Abstract
Here, we provide evidence that YqjD, a hypothetical protein of Escherichia coli, is an inner membrane and ribosome binding protein. This protein is expressed during the stationary growth phase, and expression is regulated by stress response sigma factor RpoS. YqjD possesses a transmembrane motif in the C-terminal region and associates with 70S and 100S ribosomes at the N-terminal region. Interestingly, E. coli possesses two paralogous proteins of YqjD, ElaB and YgaM, which are expressed and bind to ribosomes in a similar manner to YqjD. Overexpression of YqjD leads to inhibition of cell growth. It has been suggested that YqjD loses ribosomal activity and localizes ribosomes to the membrane during the stationary phase.
INTRODUCTION
In nature, bacteria survive in various environments, usually under stress conditions including nutrient starvation, temperature shock, osmolarity changes, and sudden changes in pH. For prolonged survival under such conditions, ordered expression of many stringent-response genes is required. The expression patterns of a large fraction of genes on the genome are altered by turning off or reducing expression of growth-related genes while switching on a set of genes that are required for adaptation to the specific stress condition. Changes in global patterns of gene expression in the stationary growth phase in organisms such as Escherichia coli involve drastic changes in cellular machineries, including changes in nucleoid conformation, transcription apparatus, and translation machinery (5, 9). From the variety of changes in the bacterial cell, we focused our attention on ribosomal changes in E. coli cells during transition from the exponential to the stationary phase (1, 14). Ribosomes are universally conserved ribonucleoproteins and are comprised of two asymmetric subunits. In bacteria, large (50S) and small (30S) subunits associate to form functional 70S ribosomes. Ribosomes can account for as much as 45% of the total mass of bacterial cells in organisms such as E. coli during the exponential phase and actively synthesize all of the cellular proteins required. However, in cells under stress conditions, such as starvation, it has been shown that ribosomal biosynthesis is repressed and that protein synthesis is also suppressed. These systems, which allow translational regulation, are very important for bacteria to survive in harsh environments.
In eukaryotes, it is known that phosphorylation of initiation factor 2α (eIF2α) is an adaptive mechanism for downregulating protein synthesis under stress conditions (7). In the Gammaproteobacteria, which includes E. coli, protein synthesis is mainly suppressed by the formation of 100S ribosomes (21). The 100S ribosome is a dimer of 70S ribosomes which is formed by the binding of ribosome modulation factor (RMF) to ribosomes (13). RMF is a small (Mr of 6,507), basic (pI 11.3) protein, and its expression increases remarkably during transition from the exponential phase to the stationary phase. Another protein factor expressed during the stationary phase, hibernation promoting factor ([HPF] also known as YhbH), also binds to ribosomes and promotes 100S ribosome formation (14, 18). In previous studies, it has been shown that RMF inactivates ribosomes by covering the peptidyl transferase center and entrance of the peptide exit tunnel (22, 23). This interconversion system between 70S and 100S ribosomes is an important strategy for bacterial survival under stress conditions. The ribosomal resting stage, the stage of 100S ribosome formation, is incorporated into the ribosome cycle and is called a hibernation stage (24). As seen above, the activity and conformation of ribosomes are altered by several protein factors in response to environmental stresses. However, the whole picture of ribosomal stress responses has not yet been elucidated.
In this study, we diligently search for novel E. coli proteins which associate to ribosomes during the stationary phase using two-dimensional gel electrophoresis. From the results, it was found that the YqjD protein (101 amino acids; Mr of 11,052, pI 9.1), with unknown physiological functions, associates to ribosomes during the stationary phase. YqjD associates with 70S and 100S ribosomes at the N-terminal region, and expression is regulated by RNA polymerase sigma factor RpoS (σs) for transcription of stationary-phase-specific genes. Interestingly, overexpression of YqjD leads to growth inhibition, in a manner similar to that of RMF; hence, there is a possibility that this protein inactivates ribosomes. A predicted secondary structure for YqjD shows that this is a membrane binding protein having a transmembrane helix in the C-terminal region. In fact, YqjD was found in the inner membrane fraction separated by centrifugation. The experimental results of this study indicate that the E. coli YqjD protein is an inner membrane protein associated with stationary-phase ribosomes, which may localize a part of the ribosome to the membrane during the stationary phase.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli W3110, a wild-type K-12 strain, was used in the main experiments. For mutational analysis, the deletion mutants of yqjD (ΔyqjD) and rpoS (ΔrpoS) (BW25113-derived strains) were obtained from the Keio collection (systematic knockout strain of E. coli K-12) (2). The coding sequences for full-length YqjD (full-YqjD; residues 1 to 101), YqjD with a deletion of the C terminus (ΔC-YqjD; residues 1 to 76), and YqjD with a deletion of the N terminus (ΔN-YqjD; residues 13 to 101), with and without N-terminal hexahistidine (His6) tags, were inserted into the E. coli expression vector pET30 (Novagen), which was transformed into E. coli BL21 cells. For prolonged culture, E. coli cells were grown in medium E containing 2% polypeptone and supplemented with 0.5% glucose at 37°C with shaking at 120 cycles per minute (19). Cells in the stationary phase were harvested after 2 days of incubation because the amounts of YqjD in cells peaked at 2 days. For purification of overexpressed proteins, cells with plasmids were grown in Luria broth at 37°C and harvested 1 h after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). His-tagged proteins were purified by a column filled with 1 ml of nickel-nitrilotriacetic acid-agarose (Ni-NTA Hi-Trap column; GE Healthcare) and then dialyzed against association buffer [100 mM CH3COONH4, 15 mM (CH3COO)2Mg · 4H2O, 20 mM Tris-HCl at pH 7.6, and 6 mM 2-mercaptoethanol] for use in an in vitro assay and stored at −80°C until use.
Preparation of ribosomes.
Each pellet of harvested cells was ground with an approximately equal volume of quartz sand and then suspended in association buffer. Homogenate was centrifuged at 15,000 rpm for 10 min at 4°C. The pellet was saved for use as a fraction of cell debris (CD). The supernatant was centrifuged at 4°C in a 55.2Ti rotor (Beckman) at 50,000 rpm for 1.5 h. The supernatant was saved for use as a fraction of the postribosomal supernatants (PRS). The pellet was resuspended in association buffer and saved for a fraction of the crude ribosomes (CR). The CR solution was layered onto a 5 to 20% linear sucrose density gradient in association buffer and centrifuged in an SW41Ti (Beckman) rotor at 40,000 rpm for 1.5 h at 4°C. The solution after centrifugation was fractionated, and absorbance at 260 nm was measured with a UV-1800 spectrophotometer (Shimadzu, Japan). The fraction containing ribosomes was passed through a filter (Amicon Ultracel-100K; Millipore) to eliminate sucrose, and the upper solution was saved for use as a ribosome fraction (RF). To obtain ribosomal 30S and 50S subunits, the buffer solution of RF was changed to dissociation buffer [100 mM CH3COONH4, 1 mM (CH3COO)2Mg · 4H2O, 20 mM Tris-HCl at pH 7.6, and 6 mM 2-mercaptoethanol] and incubated at 37°C for 20 min; the solution was layered onto a 5 to 20% linear sucrose density gradient in dissociation buffer and centrifuged in the SW41Ti rotor at 40,000 rpm for 3 h at 4°C. The collection procedures of each fraction containing 30S and 50S were the same as described above.
2D gel electrophoresis (RFHR method).
Proteins in each fraction (CD, CR, PRS, and RF) were extracted by the acetic acid method (6). One-tenth volume of 1 M MgCl2 and two volumes of acetic acid were added to each fraction, and the mixture was stirred for 1 h at 4°C. After centrifugation at 20,000 × g for 10 min, the supernatant was dialyzed against 2% acetic acid three times. Proteins were lyophilized and stored at −80°C until use. Eighty micrograms of the lyophilized protein was analyzed by radical-free and highly reducing (RFHR) two-dimensional PAGE (2D-PAGE) (20). Protein spots on gels were scanned with a GS-800 calibrated densitometer (Bio-Rad Laboratories Inc.). The optical density values of proteins (YqjD, YfiA, HPF, RMF, and the stationary-phase-induced ribosome-associated protein [SRA]) were calculated as a function of their molecular weights, and values were normalized against the value for ribosomal protein S2 on the gel. Ribosomal protein S2 was previously defined to exist as a unit copy protein (17), so it was used as a marker to estimate the copy number of each protein. Here, “copy number” means the molar ratio of the molecule to the 70S ribosome, 50S subunit, or 30S subunit.
Western blotting.
The sample of ribosomes was fractionated by 5 to 20% linear sucrose density gradient centrifugation (SDGC). Proteins in each fraction were precipitated with 10% trichloroacetic acid (TCA), separated by Tricine-SDS-PAGE on 15% gels (16), and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-FL transfer membrane; Millipore). YqjD was recognized by rabbit polyclonal antibodies against the synthetic peptide corresponding to residues 25 to 54 of YqjD or the hexahistidine tag, which were detected with ECF substrate (GE Healthcare) using a Typhoon FLA 9000 imager (GE Healthcare).
Fractionation of proteins containing inner and outer membranes.
To determine the localization of YqjD in cells, proteins containing inner and outer membranes were fractionated by differential centrifugation (4). Cells of the W3110 strain cultured for 2 days were suspended in TE buffer (50 mM Tris-HCl at pH 8.0, 5 mM EDTA) containing protease inhibitor cocktail (P8465; Sigma-Aldrich) and DNase (D5025; Sigma-Aldrich), and cells were then lysed with lysozyme (122-02673; Wako). Unlysed cells and cell debris were removed by centrifugation at 4,000 rpm for 5 min at 4°C. Supernatant was centrifuged at 4°C in a 55.2Ti rotor at 50,000 rpm for 1.5 h. Membrane proteins (pellet) were resuspended in 2 ml of TE buffer, and the suspension was loaded on a 20-ml sucrose step gradient prepared with 10 ml of 70% (wt/vol) sucrose in TE buffer layered with 10 ml of 53% (wt/vol) sucrose in TE buffer. The gradient was then centrifuged at 4°C in a 55.2Ti rotor at 55,000 rpm for 5 h. The upper band (inner membrane proteins) and lower band (outer membrane proteins) were collected from the top of the gradient. The material from each band was concentrated, and sucrose was removed using a filter (Amicon Ultracel-3K; Millipore), which was then solubilized in sodium dodecyl sulfate (SDS) sample buffer.
RESULTS
YqjD is expressed during the stationary growth phase and is contained in fractions of ribosomes and cell debris.
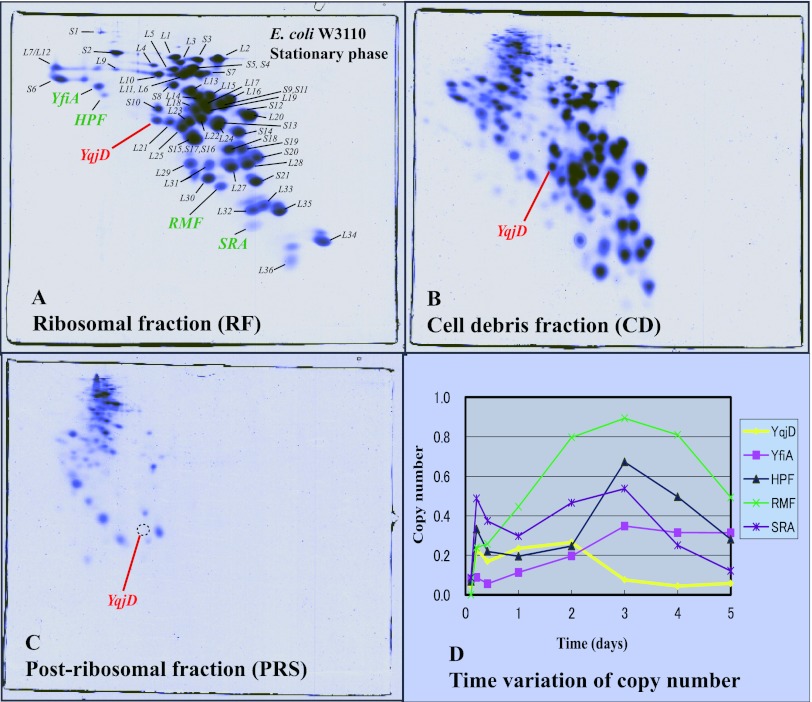
In order to search for proteins expressed during the stationary phase in E. coli, proteins in some fractions were analyzed by two-dimensional gel electrophoresis (RFHR method) (see Materials and Methods). Figure 1A shows a gel in which proteins extracted from the ribosome fraction (RF) of the W3110 strain during the stationary phase were separated. RFHR 2D-PAGE has an excellent analytical capability for basic proteins, and all E. coli ribosomal proteins including L34 (pI 13.1) are shown on the gel. Also, ribosome binding proteins expressed during the stationary phase, YfiA (Mr of 12,785, pI 6.5), HPF (Mr of 10,751, pI 6.9), RMF, and SRA (Mr of 5,095, pI 11.1), existed in this fraction. YfiA is a paralogous protein of HPF, which inhibits the formation of 100S ribosomes (14, 18). The function of SRA (stationary-phase-induced ribosome-associated protein) is as yet unknown (10). In this study, we noticed that a protein existed under a spot of S10 in this gel with stationary-phase-specific expression. This protein was identified as YqjD by mass spectrometry, which was functionally unknown. The existence of YqjD in the RF (Fig. 1A) suggests the possibility that this is a ribosome binding protein similar to RMF. YqjD also existed in a fraction of cell debris (CD), as shown in Fig. 1B. Ribosomal proteins contained in CD are generally regarded as those from generating or degrading ribosomes because the amounts of ribosomal proteins on the gel were atypical. Insoluble and membrane proteins were also contained in this fraction, which suggests the possibility that YqjD is a membrane binding protein. YqjD was not contained in a fraction of the postribosomal supernatants (PRS), proteins in the cytoplasm without ribosomal proteins, as shown in Fig. 1C. This means that YqjD does not exist in a free state. Figure 1D shows time variations of copy numbers (the molar ratio of the molecule binding to ribosomes) of YqjD, YfiA, HPF, RMF, and SRA. These proteins were not expressed during the exponential phase (cultured for 2.5 h) but increased concomitantly with the transition from the exponential phase to the stationary phase. The expression of YqjD reached a maximum at 2 days, but expression levels of other proteins reached a maximum at 3 days. It is well known that the expression levels of many proteins that increase from the early stationary phase are regulated by transcription sigma factor RpoS (8). Therefore, expression of YqjD in a mutant with a deletion of the rpoS gene was examined by a comparison of 2D-PAGE gels (see Fig. S1 in the supplemental material). From the results, YqjD was not expressed in the rpoS mutant. The above results suggest that E. coli YqjD is a ribosome and membrane binding protein and that expression is regulated by RpoS.
Fig 1.
Proteomic analysis of proteins extracted from the E. coli W3110 strain of the stationary growth phase (cultured for 2 days) separated by RFHR 2D-PAGE. (A) Proteins contained in the ribosomal fraction (RF). All ribosomal proteins are shown in the gel, and the positions of spots are indicated. The well-known proteins that bind to ribosomes during the stationary phase, YfiA, HPF, and SRA, exist in this fraction. A protein found in this study, YqjD, is shown in red. (B) Proteins in the cell debris (CD) fraction. YqjD also exists in this fraction. (C) Proteins in the postribosomal fraction (PRS). There is no YqjD in this fraction. (D) Time variations of copy numbers of ribosome binding proteins expressed during the stationary phase. Cells were harvested after 2.5 h, 5 h, 10 h, 1 day, 2 days, 3 days, 4 days, and 5 days of incubation.
YqjD has a transmembrane motif in the C-terminal region.
Homologous proteins of YqjD were searched for using the BLAST program (http://www.genome.jp/tools/blast/). From the results, only YqjD of Shigella flexneri was hit. It has been reported that S. flexneri is closely related to E. coli and belongs to the same genus (12). It has been speculated that YqjD is a protein that closely related organisms of E. coli possess. On the other hand, two paralogous proteins of YqjD, ElaB (101 amino acids; Mr of 11,306, pI 5.3) and YgaM (113 amino acids; Mr of 12,288, pI 8.0), were picked up as paralogous proteins by the BLAST search, as shown in Fig. S2 in the supplemental material. Similar to YqjD, the functions of these paralogous proteins are unknown. The gene expression levels of these proteins were checked using a real-time PCR method. The results showed that the expression patterns of elaB and ygaM were found to be the same as expression of yqjD (see Fig. S3). Although the spots of these proteins were not found on the 2D-PAGE gel, these paralogous proteins may start to be expressed from the early stationary phase in a similar manner to YqjD.
Interestingly, a program for the prediction of the secondary structure of proteins, the SOSUI system (http://bp.nuap.nagoya-u.ac.jp/sosui/), shows that YqjD has a transmembrane motif in the C-terminal region (residues 77 to 98) (marked with an asterisk Fig. S2 in the supplemental material). ElaB and YgaM also have transmembrane motifs in the C-terminal region (residues 78 to 99 and 89 to 110, respectively). Although the possibility that YqjD is a membrane binding protein is shown in Fig. 1C, the C-terminal region of YqjD may relate to a membrane binding function.
YqjD binds to 70S and 100S ribosomes.
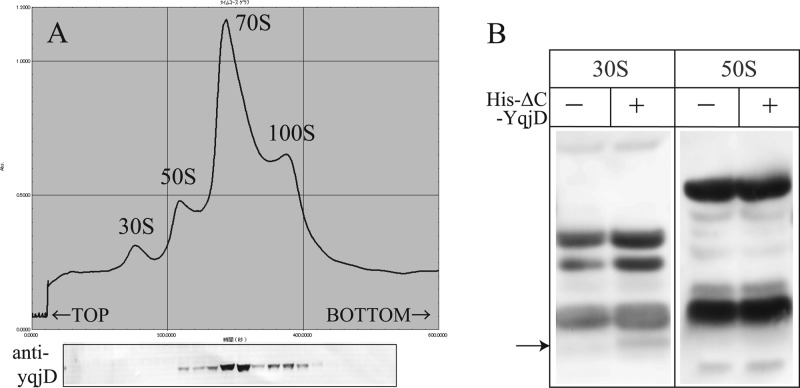
Figure 2A shows a ribosome profile of CR extracted from the W3110 strain during the stationary phase (cultured for 2 days) by sucrose density gradient centrifugation (SDGC). A peak of the 100S ribosome is seen in addition to the typical peaks of 30S, 50S, and 70S. The gradient was fractionated, and the proteins in each fraction were separated by Tricine-SDS-PAGE. YqjD in each fraction was detected by Western blotting using an antibody against YqjD, as shown in Fig. 2B (arrow). From the results, we see that YqjD existed in the fractions including the 70S and 100S ribosomes. This result indicates that YqjD binds to ribosomes.
Fig 2.
(A) Ribosome profile of E. coli during the stationary phase. YqjD in each fraction is detected by Western blotting using an antibody against YqjD. (B) Western blots of His-ΔC-YqjD in the fractions of 30S and 50S subunits. Loaded samples are proteins extracted from the 30S and 50S fractions of each subunit (−) and mixtures of each subunit and His-ΔC-YqjD (+) separated by SDGC. The arrow indicates the position of His-ΔC-YqjD.
Next, in order to examine whether YqjD binds to the 30S or 50S subunit, binding assays of YqjD to each subunit in vitro were performed. Ribosomes prepared from the deletion mutants of yqjD (ΔyqjD) were dissociated to 30S and 50S subunits by SDGC in dissociation buffer. His-ΔC-YqjD (His6-tagged YqjD truncated in the C-terminal region [residues 77 to 101]) was purified and used in this assay because the full-length YqjD was fairly insoluble (data not shown). Each of the 30S and 50S subunits was mixed with His-ΔC-YqjD in vitro and incubated at 37°C for 20 min. The reactants were divided into fractions of 30S and 50S subunits by SDGC again, and His-ΔC-YqjD in each fraction was detected by Western blotting using an antibody against His6. From the results, His-ΔC-YqjD existed in the 30S subunit fraction, as shown in Fig. 2B. Moreover, His-ΔN-YqjD (His6-tagged YqjD truncated in the N-terminal region [residues 1 to 12]) could not bind to ribosomes in the same assay (data not shown). This means that the N-terminal region of YqjD relates to the function of ribosome binding. The above results suggest that YqjD binds to the 30S subunit in 70S and 100S ribosomes at the N-terminal region.
It is well known that the behavior of protein factors binding to ribosomes depends on the ionic strength of the buffer solution. For example, RMF is released from ribosomes in a high-salt buffer [1 M CH3COONH4, 15 mM (CH3COO)2 Mg · 4H2O, 20 mM Tris-HCl at pH 7.6, and 6 mM 2-mercaptoethanol], while it associates with the 50S subunit in dissociation buffer (21). YfiA and HPF can bind to ribosomes in a high-salt buffer, while they are released from ribosomes in dissociation buffer (14). SRA can bind to ribosomes in both buffers (10). YqjD can bind to the 30S subunit in dissociation buffer, as shown in Fig. 2B, but it is released from ribosomes in a high-salt buffer (data not shown). These findings suggest that the affinity of YqjD, as well as RMF, to ribosomes is relatively low.
There is a possibility that ElaB and YgaM, paralogous proteins of YqjD, also bind to ribosomes. In order to confirm this assumption, we examined whether His6-tagged ElaB and YgaM bind to ribosomes. The overexpressing cells of these proteins were provided by the E. coli ASKA library (http://www.shigen.nig.ac.jp/ecoli/strain/top/top.jsp), and expression was induced by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The proteins contained in the RF of overexpressed cells were analyzed by RFHR 2D-PAGE. Results show that these proteins exist in the RF (see Fig. S4 in the supplemental material), which indicates that ElaB and YgaM bind to ribosomes in a similar manner as YqjD.
YqjD is an inner membrane binding protein.
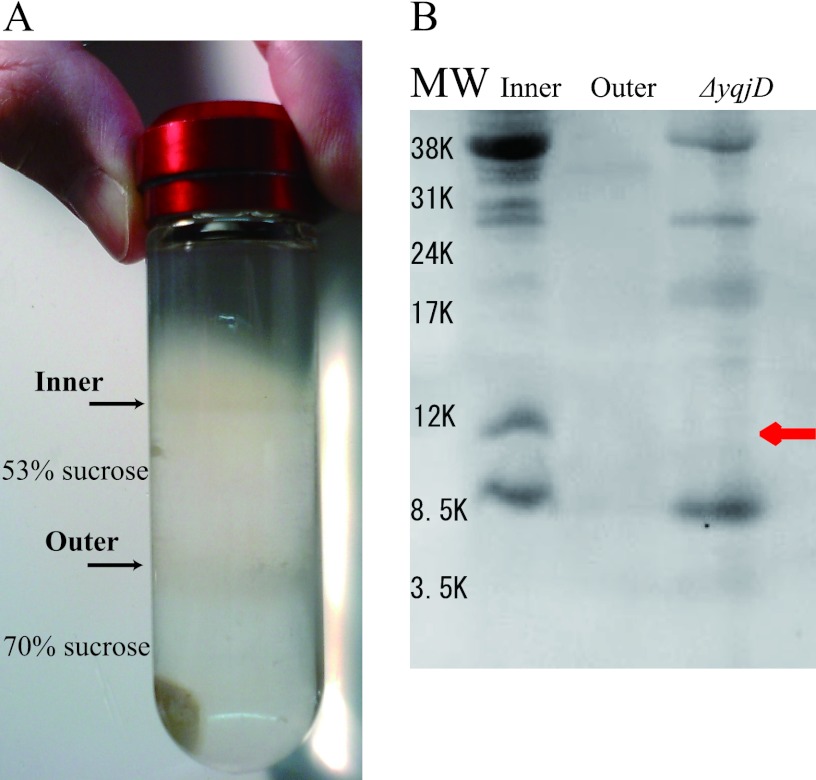
It has been speculated that YqjD is a membrane binding protein because YqjD exists in the CD fraction, as shown in Fig. 1B, and possesses the transmembrane motif in the C-terminal region, as shown in Fig. 2. In order to confirm this speculation, we examined whether YqjD was contained in an inner or outer membrane fraction. Membrane proteins prepared from the W3110 strain cultured for 2 days were separated into the inner and outer membrane fractions by successive centrifugation steps (see Materials and Methods). Figure 3A shows that inner and outer membrane proteins were contained in the upper and lower bands, respectively (4). Proteins contained in each fraction were resolved by Tricine-SDS-PAGE, and YqjD was detected by Western blotting using anti-YqjD, as shown in Fig. 3B. Membrane proteins extracted from ΔyqjD cells were also analyzed as controls. The results show that YqjD existed in the inner membrane fraction, as indicated by a red arrow, but not in the outer membrane fraction and in ΔyqjD cells. These results demonstrate that YqjD is an inner membrane binding protein.
Fig 3.
Localization of YqjD in the cell. (A) Sucrose step gradient after centrifugation, which was prepared with 70% (wt/vol) sucrose in TE buffer layered with 53% (wt/vol) sucrose in TE buffer. Inner membrane proteins are contained in the upper band on 53% sucrose, and outer membrane proteins are contained in the lower band on 70% sucrose. (B) Western blotting using polyclonal anti-YqjD for inner and outer membrane fractions. As a control, the membrane proteins of ΔyqjD cells are used. The band of YqjD is indicated by a red arrow. MW, molecular weight.
YqjD inhibits cell growth by binding to ribosomes.
To investigate the role of YqjD in the cell, we compared the features of the ΔyqjD mutant and the parental strain cultured for 2 days. The results of 2D-PAGE show that there were no significant differences in their growth rates, the shapes of cells observed by light microscopy, the ribosomal profiles by SDGC, or protein profiles except for the spot of YqjD (data not shown).
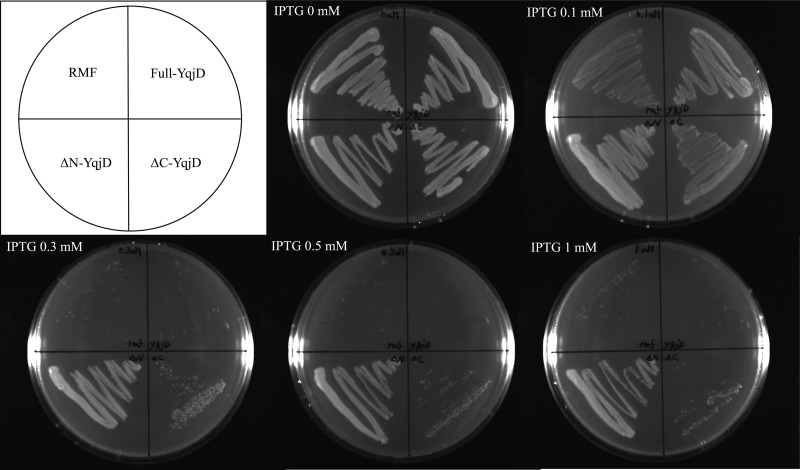
Next, cells that overexpressed the full-length YqjD protein (residues 1 to 101), ΔC-YqjD (residues 1 to 76), or ΔN-YqjD (residues 13 to 101) were cultivated on agar plates with several IPTG concentrations, as shown in Fig. 4. As a control, cells that overexpressed RMF were also cultivated. The results show that the growth of cells that overexpressed full-YqjD, ΔC-YqjD, and RMF was inhibited with an increase in IPTG concentrations but that of ΔN-YqjD was not inhibited. From the results shown in Fig. 2 and in Fig. S2 in the supplemental material, it can be suggested that the C-terminal and N-terminal regions of YqjD are involved in the functions of membrane binding and ribosome binding, respectively. Therefore, it can be interpreted that cell growth is inhibited by binding of YqjD to ribosomes at the N-terminal region. On the other hand, it has been reported that the overexpression of RMF inhibits E. coli cell growth by binding to the peptidyl transferase center in ribosomes (22, 23). The phenomenon of growth inhibition by overexpression of full-length YqjD or ΔC-YqjD, as shown in Fig. 4, is very similar to that of RMF. This fact supports a hypothesis that YqjD inactivates ribosomes by binding at the N-terminal region.
Fig 4.
Effects of YqjD overexpression on cell growth. Cells that overexpress full-YqjD (residues 1 to 101), ΔC-YqjD (residues 1 to 76), ΔN-YqjD (residues 13 to 101), or RMF were cultured overnight in Luria broth medium at 37°C, dispersed on agar plates with several IPTG concentrations (0, 0.1, 0.3, 0.5, and 1 mM), and incubated overnight at 37°C.
DISCUSSION
In this study, we provided the characterization of YqjD defined as an E. coli hypothetical protein as follows. (i) E. coli starts to express YqjD from the early stationary phase (Fig. 1D). The amount of YqjD peaks at 2 days of cell incubation and then decreases. In a previous study, it has been reported that yqjD was one of the RpoS-regulated genes identified by microarray analysis (15). In this study, it was demonstrated that the expression of this protein was regulated by the stress response sigma factor RpoS (see Fig. S1 in the supplemental material). (ii) It is suggested that YqjD binds to the 30S subunit in 70S and 100S ribosomes at the N-terminal region (Fig. 1A, 2, and 4). In a previous study, this protein was identified in proteomic data using mass spectrometric analysis as one of the proteins existing in the 30S fraction (11). In this study, it was revealed that the paralogous proteins of YqjD, ElaB and YgaM, also bind to ribosomes (see Fig. S4). (iii) It was predicted that YqjD, ElaB, and YgaM possessed transmembrane motifs in their C-terminal regions (residues 77 to 98, 78 to 99, and 89 to 110, respectively) (see Fig. S2). It was demonstrated that YqjD bound to the inner membrane (Fig. 1B and 3). (iv) Overexpression of YqjD caused the inhibition of cell growth (Fig. 4). This phenomenon can be explained by assuming that YqjD inactivates ribosomes by binding at the N-terminal region.
The peak of binding numbers of YqjD per ribosome was about 0.2 copies, as shown in Fig. 1D. However, the amount of YqjD in cells must be quite large because YqjD in CD existed in relatively large quantities, as shown in Fig. 1B. Ribosomes probably cannot bind to all of the YqjD in the inner membrane because the affinity of YqjD to ribosomes is low enough that the protein is released from them in a high-salt buffer. Although YqjD bound to free ribosomes, as shown in Fig. 1A, this binding may have been caused by excess cell breakage.
To elucidate the function of YqjD, the features of the ΔyqjD mutant and the parental strain have been examined. The results show that there were no significant differences in their phenotypes. The paralogous proteins of YqjD, ElaB and YgaM, may function complementarily because these proteins can be expressed during the stationary phase, can bind to the ribosome, and probably bind to the membrane in a manner similar to that of YqjD (see Fig. S3 and S4 in the supplemental material). If the assumption that the paralogous protein works as a backup is correct, the function of YqjD may be very important for cells. One of its important functions may be the inactivation of ribosomes. It is thought that inhibition of ribosomal activity by YqjD is reasonable during the stationary phase, the same as 100S ribosome formation. However, it is not clear why YqjD has to anchor a part of the ribosome to the inner membrane. The keywords “ribosome binding” and “membrane binding” remind us of the signal recognition particle (SRP). The SRP conveys ribosomes synthesizing inner membrane proteins to the SecYEG translocon in the plasma membrane (3). This raises the possibility that YqjD may be involved in the synthesis of some membrane proteins during the early stationary phase although YqjD itself may inactivate ribosomes.
In conclusion, YqjD is expressed under the regulation of RpoS during the stationary phase and is set in the inner membrane at the C-terminal region. A part of the ribosome (70S and 100S) may be localized to the membrane by binding to the N-terminal region of YqjD. It is suggested that the ribosome binding with YqjD loses its translational activity. Future studies should aim to elucidate details of the biological function of YqjD.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by a Grant-in-Aid for Scientific Research to H.Y. (20570167) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aiso T, Yoshida Wada HA, Ohki R. 2005. Modulation of mRNA stability participates in stationary-phase specific expression of ribosome modulation factor. J. Bacteriol. 187: 1951–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 2006.0008 doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bornemann T, Jockel J, Rodnina MV, Wintermeyer W. 2008. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15: 494–499 [DOI] [PubMed] [Google Scholar]
- 4. Castanie-Cornet MP, Cam K, Jacq A. 2006. RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 188: 4264–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frenkiel-Krispin D, et al. 2004. Nucleoid restructuring in stationary-state bacteria. Mol. Microbiol. 51: 395–405 [DOI] [PubMed] [Google Scholar]
- 6. Hardy SJ, Kurland CG, Voynow P, Mora G. 1969. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry 8: 2897–2905 [DOI] [PubMed] [Google Scholar]
- 7. Hinnebusch AG. 1994. Translational control of GCN4: an in vivo barometer of initiation factor activity. Trends Biochem. Sci. 19: 409–414 [DOI] [PubMed] [Google Scholar]
- 8. Hirsch M, Elliott T. 2005. Stationary-phase regulation of RpoS translation in Escherichia coli. J. Bacteriol. 187: 7204–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ishihama A. 1999. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells 4: 135–143 [DOI] [PubMed] [Google Scholar]
- 10. Izutsu K, et al. 2001. Escherichia coli ribosome-associated protein SRA, whose copy number increases during stationary phase. J. Bacteriol. 183: 2765–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang M, et al. 2007. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J. Bacteriol. 189: 3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jin Q, et al. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K-12 and O157. Nucleic Acids Res. 30: 4432–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kato T, et al. 2010. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18: 719–724 [DOI] [PubMed] [Google Scholar]
- 14. Maki Y, Yoshida H, Wada A. 2000. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5: 965–974 [DOI] [PubMed] [Google Scholar]
- 15. Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272: 580–591 [DOI] [PubMed] [Google Scholar]
- 16. Schaggar H, von Jagow G. 1987. Tricine-sodium dodecylsulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379 [DOI] [PubMed] [Google Scholar]
- 17. Tal M, Weissman I, Silberstein A. 1990. A new method for stoichiometric analysis of proteins in complex mixture. Re-evaluation of the stoichiometry of. E. coli ribosomal proteins. J. Biochem. Biophys. Methods 21: 247–266 [DOI] [PubMed] [Google Scholar]
- 18. Ueta M, et al. 2005. Ribosome binding proteins YfiA and YhbH have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10: 1103–1112 [DOI] [PubMed] [Google Scholar]
- 19. Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218: 97–106 [PubMed] [Google Scholar]
- 20. Wada A. 1986. Analysis of Escherichia coli ribosomal proteins by an improved two dimensional gel electrophoresis. I. Detection of four new proteins. J. Biochem. 100: 1583–1594 [DOI] [PubMed] [Google Scholar]
- 21. Wada A, Yamazaki Y, Fujita N, Ishihama A. 1990. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary phase Escherichia coli cells. Proc. Natl. Acad. Sci. U. S. A. 87: 2657–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshida H, et al. 2002. The ribosome modulation factor (RMF) binding site on the 100S ribosome of Escherichia coli. J. Biochem. 132: 983–989 [DOI] [PubMed] [Google Scholar]
- 23. Yoshida H, Yamamoto H, Uchiumi T, Wada A. 2004. RMF inactivates ribosomes by covering the peptidyl transferase center and entrance of peptide exit tunnel. Genes Cells 9: 271–278 [DOI] [PubMed] [Google Scholar]
- 24. Yoshida H, Ueta M, Maki Y, Sakai A, Wada A. 2009. Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase. Genes Cells 14: 271–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.