Abstract
Recombination between insertion sequence copies can cause genetic deletion, inversion, or duplication. However, it is difficult to assess the fraction of all genomic rearrangements that involve insertion sequences. In previous gene duplication and amplification studies of Acinetobacter baylyi ADP1, an insertion sequence was evident in approximately 2% of the characterized duplication sites. Gene amplification occurs frequently in all organisms and has a significant impact on evolution, adaptation, drug resistance, cancer, and various disorders. To understand the molecular details of this important process, a previously developed system was used to analyze gene amplification in selected mutants. The current study focused on amplification events in two chromosomal regions that are near one of six copies of the only transposable element in ADP1, IS1236 (an IS3 family member). Twenty-one independent mutants were analyzed, and in contrast to previous studies of a different chromosomal region, IS1236 was involved in 86% of these events. IS1236-mediated amplification could occur through homologous recombination between insertion sequences on both sides of a duplicated region. However, this mechanism presupposes that transposition generates an appropriately positioned additional copy of IS1236. To evaluate this possibility, PCR and Southern hybridization were used to determine the chromosomal configurations of amplification mutants involving IS1236. Surprisingly, the genomic patterns were inconsistent with the hypothesis that intramolecular homologous recombination occurred between insertion sequences following an initial transposition event. These results raise a novel possibility that the gene amplification events near the IS1236 elements arise from illegitimate recombination involving transposase-mediated DNA cleavage.
INTRODUCTION
Acinetobacter baylyi ADP1 serves as a model bacterium for investigating gene duplication and amplification (GDA) (40). GDA plays a critical role in adaptation and evolution, and it also underlies a range of health issues, such as drug resistance, cancer, and various human genetic disorders (1, 3, 4, 9, 13, 43). One mechanism for generating duplications involves homologous recombination between stretches of directly repeated DNA, such as closely spaced rRNA operons or different copies of insertion sequences (IS) (2, 10, 23, 27, 30). Therefore, it was surprising that few examples of IS-mediated rearrangement were identified during previous studies of chromosomal GDA in A. baylyi (34, 35, 40).
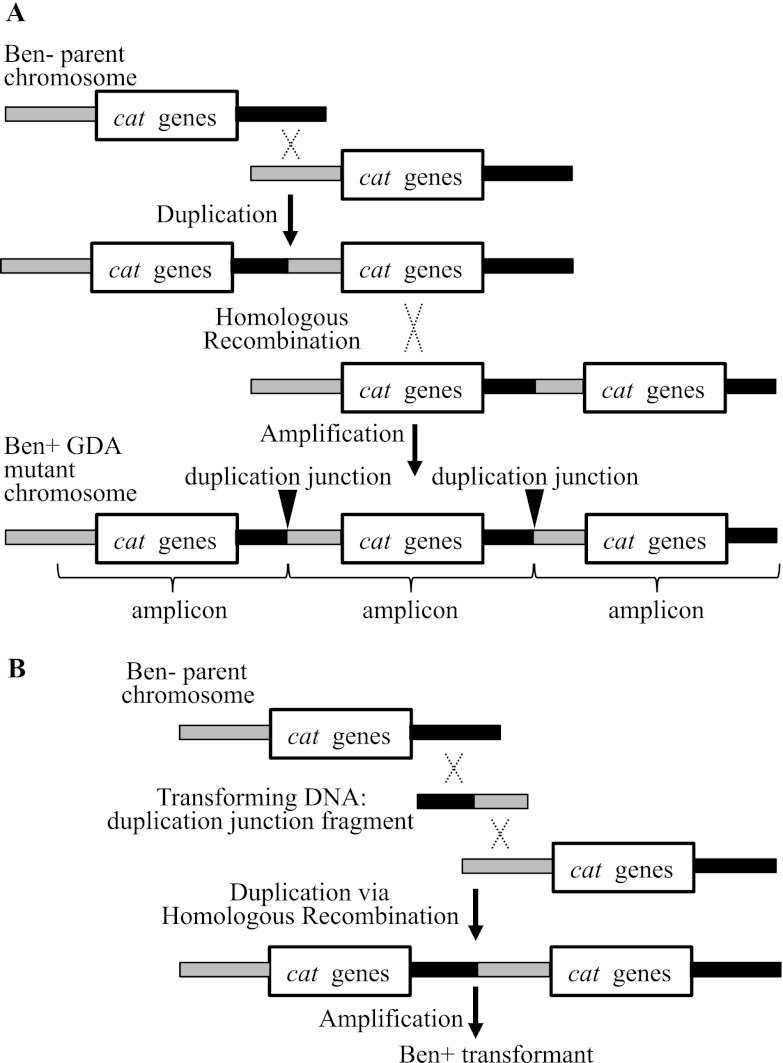
Gene amplification mutants are readily selected from an A. baylyi parent strain that is unable to consume benzoate (Ben−) (12, 16, 34, 40). This parent lacks two regulators that normally activate transcription from multiple promoters in a clustered group of genes for benzoate catabolism. Spontaneous Ben+ derivatives that acquire the ability to grow on benzoate carry tandem head-to-tail arrays of a chromosomal segment (amplicon) encompassing genes needed to degrade benzoate (cat genes) (Fig. 1A) (16, 34, 40). These arrays increase gene expression by providing multiple copies of genes with weak promoters and thereby compensate for the absence of the transcriptional regulators (34, 35, 40). The selection demands increased expression of genes controlled by at least two different promoters, and it yields gene amplification mutants almost exclusively. Furthermore, continued selection maintains the amplified DNA and permits the analysis of amplicon size and copy number. The precise site of duplication, termed the duplication junction, can be identified using an assay that exploits the high efficiency of natural transformation in A. baylyi (Fig. 1B) (34).
Fig 1.
Selection and analysis of Ben+ amplification mutants. (A) Two-step model for the formation of Ben+ amplification mutants. In the first step, a chromosomal region is duplicated by recombination (X) between DNA that is normally downstream of the cat genes (black area) and DNA normally upstream of these genes (gray area) on the Ben− parent chromosome. This step does not depend on homology and may be mediated by illegitimate recombination (34, 35, 40). The “cat genes” label denotes an approximately 10-kbp region, including the catA gene and the catBCIJFD operon, which are transcribed from separate promoters and expressed weakly due to the absence of two transcriptional activators. In the second step of the amplification process, identity between the repeated segments of the duplication enables further increases in copy number via homologous recombination. Multiple copies of the repeated segment, the amplicon, can increase gene expression and thereby generate a Ben+ mutant. The duplication junction defines the endpoints of the amplicon. Its sequence allows inferences about the recombination event that generated the duplication. (B) To identify the duplication junction, fragments of DNA from a Ben+ GDA mutant are used to transform a Ben− parent strain. A Ben+ phenotype generated by homologous recombination signals the presence of the duplication junction DNA fragment.
The A. baylyi system was expanded for genome-wide studies using the 10-kbp cat gene region, which encompasses catA and the catBCIJFD operon, as a selectable cassette for relocation into any nonessential chromosomal locus (14, 40). The cassette is used to generate diverse Ben− parent strains from which Ben+ amplification mutants are selected and analyzed. The frequency of spontaneous duplications that encompass catA can also be assessed regardless of the chromosomal location of this gene (40). The duplication frequency assay detects a small fraction of cells that retain a functional catA after one copy is inactivated by allelic replacement. When grown under conditions that do not select for duplications, 10−4 cells carry a duplication of catA in its native locus (40).
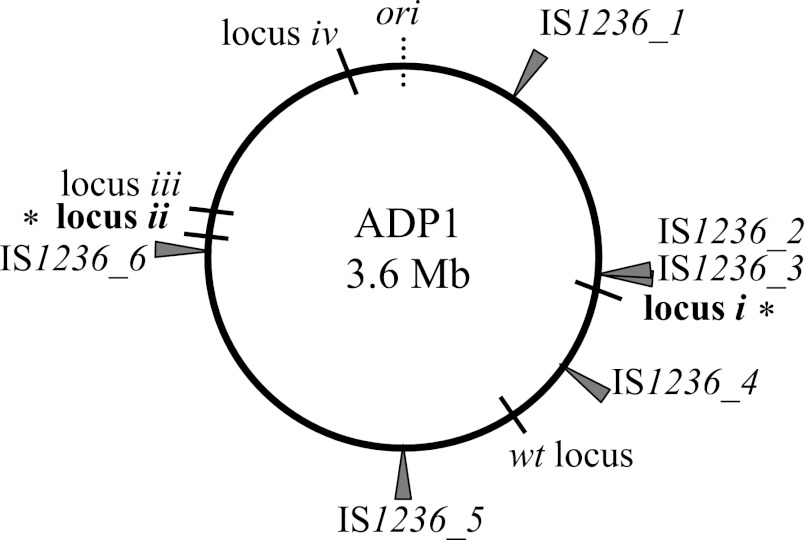
In A. baylyi ADP1, there is a single type of IS element, IS1236 (5, 18, 19, 25). In the chromosome, there are six copies of IS1236, five of which are identical. In one location (near locus i in Fig. 2), two adjacent copies (IS1236_2 and IS1236_3) can function as a composite transposon (41). IS1236 belongs to the large IS3 family, members of which transpose using a replicative excision and conservative integration mechanism, also known as the copy-and-paste method of transposition (15, 18). As a result, the element is retained in its original locus as well as being inserted at a new location (8, 15, 30, 37, 38). Transposition can insert IS1236 elements at novel chromosomal positions that might enable gene duplication via homologous recombination, as depicted in Fig. 3A. This type of scenario was inferred to account for the involvement of IS1236 in two of 91 GDA events in our initial studies of gene amplification in the vicinity of the native cat gene locus (34, 40). This locus is 190 kbp away from the nearest IS element (Fig. 2).
Fig 2.
Position of IS elements on a circular map of the ADP1 chromosome. Six genomic copies of IS1236 reside in the locations shown relative to the origin of replication (ori). IS1236_1 is oriented with the coding sequences of the transposases in a clockwise direction, and all the other copies are oppositely oriented. The native position of the cat genes (wt locus) is shown relative to where these genes were repositioned (loci i to iv) in engineered parent strains, as described in the text. Mutants described in this study arose with the cat genes located in the regions marked by bold text (*): locus i (ACIAD0982), which is 24 kbp from IS1236_3, and locus ii (ACIAD2822), which is 32 kbp from IS1236_6. The wt locus is 190 kbp from IS1236_4.
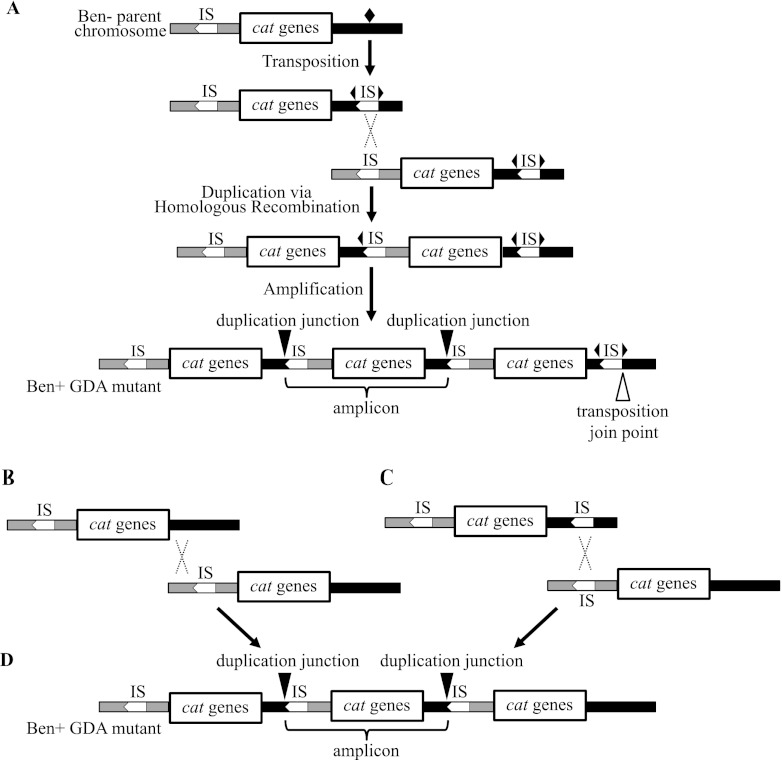
Fig 3.
Roles for IS1236 in gene amplification. (A) Model for the formation of Ben+ GDA mutants following the transposition of IS1236 (IS) to a new target site (marked by a black diamond). After the IS element is inserted in this site (depicted by two outward-facing triangles of a split diamond), homologous recombination between IS elements can generate a duplication. Further amplification can occur by homologous recombination. According to this model, there should be an IS element present downstream of the final amplicon in which the connection to downstream DNA creates a unique junction (the transposition join point). Other labels and representations are the same as those described for Fig. 1. (B) In an alternative model, illegitimate recombination could generate a duplication (not shown) that undergoes further amplification by homologous recombination. (C) In a different alternative model, there is transposition of an IS element to a position downstream of the cat genes on one copy of the chromosome (top). Homologous recombination could then occur between this transposed IS element and an IS element upstream of the cat genes on a different copy of the chromosome (bottom). (D) Homologous recombination between duplications formed by the methods shown in panels B and C could generate a Ben+ GDA mutant with the depicted chromosomal configuration. This configuration lacks a terminal IS element downstream of the cat genes.
In our current study, we investigated whether proximity of the cat gene cassette to an IS1236 element influences the percentage of selected GDA events that are IS mediated. We analyzed gene amplification mutants derived from parent strains in which the relocated catA gene was within 24 or 32 kbp of the nearest IS1236 element (locus i or ii, respectively, in Fig. 2). As described here, duplication junction sequences were determined in order to identify IS involvement. When IS-mediated rearrangements were detected, the genomic configurations of the GDA mutants were further characterized.
MATERIALS AND METHODS
Strains and growth conditions.
A. baylyi strains derived from the wild type, ADP1, were given ACN designations (Table 1). Escherichia coli strains, DH5α and Top10 (Invitrogen), were used as plasmid hosts and were grown at 37°C in Luria-Bertani (LB) medium, also known as lysogeny broth (10 g of Bacto tryptone, 5 g of yeast extract, and 10 g of NaCl per liter [39]). A. baylyi strains were grown at 30 or 37°C in LB or in minimal salts medium (42) with 10 mM succinate or 1 mM benzoate as the sole carbon source. Antibiotics were used as necessary at the following final concentrations: kanamycin, 25 μg/ml; spectinomycin and streptomycin, 12.5 μg/ml; ampicillin, 150 μg/ml.
Table 1.
Strains and plasmids used in this study
| Strain(s) or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| A. baylyi strains | ||
| ADP1 | Wild type (BD413) | 24 |
| ACN1039 | benMΩS4036 benA5147 Δcat5825 (catA-catD); cat region (1439430–1449722)b in locus i (ACIAD0982, vanK), catMΩK5541; Ben− parent strain | 40 |
| ACN1040, ACN1101, ACN1102, ACN1127, ACN1128, ACN1130, ACN1131, ACN1140, ACN1141 | Benzoate+ mutants derived from ACN1039 | 40 |
| ACN1050 | benMΩS4036 benA5147 Δcat5825; cat region (1439430–1449722)b in locus ii (ACIAD2822), catMΩK5541; Ben− parent strain | 40 |
| ACN1056–ACN1058, ACN1105, ACN1106, ACN1161–ACN1165 | Benzoate+ mutants derived from ACN1050 | 40 |
| ACN1222–ACN1223 | Benzoate+ mutants derived from ACN1039 | This study |
| Plasmids | ||
| pZErO-2 | Kmr; zero background cloning vector | Invitrogen |
| pUC19 | Apr; cloning vector | 40 |
| pBAC941 | ACN1056 junction plasmid; pZErO-2 with EcoRI insert (2779591–2783351; 2726455–2728826)b | This study |
| pBAC942 | ACN1057 junction plasmid; pZErO-2 with EcoRI insert (2779591–2783617; 2726455–2728826)b | This study |
| pBAC943 | ACN1058 junction plasmid; pZErO-2 with EcoRI insert (2779591–2781669; 2726455–2728826)b | This study |
| pBAC956 | ACN1105 junction plasmid; pZErO-2 with EcoRI insert (2779591–2784649; 2726455–2737877)b | This study |
| pBAC957 | ACN1106 junction plasmid; pZErO-2 with EcoRI insert (2762288–2763209; 2726455–2728826)b | This study |
| pBAC993 | ACN1161 junction plasmid; pZErO-2 with EcoRI insert (2779591–2783206; 2726455–2728826)b | This study |
| pBAC994 | ACN1162 junction plasmid; pZErO-2 with EcoRI insert (2801423–2802664; 2755061–2761112; 1449722–1444251)b | This study |
| pBAC995 | ACN1163 junction plasmid; pZErO-2 with EcoRI insert (2779591–2783778; 2726455–2728826)b | This study |
| pBAC996 | ACN1164 junction plasmid; pZErO-2 with EcoRI insert (2763053–2763242; 2696984–2706500)b | This study |
| pBAC999 | ACN1101 junction plasmid; pUC19 with EcoRI insert (975200–977490; 943621–946613)b | This study |
| pBAC1000 | ACN1102 junction plasmid; pUC19 with EcoRI insert (983094–992928; 943621–946634)b | This study |
| pBAC1008 | ACN1128 junction plasmid; pZErO-2 with EcoRI insert (969600–973723; 943621–946634)b | This study |
| pBAC1009 | ACN1140 junction plasmid; pZErO-2 with EcoRI insert (975200–981221; 943621–946634)b | This study |
| pBAC1010 | ACN1127 junction plasmid; pUC19 with EcoRI insert (969600–970659; 943621–946634)b | This study |
| pBAC1011 | ACN1040 junction plasmid; pUC19 with EcoRI insert (975200–980963; 943621–946634)b | This study |
| pBAC1012 | ACN1131 junction plasmid; pZErO-2 with EcoRI insert (985699–992893; 943621–946634)b | This study |
| pBAC1013 | ACN1141 junction plasmid; pZErO-2 with EcoRI insert (985899–989426; 943621–946634)b | This study |
| pBAC1014 | ACN1130 junction plasmid; pZErO-2 with EcoRI insert (983092–983865; 943621–946634)b | This study |
| pBAC1015 | ACN1165 junction plasmid; pZErO-2 with EcoRI insert (2779591–2785586; 2702408–2706593)b | This study |
| pBAC1039 | ACN1223 junction plasmid; pZErO-2 with EcoRI insert (969000–972203; 943621–946634)b | This study |
| pBAC1063 | ACN1222 junction plasmid; pZErO-2 with EcoRI insert (975200–975936; 943621–946634)b | This study |
DNA purification, plasmid construction, and the generation of plasmid libraries.
Standard molecular techniques were used for restriction digests, ligations, E. coli transformations, and plasmid isolation (39). Genomic DNA was isolated from A. baylyi strains with the Easy-DNA kit (Invitrogen). Based on previously described methods (35), libraries of DNA from amplification mutants were generated by ligation of EcoRI-cleaved genomic DNA from ACN1040, ACN1101, ACN1102, or ACN1127 into the plasmid vector pUC19. For all other strains, including the newly isolated mutants ACN1222 and ACN1223, a different plasmid vector was used, pZErO-2 (Invitrogen). Libraries were maintained in E. coli DH5α for pUC19 constructs and E. coli Top10 (Invitrogen) for pZErO-2 constructs. Plasmids were screened for their ability to transform a Ben− recipient to allow growth on benzoate as a sole carbon source (35). Plasmids that conferred the ability to consume benzoate were sequenced to determine the duplication junction site. DNA sequencing was performed at Genewiz, Inc.
Selection of Ben+ GDA mutants, chromosomal analysis by PFGE, and quantitative PCR (qPCR).
The Ben− parent strain ACN1039 was grown on succinate medium, washed, concentrated, and plated on solid medium with benzoate as the sole carbon source, as described previously (35, 40). Two individual colonies that arose independently from different starting cultures after 7 and 10 days of incubation were streak purified, and, respectively, designated ACN1222 and ACN1223. They were further characterized by pulsed-field gel electrophoresis (PFGE) analysis (CHEF-DR II; Bio-Rad). Genomic DNA was prepared in agarose plugs, and the embedded DNA was digested with the restriction enzyme NotI as previously described (20, 35, 40). PFGE was used to separate the NotI-cleaved chromosomal fragments using a 24-h protocol with a constant 6-V cm−2 current and pulse switch times that increased from 1 to 150 s (ACN1039-derived mutants) or 10 to 200 s (ACN1050-derived mutants).
To evaluate the amplicon copy numbers in ACN1222 and ACN1223, the amount of catA DNA was compared to that of a control gene, antA, assumed to be in the genome in single copy. A real-time qPCR assay was used with TaqMan 5′-exonuclease (Applied Biosystems). The primers and method were previously described (40).
Analysis of the chromosomal configurations of GDA mutants by PCR and Southern hybridization.
Strain-specific oligonucleotides (see Table S1 in the supplemental material) were used for PCR analysis to determine the genomic rearrangements present in individual GDA mutants. LongAmp polymerase (New England BioLabs) and extension times of 2 min 30 s were used to ensure the amplification of long products. Concentrations of reagents in the 25-μl reaction mixture were as follows: template DNA, ∼10 ng; dNTPs, 300 μM; oligonucleotides, 0.4 μM each; 1× LongAmp reaction buffer; 2.5 units LongAmp polymerase. PCRs were run on agarose gels at the appropriate concentrations for differentiating products of expected sizes (39) (Table 2).
Table 2.
Predicted fragment sizes for PCR analysisa
| Strain | Fragment size (bp) |
|
|---|---|---|
| x-bp fragment | x + 1,237-bp fragment | |
| ACN1056 | 818 | 2,055 |
| ACN1057 | 818 | 2,055 |
| ACN1058 | 859 | 2,096 |
| ACN1105 | 846 | 2,083 |
| ACN1106 | 866 | 2,103 |
| ACN1161 | 819 | 2,056 |
| ACN1163 | 819 | 2,056 |
| ACN1040 | 745 | 1,982 |
| ACN1101 | 742 | 1,979 |
| ACN1102 | 1,018 | 2,255 |
| ACN1127 | 817 | 2,054 |
| ACN1128 | 968 | 2,205 |
| ACN1130 | 881 | 2,118 |
| ACN1131 | 1,018 | 2,255 |
| ACN1140 | 745 | 1,982 |
| ACN1141 | 976 | 2,033 |
| ACN1222 | 883 | 2,120 |
| ACN1223 | 1,113 | 2,350 |
Southern hybridizations were performed on select amplification mutants. A digoxigenin (DIG)-labeled DNA probe was generated by a random-primed labeling method following the Roche Molecular Biochemicals DIG Application Manual using the primers listed in Table S1 in the supplemental material. Briefly, 10 ng ADP1 genomic DNA, 200 μM DIG-dNTPs, 0.5 μM each primer, and 2.6 units of Expand High Fidelity polymerase were used in a 30-cycle PCR. The labeled probe was stored at −20°C in hybridization buffer (5× SSC [0.75 M NaCl, 0.075 M Na-citrate, pH 7.0], 50% formamide, 0.02% SDS, 0.1% N-laroyl sarcosine, 2% Bio-Rad blocking reagent). PCR products were generated as a size ladder for visualization of band sizes following hybridization with the DIG-labeled probe using the primers in Table S1 in the supplemental material. DNA from the amplification mutants (2 μg) and 4 μg of DNA from wild-type ADP1 and the parent strain were digested with PstI (ACN1050, ACN1056, ACN1057, ACN1058) or EcoRV (ACN1039, ACN1101). The digested DNA was separated on a standard (1.0%) agarose gel and transferred to a nitrocellulose membrane using a TurboBlotter system (Schleicher and Schuell). Following transfer, DNA was cross-linked to the membrane by a total dose of UV light (254 nm) of 120 m J cm−2. Prehybridization, hybridization for 16 h with the DIG-labeled probe at 42°C, and low- and high-stringency washes were carried out according to the manufacturer's instructions (Roche). Following the washes, incubation of the membrane with an anti-DIG antibody (Roche) was immediately followed by incubation with a chemiluminescent substrate of alkaline phosphatase to visualize the band sizes on X-ray film according to the manufacturer's instructions and as previously described (20, 35).
RESULTS AND DISCUSSION
Selection and characterization of GDA mutants.
In a previous study, we described the construction of two Ben− strains, ACN1039 and ACN1050, in which the cat gene cassette was deleted from its native locus and relocated to the chromosomal positions indicated in Fig. 2 as locus i and locus ii, respectively (40). Here, we chose 19 of the Ben+ amplification mutants that were derived from ACN1039 or ACN1050 for further analysis. In addition, we selected two new Ben+ mutants from ACN1039, designated ACN1222 and ACN1223 (Table 1). These mutants arose similarly to those that were previously isolated, each appearing spontaneously as a colony after ACN1039 cells were spread on solid medium with benzoate as the sole carbon source.
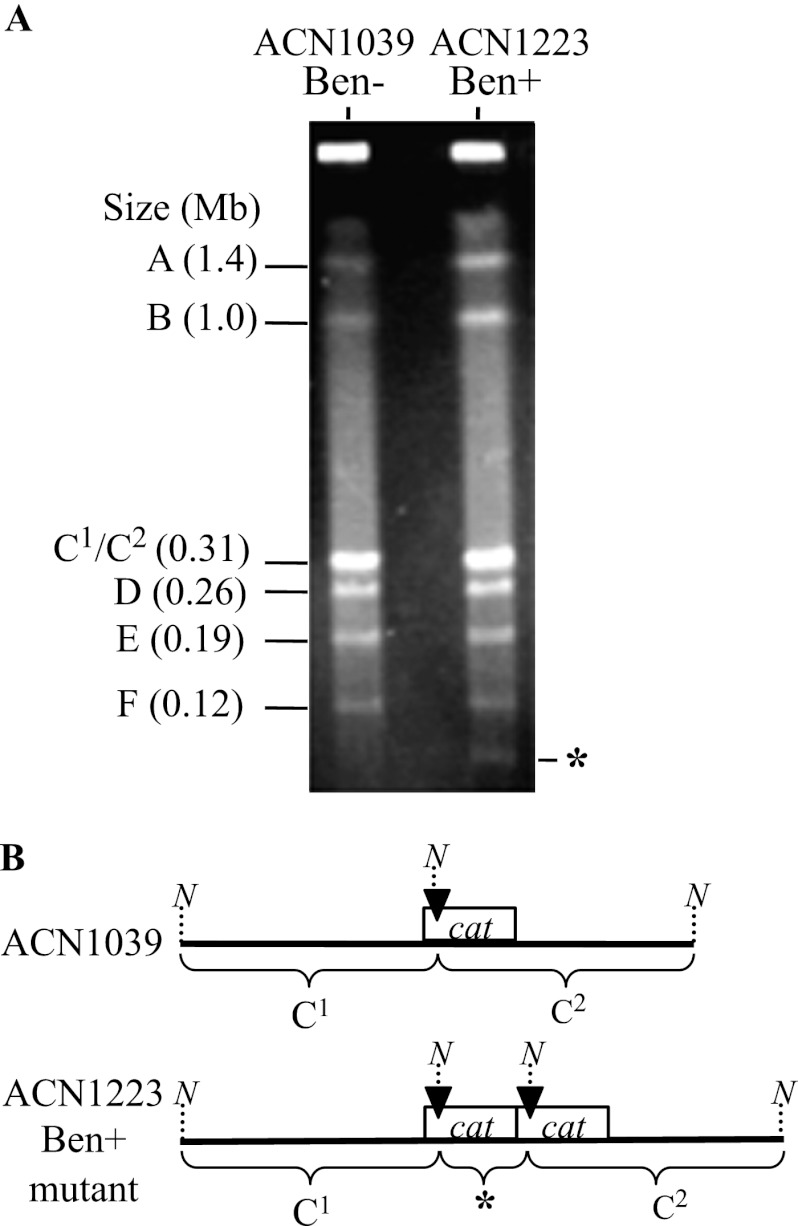
Characterization of NotI-digested ACN1223 DNA by PFGE revealed one fragment in the mutant that is not in its parent (marked by an asterisk in Fig. 4A). When the wild-type chromosome is digested with NotI, its 3.6-Mb circular chromosome is cut into 6 fragments. As depicted in Fig. 4B, a NotI site in the cat gene region was introduced when the cassette was placed in locus i of ACN1039. Introduction of this site results in cleavage of the wild-type fragment into two similarly sized fragments that are not resolved with PFGE (Fig. 4B). In GDA mutants, multiple copies of the cat gene region result in a novel NotI-generated fragment whose size equals that of its amplicon (Fig. 4B). Thus, the banding pattern of ACN1223 indicates the presence in this strain of an amplicon of ∼41 kbp. Similar analysis of ACN1222 indicated the presence of an ∼44-kbp amplicon (data not shown).
Fig 4.
Amplicon size analysis by PFGE. (A) Separation of NotI-digested genomic DNA from a Ben+ GDA mutant (ACN1223) and its Ben− parent strain (ACN1039). Labels A, B, D, E, and F correspond to wild-type NotI-generated chromosomal fragments. The wild type additionally has a fragment (C; ∼0.62 Mb) that is cleaved in both ACN1039 and ACN1223 by a NotI site in the relocated cat genes. The resulting fragments (C1 and C2) are nearly equal in size and cannot be separated by PFGE. One fragment, which is present in ACN1223 (*) but not in ACN1039, corresponds to an amplicon of ∼41 kbp, as discussed in the text. (B) Illustration of restriction pattern differences between ACN1039 and ACN1223 in the vicinity of the relocated cat gene cassette. A black arrowhead marks the position of a NotI restriction site (N) that generates two nearly equally sized fragments in ACN1039 (C1 and C2). When there is gene amplification of the cat region, multiple copies of the marked NotI site generate a fragment whose size corresponds to that of its amplicon, as depicted for ACN1223 (*).
ACN1039-derived mutants were found to have a relatively narrow range of amplicon size (39 to 61 kbp), with copy numbers of the amplicon ranging from 9 to 35 relative to the rest of the genome (40) (see Table S2 in the supplemental material). Therefore, the amplicon sizes of ACN1222 and ACN1223 are consistent with previous observations. When amplicon copy number was evaluated (see Table S2), that of ACN1222 was determined to be 23, within the range previously observed for ACN1039-derived mutants. While ACN1223 was found to have only 2 copies of its amplicon, cat gene duplications in other genomic regions have previously been shown to be sufficient for slow Ben+ growth (40). The ACN1050-derived GDA mutants have amplicon sizes spanning from 49 to 95 kbp, with copy numbers that range from 7 to 26 (40) (see Table S2 in the supplemental material).
Determination of DNA junction sequences.
The 21 GDA mutants chosen for this study resulted from amplification events in which the cat genes were closer to an IS element than they are in their native locus. The cat gene cassette in ACN1039 (locus i) is 24 kbp from IS1236_3, and the cassette in ACN1050 (locus ii) is 32 kbp from IS1236_6 (Fig. 2). We sought to evaluate whether proximity to a transposable element affects the likelihood of selecting mutants with IS-mediated rearrangements. Since DNA from the precise duplication site provides critical information about the underlying recombination event, such duplication junction sequences were analyzed in the ACN1039- and ACN1050-derived GDA mutants.
To identify the duplication junction sequences, we used a previously developed assay that exploits the high efficiency of natural transformation and homologous recombination in A. baylyi (35). The first step in this method involves construction of a recombinant plasmid library in E. coli carrying random chromosomal DNA fragments from an amplification mutant. The presence of a plasmid-borne duplication junction sequence can be identified by its ability to enable a Ben− parent strain to grow on benzoate after transformation and homologous recombination. Individual plasmid-containing E. coli colonies are patched onto a lawn of the Ben− parent strain on succinate medium, on which the E. coli strain cannot grow. Plasmid DNA that is released by the lysis of E. coli is taken up by the neighboring Ben− parent strain. If the plasmid DNA includes a duplication junction, homologous recombination between this DNA and the chromosome can generate a duplication identical to the one found in the original amplification mutant from which the plasmid library was constructed (Fig. 1B). When transferred to benzoate medium, Ben+ transformants arise from cells in which the chromosomal duplication has undergone further amplification. Plasmids are then isolated from the E. coli donors that are able to generate the Ben+ phenotype in A. baylyi, and the duplication junction is identified by DNA sequence analysis.
In this fashion, the duplication junctions of 21 amplification mutants were isolated and sequenced (Fig. 5). They reveal the precise point of recombination between DNA that had been downstream of the cat gene cassette and DNA that was previously upstream of it. Inverted black triangles mark these sites in Fig. 1A and Fig. 3A. As detailed below, the majority of these duplication junctions involved the same sequence upstream of the cat genes. The DNA sequence of the duplication junction defines the endpoints of an amplicon and therefore also the amplicon size. In all cases, the sizes inferred from junction sequence analyses (Fig. 5) are in good agreement with those determined by PFGE (Fig. 4A and data not shown).
Fig 5.
IS1236 sequences identified in duplication junctions. (A) Diagram of the IS1236 element, with red arrows representing inverted repeats. Ten nucleotides are shown at each end of these repeats. OrfA, OrfAB, and OrfB are presumed to be transposase proteins, and the coding orientations of their genes (indicated by arrows) are opposite to those of the cat genes for the IS elements closest to the cassette in locus i or ii. (B) Duplication junction sequences for Ben+ amplification mutants. The middle line shows the DNA sequence of each experimentally determined duplication junction. The top and bottom lines indicate the known chromosomal sequence of DNA that normally occurs downstream or upstream, respectively, of the relocated cat gene cassette starting with the indicated genomic coordinate (from GenBank entry CR543861). Nucleotide identity at the duplication junction is shown in bold typeface. The IS1236 sequence is shown in red text, with the first three base pairs of its inverted repeat underlined.
Predominance of IS-mediated rearrangement leading to amplification of loci i and ii.
Sequence analysis showed that 11 out of 11 duplication events from locus i and 7 out of 10 duplication events at locus ii involved a copy of IS1236 at the duplication junction (Fig. 5). The duplication junction in each of these 18 independent GDA mutants occurred exactly at the end of the IS1236 terminal inverted repeat. This observation could be readily explained if homologous recombination between two IS elements occurred as shown in Fig. 3A. In this case, the sequence identified as the duplication junction represents the target site into which the IS element had initially inserted (the leftward-pointing triangle in Fig. 3A). According to this scenario, the precise point of recombination (the duplication junction) really occurs somewhere in the 1.2-kbp region of sequence identity between two IS elements.
IS1236 sequences were evident in 86% of the duplication sites. These results differed from previous investigations, where only a small fraction (2%) of 91 characterized duplication junctions involved IS1236 (34, 40). The previous studies identified junction sequences resulting from duplication events in a different chromosomal region, the native cat gene locus, which suggests that chromosomal position influences the type of gene amplification mutants that are selected. Chromosomal position was also concluded to be a factor in the correlation between size and copy number of amplicons in different GDA mutants (40).
The effect of genomic location was also evaluated for spontaneous catA duplication frequency. This frequency was previously assessed in five chromosomal regions, the wild-type locus and the positions to which catA had been relocated, loci i to iv in Fig. 2. Interestingly, these values were similar regardless of genomic position (40). In the absence of selection, the duplication frequency of catA ranged from 10−4 to 10−5. The value for locus i was the same as for the wild-type locus (10−4), whereas that for locus ii was 10-fold lower. Therefore, the increase in the proportion of duplication sites involving IS1236 cannot be explained by an overall increase in the frequency of spontaneous duplications in locus i or ii. The relative distance of the cat genes to an IS1236 element is one difference between the wild-type locus and the two regions examined in our current study. However, further work is needed to establish if, and how, such proximity affects mechanisms underlying GDA.
IS1236 was first discovered in studies of mutations that prevent the degradation of p-hydroxybenzoate (18, 19). The spontaneous insertion of IS1236 plays a significant role in the inactivation of genes for aromatic compound catabolism in two different chromosomal regions of ADP1 (18, 19, 26, 41). One of these regions corresponds to locus i, where the cat gene cassette is inserted in vanK. This chromosomal area, which encompasses several van genes involved in the metabolism of ferulate and vanillate, undergoes frequent rearrangements, including large spontaneous deletions and the transposition of a composite element that combines IS1236_2 and IS1236_3 (20, 41). Our analysis indicates that IS1236_3 alone, not the composite IS element, was located at the duplication junctions of GDA mutants involving locus i in the current study (data not shown).
The second region that is characterized by numerous IS1236 insertions resides approximately 300 kbp away, in a clockwise direction, from the native cat genes (wt locus in Fig. 2). This region makes up an approximately 20-kbp supraoperonic gene cluster involved in aromatic compound catabolism. IS1236 transposition was shown to occur preferentially in certain genes within the cluster, although the basis of such preference remains unclear (18, 26).
GDA events near locus ii, ACN1050-derived mutants.
While all of the duplication junctions from locus i involved IS1236, three from locus ii did not (Fig. 5). These junctions were isolated from the GDA mutants ACN1162, ACN1164, and ACN1165. There were no obvious features to distinguish these amplicons from any others in terms of size or copy number (see Table S2 in the supplemental material). We examined DNA at the point of recombination in these mutants to identify short stretches of nucleotides that are identical between the sequences that recombine to form duplication junctions (bold nucleotides in Fig. 5). The lack of significant identity between the sequences generating the duplication junctions indicates that these duplications were formed by illegitimate recombination. At the precise point of recombination, there was only one nucleotide of identity between the sequences comprising the duplication junction of two mutants (ACN1164 and ACN1165), and there was no identity at the duplication junction in the other (ACN1162). This type of pattern is characteristic of most of the duplication junctions isolated in previous studies of GDA at the native cat gene locus (34, 40).
The seven remaining ACN1050-derived GDA mutants displayed IS involvement. Such involvement is intriguing in this chromosomal region because the nearest IS element to locus ii is IS1236_6 (Fig. 2). This IS1236 copy shares only 82% sequence identity with the other copies, which are all identical to one another. Moreover, the sequence variation suggests that IS1236_6 does not encode a functional transposase (see Fig. S1 in the supplemental material). Specifically, the canonical catalytic triad (DDE motif) is disrupted by a frameshift that occurs prior to the codon for the glutamate residue (8, 31, 37).
Potential role for transposition in gene amplification.
Amplification mutants could be generated by transposition of an IS1236 element followed by homologous recombination between the two IS copies, as shown in Fig. 3A. The duplication junction should identify the precise target site into which IS1236 transposed. This target site, marked by a diamond in Fig. 3A, is expected to be different in each mutant. Insertion of a copy of IS1236 into this site would create two characteristic joints where wild-type sequence is interrupted by the IS element (represented in Fig. 3A by two black triangles of a split diamond). Following duplication, homologous recombination could result in further amplification. An amplification mutant would be expected to carry multiple copies of the duplication junction, as well as a single copy of one joint that marks the downstream portion of the transposition target site (labeled as the transposition join point in Fig. 3A). This single join point highlights that the process of transposition followed by homologous recombination results in a hallmark copy of the insertion sequence downstream of the final copy of the amplicon. As described below, we failed to identify this “terminal IS element,” a result that led us to consider different possible duplication mechanisms. These possibilities are illustrated in Fig. 3B and C and are discussed later.
PCR analysis of chromosomal configurations in GDA mutants.
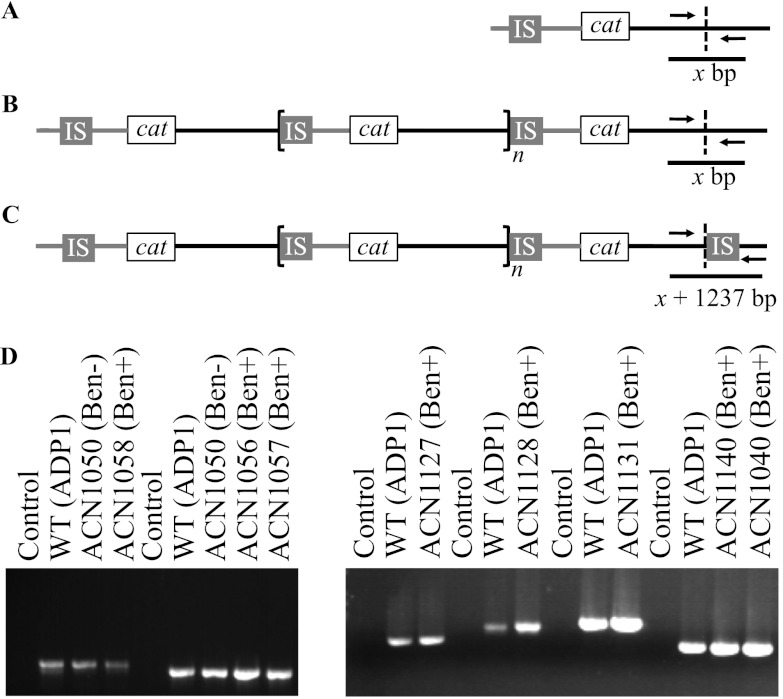
We used PCR methods to detect the presence or absence of an IS element downstream of the cat genes in each of the ACN1039- and ACN1050-derived GDA mutants. Primers were designed to amplify the target DNA where IS1236 might have transposed. The upstream portion of the target site (the leftward-pointing triangle of the split diamond in Fig. 3A) was identified experimentally by the junction sequences (Fig. 5). PCR primer pairs were designed to anneal within and immediately downstream of the final amplicon, flanking the position where a terminal insertion sequence would be located following transposition of IS1236 (see Table S1 in the supplemental material). These primers, when used with the chromosomal DNA of an amplification mutant as the template, should distinguish between the presence of an IS element inserted in the target DNA at the transposition join point (as shown in Fig. 3A) and the wild-type configuration in this region (as shown in Fig. 3D).
The expected differences between PCR products from different chromosomal templates are shown in Fig. 6A to C and Table 2. Fragment size predictions were calculated for each of the 18 amplification mutants in which an IS element was detected at the duplication junction. When no terminal IS element is located downstream of the final amplicon of a GDA mutant, its PCR product size will correspond to that of the wild type (ADP1) and the Ben− parent, x bp (Table 2, Fig. 6A and B). If there were a terminal IS element present, the PCR product size of the wild-type configuration (x bp) would be increased by the size of IS1236, 1,237 bp, yielding a total product size for the GDA mutant of x + 1,237 bp (Table 2, Fig. 6C).
Fig 6.
PCR-based analysis of chromosomal configurations. Panel A diagrams the expected PCR product in the Ben− parent strain. The wild-type chromosome is identical to this configuration except the cat gene cassette is in its native locus. The chromosomal shading (gray versus black) is as described in the legend to Fig. 1. Panels B and C depict two different chromosomal arrangements that could occur downstream of the terminal amplicon in a Ben+ GDA mutant. The region within the bracket represents n copies of an amplicon. The dashed line indicates the endpoint of the final amplicon or the equivalent nucleotide in the wild-type or parent chromosome. In panel B, the chromosomal configuration in the PCR-amplified region matches that of the wild-type strain (WT; ADP1) as well as its Ben− parent. In the configuration shown in panel C, the presence of an IS element could result from the transposition and amplification process depicted in Fig. 3A. To differentiate between these two possibilities, PCR primers (see Table S1 in the supplemental material) were designed for specific GDA mutants based on the DNA sequence from the experimentally determined duplication junction. PCR product sizes (Table 2) can be predicted from the primers used, either x bp for configurations A and B or x bp + 1,237 (the number of nucleotides in the IS element) for configuration C. (D) The results of PCR with the wild type, Ben− parent strain, Ben+ GDA mutant, or no DNA template are shown. DNA size standards (not shown) indicated in all cases that the wild-type fragments matched the expected sizes shown in Table 2. In all cases, the PCR-generated fragments from the amplification mutants were the same size as those from the wild type and the Ben− parent strain.
In ACN1039- and ACN1050-derived GDA mutants, these primer sets (see Table S1 in the supplemental material) generated a PCR product of a size corresponding to the wild-type configuration (Fig. 6D and data not shown). In no case was a larger product detected that would have indicated a terminal IS element, despite sufficient extension time and use of a polymerase mix competent for the formation of the longer product (see Materials and Methods). These results contradict the model displayed in Fig. 3A of how the gene amplification mutants could be generated by transposition of IS1236 followed by homologous recombination between identical sequences to yield duplication and further amplification.
Confirmation of results by Southern hybridization.
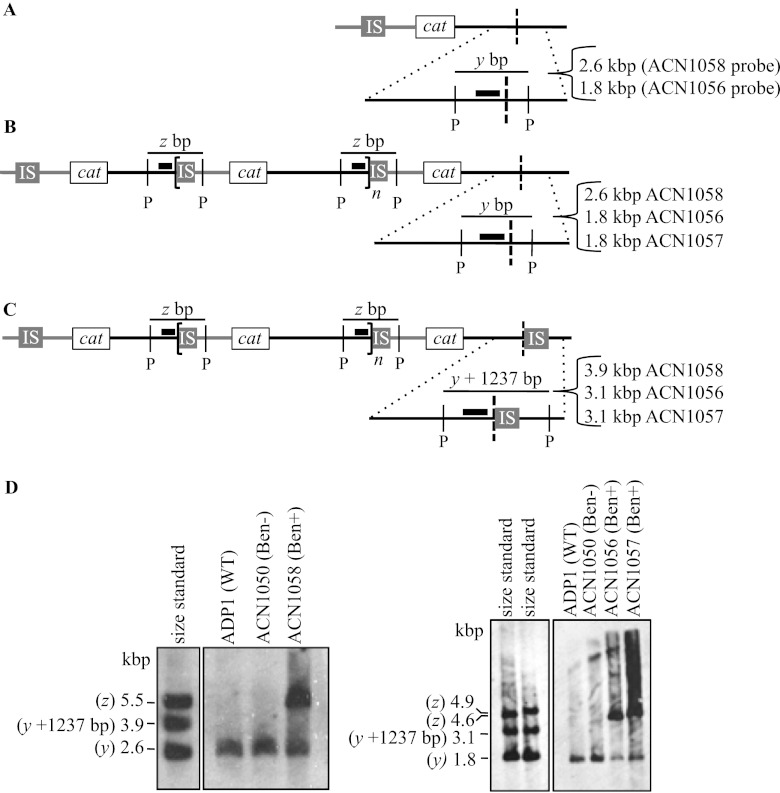
Southern hybridization was additionally used to confirm the absence of an IS element downstream of the final amplicon. In a similar fashion to the PCR method, the size of fragments detected by hybridization can distinguish the two potential configurations of an amplification mutant (Fig. 7B and C). A probe (depicted as a thick black line in Fig. 7) was designed to hybridize to DNA adjacent to the IS1236 sequence in the duplication junction. When chromosomal DNA from an amplification mutant is digested with a restriction enzyme that does not cut within IS1236, this probe should hybridize to a fragment corresponding to the junction fragment labeled z in Fig. 7B and C. Using information from the duplication junction sequence (Fig. 5), the size of this fragment can be predicted (Table 3). Because this fragment contains the duplication junction sequence, a fragment of this size would be expected only in the Ben+ GDA mutants and not in comparable samples of DNA from the wild type or Ben− parent strain. As shown in Fig. 7D, junction fragments of the expected sizes were detected in three mutants, ACN1056, ACN1057, and ACN1058 (labeled z). As expected, these fragments were not detected in the ADP1 wild-type strain or the ACN1050 parent strain (Fig. 7D).
Fig 7.
Analysis of chromosomal configurations by Southern hybridization. Panels A, B, and C depict the Ben− parent and two different possible chromosomal arrangements downstream of the terminal amplicon in a Ben+ GDA mutant. The wild-type chromosome is identical to the Ben− parent except the cat gene cassette is in its native locus. These arrangements are the same as those described in the legends to Fig. 6A, B, and C. The chromosomal shading is as described in the legend to Fig. 1. Dotted lines show an enlarged view of a region that hybridizes to a labeled probe that abuts the duplication junction (depicted as a thick black bar). When chromosomal DNA is digested with the restriction enzyme PstI (P), sequence analysis allows the size prediction of the fragment that encompasses the duplication junction to which the probe will hybridize (z). This analysis also predicts the size of the wild-type PstI-generated fragment to which the same probe will hybridize (y). For the wild type and the Ben− parent strain, the size of y depends on the probe used. For GDA mutants with the chromosomal configuration shown in panels B or C, the probe should detect a fragment of size y bp or y bp + 1,237 (the size of the IS element), respectively. The predicted fragment sizes for different mutants vary according to the specific junction and probe sequences (Table 3). (D) The results of Southern hybridizations are shown for chromosomal DNA from the wild type (ADP1) and the mutants indicated. Standards corresponding to the size of predicted fragments were generated by PCR (as described in Materials and Methods). In no case was a band of size y + 1,237 bp detected.
Table 3.
Predicted fragment sizes for Southern hybridizationa
| Strain | Fragment size (bp) |
||
|---|---|---|---|
| y fragment | y + 1,237-bp fragment | z fragment (junction) | |
| ACN1056 | 1,826 | 3,096 | 4,586 |
| ACN1057 | 1,826 | 3,096 | 4,853 |
| ACN1058 | 2,598 | 3,868 | 5,503 |
| ACN1101 | 1,501 | 2,738 | 10,807 |
The sizes of fragments y and z are calculated based on the labeled probes used for hybridizations, which are generated with the PCR primers listed in Table S1 in the supplemental material, and evaluation of the corresponding regions depicted in Fig. 7 in consideration of the genomic DNA sequence (GenBank entry CR543861).
In the region downstream of the final amplicon, this same probe should distinguish between two possible chromosomal configurations. In a GDA mutant with no terminal IS element downstream of the final amplicon (Fig. 7B), the probe would hybridize to a fragment of wild-type size, indicated as y in Fig. 7A and B, whose value is shown in Table 3. In a GDA mutant with the configuration shown in Fig. 7C, the presence of a terminal IS element would increase the size of the fragment detected by the probe. Relative to fragment y (Fig. 7B), the probe should hybridize to a fragment whose size increases by the number of nucleotides in the IS element, y + 1,237 bp (Fig. 7C, Table 3). Southern hybridization analysis of four GDA mutants (ACN1056, ACN1057, ACN1058, and ACN1101) demonstrated the presence of a wild-type-sized fragment in each strain (Fig. 7D and data not shown). These results are consistent with the PCR-based conclusion that there is no terminal IS element downstream of the final amplicon.
Contradiction of the predicted model for transposition-based gene amplification.
Our initial model proposed that an IS-mediated duplication was generated by a transposition event that places IS1236 downstream of the cat genes. Following transposition, duplication and further amplification were proposed to occur by homologous recombination (Fig. 3A). In this scheme, recombination could occur via intramolecular interactions on a single chromosome or between identical copies of the chromosome within a cell. This model is based on standard assumptions for the well-documented behavior of transposable elements and homologous recombination (11, 36). Moreover, this combination of transposition and homologous recombination has often been reported to underlie genetic duplication (21, 23, 28–30). Nevertheless, the absence of a detectable terminal IS element in our mutants indicates that this model for IS involvement is incorrect for the GDA events investigated here.
Alternative models of IS-mediated gene amplification.
Two different models, shown in Fig. 3B and 3C, might account for the observed chromosomal configuration in the GDA mutants. As illustrated in Fig. 3B, the initiating duplication event could involve some type of illegitimate recombination between the wild-type sequence downstream of the cat gene cassette and the IS1236 element that normally resides upstream of catA. The recombining DNA sequences that form each duplication junction are shown in Fig. 5. If these duplication junctions form by illegitimate recombination, then the precision of these events is clearly significant. Each duplication junction occurs at the exact base pair of the IS1236 terminal inverted repeat where the transposase would nick to yield a free 3′ hydroxyl that initiates the first strand transfer reaction in copy-and-paste transposition (15).
Illegitimate recombination has previously been considered to play a role in duplications involving an IS element. Specifically, in an experimental system involving Tn9, illegitimate recombination was proposed to initiate tandem amplification (7). In this example, high concentrations of chloramphenicol can select multiple copies of the Tn9-borne drug resistance determinant. When this process was assessed in a recA mutant, the initial step of the transposon duplication was observed to occur at a low frequency in a recA-independent fashion (7).
Because some illegitimate recombination mechanisms are mediated by short stretches of identity, we evaluated the level of identity between the DNA sequences that recombined. At the exact duplication site, all GDA mutants contain six or fewer identical nucleotides (highlighted by bold text in Fig. 5). In 10 mutants, there are no identical nucleotides at the duplication junction. Thus, the duplication mechanism does not appear to depend on sequence identity. However, the precision of the recombination site suggests that an IS1236 transposase is involved even though the model for illegitimate recombination shown in Fig. 3B does not invoke a typical transposition mechanism.
A transposase-mediated duplication process for the ACN1050-derived mutants is especially interesting. The nearest IS element to locus ii in this parent strain is IS1236_6, and, as noted earlier, IS1236_6 appears not to encode a functional transposase (see Fig. S1 in the supplemental material). At this locus, a transposase encoded by a different element may act in trans. However, trans-acting activity is presumed to occur infrequently with the IS3 family elements (11, 30).
Another explanation for the chromosomal configuration of the GDA mutants is illustrated in Fig. 3C. In this scenario, a typical transposition event places a novel IS1236 copy downstream of the cat gene cassette on one copy of the chromosome. Chromosomal duplication could then result from homologous recombination between this element and an IS1236 element upstream of the cat gene cassette on a different chromosomal copy that has not undergone transposition. Having different chromosomal copies in a bacterial cell may not be as rare as previously thought, since recent studies indicate that prokaryotic polyploidy is common (6, 22, 32, 44, 45). Unless the original copy of the chromosome that underwent the novel transposition event is replicated before homologous recombination with a sister chromosome, our Southern hybridization and PCR methods would not detect it.
To determine whether a mechanism involving transposition (Fig. 3C) could be distinguished from one without transposition (Fig. 3B), we investigated target sequence duplication. A key characteristic of IS3 family transposition is that it generates a small duplication of the target such that the IS element is typically flanked by 3-bp direct repeats (30). Unfortunately, the analysis of potential target sequence duplication is unable to distinguish between the alternative duplication mechanisms shown in Fig. 3B and C (see Fig. S2 and S3 in the supplemental material).
Copy-and-paste transposition.
The model of illegitimate recombination shown in Fig. 3B was proposed because our results could not be explained by the documented behavior of other IS elements. In the well-studied copy-and-paste transposition mechanism of the IS3 family element IS911, the transposase binds and assembles terminal inverted repeat sequences (IRL and IRR) in a synaptic complex (37). After a single end of the element is cleaved, it is transferred to the same DNA strand 3 bp from the opposite end. This intermediate is resolved by host-mediated replication, which restores the donor DNA and also yields a circular transposition molecule. This circular element can synapse with a site adjacent to an IS911 IR in the target, thereby allowing the transposase to cleave and transfer one IR strand of the mobile element to the target DNA. Host recombination enzymes complete the IS911 insertion by resolving the four-way branched DNA intermediate of this IR-targeted process (46). If the transposase brings together one IS911 IR sequence with that of another IS911 element (47) or with a chromosomal surrogate sequence (33), the resulting circular transposition intermediate includes the intervening sequence. By this method, the intervening sequence can be duplicated.
For an analogous mechanism to account for cat gene duplication, there must be a second IS1236 or a surrogate IR site downstream of the cat genes, and host replication would need to generate a >40-kbp circular transposition intermediate. However, the presence of a second appropriately placed IS1236 element was ruled out by our experiments described earlier. Moreover, sites that might serve as IR surrogates were not detected when the target sequences surrounding duplication junctions (Fig. 5) were compared to the terminal 30 bp for IRL and IRR of IS1236 (data not shown). Therefore, while the IS1236-associated duplication products are not consistent with an IR-targeted mechanism, the IS911 studies highlight the importance of assessing target sequences and proteins that might contribute to processing alternative transposition products.
Concluding remarks.
In previous studies of gene amplification in A. baylyi, IS1236 was not found to be a significant factor in GDA events and was identified in only two of 91 characterized duplication junctions (12, 34, 40). In contrast, 86% of the 21 GDA mutants that were characterized in our current study display IS-involved rearrangements to form the duplication junctions. One difference between the past and present studies lies in the chromosomal regions from which amplification mutants were selected. Here, the selection cassette resided closer to the nearest IS element than in previous studies, within 32 kbp rather than 190 kbp (Fig. 2). This proximity may be responsible for the higher proportion of IS-mediated rearrangements.
The paucity of systematic GDA studies prevents the direct comparison of these results with those from other bacteria, such as E. coli and Salmonella enterica, in which genetic rearrangements have been studied. Typically, gene amplification is difficult to study because of its transient and variable nature. The selection system developed to capture and characterize chromosomal amplicons in A. baylyi is unique and continues to reveal novel features of genomic rearrangement.
Surprisingly, PCR and Southern hybridization methods demonstrated the inability of a common model, which assumes transposition is followed by homologous recombination (Fig. 3A), to account for the formation of A. baylyi GDA mutants. The actual events responsible for generating these GDA mutants remain unknown. They may involve transposase-mediated DNA cleavage and illegitimate recombination (Fig. 3B). Alternatively, different chromosomal copies may undergo unequal homologous recombination (Fig. 3C). Aspects of the schemes shown in Fig. 3B and C differ from commonly accepted models of insertion sequence-mediated GDA. Nevertheless, the isolation of 18 independent mutants that display identical characteristics of IS-involved rearrangement suggests that a common mechanism is responsible for their formation. A greater understanding of the role of insertion sequences in gene duplication should lead to a deeper appreciation of recombination as a driving force in genomic variation.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge helpful advice from Timothy Hoover and Cory Momany.
This research was supported by National Science Foundation grants (MCB 0920893 to E.L.N. and DBI 0905829 to K.T.E.; DBI 1062589 supported M.K.I.).
Footnotes
Published ahead of print 15 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Albertson DG. 2006. Gene amplification in cancer. Trends Genet. 22: 447–455 [DOI] [PubMed] [Google Scholar]
- 2. Anderson P, Roth J. 1981. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc. Natl. Acad. Sci. U. S. A. 78: 3113–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersson DI. 2011. Evolving promiscuously. Proc. Natl. Acad. Sci. U. S. A. 108: 1199–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson DI, Hughes D. 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43: 167–195 [DOI] [PubMed] [Google Scholar]
- 5. Barbe V, et al. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic. Acids Res. 32: 5766–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breuert S, Allers T, Spohn G, Soppa J. 2006. Regulated polyploidy in halophilic archaea. PLoS One 1: e92 doi:10.1371/journal.pone.0000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandler M, de la Tour EB, Willems D, Caro L. 1979. Some properties of the chloramphenicol resistance transposon Tn9. Mol. Gen. Genet. 176: 221–231 [DOI] [PubMed] [Google Scholar]
- 8. Chandler M, Mahillon J. 2002. Insertion sequences revisited, p 305–366 In Craig NL, Craigie R, Gellert M, Lambowitz A. (ed), Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- 9. Conrad B, Antonarakis SE. 2007. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu. Rev. Genomics Hum. Genet. 8: 17–35 [DOI] [PubMed] [Google Scholar]
- 10. Coyne S, Courvalin P, Galimand M. 2010. Acquisition of multidrug resistance transposon Tn6061 and IS6100-mediated large chromosomal inversions in Pseudomonas aeruginosa clinical isolates. Microbiology 156: 1448–1458 [DOI] [PubMed] [Google Scholar]
- 11. Craig NL. 1996. Transposition, p 2339–2362 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 2 ASM Press, Washington, DC. [Google Scholar]
- 12. Craven SH. 2009. Gene expression and dosage: distinct mechanisms regulate benzoate degradation in Acinetobacter baylyi ADP1. PhD thesis. University of Georgia, Athens, GA [Google Scholar]
- 13. Craven SH, Neidle EL. 2007. Double trouble: medical implications of genetic duplication and amplification in bacteria. Future Microbiol. 2: 309–321 [DOI] [PubMed] [Google Scholar]
- 14. de Berardinis V, et al. 2008. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 4: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duval-Valentin G, Marty-Cointin B, Chandler M. 2004. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J. 23: 3897–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elliott KT, Neidle EL. 2011. Acinetobacter baylyi ADP1: transforming the choice of model organism. IUBMB Life 63: 1075–1080 [DOI] [PubMed] [Google Scholar]
- 17. Eraso JM, Kaplan S. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 176: 32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerischer U, D'Argenio DA, Ornston LN. 1996. IS1236, a newly discovered member of the IS3 family, exhibits varied patterns of insertion into the Acinetobacter calcoaceticus chromosome. Microbiology 142: 1825–1831 [DOI] [PubMed] [Google Scholar]
- 19. Gerischer U, Ornston LN. 1995. Spontaneous mutations in pcaH and -G, structural genes for protocatechuate 3,4-dioxygenase in Acinetobacter calcoaceticus. J. Bacteriol. 177: 1336–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gralton EM, Campbell AL, Neidle EL. 1997. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology 143: 1345–1357 [DOI] [PubMed] [Google Scholar]
- 21. Gray YH. 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 16: 461–468 [DOI] [PubMed] [Google Scholar]
- 22. Griese M, Lange C, Soppa J. 2011. Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323: 124–131 [DOI] [PubMed] [Google Scholar]
- 23. Haack KR, Roth JR. 1995. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics 141: 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Juni E, Janik A. 1969. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J. Bacteriol. 98: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kichenaradja P, Siguier P, Pérochon J, Chandler M. 2010. ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res. 38: D62–D68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kok RG, D'Argenio DA, Ornston LN. 1997. Combining localized PCR mutagenesis and natural transformation in direct genetic analysis of a transcriptional regulator gene, pobR. J. Bacteriol. 179: 4270–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kugelberg E, Kofoid E, Reams AB, Andersson DI, Roth JR. 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. U. S. A. 103: 17319–17324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kusumoto M, et al. 2011. Insertion sequence-excision enhancer removes transposable elements from bacterial genomes and induces various genomic deletions. Nat. Commun. 2: 152. [DOI] [PubMed] [Google Scholar]
- 29. Ling A, Cordaux R. 2010. Insertion sequence inversions mediated by ectopic recombination between terminal inverted repeats. PLoS One 5: e15654 doi:10.1371/journal.pone.0015654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62: 725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montano SP, Rice PA. 2011. Moving DNA around: DNA transposition and retroviral integration. Curr. Opin. Struct. Biol. 21: 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pecoraro V, Zerulla K, Lange C, Soppa J. 2011. Quantification of ploidy in proteobacteria revealed the existence of monoploid, (mero-)oligoploid and polyploid species. PLoS One 6: e16392 doi:10.1371/journal.pone.0016392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Polard P, Seroude L, Fayet O, Prère MF, Chandler M. 1994. One-ended insertion of IS911. J. Bacteriol. 176: 1192–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reams AB, Neidle EL. 2004. Gene amplification involves site-specific short homology-independent illegitimate recombination in Acinetobacter sp. strain ADP1. J. Mol. Biol. 338: 643–656 [DOI] [PubMed] [Google Scholar]
- 35. Reams AB, Neidle EL. 2003. Genome plasticity in Acinetobacter: new degradative capabilities acquired by the spontaneous amplification of large chromosomal segments. Mol. Microbiol. 47: 1291–1304 [DOI] [PubMed] [Google Scholar]
- 36. Roth JR, et al. 1996. Rearrangements of the bacterial chromosome: formation and applications, p 2256–2276 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 2 American Society for Microbiology, Washington, DC [Google Scholar]
- 37. Rousseau P, et al. 2002. Transposition of IS911, p 367–383 In Craig NL, Craigie R, Gellert M, Lambowitz A. ( ed), Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- 38. Rousseau P, Tardin C, Tolou N, Salomé L, Chandler M. 2010. A model for the molecular organisation of the IS911 transpososome. Mob. DNA 1: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sambrook J, Fritsch EF, Maniatis T. 1987. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 40. Seaton SC, et al. 2012. Genome-wide selection for increased copy number in Acinetobacter baylyi ADP1: locus and context-dependent variation in gene amplification. Mol. Microbiol. 83: 520–535 [DOI] [PubMed] [Google Scholar]
- 41. Segura A, Bünz PV, D'Argenio DA, Ornston LN. 1999. Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J. Bacteriol. 181: 3494–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shanley MS, Neidle EL, Parales RE, Ornston LN. 1986. Cloning and expression of Acinetobacter calcoaceticus catBCDE genes in Pseudomonas putida and Escherichia coli. J. Bacteriol. 165: 557–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stankiewicz P, Lupski JR. 2010. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61: 437–455 [DOI] [PubMed] [Google Scholar]
- 44. Tobiason DM, Seifert HS. 2010. Genomic content of Neisseria species. J. Bacteriol. 192: 2160–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tobiason DM, Seifert HS. 2006. The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol. 4: e185 doi:10.1371/journal.pbio.0040185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turlan C, Loot C, Chandler M. 2004. IS911 partial transposition products and their processing by the Escherichia coli RecG helicase. Mol. Microbiol. 53: 1021–1033 [DOI] [PubMed] [Google Scholar]
- 47. Turlan C, Ton-Hoang B, Chandler M. 2000. The role of tandem IS dimers in IS911 transposition. Mol. Microbiol. 35: 1312–1325 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.