the transcriptional repression of HMRa1 message started to decay at ∼22 h and was completely lost by 28 h (Fig. 2B). Notably, we observed that silencing at HMR persisted at levels of Sir3 that were insufficient to promote repression under steady-state conditions; by 22 h, Sir3 protein levels decreased to the uninduced level but HMR remained fully silenced. Significant repression of a1 continued for approximately two generations after this time, with a1 message only reaching fully expressed levels at ∼28 h. This indicates that previously silenced cells are able to maintain silencing under conditions insufficient to promote silencing in previously unsilenced cells. Thus, as shown previously for HML, an epigenetic mechanism exists at HMR to promote the inheritance of the repressed state in daughter cells.
Recently derepressed cells regain silencing faster than naïve unsilenced cells.
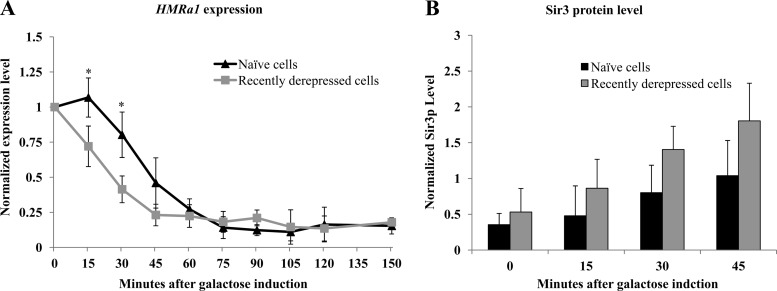
We observed that cells are able to temporarily maintain silencing when their Sir3 levels decrease to levels that are unable to support silencing under steady-state conditions. To examine whether recently derepressed cells have persistent phenotypic differences compared to long-term-derepressed cells, we compared the rate of establishment of silencing at HMRa1 in naïve cells with the rate in recently derepressed cells. We added galactose to long-term-unsilenced cells (“naïve cells”) and to cells 36 h after washing out galactose (“recently derepressed cells”) and then measured a1 mRNA levels every 15 min. As shown in Fig. 3A, silencing is established more quickly in recently derepressed cells, primarily due to the loss of a lag period in naïve cells, in which no silencing is achieved. We also compared the levels of Sir3 protein in naïve and recently derepressed cells and observed that at early time points recently derepressed cells express Sir3 at higher levels (Fig. 3B). This differential Sir3 expression may reflect an independent memory mechanism operating on the GAL10 promoter controlling SIR3 transcription (4). Thus, while we observed an increase in the rate of repression in recently derepressed cells, our experiment does not establish whether this is due to an epigenetic mechanism operating at HMR or to differences in Sir3 levels under the two conditions.
Fig 3.
Recently derepressed cells establish silencing faster than naïve cells. (A) Establishment of silencing in naïve and recently derepressed cells. Galactose was added to 2% to long-term-uninduced cells (“naïve” cells) and to recently derepressed cells. For the recently derepressed sample, cells were grown as described in the Fig. 2A legend; galactose was readded to cells 36 h after galactose was washed out. Following galactose addition, samples were collected every 15 min for 150 min. The levels of a1 mRNA were measured by qPCR analysis (see Materials and Methods). The difference in the establishment of silencing in naïve and recently derepressed cells at 15 and 30 min after galactose washout was statistically significant (*, P = 0.039 and P = 0.023, Student's t test). (B) Sir3 protein levels were measured in the cultures described for panel A as described in the legend for Fig. 2B.
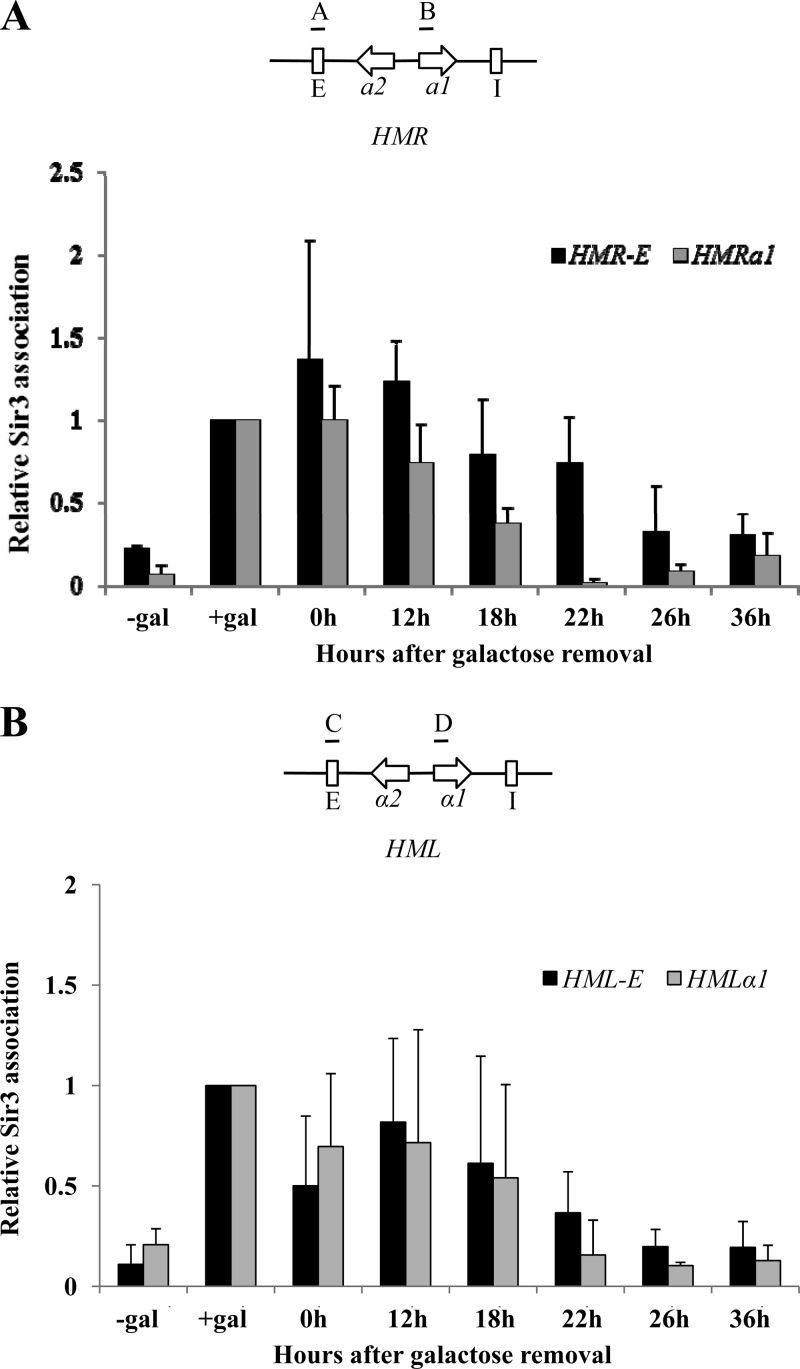
Sir3 is retained at the HMR-E silencer in an epigenetic pattern.
The ability of silenced cells to maintain silencing at HMR as Sir3 reaches limiting levels suggests that their previously silenced status gives them an advantage in recruiting or retaining limiting Sir3 protein. To study the molecular events accompanying the inheritance of silencing in low levels of Sir3 protein, we monitored the association of Sir3 at the HML and HMR loci and their silencer regions using ChIP assays. Samples were collected from the uninduced and induced control cultures and from the induced/washed culture at the 0, 12, 18, 22, 26, and 36 h time points. In long-term-induced and -uninduced cultures, we observed a galactose-dependent association of Sir3 with both silencers and the a1 and α1 genes (Fig. 4A and B). After the removal of galactose, Sir3 association with the HMRa1 gene tracks well with the overall level of Sir3 protein in the cell (Fig. 4A, gray bars). However, the association of Sir3 with the HMR-E silencer decreases with slower kinetics (Fig. 4A, black bars). In particular, at the 22 h time point Sir3 is substantially retained at HMR-E, even though Sir3 levels are at uninduced levels (Fig. 4A). As Sir3 is not enriched at HMR-E in long-term-derepressed cells, this result indicates that the Sir3 protein association with this silencer is epigenetically inherited. At the HML locus, the association of Sir3 protein at the HMLα1 gene and at HML-E tracked well with overall levels of Sir3 protein in the cell (Fig. 4B). We have not determined the minimal level of Sir3 required to repress transcription of the silent mating type loci under steady-state conditions; thus, we cannot conclude whether repression of transcription and Sir3 retention at HML also follow an epigenetic pattern. Given the clear epigenetic pattern we observed at HMR, we focused additional experiments on this locus.
Fig 4.
Association of Sir3 with the silent mating type loci. (A) (Top) The positions of the HMR-E (indicated by an “A” above the diagram) and a1 (indicated by a “B”) probes used for ChIP are shown on a diagram of the HMR locus. (Bottom) Association of Sir3 with HMR is shown, measured in galactose-induced cultures in which galactose had been removed for the indicated number of hours. Long-term-uninduced and induced controls are also shown (“−gal” and “+gal,” respectively); values are expressed relative to the galactose-induced control. The association of Sir3 at HMR-E at 22 h versus the uninduced culture (−gal) is statistically significant (P = 0.035, Student's t test). (B) Sir3 association with HML was measured as described for panel A. The association of Sir3 at HML-E and HMLα1 at the 22 h time point (and at all subsequent time points) versus the induced culture (+gal) is statistically significant (P < 0.001, Student's t test).
Decreased H4-K16 acetylation and H3-K79 dimethylation persist in recently derepressed cells.
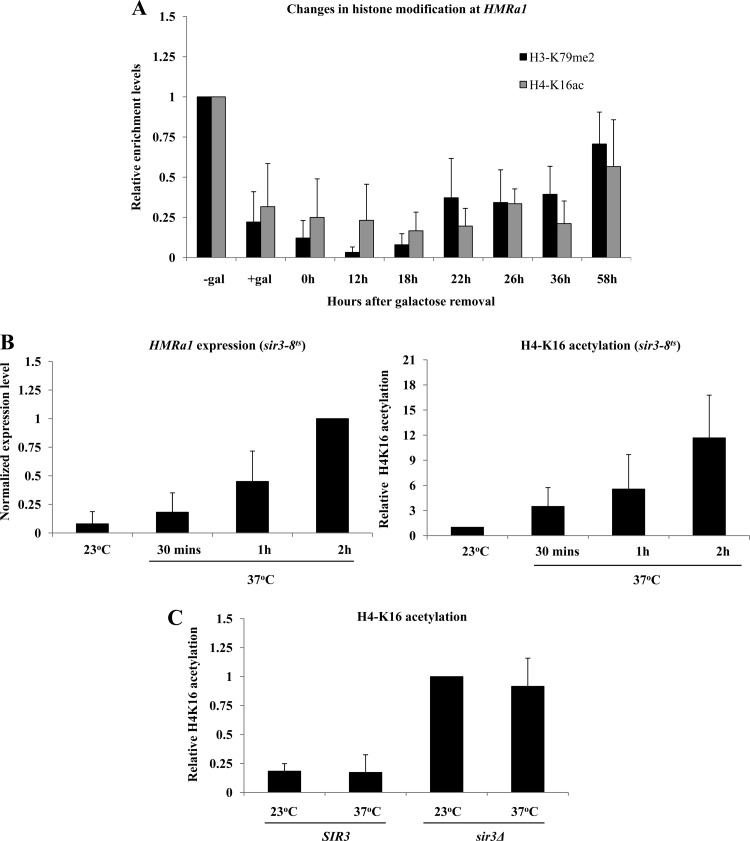
We used ChIP to investigate the pattern of histone modifications at HMR as Sir3 levels became limiting. In control experiments we observed galactose-dependent decreases in H3-K79 dimethylation and H4-K16 acetylation in long-term-induced or -uninduced cultures (Fig. 5A). In cells experiencing steadily decreasing Sir3 protein, the low levels of H3-K79 dimethylation persisted at levels significantly below those of uninduced cells for several generations, even as a1 message reached fully expressed levels (Fig. 5A, black bars). Even more strikingly, H4-K16 deacetylation remained unchanged for several generations after Sir3 reached uninduced levels (Fig. 5A, gray bars).
Fig 5.
Low levels of H3-K79 dimethylation (H3-K79me2) and H4-K16 acetylation (H4-K16ac) persist in recently derepressed cells. (A) Relative levels of H3-K79 dimethylation and H4-K16 acetylation at HMR were measured using ChIP (see Materials and Methods) with an HMRa1 probe (probe B in Fig. 4A). Association of the histone marks was measured in galactose-induced cultures in which galactose had been removed for the indicated number of hours. Long-term-uninduced and induced controls are also shown (“−gal” and “+gal,” respectively); values are expressed relative to the uninduced control. (B) Histone acetylation increases rapidly upon Sir3 inactivation. Strain YSH829 (sir3-8) was grown in YPraffinose medium to log phase at permissive temperature (23°C) and then divided into two cultures; one was kept at 23°C, and the other was shifted to nonpermissive temperature (37°C). After the temperature shift, samples were collected after 30 min, 1 h, and 2 h to measure a1 mRNA levels and H4-K16 acetylation at HMRa1. Immunoblot experiments indicated that Sir3-8p levels were similar to wild-type Sir3 levels at permissive temperature but significantly lower than wild-type levels when shifted to nonpermissive temperature (not shown), consistent with prior reports (44). a1 mRNA levels are expressed relative to the culture kept at 37°C for 2 h, and H4-K16 acetylation levels are expressed relative to the 23°C culture. (C) Relative levels of H4-K16 acetylation at HMRa1 were measured in a strain bearing a wild-type SIR3 gene (YSH831) and in a sir3Δ strain (YSH872) at permissive (23°C) and nonpermissive (37°C) temperatures. H4-K16 acetylation levels are expressed relative to the 23°C culture of the sir3Δ strain.
To determine if the slow reacquisition of these marks was a typical consequence of Sir3 loss, we monitored H4-K16 acetylation in a Sir3 temperature-sensitive strain (YSH829), shifted from permissive temperature (23°C) to nonpermissive temperature (37°C). We found that inactivation of Sir3 by temperature shift led to a rapid loss of silencing and increase in H4-K16 acetylation at HMRa1 (Fig. 5B); no increase in H4-K16 acetylation was observed in a strain bearing a wild-type strain SIR3 gene (YSH831) or in a sir3Δ strain (YSH872) following a similar temperature shift (Fig. 5C). These results suggest that the persistence of low H4-K16 acetylation under conditions of diminishing Sir3 is dependent on Sir3 activity and reflects a unique ability of previously silenced cells to utilize normally limiting levels of Sir3.
Persistence of H4-K16 deacetylation is partially dependent on Sir2 activity.
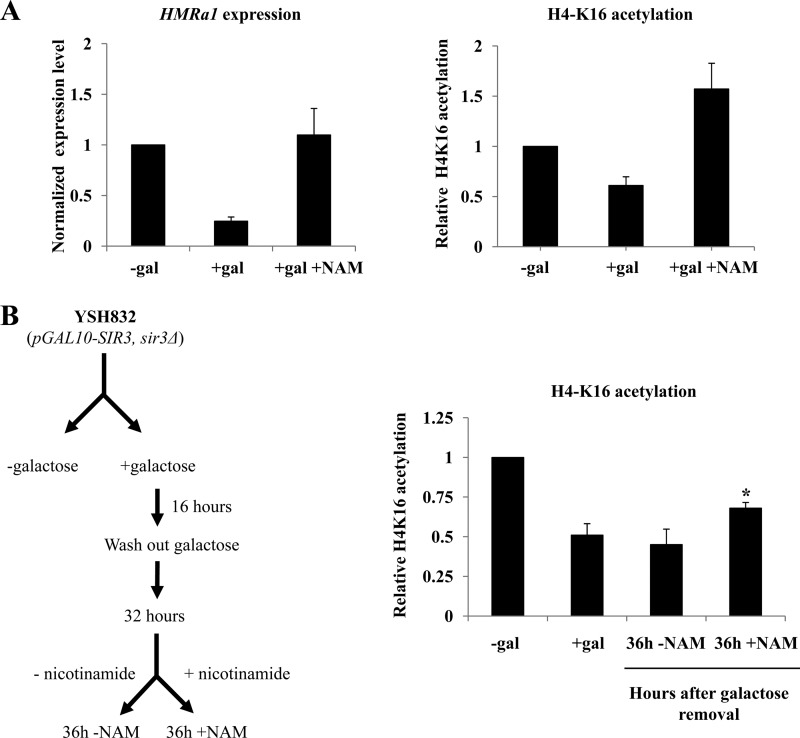
To determine if Sir2 is required to maintain the deacetylated state in recently derepressed cells, we inhibited the activity of Sir2 using nicotinamide (2). In control experiments a 4-h incubation of galactose-induced cells with nicotinamide disrupted silencing and caused an increase in H4-K16 acetylation (Fig. 6A). Next, we tested if H4-K16 deacetylation observed at HMRa1 in low levels of Sir3 protein could be reversed upon treatment with nicotinamide. Induced/washed cultures were grown in YPraffinose for 32 h. Nicotinamide was added, cells were grown an additional 4 h, and then H4-K16 acetylation was examined by ChIP. As shown in Fig. 6B, we observed a significant increase in H4-K16 acetylation at HMRa1 compared with the untreated cells (36 h +nicotinamide versus 36 h −nicotinamide), although acetylation did not return to the levels seen in the control experiment. This suggests that persistence of H4-K16 deacetylation at HMRa1 is at least partially dependent on Sir2 activity.
Fig 6.
Persistence of H4-K16 deacetylation depends on Sir2 activity. (A) Nicotinamide (NAM) causes loss of silencing and increases H4-K16 acetylation in control galactose-induced cultures. a1 mRNA levels and H4-K16 acetylation at HMRa1 were measured in cells treated for 4 h with nicotinamide at 5 mM and expressed relative to the uninduced control. (B) (Left) Cultures were grown as described in the legend to Fig. 2A; 32 h after galactose was washed out, the culture was split, with one portion receiving 5 mM nicotinamide. Four hours later (36 h time point) H4-K16 acetylation was measured at HMRa1 and expressed relative to an uninduced control culture (“−gal”). A long-term-induced control (“+gal”) is also shown. The difference between the +NAM and the −NAM cultures at the 36 h time point is significant (P = 0.019, Student's t test).
The HMR locus is peripherally localized in recently derepressed cells.
Silenced sequences and silencing proteins are nonrandomly distributed in the nucleus in yeast, with a tendency to associate with the nuclear periphery (6, 7, 11, 13, 27). To explore whether subnuclear localization of HMR sequences could explain the differences that we saw between long-term-derepressed and recently derepressed cells, we monitored the HMR position within the nucleus. We inserted ∼256 lacop binding sites approximately 2 kb telomere proximal to the HMR locus, creating YSH1089. Expression of LacI-GFP in these cells creates a single focus of GFP fluorescence, allowing the position of HMR to be determined. We also used an antibody against a component of nuclear pore complex Nsp1 to visualize the nuclear periphery. We collected three-dimensional focal stacks to measure the distance between LacI-GFP foci and the nuclear membrane in long-term-uninduced (naïve) and galactose-induced cells and in cells 22 h or 36 h following washout of galactose. Under all conditions tested, we observed a tendency to be in the outer two zones of the nucleus. However, we failed to observe a significant difference between long-term-repressed cells and naïve cells; cells at the 22 h time point also showed a pattern similar to that of these controls (Fig. 7B). Thus, to the extent of the precision of measurement possible in these assays, retention of peripheral localization does not appear to be the mechanism aiding the retention of Sir3 at HMR under conditions of limiting Sir3. Unexpectedly, we observed an increase in HMR localization to zone I in recently derepressed cells (36 h cells). This change in localization could be contributing to the retention of silent histone marks we observed at this time; however, we noted that this change in localization coincided with the onset of a1 transcription, and migration to the nuclear periphery at the onset of transcription has been observed for several other genes (4, 5, 7, 47).
DISCUSSION
The epigenetic pattern of inheritance observed for Sir-dependent silencing in budding yeast cells was established by assessing the phenotypic consequences of HML transcription in single-cell pedigree assays (26, 37). Phenotypic assays based on colony color indicated that silencing at yeast telomeres was also inherited mitotically (1). Characterization of the properties of the Sir2, Sir3, and Sir4 proteins (summarized in the introduction) revealed a potential positive-feedback loop for Sir protein recruitment and histone deacetylation consistent with basic predictions of nucleosome-based inheritance models; however, there is little direct evidence for this mode of epigenetic inheritance in this or other systems (reviewed in references 21 and 38). We tested predictions of these models by examining silencing at HML and HMR under conditions of steadily decreasing Sir3 protein.
Several prior studies have shown that HML is more sensitive than HMR to mutations that weaken silencing. For instance, specific substitution mutations or deletions of the histone H4 N terminus disrupt silencing at HML but have only minor effects on repression at HMR (19, 22, 30, 36). Similarly, deletion of the NAT1 or ARD1 genes, which code for components of an N-terminal acetyltransferase that likely acts on Sir3, also preferentially derepresses HML (33, 52, 53). Our results suggest an explanation for these prior observations: we observed that when Sir3 becomes limiting, silencing at HMR is more stable than at HML, indicating that the HMR locus is better able to sustain silencing at very low levels of Sir3. We observed that transcriptional repression at HMR persists even when Sir3 protein decreases to levels that are insufficient to support measurable silencing in long-term-derepressed cells, indicating that there exists an epigenetic mechanism to promote the persistence of silencing at HMR. HML was derepressed when Sir3 reached uninduced levels, but since the minimal level of Sir3 necessary to promote silencing is not known, we cannot determine whether HML also exhibited an epigenetic pattern in these experiments.
We note that silencing persists in cells in which Sir3 is detectable at the HMR-E silencer, but not at the HMRa1 gene subject to silencing (Fig. 4A, 22 h); at this time, silencing-specific histone modifications are still present at the a1 gene. Prior studies reported that the HMR-E and HMR-I silencers physically interact in vivo (31, 50); perhaps once these Sir-dependent interactions are established, repression is sustained even as Sir3 association diminishes. Surprisingly, we also observed that the low H4-K16 acetylation and H3-K79 methylation associated with silencing persist long after HMR is derepressed. In a control experiment we showed that H4-K16 acetylation is rapidly regained when Sir3 is inactivated using a temperature-sensitive allele. This suggests that the retention of silencing-specific histone modifications is likely still a Sir3-dependent phenomenon; in these recently derepressed cells the very low levels of Sir3 present in the cell (approximately 10% of normal levels) still have the ability to maintain low acetylation even as the genes are transcribed, an ability not observed in the long-term-uninduced cells. Persistence of low H4-K16 acetylation and H3-K79 methylation could be due to an active process of removing posttranslational modifications from histones or could be due to sequestration of the locus in a manner that inhibits access to modifying enzymes. Pertinent to these possibilities, we found that at least some of the persistent low acetylation is sensitive to nicotinamide addition, suggesting it depends on the function of Sir2 (or a Sir2 homolog). We also observed that in recently derepressed cells the HMR locus is more likely to localize to the nuclear periphery; however, this change in localization occurs coincidentally with transcription of the HMRa1 gene. In contrast to prior studies (6, 7, 11), we failed to observe a difference in HMR localization in silenced versus unsilenced cells. It was recently reported that Sir3 overexpression increased telomere clustering but decreased peripheral localization of telomeres (40); perhaps the higher-than-normal Sir3 levels during the early phases of our experiment prevented us from observing silencing-specific HMR localization.
Single-cell assays previously indicated that strains lacking the Set1 enzyme responsible for methylating lysine 4 on histone H3 converted from an unsilenced phenotype to a silenced phenotype at a higher rate than did wild-type cells but that cells lacking the Sas2 enzyme capable of acetylating lysine 16 on histone H4 were slower to acquire silencing (34, 35). We found that recently derepressed cells had lower levels of H3-K79 methylation and H4-K16 acetylation than naïve cells and also silenced more quickly when Sir3 was induced than did naïve cells. While this could reflect an absence of a lag period due to the different state of these cells' histone modifications, we also observed a somewhat quicker restoration of Sir3 levels that could also be the cause of more-rapid silencing.
If silencing epigenetics follows a self-templating pattern and involves either inheritance of modified nucleosomes to which Sir proteins subsequently bound or inheritance of Sir protein-bound nucleosomes, then silenced cells should have an advantage over derepressed cells in retaining limiting Sir3 protein. Our results show that, for HMR, this is the case; in long-term-uninduced cells, our ChIP assays were unable to detect Sir3 at the HMR locus, but when decreasing Sir3 protein reached uninduced levels in silenced cells, Sir3 was retained at the HMR-E silencer. Prior experiments using inducible recombination to remove silencer sequences in vivo demonstrated that transcriptional repression was retained in the absence of silencers but was lost as cells progressed through a single cell cycle (8, 16). These experiments showed that a chromatin state sufficient to repress transcription at HML was insufficient to promote its inheritance, indicating that a nucleosome-only inheritance model is insufficient to explain the epigenetic pattern seen in budding yeast. Consistent with this observation, at the same time that we observed retention of Sir3 at the HMR-E silencer, we failed to observe Sir3 at the HMRa1 gene, despite the presence of histone modifications conducive to Sir3 binding (Fig. 4A). These experiments indicate that neither silencer sequences nor modified nucleosomes are on their own sufficient to retain Sir3 when levels become limiting; instead, this suggests that silencer sequences act in concert with local modified nucleosomes to retain Sir3. Prior experiments reported that association of the Sir complex (Sir2, Sir3, and Sir4) with silencer sequences was reduced if Sir2's deacetylase activity was eliminated, also suggesting that stable Sir complex binding required deacetylated nucleosomes (17, 25, 41, 43). Our experiments suggest that this dual requirement for Sir complex binding may be crucial to inheriting the repressed state, consistent with a cooperative association model recently proposed (32).
ACKNOWLEDGMENTS
We are grateful to Amy MacQueen for assistance with microscopy and for comments on the manuscript, Upasna Sharma for help with ChIP assays, and members of the Holmes lab for helpful discussions.
This work was supported by a grant from the National Science Foundation (MCB-0617986) to S.G.H.
Footnotes
Published ahead of print 14 May 2012
REFERENCES
- 1. Aparicio OM, Billington BL, Gottschling DE. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279–1287 [DOI] [PubMed] [Google Scholar]
- 2. Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099–45107 [DOI] [PubMed] [Google Scholar]
- 3. Brachmann CB, et al. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 4. Brickner DG, et al. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81 doi:10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brickner JH, Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342 doi:10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bystricky K, et al. 2009. Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/Ku80 complex. Mol. Cell. Biol. 29:835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casolari JM, et al. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427–439 [DOI] [PubMed] [Google Scholar]
- 8. Cheng TH, Gartenberg MR. 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14:452–463 [PMC free article] [PubMed] [Google Scholar]
- 9. Dasgupta A, Ramsey KL, Smith JS, Auble DT. 2004. Sir Antagonist 1 (San1) is a ubiquitin ligase. J. Biol. Chem. 279:26830–26838 [DOI] [PubMed] [Google Scholar]
- 10. Fox CA, McConnell KH. 2005. Toward biochemical understanding of a transcriptionally silenced chromosomal domain in Saccharomyces cerevisiae. J. Biol. Chem. 280:8629–8632 [DOI] [PubMed] [Google Scholar]
- 11. Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119:955–967 [DOI] [PubMed] [Google Scholar]
- 12. Goldstein AL, McCusker JH. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- 13. Gotta M, Gasser SM. 1996. Nuclear organization and transcriptional silencing in yeast. Experientia 52:1136–1147 [DOI] [PubMed] [Google Scholar]
- 14. Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583–592 [DOI] [PubMed] [Google Scholar]
- 15. Hediger F, Taddei A, Neumann FR, Gasser SM. 2004. Methods for visualizing chromatin dynamics in living yeast. Methods Enzymol. 375:345–365 [DOI] [PubMed] [Google Scholar]
- 16. Holmes SG, Broach JR. 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 10:1021–1032 [DOI] [PubMed] [Google Scholar]
- 17. Hoppe GJ, et al. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin QW, Fuchs J, Loidl J. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113 (Pt 11):1903–1912 [DOI] [PubMed] [Google Scholar]
- 19. Johnson LM, Fisher-Adams G, Grunstein M. 1992. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 11:2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katan-Khaykovich Y, Struhl K. 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 24:2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaufman PD, Rando OJ. 2010. Chromatin as a potential carrier of heritable information. Curr. Opin. Cell Biol. 22:284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kayne PS, et al. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55:27–39 [DOI] [PubMed] [Google Scholar]
- 23. Knop M, et al. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963–972 [DOI] [PubMed] [Google Scholar]
- 24. Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121:515–527 [DOI] [PubMed] [Google Scholar]
- 25. Luo K, Vega-Palas MA, Grunstein M. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16:1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mahoney DJ, Marquardt R, Shei GJ, Rose AB, Broach JR. 1991. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 5:605–615 [DOI] [PubMed] [Google Scholar]
- 27. Maillet L, et al. 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10:1796–1811 [DOI] [PubMed] [Google Scholar]
- 28. Martins-Taylor K, Dula ML, Holmes SG. 2004. Heterochromatin spreading at yeast telomeres occurs in M phase. Genetics 168:65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martins-Taylor K, Sharma U, Rozario T, Holmes SG. 2011. H2A.Z (Htz1) controls the cell-cycle-dependent establishment of transcriptional silencing at Saccharomyces cerevisiae telomeres. Genetics 187:89–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Megee PC, Morgan BA, Mittman BA, Smith MM. 1990. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247:841–845 [DOI] [PubMed] [Google Scholar]
- 31. Miele A, Bystricky K, Dekker J. 2009. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 5:e1000478 doi:10.1371/journal.pgen.1000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moazed D. 2011. Mechanisms for the inheritance of chromatin states. Cell 146:510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mullen JR, et al. 1989. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Osborne EA, Dudoit S, Rine J. 2009. The establishment of gene silencing at single-cell resolution. Nat. Genet. 41:800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osborne EA, Hiraoka Y, Rine J. 2011. Symmetry, asymmetry, and kinetics of silencing establishment in Saccharomyces cerevisiae revealed by single-cell optical assays. Proc. Natl. Acad. Sci. U. S. A. 108:1209–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park EC, Szostak JW. 1990. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol. Cell. Biol. 10:4932–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pillus L, Rine J. 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59:637–647 [DOI] [PubMed] [Google Scholar]
- 38. Ptashne M. 2007. On the use of the word ‘epigenetic’. Curr. Biol. 17:R233–R236 [DOI] [PubMed] [Google Scholar]
- 39. Rockmill B. 2009. Chromosome spreading and immunofluorescence methods in Saccharomyces cerevisiae. Methods Mol. Biol. 558:3–13 [DOI] [PubMed] [Google Scholar]
- 40. Ruault M, De Meyer A, Loiodice I, Taddei A. 2011. Clustering heterochromatin: Sir3 promotes telomere clustering independently of silencing in yeast. J. Cell Biol. 192:417–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rudner AD, Hall BE, Ellenberger T, Moazed D. 2005. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 25:4514–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rusche LN, Kirchmaier AL, Rine J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481–516 [DOI] [PubMed] [Google Scholar]
- 43. Rusche LN, Kirchmaier AL, Rine J. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2207–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stone EM, Reifsnyder C, McVey M, Gazo B, Pillus L. 2000. Two classes of sir3 mutants enhance the sir1 mutant mating defect and abolish telomeric silencing in Saccharomyces cerevisiae. Genetics 155:509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Straight AF, Belmont AS, Robinett CC, Murray AW. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608 [DOI] [PubMed] [Google Scholar]
- 46. Sussel L, Vannier D, Shore D. 1993. Epigenetic switching of transcriptional states: cis- and trans-acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3919–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taddei A, et al. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441:774–778 [DOI] [PubMed] [Google Scholar]
- 48. Tanner KG, Landry J, Sternglanz R, Denu JM. 2000. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. U. S. A. 97:14178–14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tanny JC, Moazed D. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. U. S. A. 98:415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT. 2008. Long-range communication between the silencers of HMR. Mol. Cell. Biol. 28:1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vialard JE, Gilbert CS, Green CM, Lowndes NF. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X, Connelly JJ, Wang CL, Sternglanz R. 2004. Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics 168:547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Whiteway M, Freedman R, Van Arsdell S, Szostak JW, Thorner J. 1987. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol. Cell. Biol. 7:3713–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]