Abstract
Pseudomonas aeruginosa is a Gram-negative, opportunistic pathogen that utilizes polar type IV pili (T4P) for twitching motility and adhesion in the environment and during infection. Pilus assembly requires FimX, a GGDEF/EAL domain protein that binds and hydrolyzes cyclic di-GMP (c-di-GMP). Bacteria lacking FimX are deficient in twitching motility and microcolony formation. We carried out an extragenic suppressor screen in PA103ΔfimX bacteria to identify additional regulators of pilus assembly. Multiple suppressor mutations were mapped to PA0171, PA1121 (yfiR), and PA3703 (wspF), three genes previously associated with small-colony-variant phenotypes. Multiple independent techniques confirmed that suppressors assembled functional surface pili, though at both polar and nonpolar sites. Whole-cell c-di-GMP levels were elevated in suppressor strains, in agreement with previous studies that had shown that the disrupted genes encoded negative regulators of diguanylate cyclases. Overexpression of the regulated diguanylate cyclases was sufficient to suppress the ΔfimX pilus assembly defect, as was overexpression of an unrelated diguanylate cyclase from Caulobacter crescentus. Furthermore, under natural conditions of high c-di-GMP, PA103ΔfimX formed robust biofilms that showed T4P staining and were structurally distinct from those formed by nonpiliated bacteria. These results are the first demonstration that P. aeruginosa assembles a surface organelle, type IV pili, over a broad range of c-di-GMP concentrations. Assembly of pili at low c-di-GMP concentrations requires a polarly localized c-di-GMP binding protein and phosphodiesterase, FimX; this requirement for FimX is bypassed at high c-di-GMP concentrations. Thus, P. aeruginosa can assemble the same surface organelle in distinct ways for motility or adhesion under very different environmental conditions.
INTRODUCTION
Bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) is a ubiquitous bacterial second messenger that serves as a single integrated signal for many environmental inputs. Increasing levels of this small intracellular molecule are associated with bacterial transitions from motile growth to formation of adherent biofilm communities (20, 41). c-di-GMP is synthesized from GTP by diguanylate cyclases (DGC) and hydrolyzed to pGpG by phosphodiesterases (PDEs) (42). These enzymes contain conserved GGDEF and EAL (or HY-GYP) domains for making and breaking c-di-GMP, respectively. c-di-GMP DGCs and PDEs are present throughout the bacterial kingdom, and the genomes of many organisms encode large numbers of these enzymes. c-di-GMP binding to nonenzymatic protein domains modulates functions as diverse as rhamnolipid biosynthesis (2), exopolysaccharide (EPS) production (25, 31), siderophore production (27), virulence (24, 33), cell division (8), and motility organelle assembly and function (1, 19, 21). In general, most GGDEF and EAL/HY-GYP domains are associated with signal input domains, such as PAS (oxygen sensing), REC (phosphoreceiver), and GAF (cyclic nucleotide binding) domains, allowing c-di-GMP synthesis and hydrolysis to be regulated in response to multiple environmental signals (15).
The opportunistic Gram-negative human pathogen Pseudomonas aeruginosa encodes 17 GGDEF, 9 EAL, and 16 dual-domain (GGDEF/EAL)-containing proteins (http://www.ncbi.nlm.nih.gov/Complete_Genomes/SignalCensus.html). Several of these proteins have been studied in detail, including FimX, a dual-domain protein required for twitching motility (19, 21). The DGC domain of FimX is degenerate and enzymatically inactive, while the PDE domain lacks residues predicted to play roles in coordinating divalent cations important for substrate binding and catalysis. Nonetheless, FimX binds c-di-GMP with high affinity (∼100 nM) via its EAL domain and exhibits weak PDE activity in vitro (21, 35, 40). Deletion of FimX, as well as point mutations targeting the EAL or degenerate GDSIF motifs, result in a pilus assembly defect: whole-cell pilin levels are unchanged, but surface pilin expression is markedly reduced and bacterial twitching motility is impaired (19, 21).
Twitching is a surface-associated motility that is mediated by type IV pili (T4P) and exhibited by a phylogenetically diverse group of bacteria, such as Neisseria gonorrhoeae, P. aeruginosa, and Myxococcus xanthus (29). Twitching motility is important for biofilm initiation (36), a surface colonization behavior of P. aeruginosa that allows it to exploit environmental and host niches, such as the cystic fibrosis (CF) airway. Biofilm maturation also requires T4P, although these pili can be motile or nonmotile (3, 6). Biofilm formation is promoted by increased intracellular c-di-GMP, which results in elevated expression of EPS (11), surface adhesins (32, 50), and protein adhesins (4).
In this study, we carried out an extragenic suppressor screen in PA103ΔfimX to identify mutations that restored surface pilus assembly. Unexpectedly, we observed that the pilus assembly requirement for FimX could be bypassed at high intracellular concentrations of c-di-GMP. Characterization of the suppressor strains suggests that P. aeruginosa assembles T4P in distinct ways for motility and biofilm formation.
MATERIALS AND METHODS
Strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Both Escherichia coli and Pseudomonas aeruginosa were cultured in Luria broth (LB) medium with antibiotics as required at the following concentrations: gentamicin, 15 μg ml−1 for E. coli or 100 μg ml−1 for P. aeruginosa; tetracycline, 20 μg ml−1 for E. coli or 100 μg ml−1 for P. aeruginosa. M9 medium was used for cultivating P. aeruginosa for biofilms. Commercial chemically competent E. coli was used for transformations (Invitrogen), and P. aeruginosa was transformed by electroporation (7). All bacterial strains were stored at −80°C as 15% glycerol stocks.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description/relevant genotype | Source or reference |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169(Φ80dlacZΔM15) hsdR17 thi-1 relA1 recA1 | Invitrogen |
| P. aeruginosa strains | ||
| PA103 | Aflagellate virulent lung isolate | 26 |
| PAO1 | WT | 48 |
| PA103ΔfimX | PA103 containing an in frame deletion of aa 15-685 of FimX | 21 |
| PA103ΔpilA | Gmr cassette (aacC1) inserted into pilA in PA103, Gmr | 52 |
| PAO1ΔfimX | PAO1 containing an in frame deletion of aa 15-685 of FimX | 21 |
| PAO1ΔpilA | PAO1 with in-frame deletion of PilA | 10 |
| ΔfimXPA0171∷Tn5 | Tn insertion in PA0171 in PA103ΔfimX | This study |
| ΔfimXPA1121∷Tn5 | Tn insertion in PA1121 in PA103ΔfimX | This study |
| ΔfimXPA3703∷Tn5 | Tn insertion in PA3703 in PA103ΔfimX | This study |
| PA103/pMQ72 | PA103 with the arabinose-inducible plasmid | This study |
| PA103/pPA0169 | PA103 overexpressing PA0169 under PBAD | This study |
| PA103/pPA1120 | PA103 overexpressing PA1120 under PBAD | This study |
| PA103/pPA3702 | PA103 overexpressing PA3702 under PBAD | This study |
| PAO1/pPA0169 | PAO1 overexpressing PA0169 under PBAD | This study |
| PAO1/pPA1120 | PAO1 overexpressing PA1120 under PBAD | This study |
| PAO1/pPA3702 | PAO1 overexpressing PA3702 under PBAD | This study |
| ΔfimX/pMQ72 | PA103ΔfimX with the arabinose-inducible plasmid | This study |
| ΔfimX/p1120 | PA103ΔfimX overexpressing PA1120 under PBAD | This study |
| ΔfimX/p1120mutant | PA103ΔfimX overexpressing mutated PA1120 under PBAD | This study |
| ΔfimX/pPA0169 | PA103ΔfimX overexpressing PA0169 under PBAD | This study |
| ΔfimX/pPA3702 | PA103ΔfimX overexpressing PA3702 under PBAD | This study |
| ΔfimX/pPleD* | PA103ΔfimX overexpressing PleD* under PBAD | This study |
| ΔfimXPA0171 VC | ΔfimXPA0171∷Tn5 with the arabinose-inducible plasmid | This study |
| ΔfimXPA0171/pPA0171 | ΔfimXPA0171∷Tn5 overexpressing PA0171 under PBAD | This study |
| ΔfimXPA1121 VC | ΔfimXPA1121∷ Tn5 with the arabinose-inducible plasmid | This study |
| ΔfimXPA1121/pPA1121 | ΔfimXPA1121∷ Tn5 overexpressing PA1121 under PBAD | This study |
| ΔfimXPA3703 VC | ΔfimXPA3703∷ Tn5 with the arabinose-inducible plasmid | This study |
| ΔfimXPA3703/pPA3703 | ΔfimXPA3703∷ Tn5 overexpressing PA3703 under PBAD | This study |
| Plasmids | ||
| pMQ72 | Cloning vector; Gmr, URA3 PBAD; araC | 45 |
| pPleD* | PleD* under the control of the PBAD promoter in pMQ72; Gmr, URA3 | This study |
| pPA1120 | PA1120 under the control of the PBAD promoter in pMQ72; Gmr, URA3 | This study |
| pPA3702 | PA3702 under the control of the PBAD promoter in pMQ72; Gmr, URA3 | This study |
| pPA0169 | PA0169 under the control of the PBAD promoter in pMQ72; Gmr, URA3 | This study |
| pPA0171 | PA0171 under the control of the PBAD promoter in pMQ72; Gmr, URA3 | This study |
| pPA3703 | PA3703 under the control of the PBAD promoter in pMQ72; Gmr, URA3 | This study |
| pPA1121 | PA1121 under the control of the PBAD promoter in pMQ72; Gmr, URA3 | This study |
| pPA1120/topo | PA1120 cloned in pTOPO | This study |
| pPA1120mutant | Mutant PA1120 (GGDEF to AAAEF) under the control of the PBAD promoter in pMQ72; Gmr, URA3 | This study |
Extragenic suppressor screen.
Tn5<TcR> transposon (Tn) DNA (200 ng) was incubated with EZ-Tn5 transposase (Epicentre Biotechnologies) in the presence of glycerol to form transposon complexes according to the manufacturer's protocols. Electrocompetent PA103ΔfimX was prepared and electroporated with one-eighth of the transposon mixture and then incubated for 2 h at 37°C in LB and plated onto LB-tetracycline plates (7). Approximately 40,000 transformants were screened visually for the appearance of rough-edged colonies at 72 h after plating. These colonies were restreaked to LB-tetracycline to confirm colony morphology and then tested by stab agar assay for restoration of twitching motility. Colonies that displayed increased twitching motility compared to ΔfimX were further evaluated. The location of transposon insertions was mapped by inverse PCR as previously described (34).
Strain construction.
All PCR primers employed in this study are listed in Table 2 and were ordered from either Integrated DNA Technologies or Keck Facility (Yale University). PA0169, PA0171, PA1120, PA1121, PA3703, and PA3702 were amplified from P. aeruginosa PA103 genomic DNA using Phusion DNA polymerase (New England BioLabs) and then cloned in pMQ72 under an inducible pBAD promoter. PleD* His (38) was used as a template to amplify PleD*. The absence of mutations in amplified genes was confirmed by DNA sequencing. Plasmids were introduced into P. aeruginosa, and transformants were selected on LB-gentamicin. Arabinose was used at a concentration of 0.2% in all experiments to induce gene expression.
Table 2.
Primers used in this study
| Primer name | Primer sequencea (5′–3′) |
|---|---|
| 0169-F | GCGAGCTCTCGCTGGAGCAACTGATGCGGCTGGAG |
| 0169-R | GCTGCTGGTGGAGCTCAGCCGAGGCC |
| 0171-F | CCGAGCAGGCCGAGCTCTTCGTGGCGACGCTCG |
| 0171-R | CGACCCAGTCGATCACGCGGCCGCACAGTTC |
| 1120-F | CGCGCATCACCTTCGAGCTCAATCTCGACTCCATC |
| 1120-R | CCAGGCCCAGCACAAGCTTGAACAGGGCG |
| 1121-F | GGCACATCCGGTACCATTCGCCGCACACCG |
| 1121-R | GCGCAGGGTCAGCGGCCGCCCAGGGTC |
| 3703-F | CAAGGATCGTGAGGAGCTCCGTCGGCGCGGTC |
| 3703-R | CCTCGTATCAAATTGAAAGCTTTTCCACGCACCG |
| 3702-F | ATTAGCCGGGGTCGACTCCGGCCAGGCG |
| 3702-R | CTGCGAAGTATACTAAGCTTGCGCCCCACAGG |
| PleD*-F | GCGGTACCGGCAGCCTGCATGAGCGCCCGG |
| PleD*-R | GCAAGCTTTCAGGCGGCCTTGCCGACC |
| 1120 m-F | CCTGGTGGCGCGCCTGGCCGCCGCCGAATTCGCCGTCCTGC |
| 1120 m-R | GCAGGACGGCGAATTCGGCGGCGGCCAGGCGCGCCACCAGG |
| 1120-RTF | TCGCTATACGGGTTCCAATCCATC |
| 1120-RTR | TATAGCTGATGGAACGGGCGAT |
| 3702-RTF | TGCACAACCCTCATGAGAGCAAGA |
| 3702-RTR | AACAGAAATGGAAGTCGATGCCCG |
| 169-RTF | ATCTCCGACGGTTTCCAGTCGAT |
| 169-RTR | TTGAGGTCGCGCATCATCTGCT |
| rplU-RTF | CGCAGTGATTGTTACCGGTG |
| rplU-RTR | AGGCCTGAATGCCGGTGATC |
| cupA-RTF | ATGACCAGAACTTCGAACCCATGC |
| cupA-RTR | ATGTGATAGTGTTTGCCGCCATCG |
| pelA-RTR | GGCGCTCACCGTACTCGATC |
| pelA-RTR | GATGCGCCGGCCATTCTCG |
| pslD-RTR | CGCACGCCGCAGGAAATCG |
| pslD-RTR | CTTCCGAGTCCTCGCGCATG |
Restriction sites are underlined.
Subsurface stab assay for twitching motility.
Twitching motility was determined at an LB agar-plastic interface after overnight incubation at 37°C, followed by 24 h of incubation at room temperature as previously described (21). The diameter of the twitching zone on the plastic surface was measured; for noncircular zones, the longest and shortest dimensions were measured and averaged.
Surface pilin ELISA.
Surface pilin enzyme-linked immunosorbent assay (ELISA) was performed as described previously (21), using polyclonal antiserum raised against pili purified from PA103 or PAO1.
Phage sensitivity.
PO4 phage (kindly provided by Patricia Baynham, Thomas More College) was serially diluted and spotted in duplicate onto soft agar (0.7%) containing PA103, PAO1, or the indicated isogenic mutants. Plaques were counted following overnight incubation at 37°C. Efficiency of plaque formation was calculated relative to the wild-type (WT) PA103 strain.
Transmission electron microscopy (TEM).
Log-phase bacteria grown in LB (plus antibiotics and 0.2% arabinose, if required) were allowed to bind to glow-discharged carbon grids for 1 min. Grids were stained twice with 1% phosphotungstate (1 min) or once with 1% uranyl acetate (1 min) and then dried for 10 to 15 min before imaging on a Tecnai 12 Biotwin microscope (Center for Cell and Molecular Imaging, Yale University).
Determination of intracellular c-di-GMP concentrations.
Bacteria were grown overnight on LB plates (with antibiotics as indicated) at 37°C, harvested by scraping, and resuspended in 300 μl phosphate-buffered saline (PBS); a 10-μl aliquot was set aside for bicinchoninic acid (BCA) protein assay (Pierce). The remaining cells were repelleted and resuspended in 300 μl of ice-cold extraction solvent (acetonitrile-methanol-water at 40:40:20 [vol/vol/vol]) containing the internal standard cXMP (200 ng/ml). The extraction process was completed as described by Spangler et al. (46). Extracts were analyzed by liquid chromatography-tandem mass spectrometry (MS/MS) on an API 3000 triple-quadrupole mass spectrometer (Applied Biosystems, Foster City, CA), coupled with a series 200 high-performance liquid chromatography (HPLC) system (Perkin Elmer Instruments, Norwalk, CT). Liquid chromatography separation was achieved on a reverse-phase column with an ammonium acetate-methanol gradient. Final c-di-GMP concentrations were expressed as pmol/mg of bacterial protein; the molar concentration of c-di-GMP was calculated as described elsewhere (13). Biological duplicates were prepared for each sample; each strain was assayed in 2 to 3 independent experiments.
Site-directed mutagenesis.
The GGDEF motif (aa 328 to 332) of PA1120 was changed to AAAEF according to the QuikChange site-directed mutagenesis kit protocol (Stratagene) using primers 1120m-F and 1120m-R (Table 3). PA1120 cloned in pTOPO (Invitrogen) was used as a template for this reaction, and the introduction of only the desired mutations was confirmed by sequencing. PA1120m was then subcloned into pMQ72, resulting in an arabinose-inducible expression construct.
Table 3.
Efficiency of PO4 plaque formation
| Strain | Efficiency of plaque formationa |
|---|---|
| PA103 | 1.0 ± 0.26 |
| PA103ΔpilA | <1 × 10−9 |
| PA103ΔfimX | 0.01 |
| ΔfimX PA0171∷Tn5 | 1.37 ± 0.6 |
| ΔfimX PA1121∷Tn5 | 1.5 ± 0.28 |
| ΔfimX PA3703∷Tn5 | 1.19 ± 0.06 |
| ΔfimX/pMQ72 | 0.017 ± 0.005 |
| ΔfimX/pPA0169 | 0.006 ± 0.006 |
| ΔfimX/pPA1120 | 1.1 ± 0.35 |
| ΔfimX/pPA3702 | 1.0 ± 0.4 |
| ΔfimX/pPleD* | 0.85 ± 0.12 |
| PA103/pMQ72 | 1.05 ± 0.14 |
| PA103/pPA0169 | 1.25 ± 0.11 |
| PA103/pPA1120 | 0.7 ± 0.09 |
| PA103/pPA3702 | 0.88 ± 0.001 |
| PAO1/pMQ72 | 1.45 ± 0.27 |
| PAO1/pPA0169 | 1.25 ± 0.104 |
| PAO1/pPA1120 | 1.67 ± 0.29 |
| PAO1/pPA3702 | 0.95 ± 0.104 |
Efficiency of PO4 plaque formation expressed relative to the WT strain (PA103 or PAO1, as appropriate). Means ± SD of 3 to 5 independent experiments are shown.
RNA isolation and quantitative PCR.
Bacterial RNA was isolated from PA103ΔfimX and the transposon mutants grown to late exponential phase as previously described (43). RNA was checked for DNA contamination by PCR amplification of the 50S ribosomal protein gene rplU using Taq DNA polymerase and primer pair rplU-RTF and rplU-RTR. cDNA was synthesized from 1 μg RNA using an iScript cDNA synthesis kit (Bio-Rad). For quantitative reverse transcription-PCR (qRT-PCR), 1 μl of cDNA was added to SYBR green qPCR master mix (Bio-Rad) plus 500 nM the respective forward (-RTF) and reverse (-RTR) primers (Table 3). Fold change of the expression of a target gene in each extragenic suppressor strain was calculated relative to its expression level in the parent strain ΔfimX and normalized to the expression of rplU as described previously (39).
Static biofilm formation.
Biofilm formation assays were performed as previously described using 96-well plates (37). Briefly, overnight cultures grown in LB were diluted into LB (1:100); 100 μl of the dilution was used to inoculate 3 to 4 replicate wells per strain/assay. Plates were incubated for 24 h at 30°C and then gently washed to remove nonadherent bacteria. Adherent biofilm was stained with crystal violet (0.1%), followed by biofilm dissolution with glacial acetic acid (1%). The dissolved crystal violet was measured at an absorbance of 550 nm. Each experiment was repeated independently at least 3 times.
Flow-cell biofilms.
Biofilms were grown at 30°C in flow chambers with individual channel dimensions of 1 by 4 by 40 mm and supplied at a flow of 3 ml h−1 with M9 medium supplemented with 0.3 mM glucose (16). The flow system was assembled and prepared as described elsewhere (23). Cultures for inoculation of the flow channels were prepared by inoculating a single colony from a plate into test tubes containing M9 medium supplemented with 30 mM glucose and growing them at 37°C for 16 h. Cultures were diluted to an optical density at 600 nm of 0.01 in M9 medium with 0.3 mM glucose and used for inoculation. A 350-μl volume of diluted culture was injected into each flow channel with a small syringe. After inoculation, flow channels were left without flow for 1 h for cell attachment. Medium flow was then started at a constant rate of 3 ml h−1 with a Watson Marlow 205S peristaltic pump for 4 days.
Pilus labeling in biofilms.
Bacterial overnight cultures were diluted 1:100 in fresh LB medium and inoculated on polylysine-coated coverslips for 24 h at 30°C. The coverslips were washed with PBS, fixed with methanol, and labeled for surface pilin per our surface ELISA protocol. The primary antibody (anti-PilA; PAO1) was used at a dilution of 1:200, and the secondary goat anti-rabbit Alexa 488 antibody (Molecular Probes) was used at a dilution of 1:500. Primary antibody was omitted for secondary antibody-only controls. For this experiment, PAO1, PAO1ΔfimX, and PAO1ΔpilA strains were used.
Image acquisition and processing.
All microscopic observations and image acquisitions were performed by confocal laser scanning microscopy (Zeiss LSM 510). Images were obtained with a 60× water objective. Image scanning was carried out with the 488-nm laser line from an Ar-Kr laser. Simulated fluorescence projections and sections through the biofilms were generated with the Zeiss Image Browser software. Images from each experiment were analyzed by the computer program COMSTAT (17).
Statistical analysis.
Means and standard deviations were calculated using Excel (Microsoft). Data are expressed as means ± SD. P values were calculated by analysis of variance (ANOVA; one way or two way, as indicated) followed by Bonferroni's or Dunnett's multiple comparison tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001) using GraphPad Prism 5.0.
RESULTS
Isolation of fimX suppressors that restore twitching motility.
PA103ΔfimX bacteria do not assemble surface pili, resulting in colonies with a characteristic smooth appearance that is easily distinguished from the jagged colony border of WT P. aeruginosa PA103 (19, 21). We took advantage of this difference in colony morphology to visually screen a library of ca. 40,000 Tn5<TcR> transposon insertion mutants in the PA103ΔfimX background for extragenic suppressors that restored a jagged colony morphology. Forty-two mutants satisfied this screen; these were restreaked and checked for WT colony morphology, resulting in 29 mutants that had either partial or near-complete suppression of the PA103ΔfimX colony morphology phenotype. Multiple independent transposon (Tn) insertions were mapped by inverse PCR to each of three specific loci: PA0171, PA1121 (yfiR), and PA3703 (wspF) (see Fig. S1 in the supplemental material). Transposon-mediated disruption of these three genes has been previously reported to result in a wrinkly spreader phenotype, i.e., tight colonies on agar and clumping growth in liquid (9, 14). Three insertions could not be mapped with confidence, while four independent insertions mapped to mutL, a mismatch repair locus. These last seven insertions were not pursued further.
Extragenic suppressors restore surface pilus expression and function.
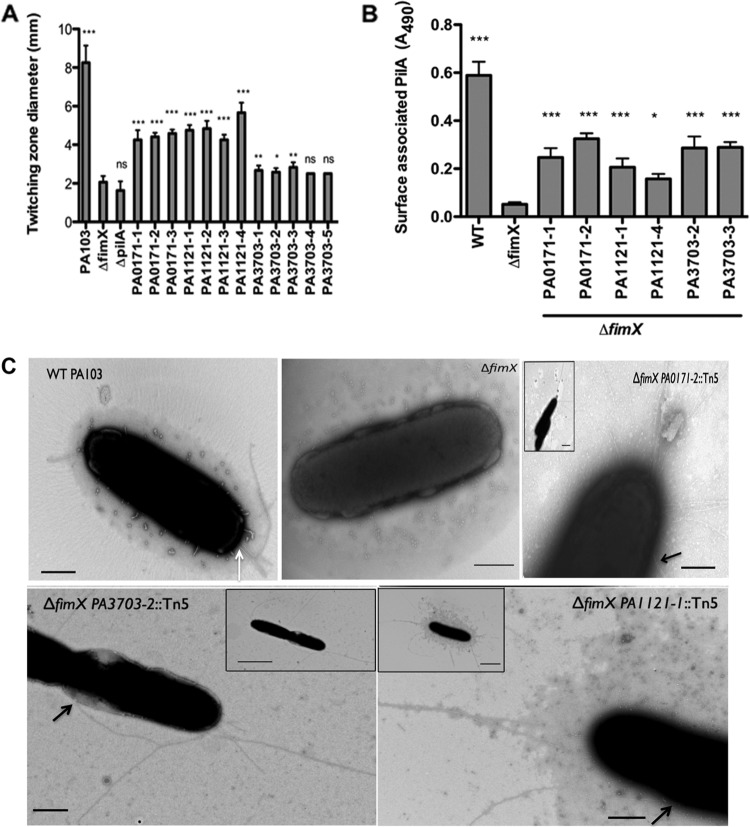
Several assays were used to assess the presence (e.g., surface pilin ELISA and transmission electron microscopy [TEM]) and function (e.g., twitching motility and pilus-specific phage sensitivity) of T4P in the PA103ΔfimX suppressors (5, 36). Multiple independent Tn insertions in PA0171, PA1121 (yfiR), and PA3703 (wspF) restored twitching motility compared to PA103ΔfimX, though no suppressor showed motility equivalent to that of WT PA103 (Fig. 1A). Surface pilin levels, measured by ELISA for two representative insertions in each gene, were also partially restored by the suppressor mutations (Fig. 1B), with all suppressor strains assembling significantly more surface pilin than the PA103ΔfimX strain. As seen by TEM, liquid-grown ΔfimX suppressors assembled surface pili at multiple sites, both polar and nonpolar, on the bacterial surface. This was in contrast to WT PA103 bacteria, which assembled only polar T4P when examined after planktonic growth (Fig. 1C; also see Fig. S2 in the supplemental material). The assembled pili appeared to be functional, as strains with insertions in PA0171, PA1121 (yfiR), and PA3703 (wspF) were as sensitive to the T4P-specific phage PO4 as WT PA103 (Table 3). Collectively, these results demonstrate that Tn insertions in PA0171, yfiR, and wspF restored surface pilin assembly to PA103ΔfimX bacteria. Assembled pili were functional, but assembly was no longer restricted to the bacterial poles.
Fig 1.
Extragenic suppressors restore surface pili. (A) Twitching zone diameter as measured by the subsurface stab assay. Strains were assayed 3 to 5 times, and means ± standard deviations (SD) (n = 4) from a representative experiment are shown. All of the strains were compared to ΔfimX using one-way ANOVA followed by Dunnett's multiple-comparison test. (B) Total surface pili as determined by surface pilin ELISA. Two representative Tn insertions were assayed for each gene. Bars indicate means ± SD from three independent experiments. All of the samples were compared to ΔfimX using one-way ANOVA followed by Dunnett's multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Visualization of T4P by transmission electron microscopy. Bacteria from early exponential growth phase were stained directly with 1% phosphotungstate. Panels show higher magnification pictures of representative cells for each strain, and insets show lower magnification pictures of the same cells. Black arrows indicate the nonpolar site of attachment of the pili with the cell body. White arrow indicates T4P on a WT surface. Scale bar, 500 nm (main panel) or 2 μm (inset).
PA103ΔfimX suppressors map to operons encoding diguanylate cyclases.
PA0171, PA1121 (yfiR), and PA3703 (wspF) each lie upstream of genes that encode proteins with predicted or confirmed DGC activity, i.e., PA0169 (siaD), PA1120 (tpbB, or yfiN), and PA3702 (wspR), raising the possibility that the observed phenotypes were due to polar effects on these downstream genes. Expression of each DGC was therefore measured by qRT-PCR in PA103ΔfimX and in the corresponding suppressor strain. We did not observe any decrease in DGC transcript levels in the suppressor strains relative to PA103ΔfimX; rather, similar or increased mRNA levels were noted. ΔfimX PA1121∷Tn5 showed an increase of 1.9- ± 0.8-fold in the mRNA of PA1120; ΔfimX PA3703∷Tn5, a 4.0- ± 0.7-fold increase in PA3702 mRNA; and ΔfimX PA0171∷Tn5, a large, 70- ± 10-fold increase in PA0169 mRNA levels.
Importantly, each suppressor mutation could be complemented by expressing WT PA0171, PA1121 (YfiR), or PA3703 (WspF) in the respective ΔfimX suppressor strain. The expression of PA1121 (YfiR) or PA3703 (WspF) completely reverted the ΔfimX suppressor back to the parental (PA103ΔfimX) phenotype, as demonstrated by the loss of twitching motility (Fig. 2A) and surface pilin expression (Fig. 2B). The expression of PA0171 in ΔfimX PA0171∷Tn5 resulted in partial complementation, as the amount of surface pilin remained higher than that observed in PA103ΔfimX (although less than that measured in ΔfimX PA0171∷Tn5) (Fig. 2B). Thus, the suppressor phenotypes appear to be due to the loss of PA0171, PA1121 (YfiR), or PA3703 (WspF) activity.
Fig 2.
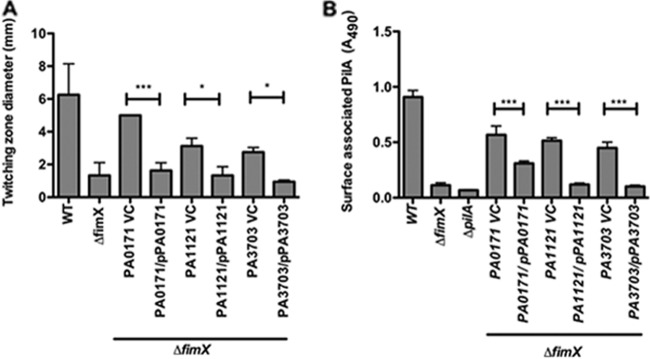
Extragenic suppressor phenotypes can be reverted by expressing PA0171, PA1121, or PA3703 in trans. Extragenic suppressor strains carrying either empty vector (VC) or PA0171, PA1121, or PA3703 cloned under the pBAD promoter were grown on LB plates plus 0.2% arabinose. Strains were assayed for surface pilin expression by ELISA (A) and twitching motility by subsurface stab assay (B). Bars represent means ± SD of triplicate samples from four independent experiments. P values were determined by ANOVA with Bonferroni's multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Intracellular cyclic-di-GMP concentrations are elevated in extragenic suppressor strains.
PA1121 (YfiR) and PA3703 (WspF) are known to negatively regulate the DGCs PA1120 (YfiN/TpbB) (27) and PA3702 (WspR) (18), respectively. It is not known whether PA0171 plays a similar role via PA0169. We measured intracellular c-di-GMP concentration in PA103, PA103ΔfimX, and each ΔfimX suppressor grown on agar plates using HPLC-MS/MS (46). PA103 and PA103ΔfimX had similar low intracellular c-di-GMP levels (∼300 nM). In contrast, each of the suppressors had significantly elevated c-di-GMP concentrations, with ΔfimX PA1121∷Tn5 having the highest (∼100-fold greater than PA103ΔfimX) followed by ΔfimX PA3703∷Tn5 (∼20-fold) and ΔfimX PA0171∷Tn5 (∼10-fold) (Fig. 3).
Fig 3.
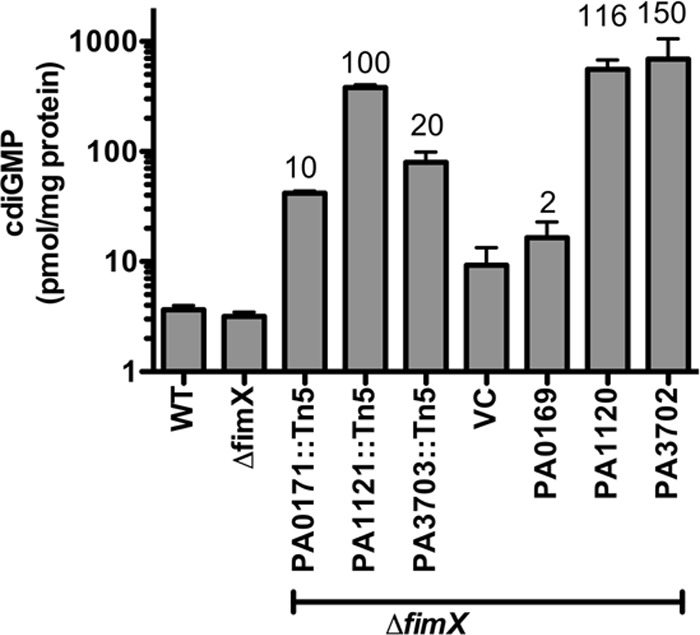
c-di-GMP levels are elevated in the extragenic suppressor strains. Bacteria were grown on LB agar plates overnight supplemented with antibiotic and 0.2% arabinose as appropriate. Each sample was prepared in biological duplicates and measured in 2 to 3 experiments; bars represent means ± SD. Values over each bar represent the c-di-GMP fold increase in the respective strains compared to the c-di-GMP level in ΔfimX or ΔfimX VC, respectively.
Type IV pilus assembly in the absence of FimX is driven by high c-di-GMP levels.
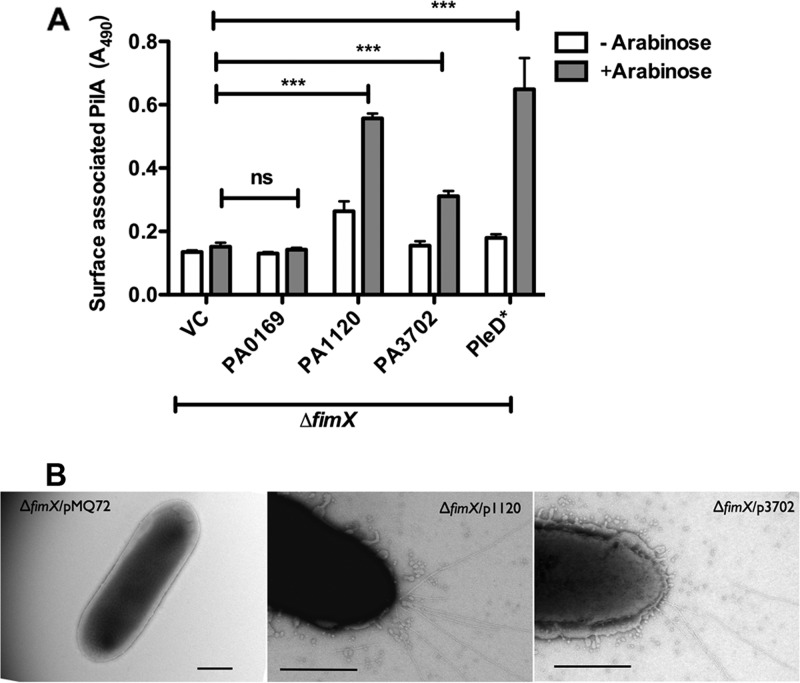
High c-di-GMP concentrations are usually associated with sessile behaviors; therefore, we found it surprising that all of the suppressor strains showed elevated levels of intracellular c-di-GMP and yet exhibited motility. We tested whether a high level of c-di-GMP was sufficient to suppress the PA103ΔfimX phenotype by expressing each DGC in PA103ΔfimX under the control of an inducible promoter. Overexpression of each DGC except PA0169 resulted in increased c-di-GMP levels as measured by HPLC-MS/MS (Fig. 3). Overexpression of either PA1120 (YfiN/TpbB) or PA3702 (WspR) led to increased surface pilin assembly (Fig. 4A) and twitching motility zones (see Fig. S3 in the supplemental material) and restored PO4 phage sensitivity compared to PA103ΔfimX (Table 3). However, PA0169 overexpression did not restore surface pilin expression (Fig. 4A), and these bacteria remained PO4 phage resistant (Table 3). Multiple pili were observed by TEM on the majority of PA103ΔfimX bacteria overexpressing PA1120 (YfiN/TpbB) or PA3702 (WspR) (Fig. 4B), whereas this was not the case for PA0169 overexpressing ΔfimX bacteria. We also tested whether the expression of an unrelated DGC could suppress the PA103ΔfimX T4P assembly defect, choosing the well-characterized Caulobacter crescentus DGC PleD for this experiment (38). Expression of PleD*, a constitutively active allele of this DGC, restored surface pilin assembly (Fig. 4A), twitching motility (see Fig. S3), and PO4 phage sensitivity (Table 3) compared to PA103ΔfimX. Similar results were obtained when we expressed the WT allele of PleD from a higher-copy-number plasmid (see Fig. S4). Thus, suppression of PA103ΔfimX phenotypes is not restricted to the DGCs identified in our genetic screen.
Fig 4.
Overexpression of diguanylate cyclases restores surface pilus assembly in ΔfimX. PA103ΔfimX carrying pMQ72 (VC) or the indicated DGCs cloned under the pBAD promoter were grown on plates with or without 0.2% arabinose. (A) Total surface pili as determined by surface pilin ELISA. Bars indicate means ± SD of triplicate samples from three independent experiments. Two-way ANOVA followed by Bonferroni's multiple comparison test was used to analyze the data. All induced (+Arabinose) samples were compared to ΔfimX/pMQ72 plus arabinose. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) T4P visualized by transmission electron microscopy after staining with 1% phosphotungstate. Panels show a higher magnification image of a representative cell. Scale bar, 500 nm. ns, not significant.
If high c-di-GMP levels are required to suppress the ΔfimX phenotype, then overexpression of an enzymatically inactive DGC should not support T4P assembly. We tested this by mutating the DGC motif of PA1120 (YfiN/TbpB) from GGDEF to AAAEF, a change predicted to disrupt DGC activity (27). Overexpression of this mutated DGC (PA1120m) in PA103ΔfimX did not suppress the T4P assembly defect (Fig. 5).
Fig 5.
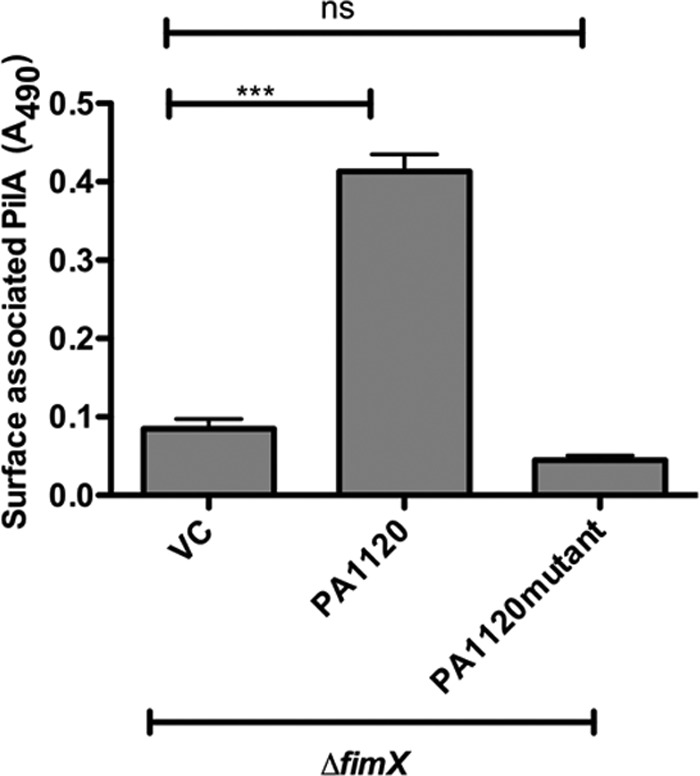
Inactive DGC does not suppress ΔfimX pilus assembly defects. ΔfimX expressing PA1120 or the enzymatically inactive allele PA1120m was harvested from plates supplemented with 0.2% arabinose. Total surface pilin was measured by ELISA. Bars indicate means ± SD of triplicate samples from three independent experiments. One-way ANOVA followed by Dunnett's comparison test was used to compare values to the vector control, ΔfimX/pMQ72 (VC). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Bacteria assemble pili in biofilms in the absence of FimX.
Biofilms likely represent a natural condition in which c-di-GMP levels are elevated, as overexpression of diguanylate cyclases promotes biofilm formation and the production of biofilm matrix components, while induction of c-di-GMP phosphodiesterases leads to biofilm dispersal (4, 12). Since experimental elevation of c-di-GMP levels in PA103ΔfimX restored surface pilus assembly, we hypothesized that this also occurred during biofilm formation and predicted that the biofilm formation defects of a ΔpilA mutant strain would not be observed for ΔfimX bacteria. Since the aflagellate WT PA103 strain produces poor biofilms, we deleted fimX in the reference strain PAO1 for these experiments, confirming that this also resulted in pilus assembly and twitching motility defects (see Fig. S5 in the supplemental material). Furthermore, the PAO1ΔfimX T4P assembly defect could be suppressed by overexpressing the DGCs PA1120 (YfiN/TbpB) and PA3702 (WspR), as we had observed for PA103ΔfimX (see Fig. S6).
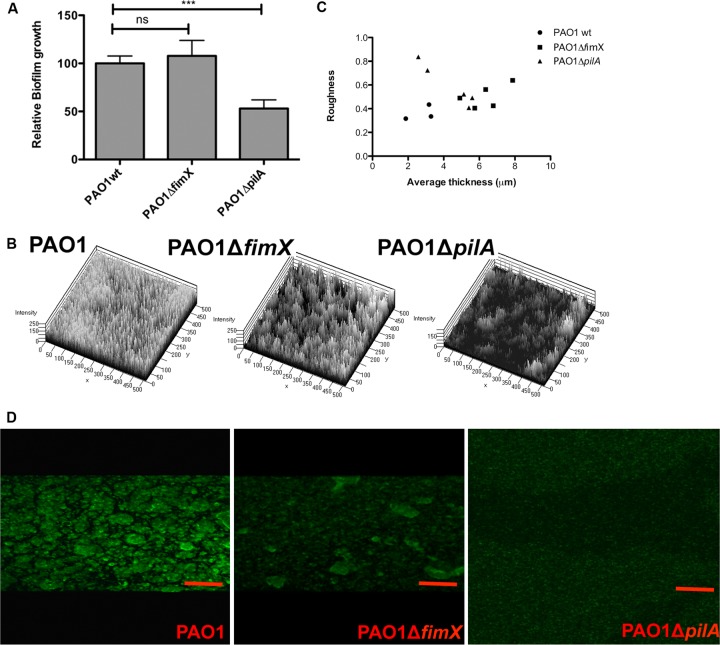
In a static biofilm model, PAO1ΔpilA had a biofilm defect, while PAO1ΔfimX formed biofilms indistinguishable from those of WT PAO1 (Fig. 6A). We also inoculated YFP-expressing PAO1, PAO1ΔfimX, and PAO1ΔpilA into flow cells and imaged biofilm structures over time by laser scanning confocal microscopy (Fig. 6B). COMSTAT analysis of day 4 biofilms showed that ΔfimX formed thicker biofilms than ΔpilA and had a roughness intermediate to that observed for PAO1 compared to PAO1ΔpilA (Fig. 6C). We confirmed that WT, ΔfimX, and ΔpilA strains had the same doubling time when grown as planktonic cultures (see Fig. S7). As bacterial spreading and colonization of surfaces in early stages of biofilm formation are thought to depend on T4P-mediated motility, we would expect increased roughness from ΔfimX bacteria. Biofilm thickness, however, is postulated to increase with T4P-dependent growth of caps on stalks during later stages of biofilm maturation. We also stained pili directly in 1-day-old static biofilms grown on coverslips, using an antibody against PAO1 pilin (Fig. 6D). WT PAO1 and PAO1ΔfimX showed qualitatively similar staining patterns, with bright staining outlining regions where microcolony structures were growing in the z axis. PAO1ΔpilA staining showed no such regions of increased signal intensity, while secondary antibody control did not give detectable fluorescence for any sample (not shown).
Fig 6.
ΔfimX bacteria form biofilms and stain for surface pilin. (A) PAO1, PAO1ΔfimX, and PAO1ΔpilA were grown as static biofilms for 24 h at 30°C in triplicate wells and then stained with crystal violet. Bars represent means ± SD from three independent experiments; values are normalized to PAO1. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Three-dimensional plot of day 4 flow biofilms formed by PAO1, PAO1ΔfimX, and PAO1ΔpilA. (C) Scatter plots showing average thickness and roughness of day 4 flow biofilms of PAO1, PAO1ΔfimX, and PAO1ΔpilA, as calculated by COMSTAT. (D) PAO1, PAO1ΔfimX, and PAO1ΔpilA were grown on polylysine-coated coverslips for 24 h under static biofilm conditions and then stained for surface pilin. All images were acquired under identical conditions. Scale bar, 10 μm.
Overexpression of DGCs in PAO1 suppresses twitching motility but not T4P assembly.
Increased activity of the DGCs PA1120 (YfiN/TbpB) (27, 51) and PA3702 (WspR) (28) results in the small-colony-variant (SCV) phenotype, characterized by wrinkly, hyperaggregative colonies that are usually reported to lack twitching motility. Although we observed no twitching motility defect for PA103 overexpressing PA1120 (YfiN/TbpB) or PA3702 (WspR), we found that overexpression of these DGCs in PAO1 did inhibit twitching motility (Fig. 7A). Since elevated c-di-GMP drives the production of exopolysaccharide and protein adhesins that might impair bacterial twitching motility (4), we explicitly tested whether the DGC-overexpressing bacteria had any defects in surface pilin assembly. No defects in pilus assembly were associated with DGC overexpression in either PAO1 or PA103 (Fig. 7B), while intact T4P function was confirmed by PO4 phage sensitivity (Table 3).
Fig 7.
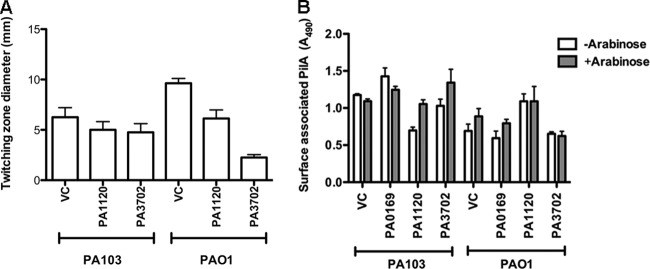
DGC overexpression in PA103 or PAO1 does not inhibit surface pilus assembly. (A) WT PA103 or PAO1 overexpressing indicated DGCs (PA1120 and PA3702) was grown on plates with 0.2% arabinose. Bars represent the twitching zone diameter as measured by the subsurface stab assay. (B) WT PA103 or PAO1 overexpressing indicated DGCs (PA0169, PA1120, and PA3702) were grown on plates with or without 0.2% arabinose. Surface pilin was measured by ELISA. Bars indicate means ± SD of triplicate samples from three independent experiments.
DISCUSSION
Alterations in c-di-GMP levels are associated with changes in bacterial behavior. In the present study, we show that the same behavior, namely, assembly of T4P on the bacterial surface, occurs over a broad range of intracellular c-di-GMP concentrations. The proteins required for assembly under low- and high-c-di-GMP conditions are not the same, however, as FimX is required only under conditions of low c-di-GMP. T4P-based motility allows movement across solid biotic and abiotic surfaces. T4P are also implicated in early stages of surface attachment and colonization, e.g., microcolony formation, which precede biofilm maturation. Recent work has demonstrated that T4P likely play roles in later stages of biofilm growth and maturation, as P. aeruginosa organisms that do not assemble T4P cannot participate in the formation of cap structures characteristic of flow-cell biofilms. However, T4P motility per se is not required for formation of cap-and-stalk, mushroom-shaped structures (3, 6, 22, 23), suggesting that the primary function of T4P in this context is as adhesins or matrix components. Thus, P. aeruginosa shows a requirement for T4P assembly in growth conditions associated with the entire spectrum of motile and sessile behaviors and, presumably, over a broad range of intracellular c-di-GMP concentrations.
FimX is a PDE that binds c-di-GMP with high affinity (equilibrium dissociation constant [KD], ∼100 nM) and that localizes to the leading pole in twitching bacteria (R. Jain and B. I. Kazmierczak, unpublished data). As most models postulate that low c-di-GMP levels are associated with motility, we initially reasoned that our screen for extragenic suppressors of PA103ΔfimX would identify proteins with DGC activity whose inactivation lowered c-di-GMP levels, perhaps locally, and restored T4P assembly. Instead, we identified many independent Tn insertions that all increased intracellular c-di-GMP in bacteria and restored T4P assembly. These Tn insertions mapped to loci that had been previously described as causing an autoaggregative growth phenotype in liquid and a wrinkled-surface colony morphology (9). Bacteria expressing this phenotype also arise spontaneously, either in directed evolution experiments (30) or from CF patients (47), as SCV. Two of the suppressor loci have been well characterized with regard to their regulation of DGC activity and c-di-GMP production. Harwood and colleagues demonstrated that the wsp operon encodes a chemosensory system whose output is the phosphorylation and activation of the DGC PA3702 (WspR) (18). PA3703 (WspF) is a homolog of the CheB methylesterase, and its inactivation is predicted to lock the Wsp signaling complex into a conformation that constantly phosphorylates and activates WspR. As expected, ΔwspF bacteria have elevated intracellular c-di-GMP, and the phenotypes associated with this mutation are no longer observed in the absence of WspR (ΔwspF ΔwspR strain) (18). Likewise, PA1121 (YfiR) negatively regulates the DGC PA1120 (YfiN/TpbB) by an unknown mechanism, and ΔyfiR bacteria have elevated levels of c-di-GMP (27, 49). Our observation of increased c-di-GMP levels in strains carrying Tn insertions in PA1121 and PA3703 is therefore consistent with their established roles as negative regulators of DGC activity.
Although mutation of PA0171 results in a wrinkly spreader phenotype (9) and twitching defects (44), the relationship between this protein and the putative DGC PA0169 (SiaD) is not established. We found that overexpression of PA0169 in ΔfimX did not phenocopy the overexpression of other DGCs, as we observed no effect on surface pilin assembly, twitching motility, PO4 phage sensitivity, or intracellular c-di-GMP levels. A prior study by Kulasakara et al. reported no increase in biofilm formation or intracellular c-di-GMP levels associated with PA0169 overexpression in P. aeruginosa PA14 (24), raising the possibility that PA0169 is not a bona fide DGC or requires some additional signal to activate it. Since PA0171 expression in trans did revert the suppressor phenotypes associated with PA0171 disruption, it is likely that PA0171 affects c-di-GMP signaling. Whether it does so via PA0169, however, is not yet established.
The genome of P. aeruginosa encodes many DGCs. Although it is possible that the three DGC-containing operons identified in our screen were targeted because these proteins have specific interactions with components of the T4P assembly apparatus, we think this is unlikely. First, the overexpression of a DGC from a phylogenetically distant alphaproteobacteria, C. crescentus PleD, suppressed the PA103ΔfimX defect in pilus assembly. Second, PA103ΔfimX suppression required expression of an enzymatically active DGC, suggesting that the reaction product, and not the specific enzyme, is important for suppression. It is more likely that the presence of negative regulators of c-di-GMP production within these operons resulted in their targeting by our insertional mutagenesis strategy.
Bacteria harboring disruptions in PA0171, PA1121 (YfiR), and PA3703 (WspF) were reported to have defective twitching motility (9, 44). As this seemingly contradicts our observation that mutations in these loci promote pilus assembly and twitching motility, we investigated this further by examining these phenotypes in the PAO1 background. Overexpression of the DGCs PA1120 (YfiN/TpbB) and PA3702 (WspR) inhibited twitching motility of PAO1 and not PA103, but T4P capable of supporting PO4 phage infection were assembled in both strains. Bacterial motility can be inhibited by the expression of adhesins, such as exopolysaccharides, and their production is upregulated by c-di-GMP to a greater extent in PAO1 than in PA103 (see Fig. S8 in the supplemental material). This may contribute to the difference in motility that we observed between PAO1 and PA103.
Taken together, our results suggest that FimX enables pilus assembly under conditions of low intracellular c-di-GMP. This role is dispensable when c-di-GMP levels are high, as we demonstrated by experimentally manipulating intracellular c-di-GMP levels. Bacteria in biofilms, a naturally occurring state characterized by high c-di-GMP, also appear to bypass the requirement for FimX for T4P assembly. Both static and flow-cell biofilms are formed differently by ΔpilA and ΔfimX strains, with the latter achieving biomass and thickness similar to that seen for the WT.
If P. aeruginosa can assemble T4P in the absence of FimX, why is this protein retained? The answer is suggested by the conditions under which T4P are assembled at polar locations for motility, i.e., when bacteria transition from liquid to surface growth. In liquid-grown P. aeruginosa, intracellular c-di-GMP concentrations are below our detection limit (<50 nM) (18, 28) but increase to about 300 nM when PA103 is cultured on a surface (Fig. 3). We expect FimX to be largely c-di-GMP bound under these conditions (KD, ∼100 nM) and observe it localizing to the pole at which pilus assembly and retraction are occurring in actively twitching bacteria (Jain and Kazmierczak, unpublished). FimX mutations that abrogate polar protein targeting result in T4P assembly at random locations on the cell envelope, leading to inefficient motility, while mutations that disrupt FimX nucleotide binding and hydrolysis result in the absence of T4P assembly altogether (21). Thus, FimX appears to spatially and temporally “license” pilus assembly at the bacterial pole through a mechanism whose molecular details are still under investigation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to G. A. O'Toole and Urs Jenal for providing plasmids, Marc Chatenay-Lapointe and Amit Kunde for assistance with mapping Tn5 insertion sites, and Ming-Jie Wu for excellent technical assistance. We thank Morven Graham for assistance with electron microscopy (Yale Center for Cellular and Molecular Imaging).
This work was supported by a Brown-Coxe Fellowship (R.J.) and R01 AI054920 from the National Institute of Allergy and Infectious Diseases (B.I.K.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print 8 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alm RA, Bodero AJ, Free PD, Mattick JS. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An S, Wu J, Zhang LH. 2010. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol. 76: 8160–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barken KB, et al. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10: 2331–2343 [DOI] [PubMed] [Google Scholar]
- 4. Borlee BR, et al. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75: 827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradley DE. 1980. A function of Pseudomonas aeruginosa PAO1 polar pili: twitching motility. Can. J. Microbiol. 26: 146–154 [DOI] [PubMed] [Google Scholar]
- 6. Chiang P, Burrows LL. 2003. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185: 2374–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64: 391–397 [DOI] [PubMed] [Google Scholar]
- 8. Christen M, et al. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328: 1295–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184: 6481–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Kerchove AJ, Elimelech M. 2007. Impact of alginate conditioning film on deposition kinetics of motile and nonmotile Pseudomonas aeruginosa strains. Appl. Environ. Microbiol. 73: 5227–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51: 675–690 [DOI] [PubMed] [Google Scholar]
- 12. Gjermansen M, Ragas P, Tolker-Nielsen T. 2006. Proteins with GGDEF and EAL domains regulate Pseudomonas putida biofilm formation and dispersal. FEMS Microbiol. Lett. 265: 215–224 [DOI] [PubMed] [Google Scholar]
- 13. Guvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66: 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haussler S. 2004. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ. Microbiol. 6: 546–551 [DOI] [PubMed] [Google Scholar]
- 15. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7: 263–273 [DOI] [PubMed] [Google Scholar]
- 16. Heydorn A, et al. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146: 2409–2415 [DOI] [PubMed] [Google Scholar]
- 17. Heydorn A, et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146: 2395–2407 [DOI] [PubMed] [Google Scholar]
- 18. Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang B, Whitchurch CB, Mattick JS. 2003. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 185: 7068–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40: 385–407 [DOI] [PubMed] [Google Scholar]
- 21. Kazmierczak BI, Lebron MB, Murray TS. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60: 1026–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50: 61–68 [DOI] [PubMed] [Google Scholar]
- 23. Klausen M, et al. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48: 1511–1524 [DOI] [PubMed] [Google Scholar]
- 24. Kulasakara H, et al. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103: 2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee VT, et al. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65: 1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Y, Davin-Regli A, Bosi C, Charrel RN, Bollet C. 1996. Epidemiological investigation of Pseudomonas aeruginosa nosocomial bacteraemia isolates by PCR-based DNA fingerprinting analysis. J. Med. Microbiol. 45: 359–365 [DOI] [PubMed] [Google Scholar]
- 27. Malone JG, et al. 2010. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 6: e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malone JG, et al. 2007. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153: 980–994 [DOI] [PubMed] [Google Scholar]
- 29. Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56: 289–314 [DOI] [PubMed] [Google Scholar]
- 30. McDonald MJ, Gehrig SM, Meintjes PL, Zhang XX, Rainey PB. 2009. Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics 183: 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65: 876–895 [DOI] [PubMed] [Google Scholar]
- 32. Monds RD, Newell PD, Gross RH, O'Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63: 656–679 [DOI] [PubMed] [Google Scholar]
- 33. Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13: 3128–3138 [DOI] [PubMed] [Google Scholar]
- 34. Murray TS, Kazmierczak BI. 2008. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J. Bacteriol. 190: 2700–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Navarro MV, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17: 1104–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30: 295–304 [DOI] [PubMed] [Google Scholar]
- 37. O'Toole GA, et al. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310: 91–109 [DOI] [PubMed] [Google Scholar]
- 38. Paul R, et al. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 282: 29170–29177 [DOI] [PubMed] [Google Scholar]
- 39. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qi Y, et al. 2011. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J. Biol. Chem. 286: 2910–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Romling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57: 629–639 [DOI] [PubMed] [Google Scholar]
- 42. Ross P, et al. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325: 279–281 [DOI] [PubMed] [Google Scholar]
- 43. Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185: 2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shan Z, et al. 2004. Identification of two new genes involved in twitching motility in Pseudomonas aeruginosa. Microbiology 150: 2653–2661 [DOI] [PubMed] [Google Scholar]
- 45. Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72: 5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spangler C, Bohm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 81: 226–231 [DOI] [PubMed] [Google Scholar]
- 47. Starkey M, et al. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191: 3492–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964 [DOI] [PubMed] [Google Scholar]
- 49. Ueda A, Wood TK. 2009. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5: e1000483 doi:10.1371/journal.ppat.1000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 98: 6911–6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei Q, et al. 2011. Phenotypic and genome-wide analysis of an antibiotic-resistant small colony variant (SCV) of Pseudomonas aeruginosa. PLoS One 6: e29276 doi:10.1371/journal.pone.0029276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Whitchurch CB, et al. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 55: 1357–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.