Abstract
The genome of Methanosarcina acetivorans encodes three homologs, initially annotated as hypothetical fused corrinoid/methyl transfer proteins, which are highly elevated in CO-grown cells versus cells grown with alternate substrates. Based only on phenotypic analyses of deletion mutants, it was previously concluded that the homologs are strictly dimethylsulfide:coenzyme M (CoM) methyltransferases not involved in the metabolism of CO (E. Oelgeschlager and M. Rother, Mol. Microbiol. 72:1260 –1272, 2009). The homolog encoded by MA4383 (here designated CmtA) was reexamined via biochemical characterization of the protein overproduced in Escherichia coli. Purified CmtA reconstituted with methylcob(III)alamin contained a molar ratio of cobalt to protein of 1.0 ± 0.2. The UV-visible spectrum was typical of methylated corrinoid-containing proteins, with absorbance maxima at 370 and 420 nm and a band of broad absorbance between 450 and 600 nm with maxima at 525, 490, and 550 nm. CmtA reconstituted with aquocobalamin showed methyl-tetrahydromethanopterin:CoM (CH3-THMPT:HS-CoM) methyltransferase activity (0.31 μmol/min/mg) with apparent Km values of 135 μM for CH3-THMPT and 277 μM for HS-CoM. The ratio of CH3-THMPT:HS-CoM methyltransferase activity in the soluble versus membrane cellular fractions was 15-fold greater in CO-grown versus methanol-grown cells. A mutant strain deleted for the CmtA gene showed lower growth rates and final yields when cultured with growth-limiting partial pressures of CO, demonstrating a role for CmtA during growth with this substrate. The results establish that CmtA is a soluble CH3-THSPT:HS-CoM methyltransferase postulated to supplement the membrane-bound CH3-THMPT:HS-CoM methyltransferase during CO-dependent growth of M. acetivorans. Thus, we propose that the name of the enzyme encoded by MA4384 be CmtA (for cytoplasmic methyltransferase).
INTRODUCTION
Most methane-producing species (methanogens) presently characterized obtain energy for growth by the CO2 reduction pathway (Table 1, reactions 1 to 3), in which CO2 is reduced stepwise with pairs of electrons derived from the oxidation of H2 (7). Although the oxidation of CO can serve as a source of electrons in the CO2 reduction pathway (Table 1, reactions 4 to 6), only three species have been investigated. Growth of Methanothermobacter thermautotrophicus (basonym, Methanobacterium thermoautotrophicum strain ΔH) with CO is extremely poor, with a rate only 1% of that of H2 (6). Although it was previously shown that Methanosarcina barkeri grows more rapidly (24), it was concluded that this species is not well evolved for growth with CO based on a poor doubling time (65 h) compared to growth with acetate (48 h) or methanol (12 h). The pathway in M. barkeri (Table 1, reactions 7 to 10) begins with the oxidation of CO to H2 followed by reduction of CO2 to methane with electrons derived from the oxidation of H2 (24). Conversely, the doubling time (20 h) for CO-dependent growth of Methanosarcina acetivorans is triple that of M. barkeri (28). M. acetivorans is incapable of metabolizing H2 (12, 35), and H2 is not detected during growth with CO (28), suggesting novel features of the CO2 reduction pathway in converting CO to CH4. Indeed, quantitative global proteomic profiling coupled with molecular and biochemical analyses of M. acetivorans grown with CO versus acetate or methanol revealed an H2-independent CO2 reduction pathway in which electron transfer reactions deviate substantially from that of M. barkeri and other H2-oxidizing, CO2-reducing species (18). Furthermore, M. acetivorans also produces acetate, formate, and dimethylsulfide (DMS) during CO-dependent growth (18, 23, 28), the only reported products other than CH4 for any methanogenic species.
Table 1.
Reactions and free energy yields of the carbon dioxide reduction pathway
| No. | Reaction | (kJ/mol) |
|---|---|---|
| 1 | 4H2 → 8e− + 8H+ | |
| 2 | CO2 + 8e− + 8H+ → CH4 + 2H2O | |
| 3 (Sum of no. 1 and 2) | CO2 + 4H2 → CH4 + 2H2O | −131 |
| 4 | 4CO + 4H2O → 8e− + 8H+ + 4CO2 | |
| 5 | CO2 + 8e− + 8H+ → CH4 + 2H2O | |
| 6 (Sum of no. 4 and 5) | 4CO + 2H2O → CH4 + 3CO2 | −210.4 |
| 7 | 4CO + 4H2O → 4H2 + 4CO2 | |
| 8 | 4H2 → 8e− + 8H+ | |
| 9 | CO2 + 8e− + 8H+ → CH4 + 2H2O | |
| 10 (Sum of no. 7, 8, and 9) | 4CO + 2H2O → CH4 + 3CO2 | −210.4 |
| 11 | 2(CH3)2S + 3H2O → 3CH4 + HCO3− + 2H2S + H+ | −49.4 |
An important step common to both M. barkeri and M. acetivorans CO2 reduction pathways is transfer of the methyl group from tetrahydrosarcinapterin (THSPT) to coenzyme M (HS-CoM), which is catalyzed by the membrane-bound N5-CH3-THSPT:HS-CoM methyltransferase complex (MtrA-H). The MtrA-H complex couples the exergonic reaction to generation of a sodium gradient (with the concentration high outside) (10) with the potential to drive ATP synthesis via the Na+/H+ ATP synthase (32). The CH3-S-CoM so produced is reductively demethylated to CH4 in the final step of the pathway. Quantitative proteomic profiling supported by quantitative reverse transcription-PCR (RT-PCR) revealed levels of MtrA-H subunits with a 9-fold mean lower abundance in CO- versus acetate-grown M. acetivorans, a result indicating downregulation of the complex in response to growth with CO (18). The analyses also revealed 50-fold elevated levels of proteins encoded by loci MA0859, MA4384, and MA4558 in cells grown with CO versus methanol or acetate (18). These proteins were first named fused corrinoid/methyltransferases (FCMT) based solely on sequence identity to methyltransferases, which transfer the methyl groups of methylamines and methanol to HS-CoM in pathways converting these growth substrates to CH4 (8). The amino-terminal domains of the FCMT homologs each contain a corrinoid-binding motif (GDVHDIGKNLV) with the conserved active-site histidine (underlined), while the C-terminal methyltransferase domains have a conserved HXCXnC motif found in several methyltransferases that binds zinc, which is required for deprotonation of HS-CoM in the catalytic mechanism (9, 17, 30). Thus, it is proposed that the FCMT homologs transfer the methyl groups of unknown substrates to HS-CoM. The reduction of MtrA-H levels, concomitant with elevated abundance of FCMT homologs in CO- versus methanol- or acetate-grown M. acetivorans, led to the proposal that the homologs function as cytoplasmic CH3-THSPT:HS-CoM methyltransferases which supplement activity of the membrane-bound sodium-pumping MtrA-H (18). However, M. acetivorans mutants, for which combinations of MA0859, MA4384, and MA4558 were deleted or disrupted, fail to produce DMS or utilize it for methanogenesis or growth (25). Furthermore, the growth phenotypes of the mutants cultured with growth-saturating CO levels are not significantly different from wild-type M. acetivorans (25). Based on these data alone, it was concluded that MA0859, MA4384, and MA4558 function exclusively in the pathway of methanogenesis from DMS, encoding DMS:HS-CoM methyltransferases that were designated MtsD, MtsF, and MtsH (methyltransferases specific for methylsulfides) (25). Remarkably, there are no reports of biochemical characterizations of any FCMT homolog validating this conclusion or investigation of the previously hypothesized CH3-THSPT:HS-CoM methyltransferase activity.
Here, we present a reexamination of the CO-dependent growth characteristics for an MA4384 deletion mutant strain of M. acetivorans and an initial biochemical investigation of the heterologously produced FCMT homolog (CmtA) encoded by MA4384. The results support the previously proposed role of cytoplasmic CH3-THSPT:HS-CoM methyltransferase for CmtA and FCMT homologs which supplement the membrane-bound CH3-THMPT:HS-CoM methyltransferase during CO-limited growth of M. acetivorans. Thus, we propose changing the name of the enzyme encoded by MA4384 to CmtA (cytoplasmic methyltransferase).
MATERIALS AND METHODS
Strains, plasmids, and chemicals.
M. acetivorans strain C2A (DSM 804) was from laboratory stocks, strain WWM1 (Δhpt) was a gift from W. W. Metcalf (University of Illinois, Urbana-Champaign), and the MA4384 deletion mutant strain DmtsF(26) (here referred to as ΔcmtA) was a gift from M. Rother (Goethe-Universität, Frankfurt, Germany). Escherichia coli strain Rosetta DE3 (pLacI) and the pET22b expression vector were from Novagen (Madison, WI). Tetrahydromethanopterin (THMPT) was a gift from R. K. Thauer (Max Planck Institute for Terrestrial Microbiology, Marburg, Germany). Preparation of CH3-THMPT from THMPT was performed as published previously (2). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and were of analytical or molecular biology grade.
Preparation of cell extracts and isolation of soluble and membrane fractions.
M. acetivorans strains were grown in high-salt medium at 37°C with 125 mM methanol or 1.0 atm of CO as previously described (18, 19). The adaptation of the wild-type and mutant strains of M. acetivorans to CO was performed as described previously (25). All steps requiring transfer of suspensions and solutions were performed under strictly anaerobic conditions in an inert-atmosphere glove bag (Coy Laboratory Products, Ann Arbor, MI), except where indicated, and at 4°C. All buffers were degassed with N2 to remove oxygen. Protein was determined by the method of Bradford (5).
Cells were harvested by centrifugation, and the cell paste was stored at −80°C. Thawed cells were resuspended in 50 mM morpholinepropanesulfonic acid (MOPS)-KOH buffer (pH 7) containing 1.5 mM phenylmethylsulfonyl fluoride and DNase I (20 μg/ml) at 2 g (wet weight) per 7 ml. Cells were disrupted by two passes through a French pressure cell at 6.9 × 103 kPa. Cell extract was obtained by centrifugation of the lysate at 1,200 × g for 12 min to pellet cell debris and unbroken cells. The membrane and soluble cellular fractions were separated by centrifugation in a discontinuous sucrose gradient as previously described (39, 41), with the following exceptions. All manipulations were performed in dim light. Cell extract (1 ml) was layered onto a gradient comprised of 70, 30, and 20% sucrose (1:2:1 ml) in 50 mM MOPS-KOH buffer (pH 7.5) and centrifuged at 150,000 × g for 2 h in a Beckman SW55 Ti swinging-bucket rotor. The membrane fraction was harvested from a tight band located at the interface of the 70 and 30% sucrose layers. The soluble protein fraction was harvested from above the 20% sucrose layer. All fractions were stored at 4°C in sealed brown bottles under a N2 atmosphere and assayed immediately.
Heterologous overproduction, purification, and reconstitution of CmtA.
The MA4384 gene sequence, obtained from the Comprehensive Microbial Resource website (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi), was amplified by PCR from genomic DNA of M. acetivorans strain C2A (DSM 804) (35) with an NdeI site at the 5′ end and an XhoI site at the 3′ end introduced into the primers (sequences of the primers are available on request). The PCR product was ligated into the pET22b expression vector to construct the recombinant plasmid pET-MA4384, with which E. coli Rosetta (DE3) cells were transformed. A stop codon (TAG) was introduced immediately after the MA4384 sequence to block expression of the His tag present in the pET22b vector to prevent possible interference with binding of corrinoid cofactors and zinc to the protein. The transformed cells were cultured at 37°C in Luria-Bertani broth (pH 7.0) containing 100 μg ampicillin/ml and 35 μg chloramphenicol/ml (final concentrations). Expression was induced by addition of isopropyl thio-β-d-galactopyranoside (1 mM final concentration) at an optical density at 600 nm (OD600) of 0.6, and the culture was incubated at 16°C for 14 h. Cells were harvested by centrifugation. Approximately 10 g (wet weight) of cells was suspended in 30 ml of 50 mM MOPS-KOH buffer (pH 7.0) containing 1.5 mM phenylmethylsulfonyl fluoride and 20 μg DNase I/ml (final concentration). Cells were disrupted by two passages through a French pressure cell at 6.9 × 103 kPa. The lysate was centrifuged at 4°C for 12 min at 1,200 × g. The supernatant solution containing cell extract was centrifuged for 30 min at 75,000 × g to pellet inclusion bodies containing CmtA and cell debris. The pellet was washed three times with 150 mM NaCl to remove remaining cell debris from inclusion bodies. The inclusion bodies were solubilized by resuspension in 5 ml of 50 mM anaerobic MOPS-KOH buffer (pH 7.0) containing 7 M urea and 10 mM dithiothreitol (DTT). From this stage onwards, all steps were performed in dim light under strictly anaerobic conditions in an inert-atmosphere glove bag.
The concentration of urea in the solution (6.5 ml) containing solubilized CmtA was reduced stepwise as follows. The solution was mixed with 153.5 ml of 50 mM MOPS-KOH buffer (pH 7.0) containing 10 mM DTT. This solution was concentrated to 60 ml using a Vivacell (Sartorius Stedim Biotech GmbH, Gottingen, Germany) pressure-operated system fitted with a 50,000-molecular-weight cutoff membrane. Buffer (102.5 ml) was added to the concentrated solution, followed again by concentration to 75 ml. Finally, buffer (75 ml) was added to this solution, followed by concentration to 1.5 ml. SDS-PAGE revealed CmtA was greater than 95% pure.
The purified protein was reconstituted with either methylcob(III)alamin or aquocobalamin by following published procedures for the MtaC corrinoid-containing protein from Methanosarcina barkeri (29) with the following modifications. The cobalamins (300 μM final concentration) and ZnCl2 (75 μM final concentration) were added to the solubilized protein and incubated at 21°C for 2 h with gentle agitation in the dark. Unbound cofactors and zinc were removed by passage through a PD-10 gel filtration column (GE Healthcare, Buckinghamshire, United Kingdom). Protein in the pass-through fraction was concentrated by filtration to 1.0 ml. The cobalt content was determined by inductively coupled plasma emission spectroscopy at the Center for Isotope Studies, University of Georgia, Athens, GA.
Methyltransferase assay.
Methylation of HS-CoM was assayed as described previously for the membrane-bound CH3-THMPT:HS-CoM methyltransferase of Methanosarcina mazei (basonym, Methanosarcina strain Gö1) (1, 3) by measuring the disappearance of HS-CoM based on thiol analysis using Ellman's reagent and an extinction coefficient for the 2-nitro-5-thiobenzoate dianion of 13,600 M−1 cm−1 (13). When activity in extracts was assayed, a final concentration of 5 mM potassium–2-bromoethanesulfonate was included to inhibit CH3-SCoM reductase activity. When DMS was the methyl donor, the methylthiol produced was removed prior to the detection of HS-CoM as follows. Protein was precipitated by the addition of cold acetone (−20°C) to 10 μl of assay mix and centrifuged. Methylthiol was removed from the supernatant solution by spinning in a Speed Vac for 20 min. Controls with a range of methylthiol (0.5 to 20 mM) added to assay mixtures confirmed complete removal.
The methylcob(III)alamin:HS-CoM methyltransferase gel activity stain was performed as previously described (36). Development of activity was dependent on both methylcob(III)alamin and HS-CoM.
RESULTS
Role in CO-dependent growth.
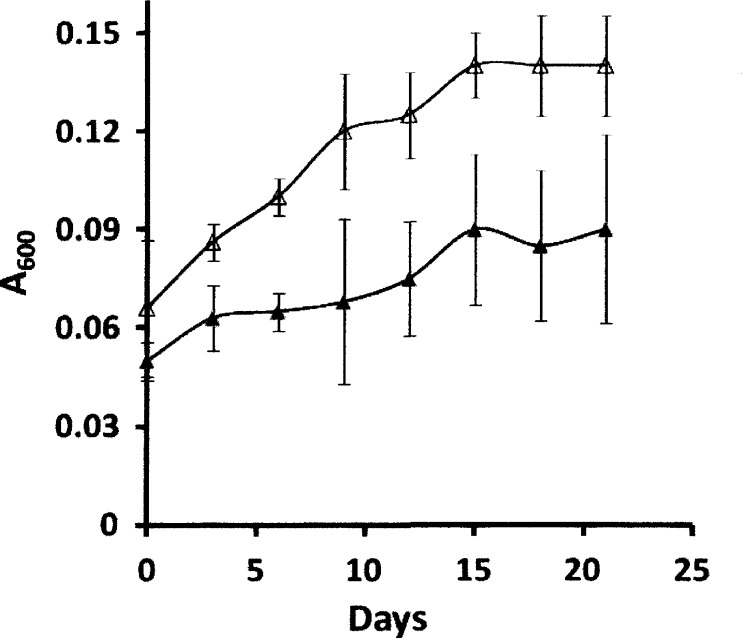
All previous investigations (4, 25) of the FCMT homologs were conducted with wild-type and mutant strains of M. acetivorans grown with a CO partial pressure of 1.5 × 105 Pa (1.5 atm), which is not growth limiting, and it is considerably greater than concentrations likely to occur in the environment. The FCMT homolog encoded by MA4384 (CmtA) is reported to be upregulated to the highest level among the three homologs (18) and was the primary focus of previous investigations (25). Thus, growth of the wild type versus the MA4384 deletion mutant ΔcmtA [DmtsF(26)] strain was undertaken with a growth-limiting partial pressure of 1.01 × 104 Pa (0.1 atm) CO (Fig. 1). Growth of the ΔcmtA strain was significantly less in terms of both doubling times (30 versus 13 days) and final yield (A600, 0.09 versus 0.14) than that of the wild type. No growth was observed in the absence of CO for either strain. The results indicate that CmtA is required for optimal growth when cultured with a growth-limiting partial pressure of CO.
Fig 1.
Growth of Methanosarcina acetivorans with a limiting partial pressure of CO. Growth was with 1.01 × 104 Pa (0.1 atm) of CO in the headspace. The inoculum was CO-grown cells. Symbols: Δ, wild-type strain WWM1; ▲, mutant strain deleted for MA4384 [DmtsF(26)].
Distribution of CH3-THMPT:HS-CoM methyltransferase activity in the soluble and membrane fractions of CO- versus methanol-grown M. acetivorans.
Previous quantitative proteomic analysis of M. acetivorans revealed a decrease in the level of several subunits belonging to the membrane-bound CH3-THSPT:HS-CoM methyltransferase (MtrA-H) complex and a substantially larger 50-fold increase in the FCMT homolog encoded by MA0859 in CO- versus methanol-grown cells (18). With translational fusions, it was also shown that CO-grown M. acetivorans contains at least 320-fold higher levels of CmtA than methanol-grown cells (4). If CmtA (MA4384) and the homolog encoded by MA0859 are soluble CH3-THSPT:HS-CoM methyltransferases as previously proposed (18), the ratio of activity in the soluble versus membrane fraction should be greater in CO-grown cells than in methanol-grown cells. The membrane and soluble fractions were cleanly isolated by discontinuous sucrose concentration gradients and assayed with THMPT, a structural and functional analog of THSPT (20, 40), from the methanogen Methanothermobacter marburgensis. The activity ratio in methanol-grown M. acetivorans was 0.16 (Table 2), similar to the ratio of 0.13 reported for methanol-grown M. mazei (3), whereas the ratio in CO-grown M. acetivorans was nearly 15-fold greater (Table 2). Furthermore, no transmembrane-spanning regions were detected in the sequences deduced from MA0859, MA4384, or MA4558 evaluated with the TMPRED program (15). These results are consistent with the previously proposed role of a soluble CH3-THSPT:HS-CoM methyltransferase for the protein encoded by MA0859 and homologs MA4384 and MA4558 (18).
Table 2.
Methyl-THMPT:HSCoM methyltransferase activity in the soluble and membrane fractions of CO- or methanol-grown Methanosarcina acetivorans
| Fraction | Total protein (mg) |
Total activitya (U) |
Sp act (U/mg) |
Total activity (%) |
||||
|---|---|---|---|---|---|---|---|---|
| CH3OH | CO | CH3OH | CO | CH3OH | CO | CH3OH | CO | |
| Extract | 50.0 | 50.0 | 6.15 ± 0.45 | 10.9 ± 1.1 | 0.12 ± 0.02 | 0.22 ± 0.03 | 100 | 100 |
| Soluble | 37.5 | 41.4 | 0.79 ± 0.15 | 7.5 ± 0.8 | 0.02 ± 0.005 | 0.18 ± 0.02 | 12.7 | 69.0 |
| Membrane | 4.5 | 3.5 | 4.77 ± 0.43 | 3.2 ± 0.2 | 1.06 ± 0.06 | 0.92 ± 0.08 | 77.5 | 29.0 |
The 100-μl assay was performed at 37°C in 50 mM MOPS-KOH (pH 7.0) containing 100 μg CmtA reconstituted with aquocobalamin, 1 mM Ti(III)-citrate, 5 mM methyl viologen, 10 mM MgCl2, 10 mM ATP, 5 mM methyl donor, 5 mM HS-CoM, and 5 mM potassium 2-bromoethanesulfonate. Unit (U) indicates μmol CH3-S-CoM/min.
Methyl-THMPT:HS-CoM methyltransferase activity of CmtA.
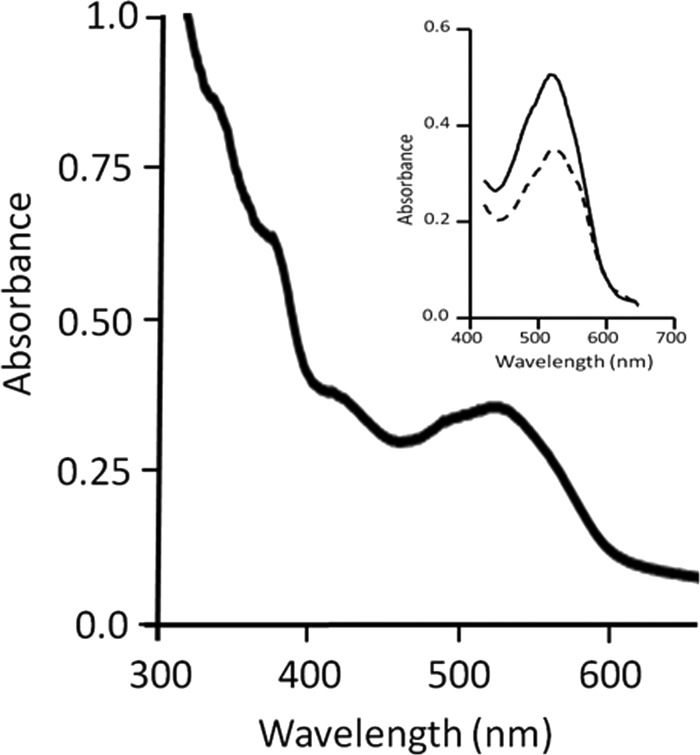
The biochemical characterization of any FCMT homolog has yet to be reported. Thus, the product of MA4384 (CmtA) was overproduced in E. coli and purified to determine the enzymatic activities. The protein was solubilized from inclusion bodies and reconstituted with methylcob(III)alamin. SDS-PAGE of the purified protein showed a single band corresponding to the predicted monomeric molecular mass of 69 kDa (see Fig. S1 in the supplemental material). Preparations were pink in color, which is typical of corrinoid-containing proteins. The UV-visible spectrum (Fig. 2) showed absorbance maxima at 370 and 420 nm with additional broad absorbance between 450 and 600 nm, containing maxima at 525 nm and shoulders at 490 and 550 nm, which is characteristic of methylated corrinoid-containing proteins of methane-producing species (14, 22, 36). Metal analysis of 6 preparations revealed 1.0 ± 0.2 mol of cobalt per mol of protein, indicating the presence of approximately one corrinoid cofactor bound to each CmtA molecule.
Fig 2.
UV-visible spectrum of CmtA. The protein (4.5 mg/ml) was reconstituted with methylcob(III)alamin. The inset shows results before (solid line) and after (dashed line) the addition of 5 mM HS-CoM (final concentration).
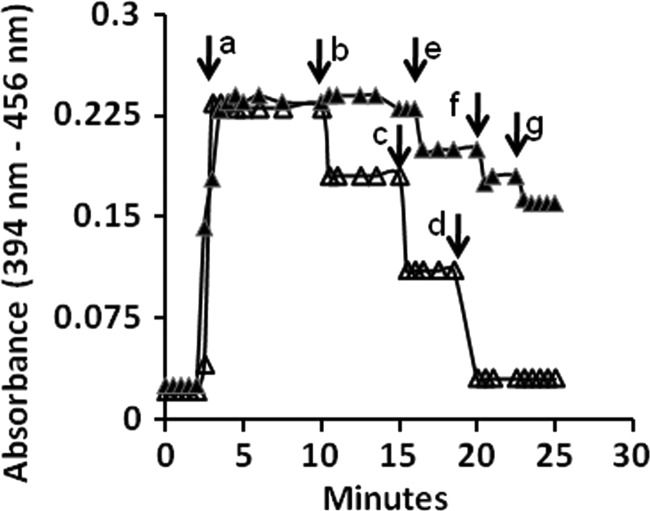
Nondenaturing gels stained positive for methylcob(III)alamin:HS-CoM methyltransferase activity with the nonphysiological substrate methylcob(III)alamin (see Fig. S1 in the supplemental material). Furthermore, incubation of methylcob(III)alamin-reconstituted CmtA with HS-CoM resulted in decreased absorbance at 525 nm (Fig. 2, inset), indicating loss of the corrinoid-containing methyl group to HS-CoM. These results indicated the potential for CH3-THSPT:HS-CoM or DMS:HS-CoM methyltransferase activity. Methyltransferase activity was further investigated by first addressing if CmtA reconstituted with aquocobalamin was competent to accept methyl groups from CH3-THMPT or DMS (Fig. 3). The reduced Co(I) cobamide form of the cofactor was first generated by reaction with 1 mM Ti(III)-citrate as revealed by the rapid development of absorbance at 394 nm (Fig. 3). Each successive addition of methyl group donor to the reduced enzyme decreased the absorbance at 394 nm relative to that at 456 nm, indicating the generation of base-off Co(III)-methyl cobamide that was observed previously for corrinoid-containing methyl transfer enzymes from species of the genus Methanosarcina (11, 16). These results suggest a mechanism in which the methyl group of CH3-THMPT is transferred to the corrinoid cofactor and subsequently transferred to HS-CoM. The transfer of methyl groups from the addition of 1 nmol CH3-THMPT was highly efficient, whereas transfer from the addition of 2 nmol of DMS was substantially less efficient.
Fig 3.
Reduction of cobamide bound to CmtA and methylation with CH3-THMPT or DMS. The reaction mixture (80-μl final volume) contained 10 μM (final concentration) CmtA reconstituted with aquocobalamin in 50 mM MOPS-KOH buffer (pH 7.0). Reactions were performed at 21°C. At the time indicated by the arrow labeled a, 2.5 μl of 32 mM Ti(III)-citrate (1.0 mM final concentration) was added to reduce aquocobalamin. Additions of methyl donors were made at time points indicated by the arrows: b and e, 1.0 nmol each of CH3-THMPT and DMS, respectively; c and d, 2.0 nmol of CH3-THMPT; f and g, 2.0 nmol of DMS. Absorbencies were corrected for dilution when adding reagents. Symbols: Δ, change in absorbance on addition of CH3-THMPT; ▲, change in absorbance on addition of DMS.
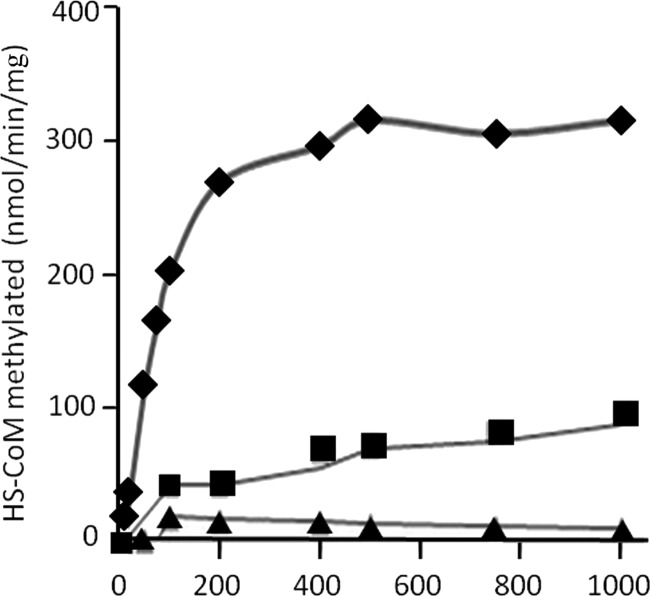
Figure 4 shows the CH3-THMPT:HS-CoM methyltransferase activity of CmtA reconstituted with aquocobalamin. The specific activity with saturating concentrations of substrates was 0.31 μmol/min/mg with CH3-THMPT and 0.18 μmol/min/mg with CH3-THF under the conditions tested. The ability of CH3-THF to substitute for CH3-THMPT is consistent with that reported for the membrane-bound CH3-THMPT:HS-CoM methyltransferase (MtrA-H) of M. mazei (2). Low levels of DMS:HS-CoM methyltransferase activity were observed early in the time course, although they were not sustained (Fig. 4). The enzyme displayed normal Michaelis-Menten kinetics, revealing apparent Km values of 135 μM for CH3-THMPT and 277 μM for HS-CoM. These results support a physiological role for CmtA in catalyzing a soluble CH3-THSPT:HS-CoM methyltransferase activity.
Fig 4.
Time course of methyl group transfer from CH3-THMPT, CH3-THF, or DMS to HS-CoM catalyzed by CmtA. The reaction was performed at 37°C in 50 mM MOPS-KOH buffer (pH 7.0) containing 100 μg of aquocobalamin-reconstituted CmtA, 1.0 mM Ti(III)-citrate, 5 mM methyl viologen, 10 mM MgCl2, 10 mM ATP, 5 mM methyl donor, and 5 mM HS-CoM in a final volume of 100 μl. Methylation of HS-CoM was determined by measuring the disappearance of HS-CoM as described in Materials and Methods. When DMS was the methyl donor, samples were processed for removal of methylthiol before determining HS-CoM as described in Materials and Methods. Symbols: ♦, CH3-THMPT; ■, CH3-THF; ▲, DMS.
Sequence analysis.
A BLASTP (http://blast.ncbi.nlm.nih.gov) search of the nonredundant databases was performed with the query sequence deduced from MA4384. The first five retrieved sequences were with full-length coverage (99%) and 54 to 57% identity to sequences deduced from FCMT homologs MA0859 and MA4558 and proteins annotated as methyltransferase cognate corrinoid proteins (see Fig. S2 in the supplemental material) from the methanogens Methanohalobium evestigatum (basonym, Methanohalobium evestigatus), Methanohalophilus mahii, and Methanosalsum zhilinae (basonym, Methanohalophilus zhilinae). The remaining 45 retrieved sequences were from diverse corrinoid-containing proteins distributed among diverse species belonging to the domains Bacteria and Archaea, having less than 35% identity and 30% coverage confined to the first 220 N-terminal residues of CmtA. The results confirm the previously reported sequence identity of the N-terminal domain to corrinoid-containing proteins (8). Since many enzymes with diverse functions contain corrinoid cofactors, the C-terminal domain of CmtA most likely determines the specific enzyme activity. Thus, the only candidate homologs of CmtA are those retrieved sequences annotated as methyltransferase corrinoid cognate proteins with identity to both the N- and C-terminal domains of CmtA. The putative homologs are from species which convert the methyl groups of methylamines or methanol to methane, and all except M. evestigatum are reported to metabolize DMS (21, 26, 27, 34, 42). However, none of these species were examined for the ability to grow with CO except M. acetivorans (18, 28), precluding assignment of the putative methyltransferase corrinoid cognate proteins to a specific pathway of methanogenesis. Nonetheless, the BLASTP search results indicate that FCMT homologs are limited to only a few methanogenic species, suggesting a novel physiological function consistent with the results documenting CH3-THSPT:HS-CoM methyltransferase activity of CmtA.
It was previously reported that M. acetivorans FCMT homologs function exclusively as DMS:HS-CoM methyltransferases based only on growth phenotypes of mutant strains in which genes encoding FCMT homologs were deleted or disrupted (25). Thus, the deduced sequence of MA4384 was aligned with the two-component DMS:HS-CoM methyltransferase of M. barkeri (see Fig. S3 in the supplemental material) comprised of two nonidentical subunits, MtsA and MtsB, for which the enzyme mechanism is well documented (37, 38). The MtsA subunit binds DMS, transferring a methyl group to the corrinoid-containing MtsB subunit, which donates the methyl group back to the MtsA subunit, where HS-CoM is bound and methylated. MtsB aligned most favorably with 31% identity to N-terminal residues 10 to 191 from the deduced sequence of MA4384, a result consistent with the N-terminal domain binding the corrinoid cofactor. MtsA aligned most favorably with C-terminal residues 270 to 620 from the deduced sequence of MA4384, although with only 19% identity. Nonetheless, the deduced sequences of MA4384 and homologs of MA4384 (see Fig. S2) contain a motif in MtsA (HXCXnC) which binds zinc, which is important for deprotonation of HS-CoM and further supports a role for the C-terminal domain of CmtA in methylation of HS-CoM. Although the low sequence identity with MtsA does not necessarily rule out DMS:HS-CoM methyltransferase activity of CmtA, the results suggest a novel function consistent with the results documenting CH3-THSPT:HS-CoM methyltransferase activity.
DISCUSSION
The results presented here establish CH3-THMPT:HS-CoM methyltransferase activity of CmtA, the FCMT homolog encoded by MA4384 in M. acetivorans. The previously reported increased levels of FCMT homologs (18) and expression of the encoding genes correlate positively with increased CH3-THMPT:HS-CoM methyltransferase activity in the soluble protein fraction of CO- versus methanol-grown M. acetivorans reported here, a result that is consistent with CH3-THMPT:HS-CoM methyltransferase activity of CmtA. The identity of CmtA to homologs encoded by MA0859 (51.7%) and MA4558 (53.7%) suggest that they also catalyze the same reaction, although biochemical analyses are necessary to draw conclusions. The results are in contrast to the previous conclusion that FCMT homologs only function in the metabolism of methylsulfides, which leads to the naming of the FCMT homologs MtsD, MtsF, and MtsH (for methyltransferases specific for methylsulfides) encoded by MA0859, MA4384, and MA4558 (25). Based on the results presented here, we propose renaming MtsF as CmtA (cytoplasmic methyltransferase).
Furthermore, the results support a role for CmtA in CO-dependent growth of M. acetivorans. Compared to the parental strain, the growth rate and final yield were significantly less for the mutant strain deleted for the gene (MA4384) encoding CmtA when cultured with a growth-limiting partial pressure of CO. This result is in contrast to growth of the same mutant in a pressurized atmosphere of 100% CO, which was previously reported to have no growth defects relative to the parental strain (25). Clearly, CO-limited growth parameters best approximate those expected in the environment from which M. acetivorans was isolated (35). The methyltransferase activity of the purified CmtA (0.31 μmol/min/mg), which was assayed with CH3-THMPT, was found to be substantially lower than that reported (7.0 μmol/min/mg) for the membrane-bound CH3-THSPT:HS-CoM methyltransferase (MtrA-H) purified from acetate-grown M. mazei and assayed with CH3-THMPT (20). Less than optimal activity of CmtA could result from substitution of the commercially available cobalamins (dimethylbenzimidazolylcobamide) for reconstitution of CmtA in place of 5-hydroxybenzimidazolylcobamide, which is present in species of the genus Methanosarcina (31). Regardless, a relatively low level of activity for CmtA is consistent with the ≥50-fold increase in levels of CmtA and FCMT homologs reported for CO- versus methanol- or acetate-grown cells to achieve the greater ratio of soluble versus membrane-bound CH3-THMPT:HS-CoM methyltransferase activities in CO-grown cells compared to methanol-grown cells reported here. Nevertheless, the results unequivocally establish the CH3-THMPT:HS-CoM methyltransferase activity of CmtA.
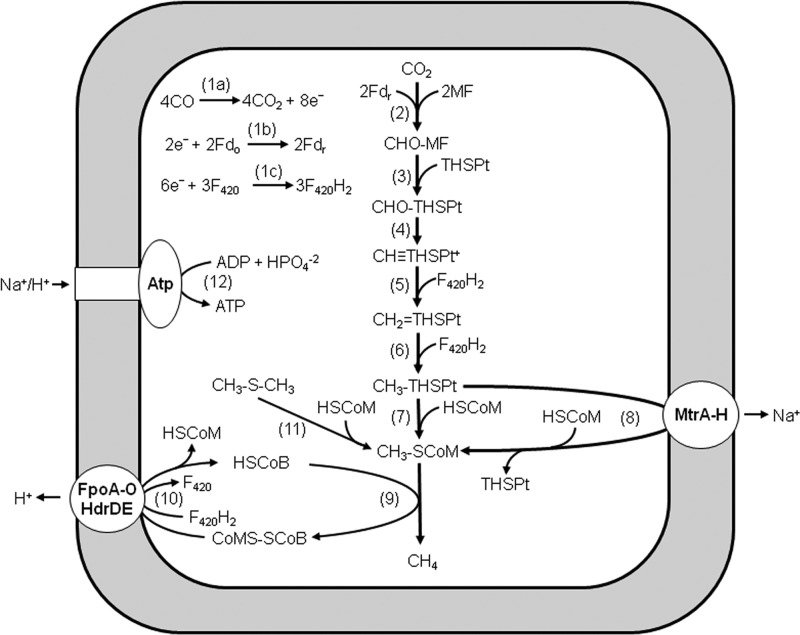
Overall, the results support the previously postulated role for CmtA and homologs in supplementing the sodium-pumping CH3-THSPT:HS-CoM methyltransferase complex MtrA-H during CO-dependent growth of M. acetivorans (18). This role for CmtA is indicated by defective growth of the mutant deleted for the encoding gene when cultured with a growth-limiting partial pressure of CO. It is conceivable that with low partial pressures of CO, the lower available free energy limits reactions coupled to the energy-requiring generation of ion gradients that drive ATP synthesis. Only two reactions in the CO-dependent CO2-reducing pathway of M. acetivorans generate ion gradients (Fig. 5) (18), the sodium-pumping CH3-THSPT:HS-CoM methyltransferase (MtrA-H) (Fig. 5, reaction 8) and the proton-pumping FpoA-O complex, which donates electrons to the heterodisulfide reductase (HdrDE), catalyzing the final reductive step (Fig. 5, reaction 10). The soluble CmtA and homologs potentially provide a mechanism for bypassing MtrA-H (Fig. 5, reaction 7), allowing growth at otherwise prohibitively low CO concentrations and equipping the cell to accommodate fluctuations in the CO concentrations that are encountered in the environment, thereby maximizing the thermodynamic efficiency for optimal ATP synthesis and growth by partitioning methyl transfer through CmtA and MtrA-H. Under laboratory conditions where cells are routinely cultured with greater than 1.0 atm of CO, CmtA and homologs would be dispensable, explaining why no growth defect was previously reported for M. acetivorans mutants cultured with 1.5 atm CO (25).
Fig 5.
Pathway proposed for the conversion of CO to CH4 by Methanosarcina acetivorans. Fdo, oxidized ferredoxin; Fdr, reduced ferredoxin; F420, coenzyme F420; MF, methanofuran; THSPt, tetrahydrosarcinapterin; HSCoM, coenzyme M; HSCoB, coenzyme B; FpoA-O, F420H2 dehydrogenase complex; HdrDE, heterodisulfide reductase; MtrA-H, CH3-THSPt:HS-CoM methyltransferase; and Atp, Na+/H+-dependent ATP synthase.
It is interesting that genes encoding M. acetivorans FCMT homologs are not present in the genomes of phylogenetically related M. barkeri or M. mazei. In contrast to M. acetivorans (12, 35), M. barkeri and M. mazei utilize H2 as a reductant in the CO2 reduction pathway, which is derived from the metabolism of fermentative and syntrophic species and is significantly more abundant than CO in the environment. Thus, M. barkeri or M. mazei is not strictly dependent on low concentrations of CO for growth, although conversion of CO to H2 could supplement abundant H2 already available from fermentative and syntrophic species in the environment. Indeed, M. barkeri grows with 0.5 atm of CO by first converting it to H2 (24). Growth of M. barkeri or M. mazei at lower partial pressures of CO as a sole energy source, as demonstrated here for M. acetivorans, has not been reported.
Mutants of M. acetivorans with deleted or disrupted genes encoding FCMT homologs, including CmtA, are defective in growth and methanogenesis with DMS, leading to the previous conclusion that FCMT homologs function exclusively in the pathway for conversion of DMS to CH4 (Table 1, reaction 11) (25). In the proposed pathway, FCMT homologs catalyze transfer of the methyl groups of DMS to HS-CoM (Fig. 5, reaction 11). Three molecules of CH3-S-CoM are reductively demethylated with three electron pairs, producing CH4 (Fig. 5, reaction 9). The methyl group from one molecule of CH3-S-CoM is transferred to THSPT for oxidation to CO2 by reversal of reactions 2 to 6, which supply the three electron pairs for reductive demethylation of CH3-S-CoM. Transfer of methyl groups from CH3-S-CoM to THSPT is accomplished by the reversal of reaction 8 (Fig. 5) (ΔG°′ = 30 kJ/mol), which is catalyzed by MtrA-H and driven by a sodium gradient. The results presented here neither support nor preclude the proposed role of FCMT homologs in the pathway. Although it is substantially less robust than CH3-THMPT, DMS served as a methyl donor to CmtA, predicting DMS:HS-CoM methyltransferase activity based on the demonstrated competency of CmtA to methylate HS-CoM. The weaker methylation with DMS and unsustained activity is explained in part by the unfavorable thermodynamics (ΔG°′ = 17.9 kJ/mol) predicted from the equilibrium constant for the DMS:cob(I)alamin methyltransferase reaction mediated by the MtsA component of the DMS:HSCoM methyltransferase from M. barkeri (38). It is also possible that the methylthiol product inhibits the enzyme. Thus, no firm conclusions can be drawn from these negative results regarding the potential for DMS:HSCoM methyltransferase activity of CmtA. However, although the thermodynamics for conversion of DMS to CH4 (33) predict good growth (Table 1, reaction 11), DMS-dependent growth of wild-type M. acetivorans is exceedingly poor, with final optical densities of ∼0.1 and maximum doubling times of 60 h (25). The poor growth suggests DMS is not a preferred substrate for M. acetivorans. The poor growth with DMS can be explained by the most plausible mechanism for CmtA. The mechanism is analogous to the three- and two-component methyltransferase systems of methanogens, wherein a subunit transfers the substrate methyl group to a corrinoid-containing subunit, which then donates the methyl group to the subunit, which binds and methylates HS-CoM. In the mechanism postulated for CH3-THMPT:HS-CoM methyltransferase activity of CmtA, the C-terminal domain binds CH3-THSPT and transfers the methyl group to the corrinoid cofactor of the N-terminal domain, which then returns the methyl group to the C-terminal domain, where HS-CoM is bound and methylated, producing CH3-S-CoM. The robust CH3-THMPT:HS-CoM methyltransferase activity demonstrated for CmtA suggests that FCMT homologs evolved with this as the primary function. The proposed mechanism for the predicted DMS:HS-CoM methyltransferase activity is similar, with the possible exception that DMS is a fortuitous methyl donor that could explain poor growth with DMS (25).
Although the reported doubling times are not significantly different between the wild-type and ΔcmtA [DmtsF(26)] strains of M. acetivorans cultured with trimethylamine, it is also reported that levels of a uidA translational fusion to cmtA [mtsF(26)] increase severalfold in wild-type M. acetivorans cultured with monomethylamine, dimethylamine, or trimethylamine versus CO, which leads to the conclusion that CmtA [MtsF(26)] is involved in the metabolism of methylotrophic substrates via an undetermined mechanism (4). Clearly, further research is needed to resolve the role of CmtA in the metabolism of methylotrophic substrates.
Conclusions.
The previous conclusion that FCMT homologs only catalyze DMS:HS-CoM methyltransferase activity was based solely on the analysis of mutants with no biochemical characterization of enzyme activity. The results reported here document the CH3-THMPT:HS-CoM methyltransferase activity of CmtA, the FCMT homolog encoded by the MA4384 gene. Thus, we propose renaming MA4384 with the designation CmtA (cytoplasmic methyl transferase), for which the methyl donor is not specified. The results also support a role for CmtA in the CO-dependent pathway of methanogenesis when cultured with growth-limiting partial pressures of CO. Finally, the results call for further biochemical investigations to determine the function of other FCMT homologs in M. acetivorans and other methanogenic species reported here.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by a grant from the Department of Energy, Energy Biosciences Program (DE-FG02-95ER20198).
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Becher B, Muller V. 1994. ΔμNa+ drives the synthesis of ATP via an ΔμNa+-translocating F1F0-ATP synthase in membrane vesicles of the archaeon Methanosarcina mazei Go1. J. Bacteriol. 176: 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Becher B, Muller V, Gottschalk G. 1992. The methyl-tetrahydromethanopterin:coenzyme-M methyltransferase of Methanosarcina strain Go1 is a primary sodium pump. FEMS Microbiol. Lett. 91: 239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Becher B, Muller V, Gottschalk G. 1992. N5methyl-tetrahydromethanopterin:coenzyme M methyltransferase of Methanosarcina strain Go1 is an Na+-translocating membrane protein. J. Bacteriol. 174: 7656–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bose A, Kulkarni G, Metcalf WW. 2009. Regulation of putative methyl-sulphide methyltransferases in Methanosarcina acetivorans C2A. Mol. Microbiol. 74: 227–238 [DOI] [PubMed] [Google Scholar]
- 5. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 6. Daniels L, Fuchs G, Thauer RK, Zeikus JG. 1977. Carbon monoxide oxidation by methanogenic bacteria. J. Bacteriol. 132: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferry JG. 2010. How to make a living exhaling methane. Annu. Rev. Microbiol. 64: 453–473 [DOI] [PubMed] [Google Scholar]
- 8. Galagan JE, et al. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12: 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gencic S, et al. 2001. Zinc-thiolate intermediate in catalysis of methyl group transfer in Methanosarcina barkeri. Biochemistry 40: 13068–13078 [DOI] [PubMed] [Google Scholar]
- 10. Gottschalk G, Thauer RK. 2001. The Na+ translocating methyltransferase complex from methanogenic archaea. Biochim. Biophys. Acta 1505: 28–36 [DOI] [PubMed] [Google Scholar]
- 11. Grahame DA, Demoll E. 1996. Partial reactions catalyzed by protein components of the acetyl-CoA decarbonylase synthase enzyme complex from Methanosarcina barkeri. J. Biol. Chem. 271: 8352–8358 [DOI] [PubMed] [Google Scholar]
- 12. Guss AM, Kulkarni G, Metcalf WW. 2009. Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri. J. Bacteriol. 191: 2826–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Habeeb AFSA. 1972. Reaction of protein sulfhydryl groups with Ellman's reagent. Methods Enzymol. 25: 457–464 [DOI] [PubMed] [Google Scholar]
- 14. Harms U, Thauer RK. 1996. The corrinoid-containing 23-kDa subunit MtrA of the energy-conserving N5-methyltetrahydromethanopterin:coenzyme M methyltransferase complex from Methanobacterium thermoautotrophicum EPR spectroscopic evidence for a histidine residue as a cobalt ligand of the cobamide. Eur. J. Biochem. 241: 149–154 [DOI] [PubMed] [Google Scholar]
- 15. Hofmann R, Stoffel W. 1993. TMbase–a database of membrane-spanning protein segments. Biol. Chem. Hoppe-Seyler 347: 166 [Google Scholar]
- 16. Jablonski PE, Lu WP, Ragsdale SW, Ferry JG. 1993. Characterization of the metal centers of the corrinoid/iron-sulfur component of the CO dehydrogenase enzyme complex from Methanosarcina thermophila by EPR spectroscopy and spectroelectrochemistry. J. Biol. Chem. 268: 325–329 [PubMed] [Google Scholar]
- 17. Kruer M, Haumann M, Meyer-Klaucke W, Thauer RK, Dau H. 2002. The role of zinc in the methylation of the coenzyme M thiol group in methanol:coenzyme M methyltransferase from Methanosarcina barkeri. New insights from X-ray absorption spectroscopy. Eur. J. Biochem. 269: 2117–2123 [DOI] [PubMed] [Google Scholar]
- 18. Lessner DJ, et al. 2006. An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. U. S. A. 103: 17921–17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li L, et al. 2007. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lienard T, Becher B, Marschall M, Bowien S, Gottschalk G. 1996. Sodium ion translocation by N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanosarcina mazei Go1 reconstituted in ether lipid liposomes. Eur. J. Biochem. 239: 857–864 [DOI] [PubMed] [Google Scholar]
- 21. Mathrani IM, Boone DR, Mah RA, Fox GE, Lau PP. 1988. Methanohalophilus zhilinae sp. nov., an alkaliphilic, halophilic, methylotrophic methanogen. Int. J. Syst. Bacteriol. 38: 139–142 [DOI] [PubMed] [Google Scholar]
- 22. Maupin-Furlow J, Ferry JG. 1996. Characterization of the cdhD and cdhE genes encoding subunits of the corrinoid iron-sulfur enzyme of the CO dehydrogenase complex from Methanosarcina thermophila. J. Bacteriol. 178: 340–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moran JJ, House CH, Vrentas JM, Freeman KH. 2008. Methyl sulfide production by a novel carbon monoxide metabolism in Methanosarcina acetivorans. Appl. Environ. Microbiol. 74: 540–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Brien JM, Wolkin RH, Moench TT, Morgan JB, Zeikus JG. 1984. Association of hydrogen metabolism with unitrophic or mixotrophic growth of Methanosarcina barkeri on carbon monoxide. J. Bacteriol. 158: 373–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oelgeschlager E, Rother M. 2009. In vivo role of three fused corrinoid/methyl transfer proteins in Methanosarcina acetivorans. Mol. Microbiol. 72: 1260–1272 [DOI] [PubMed] [Google Scholar]
- 26. Paterek J, Smith PH. 1985. Isolation and characterization of a halophillic methanogen from Great Salt Lake. Appl. Environ. Microbiol. 50: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paterek JR, Smith PH. 1988. Methanohalophilus mahii gen. nov. sp. nov. a methylotrophic halophilic methanogen. Int. J. Syst. Bacteriol. 38: 122–123 [DOI] [PubMed] [Google Scholar]
- 28. Rother M, Metcalf WW. 2004. Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc. Natl. Acad. Sci. U. S. A. 101: 16929–16934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sauer K, Thauer RK. 1998. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri. Identification of the active-site histidine in the corrinoid-harboring subunit MtaC by site-directed mutagenesis. Eur. J. Biochem. 253: 698–705 [DOI] [PubMed] [Google Scholar]
- 30. Sauer K, Thauer RK. 1997. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri. Zinc dependence and thermodynamics of the methanol:cob(I)alamin methyltransferase reaction. Eur. J. Biochem. 249: 280–285 [DOI] [PubMed] [Google Scholar]
- 31. Scherer P, Hollriegel V, Krug C, Bokel M, Renz P. 1984. On the biosynthesis of 5-hydroxybenzimidazolylcobamide (vitamin B12-factor III) in Methanosarcina barkeri. Arch. Microbiol. 138: 354–359 [Google Scholar]
- 32. Schlegel K, Leone V, Faraldo-Gomez JD, Muller V. 2012. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc. Natl. Acad. Sci. U. S. A. 109: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scholten JC, Murrell JC, Kelly DP. 2003. Growth of sulfate-reducing bacteria and methanogenic archaea with methylated sulfur compounds: a commentary on the thermodynamic aspects. Arch. Microbiol. 179: 135–144 [DOI] [PubMed] [Google Scholar]
- 34. Sowers KR. 2012. Methanogenesis, p 149–172 In Schmidt TM, Schaechter M. (ed), Topics in ecology and environmental microbiology. Academic Press, Waltham, MA [Google Scholar]
- 35. Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47: 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tallant TC, Krzycki JA. 1996. Coenzyme M methylase activity of the 480-kilodalton corrinoid protein from Methanosarcina barkeri. J. Bacteriol. 178: 1295–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tallant TC, Krzycki JA. 1997. Methylthiol:coenzyme M methyltransferase from Methanosarcina barkeri, an enzyme of methanogenesis from dimethylsulfide and methylmercaptopropionate. J. Bacteriol. 179: 6902–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tallant TC, Paul L, Krzycki JA. 2001. The MtsA subunit of the methylthiol:coenzyme M methyltransferase of Methanosarcina barkeri catalyses both half-reactions of corrinoid-dependent dimethylsulfide: coenzyme M methyl transfer. J. Biol. Chem. 276: 4485–4493 [DOI] [PubMed] [Google Scholar]
- 39. Terlesky KC, Ferry JG. 1988. Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J. Biol. Chem. 263: 4075–4079 [PubMed] [Google Scholar]
- 40. van Beelen P, et al. 1984. Derivatives of methanopterin, a coenzyme involved in methanogenesis. Eur. J. Biochem. 139: 359–365 [DOI] [PubMed] [Google Scholar]
- 41. Wang M, Tomb JF, Ferry JG. 2011. Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol. 11: 165 doi:10.1186/1471-2180-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhilina TN, Zavarzin GA. 1987. Methanohalobium evestigatus, n. gen., n. sp. The extremely halophilic methanogenic Archaebacterium. Dokl. Akad. Nauk. SSSR 293: 464–468 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.