Abstract
Previously we identified a novel component of the Staphylococcus aureus regulatory network, an extracytoplasmic function σ-factor, σS, involved in stress response and disease causation. Here we present additional characterization of σS, demonstrating a role for it in protection against DNA damage, cell wall disruption, and interaction with components of the innate immune system. Promoter mapping reveals the existence of three unique sigS start sites, one of which appears to be subject to autoregulation. Transcriptional profiling revealed that sigS expression remains low in a number of S. aureus wild types but is upregulated in the highly mutated strain RN4220. Further analysis demonstrates that sigS expression is inducible upon exposure to a variety of chemical stressors that elicit DNA damage, including methyl methanesulfonate and ciprofloxacin, as well as those that disrupt cell wall stability, such as ampicillin and oxacillin. Significantly, expression of sigS is highly induced during growth in serum and upon phagocytosis by RAW 264.7 murine macrophage-like cells. Phenotypically, σS mutants display sensitivity to a broad range of DNA-damaging agents and cell wall-targeting antibiotics. Furthermore, the survivability of σS mutants is strongly impacted during challenge by components of the innate immune system. Collectively, our data suggest that σS likely serves dual functions within the S. aureus cell, protecting against both cytoplasmic and extracytoplasmic stresses. This further argues for its important, and perhaps novel, role in the S. aureus stress and virulence responses.
INTRODUCTION
Staphylococcus aureus is an exceedingly virulent and successful pathogen, capable of causing a wide range of infections, from relatively benign skin lesions to life-threatening septicemia. With an overwhelming ability to adapt to its environment, S. aureus has become the most common cause of both hospital- and community-acquired infections and is believed to be the leading cause of death by a single infectious agent in the United States (20, 34). The threat posed by this organism to human health is further heightened by the rapid and continued emergence of multidrug-resistant isolates (1, 20, 34, 43).
Many components govern the adaptive nature of S. aureus, including complex regulatory networks, which allow it to respond to constantly changing environments via rapid shifts in gene expression. There are a number of different elements that mediate this fine-tuning, including DNA-binding proteins, two-component systems, regulatory RNAs, and alternative σ factors (10, 11, 18, 21, 22, 32, 44, 50, 51). The last class acts by binding to core RNA polymerase and redirecting promoter recognition to coordinate gene expression, bringing about expedient and wide-reaching alterations within the cell.
From a classification perspective, σ factors are divided into five discrete subfamilies, with the essential housekeeping factors (σA or σ70), which are responsible for the majority of transcription, constituting group 1. The remaining families (groups 2 to 5) contain alternative σ factors, which are important for niche-specific transcriptional regulation in response to environmental change (24, 27, 40, 41). These elements provide the ability to readily adapt to an ever-changing environment by discrete alterations in transcription profiles. As such, bacteria typically encode a number of alternative σ factors within their genome that fulfill a wide range of functions. Of the alternative families, group 4, comprising the ECF (extracytoplasmic function) σ factors, contains by far the most numerous of all such elements (27); for example, Streptomyces coelicolor contains approximately 65 σ factors, around 50 of which are of the ECF subtype (27). Interestingly, S. aureus relies on only 4 σ factors to oversee the execution of its gene expression. In addition to a primary σ factor, σA (16, 17), S. aureus, as with the majority of firmicutes, possesses a σB alternative σ factor, which controls the general stress response (18, 28, 37, 55, 64). A third σ factor, σH, has recently been reported, demonstrating homology to σH from Bacillus subtilis, and has been shown to regulate competence genes and the integration and excision of prophages (46, 68). Finally, a recent discovery in our laboratory demonstrated the existence of a fourth σ factor, σS, belonging to the ECF family (65). Unlike many other organisms, which commonly possess multiple ECF σ factors, σS is the only such element discovered in this organism thus far (27, 65).
Previous work by our group revealed a role for σS in the stress and virulence response of S. aureus (65). Specifically, we showed that σS is important in extended survival during starvation and by lysis with Triton X-100. Competitive growth analysis revealed a decreased ability of a sigS mutant to compete against its parental strain both under standard conditions and in the presence of stress. Interestingly, transcriptional analysis of sigS in the laboratory strain SH1000 revealed only baseline expression during growth in rich media over a 72-h period. Finally, using a murine model of septic arthritis, we demonstrated a role for σS in systemic infections, as mice infected with a sigS mutant displayed significantly decreased weight loss, mortality, severity of infection, systemic dissemination, and mounted immune response by the host.
In this study, we have further explored the role and regulation of σS, in an effort to understand the conditions under which S. aureus utilizes this transcriptional regulator. We show that not only is sigS transcription seemingly subject to genetic control in S. aureus cells, but it is also highly inducible in response to a variety of stresses, including those that elicit DNA damage and cell wall perturbations. Additionally, we reveal that sigS is strongly upregulated upon exposure to serum and following phagocytosis by macrophage-like cells. Finally, we present a role for σS in the response to DNA damage and cell wall stress, as well as a role in protection against components of the innate immune system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. aureus and Escherichia coli strains and plasmids are listed in Table 1. E. coli was grown in Luria-Bertani (LB) medium at 37°C. S. aureus was grown in 100 ml tryptic soy broth (TSB) (1:2.5 flask/volume ratio) at 37°C with shaking at 250 rpm, unless otherwise indicated. Synchronized cultures were obtained as described previously (65). When required, antibiotics were added at the following concentrations: ampicillin, 100 mg liter−1 (E. coli); tetracycline, 5 mg liter−1 (S. aureus); erythromycin, 5 mg liter−1 (S. aureus); lincomycin, 25 mg liter−1 (S. aureus); and chloramphenicol, 5 mg liter−1 (S. aureus). Where specified, chemically defined minimal media (CDM) and metal ion-limiting media (CL) were prepared as described previously (29, 74). Porcine serum agar was created by adding filter-sterilized porcine serum (Sigma) to preautoclaved and cooled 2% agar in distilled water.
Table 1.
Strains, plasmids, and primers used in this studya
| Strain, plasmid, or primer | Genotype or description | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | ϕ80 lacZΔM15 Δ(argF-lacZYA)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 gyrA96 relA1 | 60a |
| S. aureus strains | ||
| RN4220 | Restriction-deficient transformation recipient | Lab stocks |
| 8325-4 | Wild-type laboratory strain, rsbU mutant | Lab stocks |
| SH1000 | Wild-type laboratory strain, rsbU functional | 28 |
| Newman | Wild-type laboratory strain, human clinical isolate | Lab stocks |
| USA300 | USA300-LAC MRSA isolate cured of pUSA300-LAC-MRSA | Paul Fey, UNMC |
| LES57 | SH1000 pAZ106::sigS-lacZ sigS+ | 65 |
| HKM01 | RN4220 pAZ106::sigS-lacZ sigS+ | This study |
| HKM02 | 8325-4 pAZ106::sigS-lacZ sigS+ | This study |
| HKM03 | Newman pAZ106::sigS-lacZ sigS+ | This study |
| HKM04 | USA300 pAZ106::sigS-lacZ sigS+ | This study |
| HKM05 | RN4220 sigS::tet(lacking sigS) | This study |
| HKM06 | RN4220 sigS::tet pMK4::sigS+ | This study |
| HKM07 | USA300 sigS::tet(lacking sigS) | This study |
| HKM08 | USA300 sigS::tet pMK4::sigS+ | This study |
| Plasmids | ||
| pMK4 | Shuttle vector | 67a |
| pHKM1 | pMK4 containing a 2.1-kb sigS fragment | This study |
| Primers | ||
| OL281 | ACT GGA TCC CAG TTG CAG ATG CAT CTC TCC | |
| OL1715 | ATG CTG CAG CAA GTC TAT CTG GCG TAC | |
| OL1036 | CCG CGC ACA TTT CCC CGA AA | |
| OL1275 | ACC TTG AAG GAT ACA AGC AA | |
| OL1276 | GGC ATT TAC GCT TAA CGG AC | |
| OL1528 | GTG GTG TTT GTT GTA TAC GTC |
Primer restriction sites are underlined. Abbreviations: MRSA, methicillin-resistant S. aureus; UNMC, University of Nebraska Medical Center.
Construction of the sigS mutant and sigS-lacZ fusion strains.
All strains used in this study, other than those described below, were created via ϕ11-mediated transduction from strains previously described (Table 1).
Construction of sigS complement strains.
The complement construct generated contains approximately 1 kb of upstream and 710 bp of downstream DNA, relative to the sigS coding region. This was PCR amplified using primer pair OL281 and OL1715 (Table 1) and cloned into the Gram-positive shuttle vector pMK4, creating pHKM1. S. aureus RN4220 was transformed with this construct, with clones confirmed by PCR analysis, using a combination of gene- and vector-specific primers (OL281/OL1036). A representative clone was selected to transduce the RN4220 and USA300 sigS mutants. Clones were again confirmed by PCR analysis, creating strains HKM06 and HKM08.
β-Galactosidase assays.
Levels of β-galactosidase activity were measured as described previously (35). The results presented here are representative of three independent replicates, which showed less than 10% variability.
Real-time PCR.
Quantitative real-time PCR analysis was conducted as described previously (35) using primers listed in Table 1 specific for sigS (OL1275/OL1276). Control primers were for the 16S rRNA gene, as described previously (36). Values were calculated from three independent replicates, and the data analyzed using a Student t test with a 5% confidence limit to determine statistical significance.
Primer extension analysis.
Primer extension analysis was carried out as described previously (63) using the AMV reverse transcriptase (RT) primer extension system (Promega) according to the manufacturer's guidelines. RNA for primer extension reactions was extracted using an RNeasy kit (Qiagen) as described previously (35). For primer extension analysis, 32 μg RNA was used with primer OL1528.
Plate-based stress assays.
Plate-based assays to determine alterations in transcription resulting from external stress were performed using sigS-lacZ fusion strains as described previously (65), with the following stress chemicals: 6 M HCl, 85% phosphoric acid, 100% trichloroacetic acid (TCA), 88% formic acid, 0.2 M acetic acid, 6 M sulfuric acid, 6 M nitric acid, 6 M sodium hydroxide, 2 M NaCl, 1 M glucose, 95% ethanol, 100% methanol, 100% isopropanol, 10% SDS, 10% Triton X-100, 10% Tween 20, 1 M N-lauroyl sarcosine, 30% hydrogen peroxide, 1 M methyl viologen, 1% menadione, 2 mg ml−1 pyrogallol, 1 M sodium nitroprusside, 1 M ethyl methane sulfonate, 1 M methyl methanesulfonate (MMS), 5 mg ml−1 penicillin G, 5 mg ml−1 ciprofloxacin, 5 mg ml−1 nalidixic acid, 5 mg ml−1 cefotaxime, 5 mg ml−1 vancomycin, 2 mg ml−1 phosphomycin, 5 mg ml−1 spectinomycin, 100 mg ml−1 ampicillin, 100 mg ml−1 oxacillin, 5 mg ml−1 gramicidin, 5 mg ml−1 tetracycline, 50 mg ml−1 kanamycin, 50 mg ml−1 neomycin, 10 mg ml−1 chloramphenicol, 20 mg ml−1 puromycin, 2 mg ml−1 bacitracin, 2 mg ml−1 mupirocin, 500 mM diamide, 12.8 mg ml−1 berberine chloride, 4.21 M peracetic acid, 0.1 M EDTA, 1 M dithiothreitol (DTT). Plates were incubated for 24 h at 37°C and screened for blue halos, indicating expression.
Transcriptional analysis during growth in porcine serum.
Synchronous cultures of the sigS-lacZ fusion strains were standardized to an optical density at 600 nm (OD600) of 0.5, pelleted, and washed twice in phosphate-buffered saline (PBS) before being resuspended in 1 ml of filter-sterilized porcine serum (Sigma). The suspension was then incubated at 37°C in a rotator device for a period of 1, 5, or 24 h. At the appropriate time point 1-ml samples were pelleted and stored at −20°C for future analysis. Concomitantly, the CFU per ml for each sample was determined via serial dilution and plating on tryptic soy agar (TSA). Harvested bacterial cells were assayed for β-galactosidase production as described previously (35), with the following alterations. Arbitrary expression units were calculated as a measure of substrate cleavage (4-MUG [4-methylumbelliferyl-β-glucuronide]) by β-galactosidase into 4-MU (methylumbelliferyl), which was evaluated by measuring the fluorescence of each sample at 355/460 nm, 0.1 s, divided by the CFU ml−1. Samples collected from the initial inocula were analyzed for β-galactosidase activity and used as a measure of baseline expression to identify changes in transcription, as described previously (52, 73). The data presented were generated from 3 independent replicates and analyzed using a Student t test with a 5% confidence limit to determine statistical significance.
Macrophage cell culture and S. aureus intracellular transcriptional analysis.
Assays were carried out using the RAW 264.7 murine leukemic monocyte macrophage cell line (ATCC TIB-71) as described previously (73). Cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Sigma) supplemented with 10% fetal bovine serum (Invitrogen) and a 1% penicillin/streptomycin solution (Sigma) until infection, at which time antibiotics were used as described below. RAW 264.7 cells were seeded into 6-well plates and allowed to grow to a density of 2.5 × 106 cells per well. These were then infected with S. aureus strains resuspended in cell culture medium at 2.5 × 108 CFU per well to give a multiplicity of infection (MOI) of 100. To synchronize infections and facilitate contact between bacteria and RAW 264.7 cells, plates were centrifuged at 1,000 rpm for 10 min. Cells were subsequently incubated for 1 h at 37°C in a humidified atmosphere containing 5% CO2 to allow phagocytosis. After this time, wells were washed twice with PBS, and any remaining nonphagocytosed bacteria were killed by the addition of medium containing 30 μg ml−1 gentamicin for 1 h. This was then replaced with fresh DMEM containing 5 μg ml−1 gentamicin and incubated for 24 h. Following this, RAW 264.7 cells were washed twice with PBS and lysed using 500 μl PBS containing 0.5% Triton X-100. Samples were withdrawn to determine bacterial numbers, and the remaining bacteria were pelleted by centrifugation. Harvested bacterial cells were assayed for β-galactosidase production as described previously (35), with the modifications described above for pig serum studies. The data presented were generated from 6 independent replicates and analyzed using a Student t test with a 5% confidence limit to determine statistical significance.
DNA damage sensitivity assays.
Exponentially growing cultures were washed and resuspended in PBS before the addition of DNA-damaging agents: 150 mM H2O2, 20 mM MMS, or 2 mg ml−1 ethidium bromide (EtBr). These were placed at 37°C with shaking, and aliquots removed at the time intervals specified. Samples were then serially diluted, and CFU ml−1 determined alongside control samples that were removed prior to exposure. Percent survival was calculated by comparing initial CFU ml−1 to final CFU ml−1 from three independent assays, and the data were analyzed using a Student t test with a 5% confidence limit to determine statistical significance. Data are presented as fold change of percent survival relative to that of the wild-type strain.
UV radiation survival assay.
The UV radiation survival assay was performed as previously described (9). Briefly, strains were synchronized to an OD600 of 0.05 and allowed to grow for 4 h. Cultures were then serially diluted, and 10−2 through 10−6 dilutions plated on TSA. Dilutions 10−2 and 10−3 were subjected to UV irradiation at 4,000 μJ/cm2 using a CL-1000 UV Cross-linker (UVP). Dilutions 10−4 through 10−6 served as unexposed controls. All plates were incubated in the dark at 37°C overnight. Survival rates were calculated from three independent experiments, and the data analyzed using a Student t test with a 5% confidence limit to determine statistical significance.
MICs of cell wall-targeting antibiotics.
MIC determinations were performed using a variety of cell wall-targeting antibiotics and a broth microdilution assay, described previously (5).
Whole-blood survival assay.
The USA300 wild-type and its isogenic sigS mutant strains were subjected to analysis using a whole-human-blood model of survival as described previously (35). Pooled and deidentified whole human blood was purchased from Bioreclamation. Survival rates were calculated from three independent replicates, and the data analyzed using a Student t test with a 5% confidence limit to determine statistical significance.
Macrophage cell culture and S. aureus intracellular survival assay.
For macrophage cell culture and S. aureus intracellular survival assays, infections were carried out as described above for transcription studies, with the following alterations. RAW 264.7 cells were infected with S. aureus strains resuspended in cell culture medium at 2.5 × 106 CFU per well to give an MOI of 1. Samples were withdrawn 24 h postphagocytosis, and CFU ml−1 determined via serial dilution and plating on TSA. The data presented were generated from 3 independent replicates and analyzed using a Student t test with a 5% confidence limit to determine statistical significance.
RESULTS
σS is differentially expressed in S. aureus wild-type strains.
An unusual finding from our previous study on σS was that no expression of the sigS gene was detected in SH1000 under standard conditions (65). To assess whether this is a conserved phenomenon, transcriptional analysis using a sigS-lacZ fusion was performed in a variety of laboratory strains, including RN4220, 8325-4, SH1000, and Newman, as well as the clinical isolate USA300. Expression of sigS again exhibited only baseline activity over a 24-h period in complex liquid media in every strain, apart from RN4220 (Fig. 1A). Surprisingly, in the last strain we observed an approximate 5-fold increase in sigS expression compared to other S. aureus isolates. To ensure our findings were not an artifact of the fusion construct, we performed quantitative real-time PCR (qRT-PCR) on these wild-type strains during a window of maximal sigS expression (3 h). Using this approach, we again observed robust expression of sigS in RN4220, with minimal transcription detected in the other isolates (Fig. 1B). Curiously, while low expression was observed for the other backgrounds, transcriptional activity in Newman was almost entirely negligible. To determine if nucleotide alterations in the promoter region contributed to this variable expression, we sequenced a 945-bp region immediately 5′ of the annotated sigS start codon for each of these strains. Interestingly, all sequences were identical to each other and, where available, matched publicly available genome data for the requisite strains. As such, the differential expression of sigS is seemingly not mediated by SNPs within the promoter or regulatory regions of the sigS gene.
Fig 1.
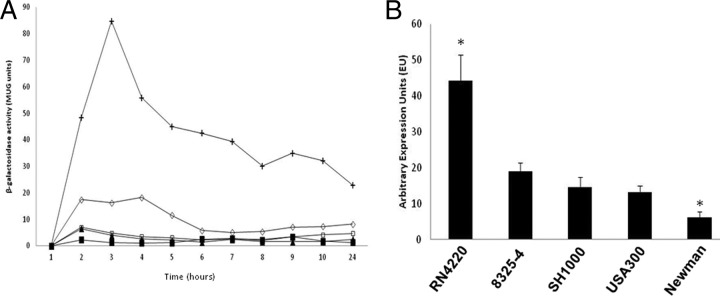
Transcription profiling of sigS in a variety of S. aureus wild-type strains. (A) sigS-lacZ fusion strains in RN4220 (+), 8325-4 (♢), SH1000 (▲), USA300 (■), and Newman (□) were grown in TSB at 37°C and sampled every hour for 10 h and again at 24 h. β-Galactosidase activity was measured to determine levels of expression. Assays were performed on duplicate samples and the values averaged. The results presented here were representative of three independent experiments that showed less than 10% variability. (B) Quantitative real-time PCR was performed on S. aureus wild-type strains grown for 3 h under the same conditions as described for panel A, with primers specific to sigS. The data presented are from at least 3 independent experiments. Error bars indicate ± standard error of the mean; *, P < 0.05 by the Student t test.
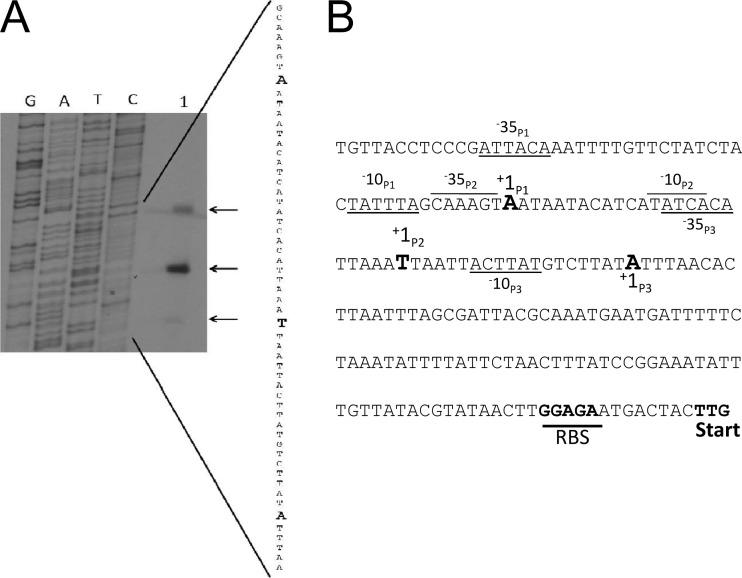
Mapping the sigS transcriptional start site.
We previously demonstrated that σS acts to upregulate itself from its own promoter region; therefore, in order to elucidate this promoter and any others, we set out to map the sigS promoters using RN4220 as a model. To this end, we performed primer extension analysis on RNA extracted from strain RN4220 from exponentially growing cultures (3 h) using a primer located 12 nucleotides (nt) downstream of the sigS initiation codon. We identified 3 unique transcription start sites (Fig. 2), the longest of which bears an adenine plus 1 residue, located 150 bp upstream of the translation start site. This is 7 nt away from a putative σA promoter, denoted P1, with a sequence of aTtACA, followed by a 17-bp spacer, and then TATtta (where lowercase typeface indicates nucleotides that differ from consensus). Promoter P2 is located 126 nt upstream of the translational start site, beginning with a thymine residue, and appears to have no notable σA or σB promoter sequences; however, a possible σS consensus was identified as CAAAGT 12 bp upstream of TATCA, the putative −10 site. The third transcription start site, positioned 107 nt upstream of the coding region, contains an adenine plus 1 residue and is 7 nucleotides away from a putative σA promoter, denoted as P3, with a sequence of aTcACA, followed by an 11-bp spacer, and then acTtAT.
Fig 2.
Primer extension analysis reveals three sigS promoters. (A) Mapping of the 5′ ends of the sigS transcripts by primer extension. RNA was extracted from RN4220 grown to exponential phase (3 h) and used in reactions (lane 1). (B) Transcriptional start sites (+1) for promoters P1, P2, and P3 are denoted with corresponding −35 and −10 regions.
sigS deletion results in a growth defect in strain RN4220.
We previously reported that the sigS mutant in the SH1000 background displayed no notable growth defect under standard conditions (65). Given the data presented above regarding differential expression of sigS in S. aureus wild-type strains, we next performed growth analysis of the sigS mutant in the RN4220, 8325-4, SH1000, and USA300 LAC (Los Angeles County clone) backgrounds. Interestingly, while mutation of σS led to no notable growth defects in the last 3 strains, we observed a significant growth defect in strain RN4220 (Fig. 3). Specifically, sigS deletion in this strain resulted in a significant defect upon exit from stationary phase, which continued through exponential growth. At hour 2, we noted a 4.4-fold decrease in optical density of the σS mutant compared to the wild type, which peaks at hour 3 with a fold change of 5.3. This trend continues through hour 4 with a fold decrease of 3. By hour 5, growth of the σS mutant is comparable to that of the parent strain. This observation is particularly interesting, as sigS expression peaks at hour 3 in RN4220, which corresponds to the time point at which we observe the highest fold decrease in growth for the mutant strain.
Fig 3.
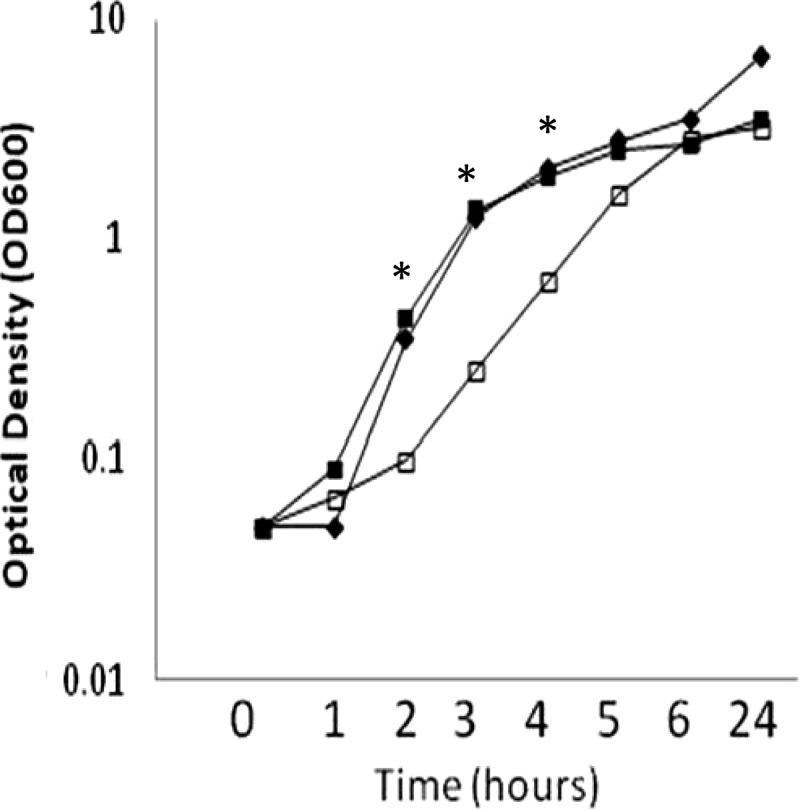
RN4220 sigS mutants have a growth defect upon exit from stationary phase. Optical density readings (OD600) of the RN4220 wild-type (■), sigS mutant (□), and sigS complement (✦) strains were taken every hour for 6 h and again at 24 h during growth at 37°C with shaking in TSB. Growth curves are representative of at least three independent experiments that showed less than 10% variability. *, P < 0.05 by the Student t test, indicating significant difference in growth between the sigS mutant and its parental and complemented strains.
sigS expression is inducible in response to external stimuli.
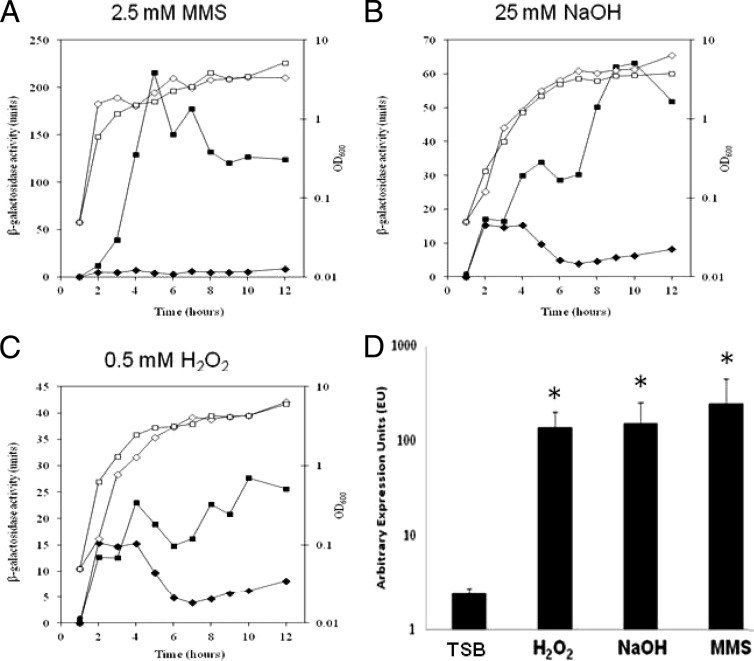
Given the differential nature of sigS expression among S. aureus strains, we next set out to explore whether transcription could be induced from this locus via external stress, as with other ECF sigma factors (27, 56, 67). This was performed using a disk diffusion assay previously described by us (35, 65), which builds on a pilot screening conducted with SH1000 (65). As such, sigS-lacZ fusion strains in 8325-4, Newman, SH1000, and USA300 were grown in the presence of a plethora of stress conditions (see Materials and Methods). While we were unable to detect upregulation of sigS in most strains, we did observe significant inducibility in 8325-4. Specifically, we noted sigS expression in the presence of a variety of chemicals (Table 2), including a number of agents that induce cell wall stress, as well as compounds known to elicit DNA damage. We also noted sigS upregulation in amino acid-limiting media, in metal ion-limiting media, and during growth on pig serum. These minimal medium studies were of particular interest, as they correlate with our previous studies demonstrating a starvation survival defect for sigS mutant strains (65). Again to rule out artifacts of the screen, we sought to verify these findings during continuous growth in liquid media. This was performed with the 8325-4 sigS-lacZ fusion strain grown in TSB containing sublethal concentrations of select chemicals (MMS, H2O2, and NaOH) shown in Table 2 and revealed increased expression of sigS in each instance (Fig. 4A to C). Specifically, in the presence of MMS, expression peaked at 5 h, with a 48.7-fold increase compared to standard conditions. Maximal expression with both NaOH and H2O2 occurred at 10 h, with fold increases of 10 and 4.4, respectively, compared to unsupplemented media. An additional qRT-PCR analysis was performed (Fig. 4D) to verify these data and again confirmed that the greatest fold increase in sigS expression was induced by exposure to MMS, resulting in a 102.6-fold increase in transcription. Transcription levels of sigS upon exposure to NaOH and H2O2 were 63.3- and 57.2-fold higher, respectively.
Table 2.
Compounds found to induce expression of a sigS-lacZ reporter fusion in strain 8325-4
| Agent/conditions | Stress/mode of action | Overall effect |
|---|---|---|
| NaOH | Alkali stress | DNA damage |
| H2O2 | Oxidative stress | DNA damage |
| MMS | Alkylates DNA | DNA damage |
| EMSa | Alkylates DNA | DNA damage |
| Ciprofloxacin | Inhibits DNA gyrase | DNA damage |
| Nalidixic acid | Inhibits DNA gyrase | DNA damage |
| Chloramphenicol | Inhibits protein synthesis | Miscellaneous |
| Pig serum | Components of the humoral immune system | Miscellaneous |
| Amino acid-limiting media | Minimal media | Miscellaneous |
| Metal-limiting media | Minimal media | Miscellaneous |
| Cefotaxime | Inhibits transpeptidation | Cell wall weakening/disruption |
| Ampicillin | Inhibits transpeptidation | Cell wall weakening/disruption |
| Oxacillin | Inhibits transpeptidation | Cell wall weakening/disruption |
| SDS | Disrupts cell walls | Cell wall weakening/disruption |
| Phosphomycin | Inhibits UDP-N-acetylglucosamine-3-enolpyruvyltransferase | Cell wall weakening/disruption |
EMS, ethyl methanesulfonate.
Fig 4.
sigS transcription is inducible in response to external stress. (A to C) The 8325-4 sigS-lacZ strain was grown in either TSB (✦) or TSB supplemented with sublethal concentrations of the indicated stress chemicals (■). Cultures were sampled every hour for 10 h, and again at 24 h, to determine β-galactosidase activity. Additionally, growth was monitored via OD600 at the times indicated for both standard (♢) and supplemented (□) growth conditions. Assays were performed on duplicate samples and the values averaged. The results presented here were representative of three independent replicates that showed less than 10% variability. (D) Quantitative real-time PCR analysis was performed with strain 8325-4 grown for 5 h under conditions identical to those described for panels A to C, using primers specific to sigS. The data presented are from at least 3 independent experiments. Error bars indicate ± standard error of the mean; *, P < 0.05 by the Student t test.
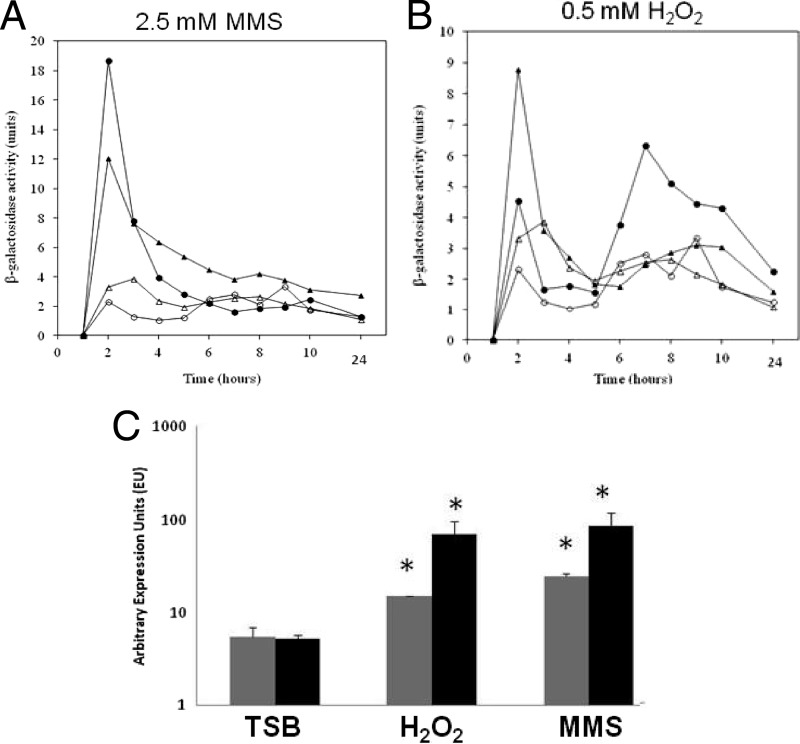
To explore if this upregulation was conserved for other S. aureus strains, but perhaps below the limit of detection for our plate-based assay, we performed experiments with MMS, NaOH, and H2O2 using SH1000 and USA300 sigS-lacZ fusion strains. Interestingly, despite a lack of blue coloration in the plate-based assay, we again detected upregulation of sigS during growth in liquid media with sublethal concentrations of MMS and H2O2 (Fig. 5A and B). Specifically, expression with MMS in both SH1000 and USA300 was highest at 2 h, with fold increases of 3.6 and 8.1, respectively, compared to standard growth conditions. In the presence of H2O2, sigS expression in SH1000 increased 2.6-fold (2 h) and 2.3-fold in USA300 (7 h). Conversely, we observed no increase in expression in the presence of NaOH when grown under these conditions, suggesting that greater, and more lethal, concentrations of this agent may be required to induce expression. We again confirmed these data by qRT-PCR in SH1000 and USA300 grown in the presence of MMS and H2O2 (Fig. 5C). We determined that SH1000 displays a 16.9-fold increase in sigS expression when cultured with MMS and a 13.5-fold increase when cultured with H2O2. Expression of sigS in USA300 grown with MMS or H2O2 displayed 4.6- and 2.8-fold increases in transcription, respectively. Collectively, these findings demonstrate a significant inducibility of sigS in response to external stimuli, which is conserved across S. aureus strains.
Fig 5.
The inducibility of sigS expression is conserved across S. aureus strains. (A and B) The SH1000 (Δ) and USA300 (○) sigS-lacZ fusion strains were grown in either TSB (open symbols) or TSB supplemented with sublethal concentrations of the indicated stress chemicals (closed symbols). Cultures were sampled every hour for 10 h, and again at 24 h, to determine β-galactosidase activity. Assays were performed on duplicate samples and the values averaged. The results presented here were representative of three independent replicates that showed less than 10% variability. (C) Quantitative real-time PCR analysis was performed with strains USA300 (gray) and SH1000 (black) grown for 2 h under conditions identical to those for panels A and B, using primers specific to sigS. The data presented are from at least 3 independent experiments. Error bars indicate ± standard error of the mean; *, P < 0.05 by a Student t test, indicating significant variation from standard conditions (TSB).
sigS is strongly upregulated during challenge by components of the innate immune system.
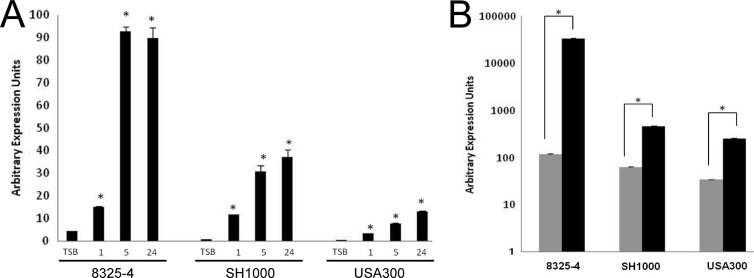
Despite limited expression under standard laboratory growth conditions, we have previously demonstrated a role for σS in the virulence of S. aureus (65). Working on the hypothesis that sigS transcription would be increased during infection, we performed expression profiling of sigS-lacZ fusion strains upon ex vivo challenge by components of the innate immune system. Indeed, our plate-based analysis already suggests that sigS expression is increased by exposure to pig serum (Table 2). In order to confirm these findings and quantify this increase across different strains, we performed transcriptional analysis using sigS-lacZ fusion strains grown in TSB and then subcultured into pig serum. We determined that after just 1 h of growth in serum, sigS expression increased 3.4-fold in strain 8325-4 compared to TSB (Fig. 6A). Expression continued to rise over time, with fold increases of 21.2 and 20.5 observed at hours 5 and 24, respectively. Additionally we observed a similar effect in strains SH1000 and USA300. Specifically, over the course of growth, we noted fold increases of 13.8, 36.2, and 44.0 in SH1000 for hours 1, 5, and 24, respectively. Finally, in USA300, sigS expression increased 6.8-, 15.6-, and 26.3-fold at hours 1, 5, and 24, respectively. We continued this line of investigation by assessing sigS expression upon phagocytosis by RAW 264.7 macrophage-like cells. Accordingly, macrophages were infected with strains 8325-4, SH1000, and USA300 bearing a sigS-lacZ fusion for a period of 24 h, before β-galactosidase activity was measured. Expression of sigS was significantly increased after phagocytosis in all strains tested (Fig. 6B), with the highest levels observed in 8325-4. In this strain we found a 286.6-fold increase in sigS expression compared to background levels. Expression in SH1000 and USA300 increased 7.5- and 7.4-fold, respectively, compared to background levels. As such, these findings support our hypothesis and suggest that σS is required during the interaction of S. aureus with its host.
Fig 6.
Profiling of sigS expression during challenge by components of the innate immune system. Fusion strains were assayed for β-galactosidase activity prior to (TSB), and during (1 h, 5 h, and 24 h), growth in pig serum (A); and prior to phagocytosis (gray bars) and 24 h postphagocytosis (black bars) by RAW 264.7 murine macrophage-like cells (B). Cells were infected at an MOI of 1:100 and incubations carried out at 37°C in a humidified atmosphere of 5% CO2. The data presented are from at least 3 independent experiments. Error bars indicate ± standard error of the mean; *, P < 0.001 by a Student t test.
σS mutants are sensitive to DNA damage stress and cell wall-targeting antibiotics.
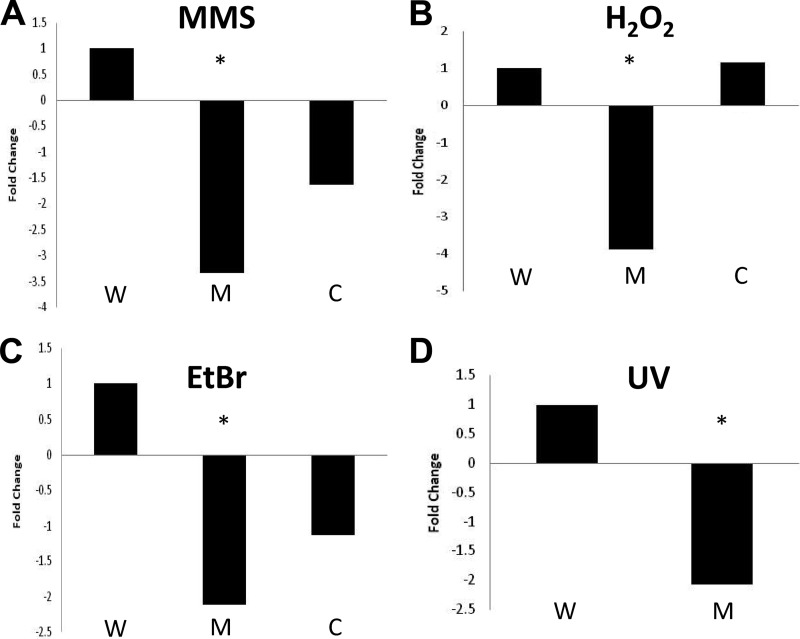
Thus far, we have demonstrated that chemicals known to induce DNA damage and cell wall stress strongly impact sigS transcription. Accordingly, we next sought to perform death-curve kill studies to examine the viability of sigS mutants during exposure to lethal concentrations of these agents in strains 8325-4, SH1000, and USA300. We found that when exponentially growing cultures were exposed to 5× the MIC of MMS for 30 min, a consistent decline in mutant cell viability was seen across all strains tested (the data for USA300, which are representative, are presented in Fig. 7A). Specifically, we recovered 3.33-fold fewer sigS mutant cells than wild-type cells when cultures were exposed to this agent. Complementation of the σS mutation reduced the observed growth impairment significantly, although not completely to wild-type levels. This lack of full complementation is likely attributed to plasmid instability in the presence of DNA-damaging conditions, as suggested by others previously (3, 26, 49, 60, 72, 75). Further to this, in order to determine whether the role of σS was limited to protection against DNA alkylation (as induced by MMS), we next examined the ability of the USA300 σS mutant to survive exposure to agents that induce other types of DNA damage. As such, analysis was carried out during exposure to oxidative stress resulting from the addition of H2O2. Following a 5-min exposure to this agent, we observed a 3.9-fold decrease in sigS mutant viability compared to the parent strain, which was fully complementable (Fig. 7B). We next used the DNA-intercalating agent ethidium bromide and found that the sigS mutant displayed a 2.1-fold decrease in viability compared to the wild-type strain after 15 min of exposure (Fig. 7C). We again saw that complementation was able to abrogate these effects, but not completely to the levels of the wild-type strain. This is likely attributed to the ability of EtBr to cure plasmids upon exposure, as observed by others previously (3). Finally, in order to determine if σS mediates protection against UV-induced lesions and double-strand breaks, we compared the survivability of the wild-type strain and its sigS mutant. Exponentially growing cultures were serially diluted on TSA and subjected to UV at a dosage of 4,000 μJ cm−2. Exposure at this level resulted in a 2.1-fold decrease in viability for the mutant (Fig. 7D). Complementation in this assay is not possible because of plasmid instability, as we observed >83% loss upon exposure (data not shown).
Fig 7.
sigS mutants are sensitive to a variety of DNA damage-inducing stresses. The USA300 wild-type (W) and sigS mutant (M) strains, along with a sigS complement strain (C), were analyzed for viability in the presence of DNA damage-inducing stressors. CFU counts were determined both pre- and postexposure, and the survivability was determined. The data are presented as fold change relative to the wild-type strain and are representative of at least three independent experiments that showed less than 10% variability. Shown are exposures of 30 min to 25 mM MMS (A), of 5 min to 150 mM H2O2 (B), of 15 min to 5 mM EtBr (C), and to UV at 4,000 μJ per cm2 (D). *, P < 0.05 by a Student t test.
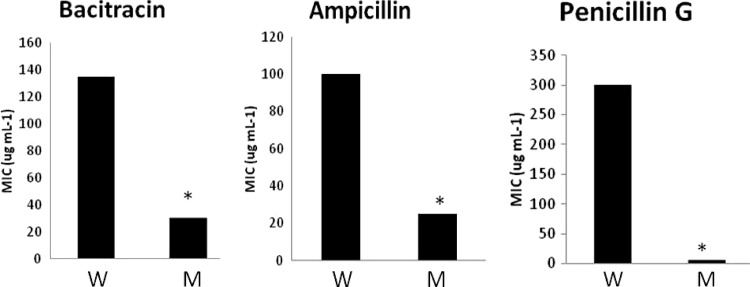
Following this, we next assessed the sensitivity of sigS mutants to a variety of cell wall-targeting antibiotics (Fig. 8). Analysis using bacitracin in the USA300 background revealed a 4.5-fold decrease in MIC for the mutant strain (30 μg ml−1) compared to the parent (135 μg ml−1). We observed a similar degree of sensitivity with ampicillin, resulting in a 4-fold decrease in MIC for the mutant (25 μg ml−1) compared to wild-type USA300 (100 μg ml−1). Finally, analysis performed using penicillin G yielded a striking 60-fold decrease in MIC for the sigS mutant (5 μg ml−1) compared to the parent (300 μg ml−1).
Fig 8.
sigS mutants are sensitive to a number of cell wall-targeting antibiotics. The USA300 wild-type (W) and sigS mutant (M) strains were grown in TSB containing increasing concentrations of the cell wall-targeting antibiotics bacitracin, ampicillin, and penicillin G in a 96-well plate format. The cultures were allowed to grow overnight at 37°C and subsequently analyzed for growth, and the MICs were determined. The data are representative of at least three independent experiments that showed less than 10% variability. *, P < 0.05 by a Student t test.
σS aids in protection of the S. aureus cell during interaction with components of the innate immune system.
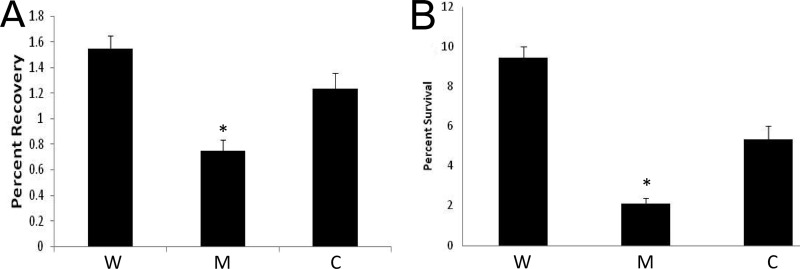
Previously we have shown a role for σS in virulence, using a murine model of septic arthritis. We have also demonstrated here that sigS expression increases not only upon exposure to serum but also during phagocytosis by macrophage-like cells. Therefore, we sought to determine the importance of σS during challenge by components of the innate immune system. This was first performed using whole human blood and the USA300 wild type, its sigS mutant, and its complemented strains. Exponentially growing cells were inoculated into whole human blood and incubated for 4 h. After this time, we recovered 2.1-fold-fewer viable cells of the sigS mutant than of the wild-type strain (Fig. 9A). Complementation analysis restored viability to levels similar to that of the wild type. Following this, we also conducted macrophage survival assays, again in the USA300 background, to assess the ability of the sigS mutant to persist upon phagocytosis. At 24 h postphagocytosis by RAW 264.7 cells, we observed a 4.5-fold decrease in survivability of the sigS mutant compared to the parent (Fig. 9B). Complementation of this finding increases survivability of the mutant cells, but not completely to levels of the wild-type strain. This is likely explained by the significant instability of the plasmid during phagocytosis, which we routinely observed when performing this assay. Collectively, these findings support our earlier work, which indicates an important role for σS in the virulence of S. aureus and confirms our expression analysis, demonstrating sigS upregulation during interaction with components of the innate immune system.
Fig 9.
σS aids in protection of the S. aureus cell during interaction with components of the innate immune system. The USA300 wild type (W) and sigS mutant (M), along with a sigS complement strain (C), were analyzed for viability 4 h after exposure to whole human blood (A) and 24 h postphagocytosis by 264.7 RAW murine macrophage-like cells (B). CFU counts were determined both pre- and postexposure, and the percent survival was determined. Error bars indicate ± standard error of the mean; *, P < 0.05 by a Student t test.
DISCUSSION
In this study, we provide new evidence for the role of σS, a novel ECF σ factor in S. aureus. In our previous works we have shown that σS is a functioning sigma factor that controls its own expression (65). Additionally, we have demonstrated a role for it in the stress and virulence responses of this organism. From a gene expression standpoint, we have previously shown that sigS expression is minimal during growth under standard laboratory conditions in SH1000 (65). In this study, we reveal that this phenomenon is conserved across a variety of S. aureus strains, including laboratory (8325-4, SH1000, and Newman) and clinical (USA300) isolates. In each case, we observed low levels of expression of sigS during growth in rich media. These results may not be entirely surprising, as the majority of ECF σ factors are employed to protect the cell during times of stress and are often transcribed only when required (27, 56, 67).
Interestingly, we did observe robust sigS expression in the highly mutated laboratory strain RN4220. Among the many mutations present in this strain are those that render the activity of other global regulators nonfunctional, including agr and sigB (15, 70). As such, the regulatory circuits in place in this strain are likely to be highly disordered, potentially explaining sigS dysregulation and therefore upregulation. Interestingly, these effects do not appear to be mediated directly through either Agr or σB, as mutations in the genes encoding these proteins alone do not affect sigS expression in either SH1000 or USA300 (data not shown). Recent sequencing of the RN4220 genome has revealed a number of single-nucleotide polymorphisms (SNPs) and deletions relative to the parental strain NCTC 8325 (2, 48). Of interest, a number of these are in genes involved in DNA metabolism, replication, recombination, and repair. Most notably, RN4220 carries an SNP in UvrC, a component of the UvrABC exonuclease, which in Escherichia coli repairs DNA damage induced by a number of mechanisms, including UV light (61). Interestingly, our analysis here demonstrates σS mutants are less able to survive exposure to UV stress. Additionally, SNPs in RN4220 are located in a putative helicase, SAOUHSC_02790, as well as a truncated resolvase, SAOUHSC_02392. Collectively, these observations suggest that RN4220 is perhaps more prone to DNA damage than other wild-type strains, as a result of mutated and nonfunctional repair pathways. This would then perhaps explain why this strain exhibits stronger sigS expression than other wild types, as we implicate σS in influencing the response of S. aureus to DNA damage in this study. Indeed, we have observed that the increasing levels of sigS transcription in different wild-type strains directly correlate with their sensitivity to DNA-damaging agents, such as MMS (data not shown).
Promoter mapping of the sigS locus reveals three discrete transcriptional start sites. Promoters P1 and P2 both appear to be under the control of the housekeeping σ factor σA; however, both are severely corrupted from consensus sequences and/or spacing. Due to the relative weakness of these promoters, it is likely that other regulatory elements must act to activate transcription from these sequences. This again likely explains the low levels of sigS expression observed in the majority of S. aureus strains and argues for a genetic regulatory network that controls expression of this regulator. Previously we demonstrated that sigS, as with other ECF sigma factors (27, 45, 65), controls its own expression via autoregulation. During promoter mapping in the present study, we reveal a likely σS-controlled transcript, P2 (CAAAGT-12 bp-TATCA). Typically, ECF sigma factor consensus sequences display a conserved AAC motif in the −35 region (27, 45); however, exceptions exist. Specifically, the ECF sigma factor of Neisseria gonorrhoeae does not recognize an AAC motif (25), while σR of Streptomyces coelicolor recognizes an AAT motif (54). More importantly, σX, an ECF sigma factor in several Pseudomonas spp., specifically recognizes an AAG motif, as seen here for σS (4, 39). ECF σ factors typically have significant divergence and decreased homology within their region 2.4 (41, 45), which specifically recognizes −10 promoter elements. Accordingly, such sites are often difficult to ascertain; however, the identified putative −10 element is strikingly similar to the TCTGA recognized sequence of σE in E. coli (13).
In addition to examining expression in wild-type strains, we have also assessed the level to which sigS is upregulated in response to external stress. We show that a variety of stressors can induce expression of sigS, ranging from those that elicit alkali stress to those that affect protein synthesis. Interestingly, this is most pronounced in strain 8325-4, which, like RN4220, lacks natural σB activity. We are able to demonstrate that the conditions of sigS inducibility in 8325-4 hold true for the σB functional strains SH1000 and USA300, although not always at the same levels. Of note, when we inactivate sigB in SH1000, we do not observe the same robust increases in sigS inducibility seen in 8325-4 (data not shown), despite these strains being very closely related. This observation is perhaps explained by the fact that 8325-4 is an rsbU mutant, which is an activator of σB activity, rather than a true sigB-null strain. Given recent findings showing a role for RsbU outside its influence of σB activity (71), it is possible that these differences are mediated by RsbU, rather than σB, mechanisms. It has also recently been shown that 8325-4 and SH1000 have genetic differences beyond the 11-bp deletion in rsbU (53), perhaps suggesting that SNPs and other genetic variations between these 2 strains influence sigS expression.
With regard to environmental influence on sigS expression, those chemicals that induce DNA damage, such as methyl methanesulfonate, appear to have the most profound effects. These findings correlate well with our phenotypic studies, showing that sigS mutants have increased sensitivity to a broad range of DNA damage-inducing stresses. These include alkylating and intercalating agents, reactive oxygen species, and UV-induced damage, each of which leads to the activation of specific and distinct repair pathways. Interestingly, when we analyzed the transcription of a number of DNA repair pathway genes (ogt, uvrB, and mutM) in both sigS mutants and S. aureus wild-type strains, we observed no alterations in expression (data not shown). As such, our findings suggest that σS is involved in mediating a comprehensive response to DNA damage by an as-yet-unknown mechanism. These findings are somewhat novel, as the majority of ECF σ factors typically respond to perturbations in the cell wall. However, reports on ECF σ factors from other organisms reveal several examples of these factors that function in sensing and responding to cytoplasmic stress. Specifically, both RpoE of Rhodobacter sphaeroides and Ecf of Neisseria gonorrhoeae respond to oxidative stress, which can in turn lead to DNA damage (6, 19, 25).
Interestingly, a number of agents that were identified as inducing sigS expression are not typically thought of as inducing DNA damage but can also induce this kind of stress. For example, H2O2 resulted in sigS upregulation and can react with intracellular iron to form hydroxyl radicals, which cause damage to DNA (8, 30, 31, 57). Additionally, SOS and DNA damage repair genes have previously been shown in Escherichia coli to be upregulated during alkali stress caused by excess NaOH, which also upregulates sigS expression (23, 62). Finally, the protein synthesis-inhibiting antibiotic chloramphenicol upregulated sigS and has been shown to lead to the degradation of double-stranded DNA and the inhibition of DNA synthesis (47).
A consideration with these DNA damage agent studies is that these agents may not be directly upregulating sigS expression but might cause mutations within the S. aureus genome, leading to SNPs. In such a scenario, this could lead to dysregulation of regulatory circuits, leading to sigS upregulation in a manner akin to that proposed for RN4220 and 8325-4. To examine this, we analyzed 8325-4 sigS-lacZ fusion strains exposed to DNA-damaging agents for a 24-h period. Upon removal of the stressor, strains were grown on agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). We found no detectable blue coloration on such plates (data not shown), indicating that DNA damage-induced upregulation of sigS does not appear to be mediated via heritable SNPs but results directly from exposure to these agents. As such, the increase in expression of sigS is due solely to exposure to agents such as MMS and suggests that σS is utilized by the cell to adapt during times of DNA damage.
We also observed substantial increases in expression of sigS following exposure to a number of cell wall-targeting chemicals, suggesting a role for σS in protection against this type of stress. This correlates well with other works presented in this study, which demonstrate sigS mutants have increased sensitivity to the cell wall-targeting antibiotics. These findings also corroborate our previous work, which demonstrates that sigS mutants are sensitive to growth in the presence of a number of cell wall-disrupting agents, including Triton X-100 and SDS (65). This suggests a role for σS in the S. aureus cell wall stress response, which is typical of ECF σ factors. For example, RpoE in Escherichia coli serves to upregulate genes involved in the heat shock response and is triggered by misfolded proteins accumulating in the periplasm and outer membrane (13). Furthermore, σW and σM of Bacillus subtilis both respond to cell wall biosynthesis-inhibiting antibiotics, with σM proving vital for survival during exposure to phosphomycin (7, 69). This information, alongside the observation that σS is the lone ECF σ factor in S. aureus, suggests it likely serves dual functions within the cell, protecting against both cytoplasmic and extracytoplasmic stresses.
We have also demonstrated here that sigS transcription is increased considerably when S. aureus is challenged by complement during growth in pig serum. We also present evidence for sigS upregulation during ex vivo infection, revealing high levels of expression upon phagocytosis by murine macrophage-like cells. Phenotypically, we show that σS is important for survival during growth in whole human blood and following phagocytosis. Collectively, this supports our previous work, which reveals a major requirement for σS during virulence (65). As part of the microbicidal mechanism employed by macrophages, reactive oxygen species and reactive nitrogen intermediates (RNI) are excreted at very high levels, leading to DNA damage in invading organisms during infection (33, 38, 42, 59, 66). Moreover, it has been observed that pathogenic organisms such as Burkholderia spp., Brucella abortus, and Vibrio cholerae defective in DNA damage repair mechanisms are attenuated in virulence, underscoring their importance during infection (12, 14, 58, 76). Together these findings suggest that, upon entry into the host, bacterial pathogens are faced with an array of DNA-damaging conditions. Given that these conditions lead to activation of σS in S. aureus, this likely goes some way toward explaining the avirulent phenotype of sigS mutants.
In summary, we present extended characterization of the lone, and novel, ECF σ factor σS in S. aureus. We reveal that, under standard conditions, its transcription remains low in a range of wild-type strains but can be upregulated in response to external stimuli. Specifically, chemicals leading to DNA damage and cell wall disruption strongly induce expression of sigS. This upregulation is seemingly of importance, as functional characterization reveals that sigS mutants are sensitive to both of these types of stress. Additionally, we reveal strong upregulation of this gene during growth in pig serum as well as upon phagocytosis by murine macrophage-like cells, which is seemingly protective to the cell. Collectively, our data suggest that σS likely serves dual functions within the cell, protecting against both cytoplasmic and extracytoplasmic stresses. This further argues for its important, and perhaps novel, role in the S. aureus stress and virulence responses.
ACKNOWLEDGMENT
This study was supported in part by grant 1R01AI080626-01A2 (LNS) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 8 June 2012
REFERENCES
- 1. Archer GL. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26: 1179–1181 [DOI] [PubMed] [Google Scholar]
- 2. Berscheid A, Sass P, Weber-Lassalle K, Cheung AL, Bierbaum G. 2012. Revisiting the genomes of the Staphylococcus aureus strains NCTC 8325 and RN4220. Int. J. Med. Microbiol. 302: 84–87 [DOI] [PubMed] [Google Scholar]
- 3. Bouanchaud DH, Scavizzi MR, Chabbert YA. 1968. Elimination by ethidium bromide of antibiotic resistance in enterobacteria and staphylococci. J. Gen. Microbiol. 54: 417–425 [DOI] [PubMed] [Google Scholar]
- 4. Brinkman FS, Schoofs G, Hancock RE, De Mot R. 1999. Influence of a putative ECF sigma factor on expression of the major outer membrane protein, OprF, in Pseudomonas aeruginosa and Pseudomonas fluorescens. J. Bacteriol. 181: 4746–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burda WN, et al. 2012. Neutral metallated and meso-substituted porphyrins as antimicrobial agents against gram-positive pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 31: 327–335 [DOI] [PubMed] [Google Scholar]
- 6. Campbell EA, et al. 2007. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell 27: 793–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao M, Wang T, Ye R, Helmann JD. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol. Microbiol. 45: 1267–1276 [DOI] [PubMed] [Google Scholar]
- 8. Chang W, Small DA, Toghrol F, Bentley WE. 2006. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188: 1648–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Z, Luong TT, Lee CY. 2007. The sbcDC locus mediates repression of type 5 capsule production as part of the SOS response in Staphylococcus aureus. J. Bacteriol. 189: 7343–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheung AL, Koomey JM, Butler CA, Projan SJ, Fischetti VA. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. U. S. A. 89: 6462–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung AL, Wolz C, Yeaman MR, Bayer AS. 1995. Insertional inactivation of a chromosomal locus that modulates expression of potential virulence determinants in Staphylococcus aureus. J. Bacteriol. 177: 3220–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuccui J, et al. 2007. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect. Immun. 75: 1186–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dartigalongue C, Missiakas D, Raina S. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276: 20866–20875 [DOI] [PubMed] [Google Scholar]
- 14. Davies BW, et al. 2011. DNA damage and reactive nitrogen species are barriers to Vibrio cholerae colonization of the infant mouse intestine. PLoS Pathog. 7: e1001295 doi:10.1371/journal.ppat.1001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Azavedo JC, et al. 1985. Expression of the cloned toxic shock syndrome toxin 1 gene (tst) in vivo with a rabbit uterine model. Infect. Immun. 50: 304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deora R, Misra TK. 1996. Characterization of the primary sigma factor of Staphylococcus aureus. J. Biol. Chem. 271: 21828–21834 [DOI] [PubMed] [Google Scholar]
- 17. Deora R, Misra TK. 1995. Purification and characterization of DNA dependent RNA polymerase from Staphylococcus aureus. Biochem. Biophys. Res. Commun. 208: 610–616 [DOI] [PubMed] [Google Scholar]
- 18. Deora R, Tseng T, Misra TK. 1997. Alternative transcription factor sigmaSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 179: 6355–6359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dufour YS, Landick R, Donohue TJ. 2008. Organization and evolution of the biological response to singlet oxygen stress. J. Mol. Biol. 383: 713–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emori TG, Gaynes RP. 1993. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6: 428–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fournier B, Klier A, Rapoport G. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41: 247–261 [DOI] [PubMed] [Google Scholar]
- 22. Giraudo AT, Martinez GL, Calzolari A, Nagel R. 1994. Characterization of a Tn925-induced mutant of Staphylococcus aureus altered in exoprotein production. J. Basic Microbiol. 34: 317–322 [DOI] [PubMed] [Google Scholar]
- 23. Goodson M. 1989. Habituation to alkali in Escherichia coli. Lett. Appl. Microbiol. 9: 71–73 [Google Scholar]
- 24. Gruber TM, Gross CA. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57: 441–466 [DOI] [PubMed] [Google Scholar]
- 25. Gunesekere IC, et al. 2006. Ecf, an alternative sigma factor from Neisseria gonorrhoeae, controls expression of msrAB, which encodes methionine sulfoxide reductase. J. Bacteriol. 188: 3463–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hashimoto H, Kono K, Mitsuhashi S. 1964. Elimination of penicillin resistance of Staphylococcus aureus by treatment with acriflavine. J. Bacteriol. 88: 261–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helmann JD. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46: 47–110 [DOI] [PubMed] [Google Scholar]
- 28. Horsburgh MJ, et al. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184: 5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69: 3744–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240: 640–642 [DOI] [PubMed] [Google Scholar]
- 31. Imlay JA, Linn S. 1988. DNA damage and oxygen radical toxicity. Science 240: 1302–1309 [DOI] [PubMed] [Google Scholar]
- 32. Janzon L, Lofdahl S, Arvidson S. 1986. Evidence for a coordinate transcriptional control of alpha-toxin and protein-a synthesis in Staphylococcus aureus. FEMS Microbiol. Lett. 33: 193–198 [Google Scholar]
- 33. Kennedy LJ, Moore K, Jr, Caulfield JL, Tannenbaum SR, Dedon PC. 1997. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem. Res. Toxicol. 10: 386–392 [DOI] [PubMed] [Google Scholar]
- 34. Klevens RM, et al. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298: 1763–1771 [DOI] [PubMed] [Google Scholar]
- 35. Kolar SL, et al. 2011. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 157: 2206–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koprivnjak T, et al. 2006. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 188: 3622–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kullik II, Giachino P. 1997. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167: 151–159 [DOI] [PubMed] [Google Scholar]
- 38. Lancaster JR., Jr 1997. A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide 1: 18–30 [DOI] [PubMed] [Google Scholar]
- 39. Lane WJ, Darst SA. 2006. The structural basis for promoter-35 element recognition by the group IV sigma factors. PLoS Biol. 4: e269 doi:10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lonetto M, Gribskov M, Gross CA. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174: 3843–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lonetto MA, Brown KL, Rudd KE, Buttner MJ. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. U. S. A. 91: 7573–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. MacMicking J, Xie QW, Nathan C. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15: 323–350 [DOI] [PubMed] [Google Scholar]
- 43. McDougal LK, et al. 2010. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob. Agents Chemother. 54: 3804–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McNamara PJ, Milligan-Monroe KC, Khalili S, Proctor RA. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182: 3197–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Missiakas D, Raina S. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28: 1059–1066 [DOI] [PubMed] [Google Scholar]
- 46. Morikawa K, et al. 2003. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells 8: 699–712 [DOI] [PubMed] [Google Scholar]
- 47. Murray TR, Downey KM, Yunis AA. 1983. Chloramphenicol-mediated DNA damage and its possible role in the inhibitory effects of chloramphenicol on DNA synthesis. J. Lab. Clin. Med. 102: 926–932 [PubMed] [Google Scholar]
- 48. Nair D, et al. 2011. Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J. Bacteriol. 193: 2332–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakamura S. 1990. Curing effects of chlorination, ozone and UV treatments on plasmid DNAs. Nihon Koshu Eisei Zasshi. 37: 745–751 ( In Japanese.) [PubMed] [Google Scholar]
- 50. Novick RP, Jiang DR. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149: 2709–2717 [DOI] [PubMed] [Google Scholar]
- 51. Novick RP, et al. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248: 446–458 [DOI] [PubMed] [Google Scholar]
- 52. O Cróinín T, Dorman CJ. 2007. Expression of the Fis protein is sustained in late-exponential- and stationary-phase cultures of Salmonella enterica serovar Typhimurium grown in the absence of aeration. Mol. Microbiol. 66: 237–251 [DOI] [PubMed] [Google Scholar]
- 53. O'Neill AJ. 2010. Staphylococcus aureus SH1000 and 8325-4: comparative genome sequences of key laboratory strains in staphylococcal research. Lett. Appl. Microbiol. 51: 358–361 [DOI] [PubMed] [Google Scholar]
- 54. Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol. Microbiol. 42: 1007–1020 [DOI] [PubMed] [Google Scholar]
- 55. Pane-Farre J, Jonas B, Forstner K, Engelmann S, Hecker M. 2006. The sigmaB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296: 237–258 [DOI] [PubMed] [Google Scholar]
- 56. Raivio TL, Silhavy TJ. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55: 591–624 [DOI] [PubMed] [Google Scholar]
- 57. Repine JE, Fox RB, Berger EM. 1981. Hydrogen peroxide kills Staphylococcus aureus by reacting with staphylococcal iron to form hydroxyl radical. J. Biol. Chem. 256: 7094–7096 [PubMed] [Google Scholar]
- 58. Roux CM, et al. 2006. RecA and RadA proteins of Brucella abortus do not perform overlapping protective DNA repair functions following oxidative burst. J. Bacteriol. 188: 5187–5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Salgo MG, Stone K, Squadrito GL, Battista JR, Pryor WA. 1995. Peroxynitrite causes DNA nicks in plasmid pBR322. Biochem. Biophys. Res. Commun. 210: 1025–1030 [DOI] [PubMed] [Google Scholar]
- 60. Salisbury V, Hedges RW, Datta N. 1972. Two modes of “curing” transmissible bacterial plasmids. J. Gen. Microbiol. 70: 443–452 [DOI] [PubMed] [Google Scholar]
- 60a. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 61. Sancar A, Rupp WD. 1983. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33: 249–260 [DOI] [PubMed] [Google Scholar]
- 62. Schuldiner S, et al. 1986. Induction of SOS functions by alkaline intracellular pH in Escherichia coli. J. Bacteriol. 168: 936–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shaw L, Golonka E, Potempa J, Foster SJ. 2004. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150: 217–228 [DOI] [PubMed] [Google Scholar]
- 64. Shaw LN, et al. 2006. Investigations into sigmaB-modulated regulatory pathways governing extracellular virulence determinant production in Staphylococcus aureus. J. Bacteriol. 188: 6070–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shaw LN, et al. 2008. Identification and characterization of sigma(S), a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One 3: e3844 doi:10.1371/journal.pone.0003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Spencer JP, et al. 1996. Base modification and strand breakage in isolated calf thymus DNA and in DNA from human skin epidermal keratinocytes exposed to peroxynitrite or 3-morpholinosydnonimine. Chem. Res. Toxicol. 9: 1152–1158 [DOI] [PubMed] [Google Scholar]
- 67. Staron A, et al. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74: 557–581 [DOI] [PubMed] [Google Scholar]
- 67a. Sullivan MA, Yasbin RE, Young FE. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29: 21–26 [DOI] [PubMed] [Google Scholar]
- 68. Tao LA, Wu XQ, Sun BL. 2010. Alternative sigma factor sigma(H) modulates prophage integration and excision in Staphylococcus aureus. PLoS Pathog. 6(5): e1000888 doi:10.1371/journal.ppat.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thackray PD, Moir A. 2003. SigM, an extracytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185: 3491–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Traber K, Novick R. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol. Microbiol. 59: 1519–1530 [DOI] [PubMed] [Google Scholar]
- 71. Truong-Bolduc QC, Hooper DC. 2010. Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus. J. Bacteriol. 192: 2525–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Voureka A. 1952. Induced variations in a penicillin-resistant staphylococcus. J. Gen. Microbiol. 6: 352–360 [DOI] [PubMed] [Google Scholar]
- 73. Walthers D, et al. 2007. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol. Microbiol. 65: 477–493 [DOI] [PubMed] [Google Scholar]
- 74. Watson SP, Clements MO, Foster SJ. 1998. Characterization of the starvation-survival response of Staphylococcus aureus. J. Bacteriol. 180: 1750–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Willetts NS. 1967. The elimination of Flac+ from Escherichia coli by mutagenic agents. Biochem. Biophys. Res. Commun. 27: 112–117 [DOI] [PubMed] [Google Scholar]
- 76. Yeager CM, Bottomley PJ, Arp DJ. 2001. Requirement of DNA repair mechanisms for survival of Burkholderia cepacia G4 upon degradation of trichloroethylene. Appl. Environ. Microbiol. 67: 5384–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]