Abstract
The differentiation of gametes involves dramatic changes to chromatin, affecting transcription, meiosis, and cell morphology. Sporulation in Saccharomyces cerevisiae shares many chromatin features with spermatogenesis, including a 10-fold compaction of the nucleus. To identify new proteins involved in spore nuclear organization, we purified chromatin from mature spores and discovered a significant enrichment of the linker histone (Hho1). The function of Hho1 has proven to be elusive during vegetative growth, but here we demonstrate its requirement for efficient sporulation and full compaction of the spore genome. Hho1 chromatin immunoprecipitation followed by sequencing (ChIP-seq) revealed increased genome-wide binding in mature spores and provides novel in vivo evidence of the linker histone binding to nucleosomal linker DNA. We also link Hho1 function to the transcription factor Ume6, the master repressor of early meiotic genes. Hho1 and Ume6 are depleted during meiosis, and analysis of published ChIP-chip data obtained during vegetative growth reveals a high binding correlation of both proteins at promoters of early meiotic genes. Moreover, Ume6 promotes binding of Hho1 to meiotic gene promoters. Thus, Hho1 may play a dual role during sporulation: Hho1 and Ume6 depletion facilitates the onset of meiosis via activation of Ume6-repressed early meiotic genes, whereas Hho1 enrichment in mature spores contributes to spore genome compaction.
INTRODUCTION
Gametogenesis is a complex, highly regulated differentiation program that is integral to the survival of higher eukaryotes. Mammalian spermatogenesis begins with DNA replication and recombination, ultimately generating four genetically unique haploid cells. The postmeiotic differentiation of spermatozoa involves a complete reorganization of the chromatin and a dramatic compaction of the nucleus that prepare the genome for passage to a new generation (16, 30). In fact, specific combinations of histone posttranslational modifications (PTMs) are found at genes encoding important developmental regulators (5, 20).
Gametogenesis in Saccharomyces cerevisiae, or sporulation, shares several chromatin features with mammalian spermatogenesis, namely, drastic compaction of the nucleus and stage-specific appearance of histone PTMs such as phosphorylation of histone H3 at serine 10 (H3S10ph), H3T11ph, and H4S1ph (17, 31). Sporulation takes place in diploid yeast deprived of nitrogen and a fermentable carbon source (15, 39). Under these starvation conditions, yeast cells enter a meiotic program similar to that found in mammalian spermatogenesis. During the postmeiotic phase, cell and nuclear volumes are substantially decreased, the genome is compacted, transcription is silenced, and a protective spore wall is synthesized. The resultant quiescent spore is able to survive unfavorable growth conditions of heat, dehydration, and chemical insult. When a fermentable carbon source is reintroduced, spores are induced to reenter the cell cycle through a process called germination, characterized by a specific transcriptional cascade and the disappearance of spore morphology (25).
The progression of sporulation is tightly controlled by transcription (28). Each stage of sporulation is induced by the temporally distinct activation of specific groups of genes, which are broadly classified into early, middle, and late genes (3, 6, 11, 28, 32, 43). Early and middle genes are involved in DNA synthesis and meiosis, while late genes control postmeiotic differentiation of the spores. Induction of sporulation is highly regulated and repressed during vegetative growth and in haploid cells. Spontaneous induction of sporulation during vegetative growth limits the growth of a diploid colony, and the meiotic process is lethal in haploids. Thus, early meiotic genes are strongly repressed by a transcriptional repressor, Ume6 (a DNA-binding transcription factor), together with two corepressor complexes, one containing Rpd3 (a histone deacetylase) and the second Isw2 (a chromatin remodeler) (14, 26). Ume6 binds to the URS1 sequence upstream of early meiotic genes and recruits these corepressors (40). Deletion of UME6 results in the derepression of early meiotic genes and severe sporulation defects (47, 54).
Chromatin architecture and cell morphology also change dramatically over the course of gametogenesis. A hallmark feature of both yeast sporulation and human spermatogenesis is a 10-fold reduction in nuclear volume, which protects the genome and involves extreme compaction of chromatin. In human sperm, this compaction is associated with the replacement of most histones with smaller, highly basic, sperm-specific proteins called protamines (2). Interestingly, yeast and certain higher eukaryotes, including some fish taxa, achieve similar compaction without protamines (9). In fact, histones are retained in the yeast spore genome, and nucleosome positioning remains largely unchanged during sporulation (17, 58). Certain histone PTMs, such as acetylation of the H4 histone tail and H4S1ph, have been shown to play a role in spore chromatin compaction (17, 31). We hypothesized that other unknown chromatin-associated factors may also help facilitate chromatin compaction in the spores of S. cerevisiae as protamines do in mammals. Indeed, here we report that the yeast H1 linker histone (Hho1) is involved in spore chromatin function.
In higher eukaryotes, in vitro studies have shown that H1 binds to the DNA entry/exit point of the nucleosome and facilitates higher-order chromatin structure such as the 30-nm fiber (21). Hho1 binds to nucleosomes and protects an extra 20 bp of nucleosomal DNA from micrococcal nuclease (MNase) digestion, similarly to higher eukaryotic linker histones (41). Chromatin immunoprecipitation-on-chip (ChIP-chip) and DNA adenine methyltransferase identification (DamID) studies in yeast and Drosophila, respectively, have demonstrated that H1 tends to be highly associated with nucleosomes and depleted at active genes; however, there is no genome-wide in vivo evidence for H1 binding to nucleosomal linker DNA (4, 46, 57).
Furthermore, the precise biological function of H1 remains unclear. One complicating factor is that several isoforms of H1 are present in humans and mice, and knockout studies have proven to be difficult due to functional redundancy (21). The linker histone represses transcription in vitro, but its transcriptional role in vivo has been the subject of debate (7, 24). Although H1 has historically been associated with chromatin compaction and general repression, several recent studies have implicated the linker histone in the repression of specific genes via recruitment by transcription factors (10, 33, 37, 45, 48, 52).
In S. cerevisiae, the function of the linker histone has also proven elusive, as deletion of HHO1 in vegetatively growing yeast shows no striking phenotype (41). Hho1 plays subtle structural and transcriptional roles. It is required for chromatin compaction during stationary phase and for higher-order chromatin organization during vegetative growth, and it represses homologous recombination in the context of DNA double-strand breaks (8, 12, 46). Hho1 has also been implicated in both transcriptional repression and activation, although the effects of HHO1 deletion on genome-wide transcription are modest (22, 35, 51, 56). Here, we investigated the role of Hho1 during the entire sporulation program, from induction through formation of mature spores and on to early germination. We provide evidence for a dual role for Hho1: first as a gene-specific factor in association with the meiotic gene repressor Ume6 and second as a genome-wide factor in the regulation of spore chromatin structure.
MATERIALS AND METHODS
Antibodies.
Hho1 antibody was obtained from Abcam (ab71833, lot 586128). H4 antibody was obtained from Active Motif (39269, lot 11908001). Ume6 antibodies were obtained from Abcam (ab37630, lot GR13708-1) and Sigma (GW22454A, lot 029K1664). H4Ac, H4S1ph, and H3S10ph antibodies have been described previously (17).
Yeast strains.
Strain genotypes can be found in Table 1. Strains used for sporulation and germination time courses were in the W303 background. The nuclear size assay was performed in the S288c background. Chromatin purification, ChIP sequencing, and meiotic transcription analysis were performed in the SK1 background. The hho1Δ and ume6Δ strains were made by replacing the HHO1 and UME6 genes with the KanMX3 marker. The dit1Δ and rpd3Δ strains were made by replacing the DIT1 and RPD3 genes with the TRP1 marker. The Hho1-3×FLAG strain used for ChIP sequencing was made by C-terminal fusion of the endogenous HHO1 gene with the 3×FLAG-KanMX3 construct. The cdc16-1 strain was a gift from Randy Strich, University of Medicine and Dentistry of New Jersey. All disruption alleles or epitope tags were constructed using PCR-based gene replacement (36).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| JB60a | MATa/MATα hho1::KanMX3/hho1::KanMX3 | This study |
| JB69b | MATa rpd3::TRP1 | This study |
| JB70b | MATα rpd3::TRP1 | This study |
| JB71b | MATa hho1::KanMX3 rpd3::TRP1 | This study |
| JB72b | MATα hho1::KanMX3 rpd3::TRP1 | This study |
| JB89a | MATa ume6::KanMX3 | This study |
| JB90a | MATα ume6::KanMX3 | This study |
| JG50b | MATa hho1::KanMX3 | This study |
| JG51b | MATα hho1::KanMX3 | This study |
| JG65 | MATa/MATα leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2-SK1/lys2-SK1 his4-N/G/his4-N/G ura3-SK1/ura3-SK1 ho::LYS2/ho::LYS2 dit1::TRP1/dit1::TRP1 | This study |
| JG186c | MATa/MATα hho1::KanMX3/hho1::KanMX3 | This study |
| JG192 | MATa/MATα his3-200/his3-200 leu2-1/leu2-1 ura3-52/ura3-52 trp1::hisG/trp1::hisG | This study |
| JG196b | MATa/MATα HHO1-3×FLAG::KanMX/HHO1-3×FLAG::KanMX | This study |
| LNY150 | MATa/MATα leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2-SK1/lys2-SK1 his4-N/G/his4-N/G ura3-SK1/ura3-SK1 ho::LYS2/ho::LYS2 | 42 |
| RSY954 | MATa/MATα cyh2R-z/cyh2R-z ho::LYS2/ho::LYS2 leu2::hisG/leu2::hisG lys2/lys2 trp1::hisG/trp::hisG ura3/ura3 cdc16-1/cdc16-1 | R. Strich |
| W1588-4C | MATa RAD5 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | R. Rothstein |
Isogenic relative to W1588-4C.
Isogenic relative to LNY150.
Isogenic relative to JG192.
Sporulation induction and spore analysis.
For sporulation, diploid yeast cells were grown in presporulation medium (2% Bacto peptone, 1% yeast extract, 2% K acetate) for approximately 15 h to an optical density at 600 nm (OD600) of approximately 1. Yeast cells were then washed with water and resuspended in sporulation medium (2% K acetate supplemented with auxotrophic amino acids) at an OD600 of 2. Samples were taken for Western blotting or reverse transcription-quantitative PCR (RT-qPCR) analysis at the indicated time points. Progression through sporulation was determined by fixing cells equivalent to 1 OD600 unit in 50% ethanol at various time points after induction of sporulation. Cells were then incubated in 100 μl of 10 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) stain (Sigma) for 10 min. The cells were washed once in water and then placed on a glass slide. A Nikon upright fluorescence microscope was used to count 200 cells for each sample. Cells with one nucleus were counted as vegetatively growing yeast asci with two nuclei were counted as dyads, and asci with three or more visible nuclei were counted as tetrads. Sporulation efficiency was determined after 48 h of incubation in sporulation medium and equals the percentage of cells induced to sporulate that became mature tetrads. Nuclear size assays were performed as previously described (31).
Purification of chromatin in yeast and spores.
The preparation of nuclei was based on published methods (18, 29). A dit1Δ mutant was used to make possible the digestion of the spore wall by Zymolyase. In brief, yeast or spores were incubated with two volumes of 2.5 mM EDTA–0.7 M β-mercaptoethanol for 30 min at 30°C. Cells were centrifuged at 3,000 × g and washed in 1 M sorbitol. Cells were resuspended in 1 M sorbitol–5 mM β-mercaptoethanol. Cells were incubated with Zymolyase 100T (0.4 mg/ml) at 30°C for 15 min to 1 h in order to achieve complete digestion of the cell wall. The digestion was monitored by adding 1% SDS to 20 μl of the digestion reaction and taking the OD at 600 nm. The digestion was considered complete when the OD600 dropped by 95% (Fig. 1B). Cells were washed in 1 M sorbitol with a protease inhibitor cocktail (Complete; Roche). Cells were lysed by osmotic shock in 7 volumes of 18% Ficoll–20 mM potassium phosphate (pH 6.8)–1 mM MgCl2–0.5 mM EDTA. Nuclei were purified by centrifugation at 30,000 × g for 30 min at 4°C. Purified nuclei were then incubated on ice for 30 min. Acid extraction (to enrich for basic proteins) was accomplished by resuspending the purified nuclei in 0.4 M H2SO4. Centrifugation at 20,000 × g for 15 min at 4°C removed precipitated debris. Proteins from the supernatant were precipitated by addition of 20% trichloroacetic acid (TCA), pelleted by centrifugation, and washed in acetone–0.05% HCl and then again in acetone alone. The pellet was briefly dried, and proteins were resuspended in water and resolved on an SDS-polyacrylamide gel.
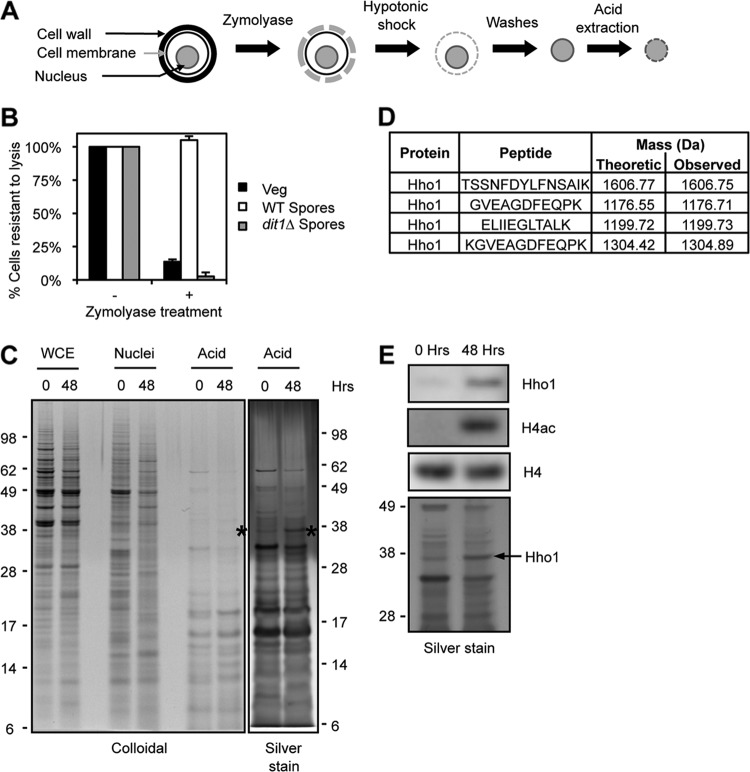
Fig 1.
Differential analysis of spore chromatin and identification of Hho1. (A) Purification scheme. The cell wall is first digested with Zymolyase. Nuclei are then released by a hypotonic shock and washed. Finally, basic proteins present in the chromatin are extracted with sulfuric acid. (B) Evaluation of the Zymolyase digestion. Cells with an intact wall are resistant to lysis in 1% SDS. Vegetative cells are sensitive to Zymolyase (black bars), but wild-type spores are resistant (white bars). Mutation of the DIT1 gene impairs the maturation of the spore wall, which can then be digested by Zymolyase (gray bars). (C) Protein profiles obtained at each purification step, stained by colloidal Coomassie blue or silver stain, in vegetatively growing cells (0 h) or in mature spores (48 h). Asterisks mark Hho1. Molecular masses are indicated on the sides of the gels (kDa). (D) Hho1 peptides identified by mass spectrometry analysis of the band indicated with asterisks in panel C. (E) Western blot analysis of acid extracts reveals that Hho1 is present at higher levels in spore chromatin (48 h) than in that from yeast in presporulation medium (0 h). H4 acetylation is detected with an antibody raised against a tetra-acetylated peptide (H4K5,8,12,16Ac). Molecular masses are indicated on the side of the silver-stained gel (kDa).
Yeast whole-cell extract preparation and Western blot analysis.
Yeast cells were harvested by centrifuging cultures at 4,000 rpm for 2 min at 4°C. The cells were washed with ice-cold water once and broken in lysis buffer (50 mM Tris-HCl [pH 7.4]; 300 mM NaCl; 0.5% NP-40; 10% glycerol; 1 mM EDTA [pH 8]; protease inhibitors that included pepstatin, leupeptin, aprotinin, and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]; phosphatase inhibitor cocktail from Sigma) using silica beads and bead beater (Bio-Spec). The lysate was sonicated (Bioruptor; Diagenode) for 5 min and cleared at 14,000 rpm at 4°C for 15 min. The supernatant was collected and stored at −80°C. Protein estimation was determined using Bradford dye. Extracts were resolved on SDS-polyacrylamide gels, and images of the resultant gels or Western blots were taken with a Fujifilm LAS-4000 imager. Quantification and contrast adjustment of images were carried out with ImageJ software.
RNA analysis.
RNA was purified using the Qiagen miRNeasy purification kit (217004) according to the manufacturer's instructions. Reverse transcription was performed using the Applied Biosystems high-capacity RNA-to-cDNA kit (4387406). cDNA was quantified using standard procedures on a 7900HT Fast-Real-Time PCR machine (ABI). During vegetative growth, mRNA levels were normalized to ACT1 or GAPDH. During sporulation, mRNA levels were normalized to NUP85 levels, which appear to be constant in three independent genome-wide transcriptome analyses (6, 11, 43). Primers are presented in Table 2. Three or four independent biological replicates were analyzed, and each data point acquired by qPCR was the average from three independent PCRs.
Table 2.
Primers used in this study
| Gene or locus | Forward/reverse primer sequence (5′→3′) |
|---|---|
| DIT1 | TTTTTTACATATTCCCACACCTTGG/TCCTCAACCTTCACGACACAAT |
| GAPDH promoter | TCCACGTGCAGAACAACATAGTC/ATGAACATGCTCCTCCCCC |
| GAPDH gene body | TTACACCGAAGATGCCGTTG/GCGTGAGTGTCACCCAAGAA |
| HHO1 | GATCATTGAAGGGCTCACGG/GGACGACTGGATCCCTTACGT |
| HOP1 promoter | AAAGCCGCTGGTCCAAGAA/TTGGGTCCTGTATATGTCTTGTAAATCA |
| HOP1 gene body | CAAATAACCCAGTCACGGGC/GAACTTCCAATCCACATTCGC |
| LEU3 | CTGTATATTCCGTGCAAGCATTTC/TTAAGGGAGGCCAGAACGTG |
| MEI4 promoter | TTTAGAAGAGTAAGGACATGAGTAGAGGC/TTTGTTCCATATCTTCCAGTTTGC |
| MEI4 gene body | CCAAAACGATCCAAAATGGCT/TTGCCATCTCCCTGGTCG |
| MEK1 | CACTCCGGAATACTGCGCTC/GCTTTTCTGTTTGCTCTGAATCC |
| MSH5 | GGCAGGGTTCTTGAGCTGG/AATCGCGCCATAAATATCGC |
| NUP85 | CAGCAAAGAGTTTTCTGCATACGTATCAGG/ATTAAACACTCTGTCATCACCTAAATCACGG |
| PCH2 promoter | GAAAGTTTCCCAAAAGACCGTTTA/TTCTTGGCCAGTTATAATCGAAGTC |
| PCH2 gene body | ACTCCTTGTCCATGGTCCTCC/GCTTTGCATAACGTTGTTTTACCTG |
| REC104 promoter | GAAGAAAAGAACATACAGCAACTTGG/TCTTCCTCCTCGATGGACATG |
| REC104 gene body | AAATCTGCAACAGTATCTCTTCCGT/CATGCATACTGGGTCGCCT |
| SEC13 promoter | AAATGGTCGTCATAGCTAATGCG/GAACAGCGTCATGGATTAATTCG |
| SEC13 gene body | GGTACTACTTCCCCAATAATCATCG/AGAGTTAACGCCAATGGCATG |
| SMK1 promoter | TTTGTTTGCCCACCGCTAA/TTTGCCGCCTATTTTTTTCG |
| SMK1 gene body | TGTGCACATTGAATGGCTGTT/CCCTAGCGAGACCAAAATCG |
| SPO11 | CTCCAACGTGGAATTGTTTCAA/TCCAGCCACTGGACTACGTTT |
| TUB1 promoter | TTCTCGCCACCCAAGATCTG/CTCTCATTGTTTGTTTGCAGTTGTAA |
| TUB1 gene body | ATCAAAGGCATTCCATGAGTCC/CAAGCGTTTGTAATTTCTGACAC |
Chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR).
Approximately 50 OD600 units of cells were cross-linked in 1% formaldehyde for 10 min at 25°C, quenched with 125 mM glycine for 5 min at 25°C, and washed with water. Extracts were made as described above with FA lysis buffer (50 HEPES mM [pH 7.5], 140 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.5 mM PMSF, aprotinin [2 mg/liter], leupeptin [2 mg/liter], pepstatin A [2 mg/liter], trichostatin A [TSA; 100 mg/liter], phosphatase inhibitor cocktail [Sigma, P2850]). Extracts were sonicated 32 times for 30 s each with intermediate incubation of 30 s (Bioruptor; Diagenode). Cellular debris was removed by centrifugation at 20,000 × g for 15 min at 4°C. The protein concentration in the supernatant was quantified by the Bradford assay. Fifty micrograms of extract was used for input samples, 500 μg of extract was used for Hho1 ChIP with 5 μl of antibody per sample, and 30 μl of protein G Dynabeads (Invitrogen, 100.02D) was used per sample. Controls with rabbit IgG were routinely performed and did not show specific signals. Primers are presented in Table 2. Three or four independent biological replicates were analyzed, and each data point acquired by qPCR was the average from three independent PCRs. Error bars in the figures are standard errors of the means (SEM).
Chromatin immunoprecipitation followed by sequencing (ChIP-seq).
Hho1-3×FLAG and wild-type (WT) (mock ChIP) diploid cells were grown in presporulation medium (“0 h”) or sporulated to maturity for 48 h. Approximately 50 OD600 units of cells were cross-linked in 1% formaldehyde for 10 min at 25°C, quenched with 125 mM glycine for 5 min at 25°C, and washed with water. Cells were resuspended in FA lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.2% SDS, CPI Mini EDTA-free protease inhibitor cocktail [Roche]). One milliliter of zirconia/silica beads was added to each tube, and cells were disrupted twice for 3 min each for the 0-h sample and four times for 3 min each for the 48-h sample at maximum speed with intermediate incubation at −20°C for 3 min (Mini-Beadbeater 96; Biospec). Chromatin was washed twice with FA lysis buffer and sonicated for 30 cycles (30 s “on” at high level and 30 s “off” per cycle) (Bioruptor; Diagenode). Cellular debris was removed by centrifugation at 14,000 rpm for 15 min at 4°C. Extracts were diluted with 3 volumes of FA lysis buffer without SDS (final SDS concentration of 0.05%) and incubated overnight with 200 μl of anti-FLAG M2 resin slurry (Sigma). The resin was washed to eliminate nonspecific binding and resuspended in 100 μl M2 elution buffer (50 ml NaCl, 10 mM Tris-HCl, 1 mM EDTA). Ten microliters of 15-mg/ml 3×FLAG peptide was added to elute Hho1-3×FLAG protein for 60 min at 4°C. Reverse cross-linking was performed at 65°C overnight (no longer than 16 h) with 2 μl proteinase K (20 mg/ml).
ChIP samples were subjected to paired-end sequencing using an Illumina GAII instrument in accordance with the manufacturer's instructions. The midpoint coordinate of pair tags demarcated the binding location of Hho1. The Hho1-3×FLAG ChIP signal was normalized to the WT (untagged) ChIP signal. For scaling the ChIP-seq data as described in reference 58, we used ImageJ to quantify Hho1 and H4 band intensities from their respective Western blots. Hho1 signal was normalized to the amount of H4 present (loading control), and we determined that there was a 5-fold increase of Hho1 between the 0-h and 48-h time points (Fig. 1E and other replicates not shown). Two biological replicates of each time point were merged to demonstrate that the resulting patterns were reproducible, and the relative tag counts at 0 h were set to be one-fifth of those at 48 h, based on the Western blot results.
ChIP-chip data analysis.
ChIP-chip data were obtained from reference 50. For all tandem affinity purification (TAP)-tagged proteins, signal intensities were normalized against an untagged control IP. The maximum value of the two promoter probes designed for each gene was used as a measurement of the promoter binding by each protein. A standard coefficient of determination was computed after removing genes with partial or missing measurements. We performed the same analysis solely for genes in cluster 4 of the transcriptional clustering performed previously (43), which are genes expressed in early meiosis.
RESULTS
Differential analysis of spore chromatin and identification of Hho1.
Over the course of sporulation, the nuclear volume of budding yeast is decreased as much as 10-fold (J. Govin and S. L. Berger, unpublished data). The extent of genome compaction in spores is similar to that in mammalian sperm, but mammals and other higher eukaryotic organisms have evolved a dedicated set of highly basic, sperm-specific proteins called protamines for this purpose (27). We failed to identify protamine homologues in an in silico search of the yeast genome. Moreover, detection of histones by Western blot analysis suggests that they are largely maintained in spores and are not generally replaced (this work and reference 17). This conclusion is confirmed by ChIP sequencing analysis showing that nucleosome positioning remains relatively constant during sporulation (58). Therefore, we hypothesized that other chromatin-associated factors may be acting to compact the spore genome.
We developed an unbiased biochemical approach to identify nuclear factors specifically enriched in spore chromatin compared to that in vegetatively growing yeast (Fig. 1A). We first used a combination of Zymolyase and mutation of the DIT1 gene to digest the highly resistant sugar polymer- and dityrosine-enriched cell wall of a >98% pure population of spores (Fig. 1B). We then purified nuclei using standard yeast protocols involving a hypotonic shock followed by high-speed centrifugation (Fig. 1A). Overall, the protein profiles were similar in nuclear extracts from vegetatively growing yeast (0 h) and spores (48 h) (Fig. 1C, Nuclei).
Highly basic chromatin-associated proteins such as histones or protamines are classically purified using acid extraction. To minimize background levels of nuclear proteins, we performed an acid extraction of nuclei from vegetatively growing yeast and spores (Fig. 1A). This acid extraction led to the identification of a protein that was highly enriched in spore chromatin, clearly revealed by silver staining (Fig. 1C, asterisk). Mass spectrometry analysis of this band revealed several peptides corresponding to a single yeast protein, Hho1, the yeast H1 linker histone (Fig. 1D). We confirmed enrichment of Hho1 in spore chromatin by comparing chromatin extracts from vegetatively growing yeast and spores by Western blot analysis (Fig. 1E). Hho1 protein levels are much higher in the chromatin of mature spores, which is also known to be marked by acetylation of H4K5, K8, K12, and K16 (Fig. 1E) (17).
Hho1 is depleted in meiotic yeast and enriched in mature spores.
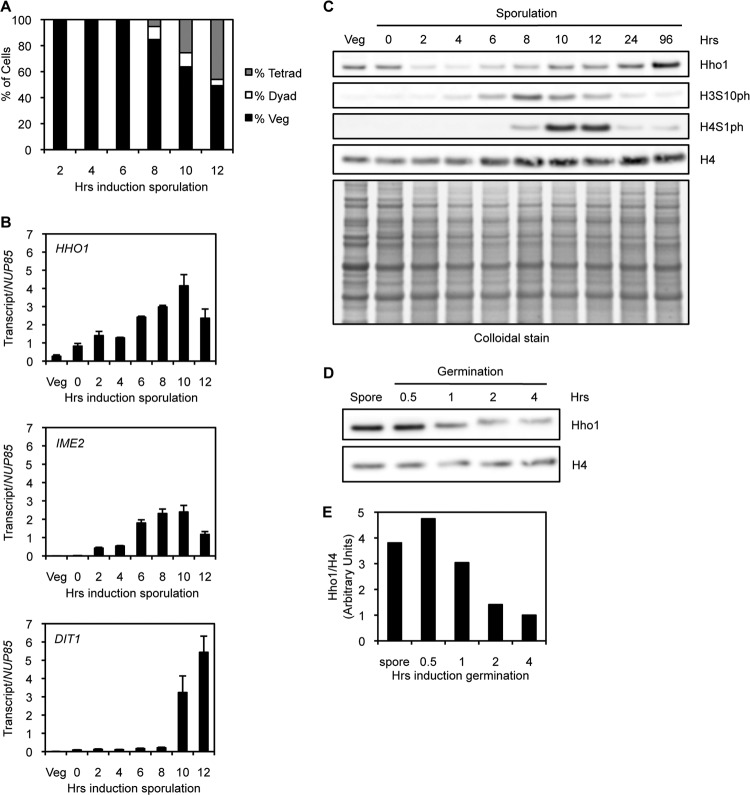
To characterize Hho1expression patterns during the sporulation process, we performed a sporulation time course and measured HHO1 transcript and protein levels by reverse transcription followed by quantitative PCR (RT-qPCR) and Western blotting. Diploid yeast cells were induced to enter sporulation synchronously, samples were taken at the indicated time points, and progression of meiosis was monitored by DAPI staining (Fig. 2A). HHO1 transcript levels are lowest in vegetatively growing yeast, slowly increase during meiosis (2 to 8 h postinduction [p.i.] of sporulation), peak near the end of meiosis (10 h p.i. of sporulation), and then decrease after meiosis (12 h p.i. of sporulation), a time when transcription of most genes decreases as the nucleus is compacted (Fig. 2B, top panel). The transcription pattern of HHO1 is similar to that of a well-characterized early meiotic gene, IME2, and is distinct from that of a late meiotic gene, DIT1 (Fig. 2B, middle and bottom panels).
Fig 2.
Hho1 is depleted in meiotic yeast and enriched in mature spores. (A) A sporulation time course indicates the percentage of cells/asci with a single nucleus (vegetatively growing), two nuclei (dyad), or three or more nuclei (tetrad). Diploid yeast cells are deprived of nutrients, induced to enter sporulation synchronously, and stained with DAPI at different times postinduction. Meiotic divisions begin at approximately 8 h after induction of sporulation. (B) RT-qPCR of RNA from yeast harvested at different times over the course of sporulation. HHO1 is transcribed at a basal level in vegetatively growing yeast (Veg), but its transcription increases over the course of meiosis. The HHO1 transcription pattern is similar to that of an early meiotic gene (IME2) and temporally distinct from that of a late meiotic gene (DIT1). NUP85 is used as a control transcript for all time points. (C) Western blot analysis of protein extracts from yeast harvested at different times over the course of sporulation. Hho1 protein levels decrease dramatically during early meiosis and peak in mature spores. H3S10ph marks meiotic divisions, and H4S1ph marks the beginning of postmeiotic compaction. Total histone H4 and a colloidal stain of total protein are used as loading controls. (D) Western blot analysis of protein extracts from yeast harvested at different times over the course of germination. Cell division begins at 4 h after induction of germination. Hho1 protein levels decrease during germination as spores reenter the cell cycle. Total histone H4 is used as a loading control. (E) Quantification of the Western blot in panel D. Hho1 protein levels are normalized to H4 protein levels. The ratio in vegetative yeast (4 h after induction of germination) is set to 1.
Hho1 protein levels show a distinct pattern compared to HHO1 transcript levels. The protein is present at a basal level in vegetatively growing yeast, but levels decrease dramatically during the onset of meiosis (2 to 4 h p.i. of sporulation) (Fig. 2C). Interestingly, Hho1 protein levels are at their lowest when DNA is undergoing replication and crossing-over. Moreover, HHO1 transcript levels are relatively stable during the onset of meiosis (Fig. 2B), suggesting that Hho1 protein levels are controlled posttranscriptionally. Hho1 protein then rises to the basal level after meiosis is complete (10 to 12 h p.i. of sporulation), a stage that is also marked by the attenuation of H3S10ph, a marker of meiotic divisions, and rising levels of H4S1ph, which is involved in postmeiotic chromatin compaction, (Fig. 2C) (31). Finally, Hho1 levels peak in mature spores (96 h p.i. of sporulation), where the nucleus is most compact, suggesting a role for Hho1 during genome compaction at the end of sporulation (see below). Hho1 protein returns to basal levels as spores are given nutrients and induced to reenter the cell cycle through germination (Fig. 2D and E).
Hho1 helps to compact the spore nucleus late in sporulation.
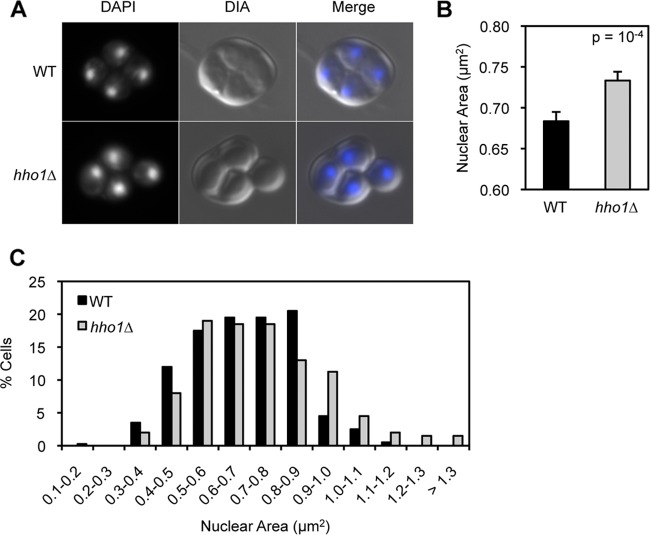
Because Hho1 is enriched in mature spore chromatin, we hypothesized that it may be acting to compact the genome in mature spores. To determine if Hho1 is required for spore genome compaction, we compared the nuclear areas of WT and hho1Δ spores. Cells were collected after 48 h in sporulation medium, fixed with formaldehyde, adhered to slides, and stained with DAPI. Images were taken using an E600 Nikon upright microscope (an example is shown in Fig. 3A). Quantitative immunofluorescence of spore nuclei revealed that hho1Δ spores have a significantly larger nuclear area (0.73 μm2) than WT spores (0.68 μm2) (Fig. 3B). Indeed, a histogram demonstrating the range of nuclear area measurements from WT and hho1Δ spores shows that a greater percentage of hho1Δ spores fall into larger nuclear size categories, while a greater percentage of WT spores fall into smaller nuclear size categories (Fig. 3C). Thus, the results suggest that Hho1 is required for normal compaction of spore nuclei. Interestingly, WT and hho1Δ spores germinate with the same efficiency (data not shown), suggesting that hho1Δ yeast that successfully completes meiosis and progresses to the mature spore stage is viable despite the defect in nuclear compaction.
Fig 3.
Hho1 helps to compact the nucleus late in sporulation. (A) WT and hho1Δ spores stained with DAPI demonstrate an increase in nuclear size of hho1Δ compared to WT spores. (B) hho1Δ spores show a statistically significant increase in nuclear size compared to WT spores. Six hundred spore nuclei were counted per strain. Error bars represent standard deviations. (C) The measurements from panel B were clustered into 0.1-μm2 intervals and are presented as a distribution across the intervals. A higher percentage of hho1Δ spores than of WT spores show larger nuclear areas.
Hho1 binds linker DNA and helps to compact the spore nucleus by increased binding genome-wide.
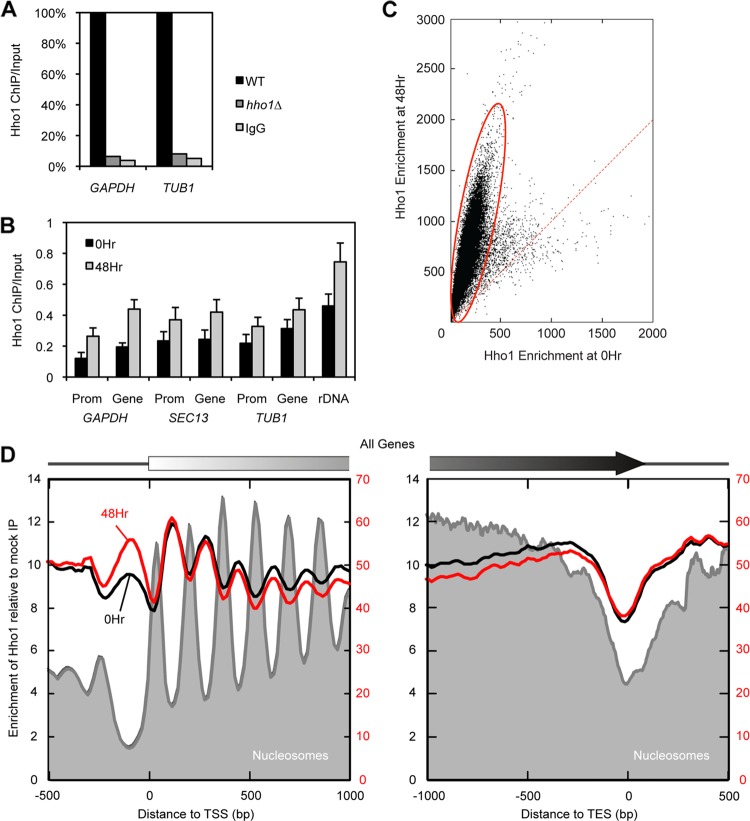
It is possible that increased genome-wide Hho1 binding is responsible for spore genome compaction. Hho1 protein levels are much higher in spores than in vegetatively growing yeast (Fig. 1E), while the number of nucleosomes associated with the DNA does not change appreciably (58). Thus, the increased ratio of linker histone to nucleosome may contribute to the compaction of the genome. In order to uncover differences in Hho1 binding patterns during sporulation, we performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) in growing yeast and spores. To demonstrate the specificity of the Hho1 antibody, we carried out ChIP-qPCR with an antibody against endogenous Hho1 in WT and hho1Δ cells (Fig. 4A). The ChIP enrichment obtained with the Hho1 antibody in WT cells was approximately 20 times greater than that obtained with the Hho1 antibody in hho1Δ cells or with IgG in WT cells, as measured by qPCR of housekeeping gene loci. These results confirm the specificity of the Hho1 antibody which we used to perform ChIP-qPCR in growing yeast and spores. Hho1 binding increases over the course of sporulation at rDNA and at promoters and gene bodies of the GAPDH, SEC13, and TUB1 housekeeping genes (Fig. 4B).
Fig 4.
Hho1 binds linker DNA and helps to compact the spore nucleus by increased binding genome-wide. (A) ChIP-qPCR of Hho1 within the GAPDH and TUB1 gene bodies shows the specificity of the antibody against endogenous Hho1. Hho1 ChIP was performed in vegetatively growing WT (set to 100%) and hho1Δ cells. An IgG ChIP control was performed in WT cells. (B) ChIP-qPCR shows that Hho1 binding increases at various housekeeping genes (Gene) and their promoters (Prom) in mature spores compared to yeast cells before the onset of sporulation. Cells were cross-linked in presporulation medium (0 h) and after 48 h in sporulation medium. (C) Comparison of Hho1 enrichment genome-wide before (0 h) and after (48 h) sporulation. The majority of loci show increased enrichment in spores (circled in red). A small population of loci shows constant Hho1 enrichment and overlaps significantly with genes expressed in the spore (P < 0.01). Each dot corresponds to a 100-bp section of the genome. The x and y coordinates represent the total normalized tag counts. Total tag counts between the two conditions were scaled to reflect Hho1 protein levels as determined by Western blotting. The dotted red line marks the 1:1 enrichment ratio. (D) Hho1 binding pattern averaged over all genes (5′ and 3′ ends) before (0 h) and after (48 h) sporulation. Scaled tag counts for Hho1-3×FLAG ChIP and mock-IP data sets were binned in 15-bp intervals relative to the TSS (left panel) or TES (right panel) of 6,576 genes. The ChIP values were then normalized to the mock-IP values and plotted with a 3-bin moving average. Also shown are occupancy levels of nucleosomal midpoints measured in the same genetic background (58).
To gain insight into the changes occurring in the genome-wide Hho1 binding pattern during sporulation, we performed chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq) using yeast in which HHO1 was tagged with the FLAG epitope. Cells were cross-linked and harvested before sporulation was induced (0 h) and 48 h after sporulation was induced (48 h). The Hho1-3×FLAG ChIP signal was normalized to a mock ChIP signal obtained from yeast with untagged Hho1. To compare the 0-h and 48-h data sets, we scaled the data (multiplied by a constant) to reflect the ratio of Hho1 protein present at 0 h and 48 h, as measured by Western blotting (Fig. 1E).
A scatter plot comparison of Hho1 enrichment genome-wide at 0 h versus 48 h revealed two trends of Hho1 occupancy over sporulation (Fig. 4C). The vast majority of genomic loci increased in Hho1 occupancy (highlighted by a red circle in Fig. 4C); however, a second population, encompassing approximately 260 genes, showed a fairly constant Hho1 occupancy between 0 h and 48 h. These genes were enriched in the Gene Ontology categories of gluconeogenesis (GO:0006094; P value, 1.7 × 10−6) and stress response (GO:0006950; P value, 3.4 × 10−6), both of which include genes that are expressed in the later stages of sporulation (25). These results suggest that Hho1 may compact the mature spore genome through a general increase in genome-wide occupancy, with the exception of genes that are expressed late in sporulation.
To obtain a high-resolution view of Hho1 occupancy over genes, we generated a composite plot for Hho1 enrichment relative to the transcriptional start sites (TSS) and transcriptional end sites (TES) of all genes (Fig. 4D). This genome-wide data set revealed a striking enrichment of Hho1 binding in nucleosomal linker regions in both the 0-h and 48-h data sets, having the same periodicity but opposite phase as nucleosomes mapped in a previous study (58). Interestingly, Hho1 was also detected in 5′ nucleosome-depleted promoter regions (NDRs), but not at 3′ NDRs, at the ends of genes. Indeed, Hho1 enrichment increases most significantly at the 5′ NDRs between the 0-h and 48-h time points. These findings suggest that Hho1 may contribute to spore genome compaction and to stabilization of the transcriptionally silent chromatin state by increased binding over both the promoters and bodies of genes.
Hho1 is required for efficient sporulation and plays a role in the progression of meiosis.
Hho1 has been implicated in sporulation, although its function in this context remains unclear (41). To investigate this potential role, we created hho1Δ diploid yeast, induced WT and hho1Δ yeast to sporulate, and compared their sporulation efficiencies (percentage of cells induced to sporulate that become mature spores) after 2 days in sporulation medium. Interestingly, hho1Δ yeast shows a statistically significant decrease in sporulation efficiency compared to that of WT yeast, where 53% of hho1Δ yeast cells become mature tetrads, compared to 64% of WT yeast cells (Fig. 5A). Thus, Hho1 is necessary for efficient sporulation in S. cerevisiae.
Fig 5.
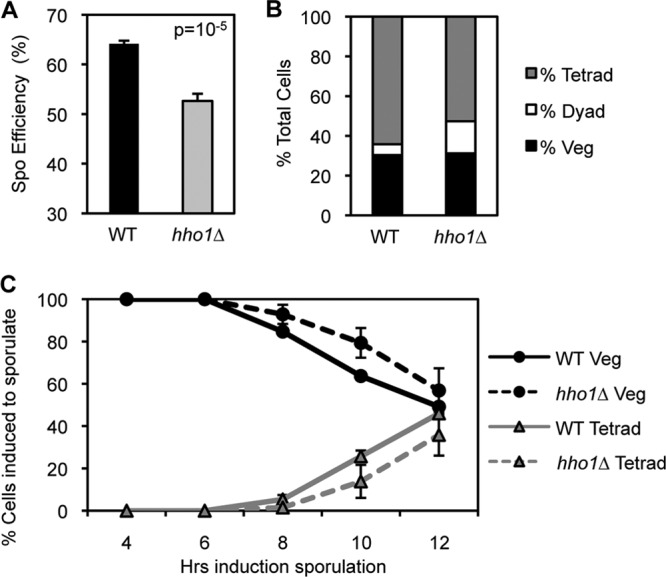
Hho1 is required for efficient sporulation and plays a role in the progression of meiosis. (A) hho1Δ yeast shows a decrease in sporulation efficiency compared to WT yeast. WT and hho1Δ diploid yeasts were incubated in sporulation medium for 48 h and stained with DAPI. Sporulation efficiency is the percentage of cells induced to sporulate that become tetrads. (B) hho1Δ yeast shows a 3-fold increase in the percentage of cells that become dyads compared to WT yeast after 48 h in sporulation medium. (C) hho1Δ yeast is delayed 2 h in its progression through meiotic divisions compared to WT yeast. A sporulation time course indicates the percentage of cells/asci with a single nucleus (Veg) or three or more nuclei (Tetrad). Diploid yeast cells are deprived of nutrients, induced to enter sporulation synchronously, and stained with DAPI at different times postinduction. Meiotic divisions begin at approximately 8 h after induction of sporulation.
In order to better characterize the sporulation defect seen in hho1Δ strains, we compared the meiotic progression of WT and hho1Δ diploids. Both were induced to enter sporulation synchronously, and samples were taken at the indicated time points. Cells were stained with DAPI, and the number of nuclei per cell/ascus was counted. After 48 h in sporulation medium, the hho1Δ culture contained three times as many dyads (asci with two nuclei) as the WT culture, indicating that a percentage of hho1Δ yeast cells arrest between the first and second meiotic divisions (Fig. 5B). In addition, hho1Δ yeast shows a 2-h lag in timing of meiotic divisions compared to WT yeast (Fig. 5C). These results suggest that Hho1 may play a role in the progression of meiosis.
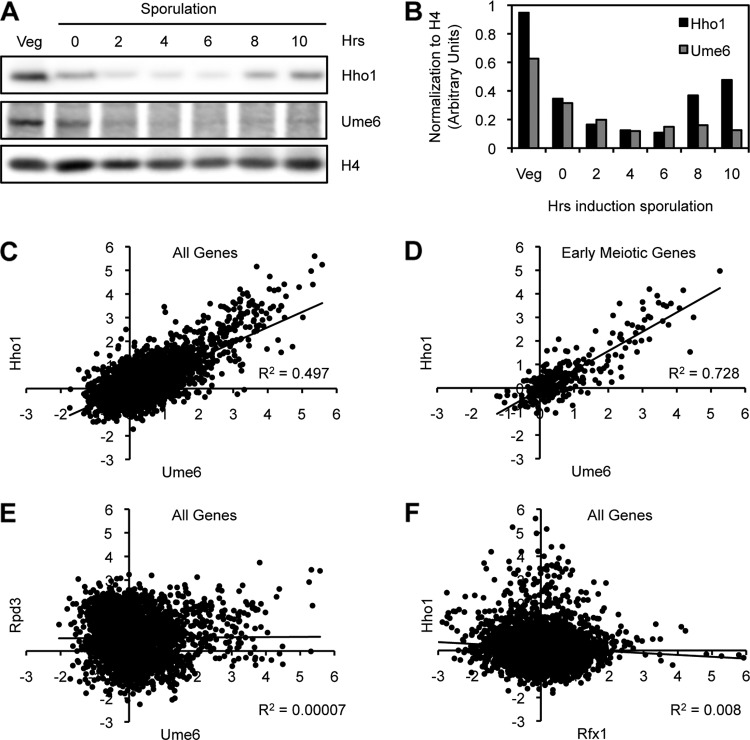
Hho1 and Ume6 are depleted during meiosis and show a high genome-wide binding correlation.
Hho1 depletion may be required during meiosis if it represses the transcription of genes that are expressed specifically during meiosis. It was previously shown that the master repressor of early meiotic genes, Ume6, is actively degraded during meiosis to permit the activation of early meiotic genes (38). Since Hho1 protein levels are also reduced early in meiosis (Fig. 2C), we compared the profiles of Ume6 and Hho1 protein expression in the same sporulation time course using Western blot analysis (Fig. 6A). A quantitative analysis of the Western blot revealed that Hho1 and Ume6 are depleted at the onset of meiosis with similar kinetics when normalized to H4 levels, which remain relatively constant over the course of sporulation (Fig. 6B).
Fig 6.
Hho1 and Ume6 are depleted during meiosis and show a high genome-wide binding correlation. (A) Hho1 and Ume6 proteins are depleted with the same kinetics at the onset of meiosis (2 to 4 h after induction of sporulation). Western blot analysis of protein extracts from yeast harvested at different times over sporulation was carried out for Hho1 and Ume6. (B) Quantification of the Western blot in panel A. Hho1 and Ume6 levels are normalized to H4. (C to F) A ChIP-chip study of many chromatin-associated proteins in vegetatively growing yeast (50) was analyzed for binding correlations between various factors. Hho1 and Ume6 binding is well correlated at the promoters of all genes (C) but is most highly correlated at the promoters of early meiotic genes (D). Such a high correlation is not seen for Ume6 and Rpd3, a known corepressor in the Ume6 complex (E), or for Hho1 and Rfx1, a strong transcriptional repressor of DNA damage-regulated genes (F).
During vegetative growth, Ume6 binds to a specific sequence upstream of early meiotic genes and recruits corepressors (1, 14, 26, 40). Upon entry into meiosis, Ume6 is degraded by the anaphase-promoting complex (APC), and early meiotic genes are then activated (38). Moreover, ume6Δ yeast shows major meiotic defects due to the deregulation of the tightly controlled meiotic transcriptional program (47, 54). Similar to Ume6, linker histones in other eukaryotes have generally been associated with transcriptional repression (7).
Recently, a large-scale ChIP-chip study of many chromatin-associated proteins in vegetatively growing yeast revealed a strong binding correlation between Hho1 and Ume6 genome-wide (Fig. 6C) (50). Further analyses of these data revealed that the correlation in binding between Hho1 and Ume6 is strongest at the promoters of genes expressed specifically during early meiosis (Fig. 6D) (43). Indeed, the binding correlation between Hho1 and Ume6 has an r2 value of 0.728 at the promoters of these early meiotic genes, compared to an r2 value of 0.497 at the promoters of all genes in the genome. These binding correlations are much higher than that seen for Ume6 and Rpd3 (r2 = 10−4), a known corepressor of Ume6 (Fig. 6E) (26). Moreover, Hho1 shows a very low genome-wide binding correlation with other strong transcriptional repressors such as Rfx1 (r2 = 10−2), a major mediator of DNA damage-regulated genes (Fig. 6F) (23). These results suggest that the association between Hho1 and Ume6 is highly specific, possibly more specific than the association of Ume6 with its previously established corepressors.
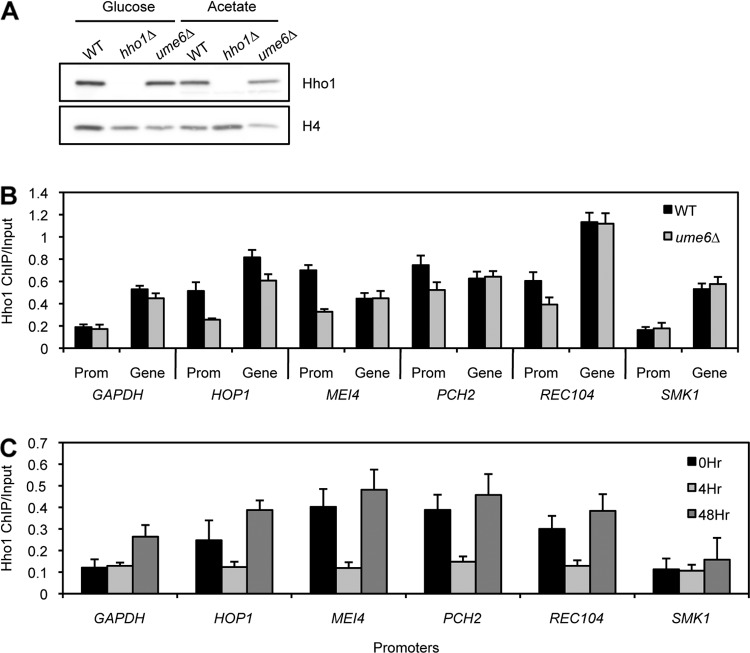
Hho1 enrichment at meiotic gene promoters is Ume6 dependent.
Based on the ChIP-chip data analysis, we hypothesized that Hho1 may be recruited to early meiotic genes by Ume6 to aid in their repression during vegetative growth. To determine if Ume6 recruits Hho1 to the promoters of meiotic genes, we first confirmed that Hho1 protein levels are similar in WT and ume6Δ strains (Fig. 7A) and then performed ChIP-qPCR with an antibody against endogenous Hho1 in these strains grown in presporulation medium (Fig. 7B). Hho1 binding is comparable in WT and ume6Δ strains at the promoters and gene bodies of controls GAPDH, a constitutively active housekeeping gene, and SMK1, a strongly repressed middle sporulation gene that is not controlled by Ume6 (54). Interestingly, in the absence of Ume6, Hho1 is depleted specifically at promoters, but not at gene bodies, of several early meiotic genes that are repressed by Ume6: HOP1, MEI4, PCH2, and REC104. The finding that Hho1 remains bound to gene bodies in the ume6Δ strain serves as a control to show that Hho1 is not evicted from the promoters of these early meiotic genes due to their aberrant activation in the absence of Ume6. These data suggest that the binding of Ume6 to the promoters of early meiotic genes is necessary for Hho1 enrichment at or recruitment to these loci, although we have not detected a physical interaction between Hho1 and Ume6 (data not shown).
Fig 7.
Hho1 enrichment at meiotic gene promoters is Ume6 dependent. (A) Western blot analysis of protein extracts from WT, hho1Δ, and ume6Δ strains shows that Hho1 protein levels are comparable in WT and ume6Δ yeasts growing in glucose or acetate medium. Total histone H4 is used as a loading control. (B) ChIP-qPCR of Hho1 shows a decrease in Hho1 binding in ume6Δ cells specifically at promoters (Prom) of early meiotic genes repressed by Ume6 (HOP1, MEI4, PCH2, and REC104). GAPDH is a constitutively active housekeeping gene, and SMK1 is a middle sporulation gene that is not repressed by the Ume6 complex. (C) ChIP-qPCR of Hho1 shows that it is enriched at the promoters of early meiotic genes before, but not during, meiosis. Cells were cross-linked in presporulation medium (0 h), during meiosis (4 h), and in mature spores (48 h).
Hho1 enrichment at the promoters of early meiotic genes is Ume6 dependent, suggesting that Hho1 may help to repress early meiotic genes in vegetatively growing yeast. It has been demonstrated that Ume6 recruits multiple corepressors, including Rpd3 (14). Thus, we created hho1Δ, rpd3Δ, and hho1Δrpd3Δ mutants and analyzed the levels of early meiotic gene transcription during vegetative growth by RT-qPCR. While the rpd3Δ mutant shows clear derepression of early meiotic genes (MEK1, MSH5, REC104, and SPO11), there is a slight derepression in hho1Δ cells and a trend toward additive derepression in hho1Δrpd3Δ cells (see Fig. S1 in the supplemental material). This derepression was specific to early meiotic genes controlled by Ume6 and not to a middle sporulation gene, SMK1, or a constitutively transcribed gene, LEU3. These data suggest that Hho1 plays a subtle role in the repression of Ume6-controlled early meiotic genes, perhaps stabilizing the repressive chromatin structure created by Rpd3 and other corepressors.
Since Hho1 binding to early meiotic gene promoters is Ume6 dependent, we also hypothesized that Hho1 occupancy specifically at these genes would be reduced during meiosis, when Ume6 is degraded. To investigate the binding pattern of Hho1 over the course of sporulation, we performed ChIP with an antibody against endogenous Hho1 at various points after induction of sporulation (Fig. 7C). WT diploid yeast cells were induced to sporulate synchronously, and cells were harvested and cross-linked in presporulation medium (0 h), during meiosis (4 h), and at the mature spore stage (48 h).
In presporulation medium (0 h), Hho1 is enriched at the promoters of early meiotic genes that are repressed by Ume6 (HOP1, MEI4, PCH2, and REC104) relative to the promoter of GAPDH, a constitutively active housekeeping gene, or SMK1, a strongly repressed middle sporulation gene that is not controlled by Ume6 (Fig. 7C) (54). During meiosis (4 h), Hho1 occupancy drops to a basal level at all loci tested (Fig. 7C). In mature spores, Hho1 binding increases at all loci but most notably at the promoters of meiotic genes controlled by Ume6 (Fig. 7C). These data suggest that Hho1 is evicted from most of the genome during meiosis (possibly because its protein levels are depleted at this time) but is enriched specifically at the promoters of early meiotic genes controlled by Ume6 at all other times during sporulation.
DISCUSSION
The existence of the linker histone in budding yeast was controversial until the sequencing of the genome revealed a gene, HHO1, that showed clear homology to linker histone genes in higher eukaryotes (49). Since the discovery of Hho1, however, its biological function has been unclear, as deletion of the HHO1 gene shows no striking phenotype in vegetatively growing cells (41). Here we characterized the linker histone function during S. cerevisiae sporulation, and we propose a dual role for Hho1: Hho1 depletion early in sporulation may promote meiosis, while Hho1 enrichment late in sporulation helps to facilitate dramatic genome compaction in mature spores.
In agreement with in vitro studies of linker histones in higher eukaryotes, Hho1 plays a role in compacting chromatin (12, 46). Interestingly, the sporulation process does not involve changes in genome-wide nucleosome occupancy and positioning, suggesting that there are other factors involved in compacting the spore genome (58). We found Hho1 to be highly enriched in spore chromatin, and yeast lacking Hho1 shows an increase in the size of the spore nucleus (Fig. 1 and 3). Thus, Hho1 plays a role in genome compaction in the later stages of sporulation. Indeed, our Hho1 ChIP-seq data demonstrate a general increase in Hho1 binding genome-wide in mature spores (Fig. 4).
Additionally, we provide the first in vivo high-resolution, genome-wide evidence that the linker histone binds to nucleosomal linker DNA. Hho1 enrichment at the 5′ ends of genes has the same periodicity, but opposite phase, as nucleosomes mapped in a previous study (Fig. 4D) (58). It has been suggested that in vegetatively growing yeast, Hho1 binds dynamically to the genome and is displaced from genes by the transcriptional machinery (46, 57). Thus, we propose that in the transcriptionally silent genome of the spore, Hho1 may fill in the promoters and open reading frames from where it is evicted by active transcription during the normal cell cycle.
In the sperm of many higher eukaryotes, postmeiotic genome compaction is achieved by the near-complete replacement of histones with highly basic, sperm-specific proteins called protamines. Yeast does not appear to have protamines to facilitate compaction of the genome during sporulation, but our data show that the linker histone is involved in the compaction of the yeast spore genome. Interestingly, evolutionary studies have suggested that protamines may be evolutionarily derived from linker histones (9). In agreement with our findings in yeast, several higher eukaryotic organisms, such as zebrafish, have no protamines and similarly have a higher level of linker histone in their spermatozoa (55).
While hho1Δ yeast shows an increase in the size of the spore nucleus (Fig. 3), it is not a complete decompaction to the size of the nucleus during vegetative growth. Moreover, WT and hho1Δ spores show similar abilities to reenter the cell cycle through germination (data not shown). Thus, Hho1 may have a redundant compaction function in the mature spore with other proteins or postmeiotic chromatin features such as H4Ac and H4S1ph, although the H4S1ph and Hho1 compaction pathways seem to be distinct (references 17 and 31 and data not shown). Interestingly, H4S1ph, H4Ac, and sperm-specific linker histones are all found in spermatogenesis of higher eukaryotes (13, 17, 31), showing conservation of these features.
The second role of Hho1 in sporulation is more complex and appears to be related to gene-specific repression. It is interesting that Hho1 plays a role in chromatin compaction in the late stages of sporulation yet is depleted during meiosis (Fig. 2C), when chromatin is also condensed to a relatively high degree. Thus, we propose a more specific role for Hho1 in two potential pathways that require its depletion during meiosis. First, Hho1 may suppress homologous recombination in meiosis, as previously shown in the context of double-strand break repair (8). Second, Hho1 may contribute to compaction of repressed genes that are specifically transcribed during meiosis. We are currently investigating a role for Hho1 in regulating meiotic recombination; however, we have gained insight into the second hypothesis.
We uncovered several links between Hho1and Ume6, the master repressor of early meiotic genes. Ume6 binds to the promoters of early meiotic genes and recruits corepressor complexes containing Rpd3 and Isw2 to repress these genes until early meiosis, when Ume6 is degraded via action of the APC (14, 26, 38). First, we observed that both Ume6 and Hho1 protein levels drop with similar kinetics during meiosis (Fig. 6A and B). In addition, Ume6 and Hho1 show a high degree of binding correlation at early meiotic genes (Fig. 6C to F), and Ume6 promotes Hho1 binding specifically to the promoters of these genes (Fig. 7B). Finally, Hho1 binding to these Ume6-repressed early meiotic genes is enriched at all times during sporulation except during meiosis (Fig. 7C).
These data suggest that Ume6 recruits Hho1 to early meiotic genes along with the Rpd3 and Isw2 complexes to repress the transcription of these genes until meiosis. The derepression of early meiotic genes in ume6Δ cells leads to severe meiotic defects, which are seen to a lesser extent in rpd3Δ yeast (47, 53, 54). When induced to sporulate, hho1Δ yeast shows a 3-fold increase in cells that are arrested between the first and second meiotic divisions (Fig. 5B), a phenotype similar to, though less severe than, that seen in ume6Δ yeast. The slight derepression of early meiotic genes that we observed in hho1Δ yeast, although less pronounced than that in ume6Δ or rpd3Δ yeast, may contribute to deregulation of the meiotic transcriptional program, resulting in meiotic arrest (see Fig. S1 in the supplemental material) (14). This arrest is not seen in the majority of hho1Δ cells induced to sporulate, likely because the other corepressor complexes recruited by Ume6 compensate for the lack of Hho1 in many cells. Indeed, when combined with the rpd3Δ mutation, hho1Δ shows a slight increase in derepression of meiotic genes over that seen in the rpd3Δ mutant alone (see Fig. S1 in the supplemental material).
We have shown that Ume6 helps to recruit Hho1 to chromatin; however, we have not detected a physical interaction between Hho1 and Ume6 by coimmunoprecipitation (data not shown). Thus, we are still investigating the basis of their interaction, which may be through other corepressors associating with Ume6 or an unknown scaffolding protein. Alternatively, Hho1 may be phosphorylated to affect the interaction, as linker histone association to chromatin is dynamic and affected by phosphorylation in higher eukaryotes (34). Moreover, the heterochromatin protein HP1 has been shown to recruit specifically methylated H1 to the promoters of genes involved in the immune response in Caenorhabditis elegans (48). Thus, it is possible that in S. cerevisiae, posttranslational modification of Hho1 or Ume6 may alter their association.
Additional insight may be gained into the link between Hho1 and Ume6 by investigating the mechanism by which Hho1 is depleted during meiosis. Our study was initially triggered by detection of an overall increase in Hho1 protein levels in mature spores relative to vegetatively growing yeast (Fig. 1); however, further investigation revealed that Hho1 protein levels are strongly reduced during meiosis (Fig. 2C and 6A), a time when new H2A, H2B, H3, and H4 are synthesized and loaded onto replicated DNA. Hho1 protein levels begin increasing after meiosis is completed, peaking in mature spores. Comparison to transcription showed that, unlike the S-phase-specific transcription of the other core histone genes (19), HHO1 transcription appears to take place throughout sporulation, peaking as meiosis ends. These data suggest that Hho1 depletion during meiosis is controlled posttranscriptionally by a pathway yet to be determined, as we have seen Hho1 depletion in the absence of a functional APC (see Fig. S2 in the supplemental material).
Although Hho1 levels are largely depleted during meiosis, a basal level of Hho1 protein is detectable and binds to the genome (Fig. 2C and 7C). Hho1 has been shown to prevent homologous recombination in the context of double-strand DNA breaks (8). Some Hho1 may remain bound to the genome during meiosis in order to prevent excessive homologous recombination, resulting in meiotic defects if Hho1 is absent. Interestingly, linker histones have also been shown to be essential for meiotic progression in plants (44).
Overall, our data reveal Hho1 to be a versatile protein subject to intricate regulation. Its Ume6-dependent recruitment to early meiotic genes supports a growing body of evidence that linker histones, although ubiquitous throughout the genome, may play specific roles in the repression of related genes via transcription factor recruitment (10, 33, 37, 48, 52). In addition, Hho1 plays a role in genome compaction specifically in the late stages of sporulation. Moreover, its role in spore genome compaction is consistent with an evolutionary link to the protamines of higher eukaryotic gametogenesis. Future studies will further unravel physiological roles of the once elusive yeast linker histone in the complex process of sporulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Berger lab for all their support and advice. We thank Alan Fox and Jonathan Schug of the University of Pennsylvania Functional Genomics Core for their help with ChIP sequencing and the Wistar Proteomics Core for help with mass spectrometry. We thank Randy Strich for helpful insight and yeast strains. We thank Pierre-Antoine Defossez for sharing his antibody against Hho1 in the initial step of this project.
This research was supported by NIH grants GM055360 to S.L.B. and HG004160 to B.F.P. J.M.B. was supported by the T32 Genetics Training Grant at the University of Pennsylvania (GM008216). J.G. was supported by several programs of the French Fondation pour la Recherche Medicale (Program Post Doc and Retour de Post Doc) and by the Philippe Foundation.
Footnotes
Published ahead of print 14 May 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Anderson SF, Steber CM, Esposito RE, Coleman JE. 1995. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a zinc-dependent manner. Protein Sci. 4:1832–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balhorn R. 2007. The protamine family of sperm nuclear proteins. Genome Biol. 8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brar GA, et al. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335:552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braunschweig U, Hogan GJ, Pagie L, van Steensel B. 2009. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 28:3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brykczynska U, et al. 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17:679–687 [DOI] [PubMed] [Google Scholar]
- 6. Chu S, et al. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699–705 [DOI] [PubMed] [Google Scholar]
- 7. Croston GE, Lira LM, Kadonaga JT. 1991. A general method for purification of H1 histones that are active for repression of basal RNA polymerase II transcription. Protein Expr. Purif. 2:162–169 [DOI] [PubMed] [Google Scholar]
- 8. Downs JA, Kosmidou E, Morgan A, Jackson SP. 2003. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol. Cell 11:1685–1692 [DOI] [PubMed] [Google Scholar]
- 9. Eirin-Lopez JM, Ausio J. 2009. Origin and evolution of chromosomal sperm proteins. Bioessays 31:1062–1070 [DOI] [PubMed] [Google Scholar]
- 10. El Gazzar M, et al. 2009. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol. Cell. Biol. 29:1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedlander G, et al. 2006. Modulation of the transcription regulatory program in yeast cells committed to sporulation. Genome Biol. 7:R20 doi:10.1186/gb-2006-7-3-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Georgieva M, Roguev A, Balashev K, Zlatanova J, Miloshev G. 2012. Hho1p, the linker histone of Saccharomyces cerevisiae, is important for the proper chromatin organization in vivo. Biochim. Biophys. Acta 1819:366–374 [DOI] [PubMed] [Google Scholar]
- 13. Godde JS, Ura K. 2009. Dynamic alterations of linker histone variants during development. Int. J. Dev. Biol. 53:215–224 [DOI] [PubMed] [Google Scholar]
- 14. Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423–433 [DOI] [PubMed] [Google Scholar]
- 15. Govin J, Berger SL. 2009. Genome reprogramming during sporulation. Int. J. Dev. Biol. 53:425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. 2004. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur. J. Biochem. 271:3459–3469 [DOI] [PubMed] [Google Scholar]
- 17. Govin J, et al. 2010. Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev. 24:1772–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gregory PD, Horz W. 1999. Mapping chromatin structure in yeast. Methods Enzymol. 304:365–376 [DOI] [PubMed] [Google Scholar]
- 19. Gunjan A, Paik J, Verreault A. 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie 87:625–635 [DOI] [PubMed] [Google Scholar]
- 20. Hammoud SS, et al. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Happel N, Doenecke D. 2009. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 431:1–12 [DOI] [PubMed] [Google Scholar]
- 22. Hellauer K, Sirard E, Turcotte B. 2001. Decreased expression of specific genes in yeast cells lacking histone H1. J. Biol. Chem. 276:13587–13592 [DOI] [PubMed] [Google Scholar]
- 23. Huang M, Zhou Z, Elledge SJ. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595–605 [DOI] [PubMed] [Google Scholar]
- 24. Izzo A, Kamieniarz K, Schneider R. 2008. The histone H1 family: specific members, specific functions? Biol. Chem. 389:333–343 [DOI] [PubMed] [Google Scholar]
- 25. Joseph-Strauss D, Zenvirth D, Simchen G, Barkai N. 2007. Spore germination in Saccharomyces cerevisiae: global gene expression patterns and cell cycle landmarks. Genome Biol. 8:R241 doi:10.1186/gb-2007-8-11-r241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kadosh D, Struhl K. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365–371 [DOI] [PubMed] [Google Scholar]
- 27. Kasinsky HE, Eirin-Lopez JM, Ausio J. 2011. Protamines: structural complexity, evolution and chromatin patterning. Protein Pept. Lett. 18:755–771 [DOI] [PubMed] [Google Scholar]
- 28. Kassir Y, et al. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224:111–171 [DOI] [PubMed] [Google Scholar]
- 29. Kizer KO, Xiao T, Strahl BD. 2006. Accelerated nuclei preparation and methods for analysis of histone modifications in yeast. Methods 40:296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kota SK, Feil R. 2010. Epigenetic transitions in germ cell development and meiosis. Dev. Cell 19:675–686 [DOI] [PubMed] [Google Scholar]
- 31. Krishnamoorthy T, et al. 2006. Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis. Genes Dev. 20:2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lardenois A, et al. 2011. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc. Natl. Acad. Sci. U. S. A. 108:1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee H, Habas R, Abate-Shen C. 2004. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304:1675–1678 [DOI] [PubMed] [Google Scholar]
- 34. Lever MA, Th'ng JP, Sun X, Hendzel MJ. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873–876 [DOI] [PubMed] [Google Scholar]
- 35. Levy A, et al. 2008. Yeast linker histone Hho1p is required for efficient RNA polymerase I processivity and transcriptional silencing at the ribosomal DNA. Proc. Natl. Acad. Sci. U. S. A. 105:11703–11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Longtine MS, et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 37. Mackey-Cushman SL, et al. 2011. FoxP3 interacts with linker histone H1.5 to modulate gene expression and program Treg cell activity. Genes Immun. 12:559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mallory MJ, Cooper KF, Strich R. 2007. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol. Cell 27:951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neiman AM. 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189:737–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park HD, Luche RM, Cooper TG. 1992. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 20:1909–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patterton HG, Landel CC, Landsman D, Peterson CL, Simpson RT. 1998. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem. 273:7268–7276 [DOI] [PubMed] [Google Scholar]
- 42. Pierce M, et al. 1998. Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5970–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Primig M, et al. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415–423 [DOI] [PubMed] [Google Scholar]
- 44. Prymakowska-Bosak M, et al. 1999. Linker histones play a role in male meiosis and the development of pollen grains in tobacco. Plant Cell 11:2317–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sancho M, Diani E, Beato M, Jordan A. 2008. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 4:e1000227 doi:10.1371/journal.pgen.1000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schafer G, McEvoy CR, Patterton HG. 2008. The Saccharomyces cerevisiae linker histone Hho1p is essential for chromatin compaction in stationary phase and is displaced by transcription. Proc. Natl. Acad. Sci. U. S. A. 105:14838–14843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Steber CM, Esposito RE. 1995. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl. Acad. Sci. U. S. A. 92:12490–12494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Studencka M, et al. 2012. Novel roles of Caenorhabditis elegans heterochromatin protein HP1 and linker histone in the regulation of innate immune gene expression. Mol. Cell. Biol. 32:251–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ushinsky SC, et al. 1997. Histone H1 in Saccharomyces cerevisiae. Yeast 13:151–161 [DOI] [PubMed] [Google Scholar]
- 50. Venters BJ, et al. 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 41:480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Veron M, et al. 2006. Histone H1 of Saccharomyces cerevisiae inhibits transcriptional silencing. Genetics 173:579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vicent GP, et al. 2011. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 25:845–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vidal M, Gaber RF. 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:6317–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Williams RM, et al. 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. U. S. A. 99:13431–13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu SF, Zhang H, Cairns BR. 2011. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 21:578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu Q, et al. 2009. Saccharomyces cerevisiae linker histone Hho1p functionally interacts with core histone H4 and negatively regulates the establishment of transcriptionally silent chromatin. J. Biol. Chem. 284:740–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zanton SJ, Pugh BF. 2006. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 20:2250–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang L, Ma H, Pugh BF. 2011. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 21:875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.