Abstract
Most antibodies that broadly neutralize HIV-1 are highly somatically mutated in antibody clonal lineages that persist over time. Here, we describe the analysis of human antibodies induced during an HIV-1 vaccine trial (GSK PRO HIV-002) that used the clade B envelope (Env) gp120 of clone W6.1D (gp120W6.1D). Using dual-color antigen-specific sorting, we isolated Env-specific human monoclonal antibodies (MAbs) and studied the clonal persistence of antibodies in the setting of HIV-1 Env vaccination. We found evidence of VH somatic mutation induced by the vaccine but only to a modest level (3.8% ± 0.5%; range 0 to 8.2%). Analysis of 34 HIV-1-reactive MAbs recovered over four immunizations revealed evidence of both sequential recruitment of naïve B cells and restimulation of previously recruited memory B cells. These recombinant antibodies recapitulated the anti-HIV-1 activity of participant serum including pseudovirus neutralization and antibody-dependent cell-mediated cytotoxicity (ADCC). One antibody (3491) demonstrated a change in specificity following somatic mutation with binding of the inferred unmutated ancestor to a linear C2 peptide while the mutated antibody reacted only with a conformational epitope in gp120 Env. Thus, gp120W6.1D was strongly immunogenic but over four immunizations induced levels of affinity maturation below that of broadly neutralizing MAbs. Improved vaccination strategies will be needed to drive persistent stimulation of antibody clonal lineages to induce affinity maturation that results in highly mutated HIV-1 Env-reactive antibodies.
INTRODUCTION
Development of an effective vaccine against HIV-1 is a global priority (22). While many candidate vaccines have been developed, to date only four efficacy trials in humans have been carried out, and only one, the ALVAC/HIV AIDSVax B/E RV144 trial, demonstrated an estimated vaccine efficacy of 31% (42). One hypothesis is that vaccine-induced antibodies (Abs) mediated a component of the protective response in the RV144 trial (17). Thus, strategies that induce protective antibody responses of greater breadth and magnitude than those found in RV144 are needed.
Broadly neutralizing antibodies are induced in approximately 15 to 20% of chronically HIV-1-infected subjects (31, 47), and these antibodies are generally highly mutated compared to antibodies induced by most viral infections (34, 55). These observations have led to a hypothesis that prolonged antigenic stimulation of persistent clonal lineages is required to induce broadly neutralizing antibodies against HIV-1 (17a, 55, 62; also W. Chen et al., presented at the AIDS Vaccine 2008 Conference, Cape Town, South Africa, 13 to 16 October 2008). In animal models, repeated immunization has been shown to recruit both previously stimulated memory B cells and naïve B cells in response to antigens (13), but in humans it is unknown whether repeated immunization recruits new clonal lineages or continues to mature previously recruited clonal lineages. It is known that with repeated stimulation, somatic hypermutation of germinal center B cell lineages can result in the development of populations of autoreactive B cells (30, 44, 48) while other B cells may lose affinity for antigen or even the capacity to express antibody (4, 20, 65); both of these processes can lead to extinction of developing clonal lineages (4, 20, 65). That there is a limit to the level of somatic hypermutation achievable by vaccination is suggested by studies of influenza vaccination where the mean mutation frequency of antibodies isolated from humans is ∼6% (34). This suggests that in the setting of vaccination, sequential recruitment rather than persistence of B cell clones also occurs in humans (34).
The technology to isolate antigen-specific human memory B cells and express recombinant monoclonal antibodies (rMAbs) provides a means by which to study HIV-1 antigen-induced affinity maturation (14, 26, 36). To date, this technology has not been used to study an HIV-1 vaccine trial. We report here the analysis of HIV-1 envelope gp120 vaccine-induced antibodies from a phase I/II trial, the GSK PRO HIV-002 trial, that used clade B HIV-1 clone W6.1D gp 120 (gp120W6.1D) envelope and a Nef-Tat fusion protein formulated with the Adjuvant System AS01B (24). This candidate vaccine induced serum antibodies that showed autologous HIV-1 Env pseudovirus neutralization and cellular immune responses. We have used single-cell memory B and plasma cell sorting with rMAb technology for the first time in the evaluation of an HIV-1 vaccine trial to isolate 34 rMAbs reflective of the vaccine-induced anti-HIV-1 serum antibody activity. We show that the gp120W6.1D immunogen stimulated persistent B cell clonal lineage maturation and, as well, recruited new B cell clonal lineages over the four vaccine immunizations.
MATERIALS AND METHODS
Ethics statement.
The clinical material used for the present study was obtained from GlaxoSmithKline Biologicals (GSK, Rixensart, Belgium) out of remaining study specimens. The present work was performed under a protocol approved by the Duke University Health System Institutional Review Board for Clinical Investigations. Subjects were recruited into the clinical study at the Center for Vaccinology in Ghent, Belgium, and the study was approved by the local independent ethics committee and conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines. Written informed consent was obtained from all subjects prior to study entry.
Subjects.
Subjects were immunized with gp120/Nef-Tat protein vaccine antigens formulated in AS01B in a study performed at the Center for Vaccinology in Ghent, Belgium, as described previously (24). Recombinant gp120W6.1D is a truncated form of a clade B HIV-1 envelope protein produced in a CHO cell line. The lyophilized pellet comprising 20 μg of Nef-Tat and 20 μg of gp120 prepared as described previously (12) was reconstituted with 0.5 ml of AS01B (liposome-based adjuvant system with 50 μg of monophosphoryl lipid A [MPL] and 50 μg of QS21) prior to administration. Subjects were immunized at 0, 1, 3, and 6 months and were followed for 24 months per protocol. Peripheral blood mononuclear cells (PBMC) and/or serum isolated at day 0, day 42, day 98, day 182, and day 672 was available for the present study.
Flow cytometry panel antibodies, recombinant proteins, and assay control antibodies.
HIV-1 Env gp120W6.1D identical to the protein used in the vaccine was obtained from GSK Biologicals (Rixensart, Belgium). Recombinant group M consensus gp140 protein (gp140ConS) was produced in recombinant vaccinia virus as a secreted protein as described previously (27). Flow cytometry was performed using a panel of antibodies reactive with the following cell surface molecules: CD138 (fluorescein isothiocyanate [FITC]), surface IgM (FITC), surface IgD (phycoerythrin [PE]), CD3 (PE-Cy5), CD16 (PE-Cy5), CD235a (PE-Cy5), CD20 (PE-Cy7), CD19 (allophycocyanin [APC]-Cy7), and CD27 (Pacific Blue) (all, BD Biosciences, San Jose, CA); CD14 (PE-Cy5) (Invitrogen, Carlsbad, CA); CD27 (PE-Cy7) and CD38 (APC-Alexa Fluor 700) (Beckman Coulter, Brea, CA).
HIV-1 Env gp120W6.1D or gp140ConS was labeled with Pacific Blue and Alexa Fluor 647 using fluorochrome labeling kits (Invitrogen, Carlsbad, CA). The proteins were quality controlled after conjugation for their ability to bind to CD4 expressed on the surface of the H9 T cell line.
Hyperimmune HIV-1 globulin (HIVIG) was obtained from the NIH AIDS Research and Reference Reagent Repository. Palivizumab, a humanized monoclonal antibody against the F protein of respiratory syncytial virus, was purchased from MedImmune, LLC (Gaithersburg, MD).
Antibody reactivity by binding antibody multiplex assay, enzyme-linked immunosorbent assay (ELISA), and indirect immunofluorescence.
Serum samples and expressed rMAbs were studied for reactivity to HIV-1 antigens using a standardized custom binding antibody multiplex assay using Luminex (51). This assay was also used to determine isotype-specific binding antibody titers in serum samples from days 0, 42, 98, and 182. IgG subclass responses specific for HIV-1 Nef (GSK Biologicals, Rixensart, Belgium), HIV-1 Tat (GSK Biologicals, Rixensart, Belgium), and HIV-1 Env gp120W6.1D were determined as described previously (63). For enhanced detection of IgA, serum samples were depleted of IgG using protein G (49, 51). Bound IgG was then eluted and used for antibody-dependent cellular cytotoxicity (ADCC) assays. IgA antibodies were detected with goat anti-human IgA (Jackson ImmunoResearch, West Grove, PA). A constant HIV-positive (HIV+) serum titration was utilized as a positive control, and negative controls were included in every assay. All assays were run under conditions compliant with Good Clinical Laboratory Practice, including tracking of positive controls by Levy-Jennings charts. Positivity criteria for antibody-antigen pairs were determined using preimmune samples from study subjects. FDA-compliant software, Bio-Plex Manager, version 5.0 (Bio-Rad, Hercules, CA), was utilized for the analysis of specimens. For rMAb screening by binding antibody multiplex assays, rMAbs were screened against a panel of HIV-1 antigens (gp14000MSA_4076, gp12089.6, gp120A244, gp120W6.1D, Nef, Tat, gp41MN, gp120MN, p24, p31, p66); rMAbs that had a blank-bead-subtracted value greater than 20 units and greater than 10 times the rMAb IgG concentration in μg/ml were selected for further evaluation. Those rMAbs with blank-bead-subtracted values for Env antigens greater than 2,000 units and greater than 650 times the rMAb IgG concentration in μg/ml were considered positive; rMAbs with values below 2,000 were tested in a titration series to confirm Env reactivity.
ELISA screening of rMAbs was performed as described previously (3) against a panel of HIV-1 Envs (gp41MN, gp140JRFL, and gp120W6.1D). ELISA testing for antigens was considered positive if the optical density reading was above 0.3 units and greater than 4-fold over background. For epitope mapping, ELISA was performed in the same manner using a peptide array obtained from the NIH AIDS Reagent Repository (HIV-1 MN Env 15-mer complete set); binding was considered positive if the optical density reading was above 0.3 and was 4-fold over background.
Indirect immunofluorescence binding of rMAbs to HEp-2 cells (Inverness Medical Professional Diagnostics, Princeton, NJ) was performed as described previously (16). Briefly, 20 μl of antibody at 25 μg/ml was placed on a predetermined spot on the surface of an ANA HEp-2 kit slide, incubated for 25 min at room temperature, washed, and developed with 20 μl of goat anti-human Ig-FITC at 20 μg/ml (Southern Biotech, Birmingham, AL) for 25 min. For images of murine MAb 13D5, goat anti-mouse Ig-FITC at 20 μg/ml (Jackson ImmunoResearch, West Grove, PA) was used. Incubations were performed in humid chambers in the dark. Slides were washed and dried; a drop of 33% glycerol was placed on each spot prior to the fixing of coverslips. Images were taken on an Olympus AX70 with SpotFlex FX1520 charge-coupled device (CCD) with a UPlanFL 40× (numerical aperture, 0.75) objective at 25°C in the FITC channel using SPOT software. All images were acquired for the time specified in the figure legend. Image layout and scaling were performed in Adobe Photoshop without image manipulation.
Flow cytometric analysis and single-cell sorting.
Single-cell sorting was performed using a BD FACSAria or a BD FACSAria II (BD Biosciences, San Jose, CA), and the flow cytometry data were analyzed using FlowJo (Treestar, Ashland, OR). Plasma cells/plasmablasts were sorted as previously described (26) by gating on CD3− CD14− CD16− CD235a− (CD3/14/16/235a−) CD19+ CD20−/lo (CD20− or expressing low levels of CD20) CD27hi (expressing high levels of CD27) CD38hi cells. Antigen-specific memory B cells were identified by using antibodies in combination with gp120W6.1D or gp140ConS labeled with Alexa Fluor 647 and Pacific Blue; cells were gated on CD3− CD14− CD16− CD235a− CD19+ surface IgD− gp120W6.1D+/+. For both plasma cells and memory B cells, single cells were directly sorted into 96-well plates containing 20 μl per well of reverse transcription (RT) reaction buffer (5 μl of 5× first-strand cDNA buffer, 0.5 μl of RNaseOUT [Invitrogen, Carlsbad, CA], 1.25 μl of dithiothreitol, 0.0625 μl Igepal CA-630 [Sigma, St. Louis, MO], 13.25 μl of distilled H2O [dH2O; Invitrogen, Carlsbad, CA]); plates were stored at −80°C until use and again stored at −80°C until PCR was performed.
PCR isolation of Ig VH, Vκ, and Vλ Genes.
Single-cell PCR was performed as described previously (25, 26, 34, 45, 60). Briefly, reverse transcription (RT) was performed using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) and human constant region primers for IgG, IgA1, IgA2, IgM, IgD, Igκ, and Igλ. Separate reactions were used to amplify individual variable region heavy chain, (VH), Vκ, and Vλ families from the cDNA template using two rounds of PCR. Products were evaluated for the presence of PCR amplicands using agarose gels (1.2%) and purified using PCR purification kits (Qiagen, Valencia, CA).
PCR amplicons were sequenced in forward and reverse directions using a BigDye sequencing kit on an ABI 3700 (Applied Biosystems, Foster City, CA). Sequence base calling was performed using Phred (8, 9); forward and reverse strands were assembled using an assembly algorithm based on the quality scores at each position (21). The estimated PCR artifact rate was 0.28 or approximately one PCR artifact per five genes amplified (25, 34). Ig isotype was determined by local alignment with genes of known isotype (46); V, D, and J region genes, complementarity-determining region 3 (CDR3) lengths, and mutation rates were identified using SoDA (56). All data were annotated so that matching subject data and sort information were linked to the cDNA sequence and analysis results.
Clonal lineage determination.
Antibody gene sequences from individual subjects were grouped and analyzed using the following criteria: (i) matching of variable and joining region gene segments, (ii) matching of CDR3 loop lengths, and (iii) ≥70% homology in CDR3 nucleotide sequence. Potential clonal lineages were only identified if both heavy and light chains for a given group satisfied all three criteria. For heavy chain (HC) alignments, D region usage was not considered; i.e., heavy chain genes could be considered part of a potential clonal lineage if they satisfied the above three criteria but had different predicted D region gene usage. Clonal lineages were confirmed by alignment of the complete V(D)J sequence, and maximum-likelihood trees for clonal lineages were generated using V(D)J regions (excluding constant region sequences); trees were constructed (dnaml), reorganized (retree), and plotted (drawgram) with the PHYLIP package, version 3.69 (10).
Expression of VH and VL as full-length IgG1 rMAbs.
Isolated Ig VH and VL gene pairs were assembled by PCR into linear full-length Ig heavy- and light-chain gene expression cassettes using methods described previously (26). Human embryonic kidney cell line 293T (ATCC, Manassas, VA) was grown to near confluence in six-well tissue culture plates (Becton, Dickinson, Franklin Lakes, NJ) and transfected with 2 μg per well of both IgH and IgL purified PCR-produced cassettes using PolyFect or Effectene (Qiagen, Valencia, CA). For PolyFect-transfected cells, 6 to 8 h after transfection, cells were fed with fresh culture medium supplemented with 2% fetal bovine serum and were incubated at 37°C in a 5% CO2 incubator. For Effectene-transfected cells, this wash step was omitted. Culture supernatants were harvested 3 days after transfection and concentrated 4-fold using centrifugal concentrators; expressed IgG was quantitated by ELISA (15); tested rMAbs were expressed at 10 μg/ml up to 20 mg/ml. For larger-scale production of rMAbs, some linear IgH and IgL gene constructs were cloned into pcDNA 3.3 using standard molecular protocols or were synthesized (GeneScript, Piscataway, NJ). Transiently expressed rMAbs with low IgG concentration were retransfected and reassayed. In cases where multiple tests of the same antibody were available, the final data set contained data from the assay with the highest concentration of MAb.
Neutralization assay in TZM-bl cells.
Neutralizing antibody assays in TZM-bl cells were performed as described previously (33). Antibodies were tested at concentrations starting at 50 μg/ml using serial 3-fold dilutions for a total of 8 concentrations tested. Env-pseudotyped viruses were added to the antibody dilutions at a predetermined titer to produce measurable infection and incubated for 1 h. TZM-bl cells were added and incubated for 48 h. Firefly luciferase (Luc) activity was measured as a function of relative luminescence units (RLU) using a Britelite Luminescence Reporter Gene Assay System as described by the supplier (PerkinElmer Life Sciences, Waltham, MA). Neutralization was calculated as a reduction in RLU in test wells compared with control wells after subtraction of background RLU in cell control wells and reported as MAb 50% inhibitory concentration (IC50) in μg/ml (33). Env-pseudotyped viruses were prepared in 293T cells and titrated in TZM-bl cells as previously described (33).
ADCC assay.
Antibody-dependent cell-mediated cytotoxicity (ADCC) assays were performed with purified serum IgG fractions and with rMAbs using the HIV-1 A1953.B chronically infected CEM.NKRCCR5 cell line as described previously (41).
SPR analysis of antibody reactivity.
Surface plasmon resonance (SPR) binding assays were performed on a BIAcore 3000 (BIAcore Inc., Piscataway, NJ) maintained at 20°C. HIV-1 Env proteins were immobilized on a CM5 sensor chip by standard amine coupling as described previously (2, 3). Additional tests were performed by capturing human rMAbs on anti-human Fc antibody-coupled surfaces; each human rMAb was captured to about 200 to 500 resonance units (RU). Specific binding responses of rMAb binding were obtained following subtraction of nonspecific binding on control surfaces. Rate constants were measured using the bivalent analyte model (to account for the avidity of bivalent Ig molecules), and global curve fitting to binding curves was obtained from rMAb titrations. Antibodies were injected at 30 μl/min for 2 to 6 min; glycine-HCl, pH 2.0, was used as the regeneration buffer.
Statistical analysis.
Statistical tests were performed in SAS, version 9.2 (SAS Institute, Cary, NC). Comparisons for multiple groups were performed using multiple degree of freedom F-tests using PROC GLM in SAS, version 9.2, with subsequent pairwise comparisons. For percentage data from individuals across multiple time points, a Kruskal-Wallis test was performed with subsequent pairwise comparisons. For data consisting of two groups only, t tests using the Satterthwaite correction (for continuous variables), the Kolmogorov-Smirnov test (for comparison of probability distributions), and Pearson's chi-square tests (for 2-by-2 category tables) were performed using the appropriate SAS PROC in SAS, version 9.2; the statistical test used is noted when P values are presented. Graphs of the data were created using GraphPad Prism (GraphPad Software, La Jolla, CA) with layout in Illustrator CS5 (Adobe, San Jose, CA).
RESULTS
Anti-gp120 vaccine-induced serum antibody is primarily IgG1, IgG3, and IgA.
This vaccine was previously reported to produce robust antibody responses to the three vaccine proteins (Env, Nef, and Tat) in addition to cellular responses (24). To further define this antibody response, we tested for isotype-specific responses against the three vaccine proteins following vaccination (Table 1). Prevaccination serum samples were negative (Fig. 1A to C). After vaccination, all subjects had detectable IgG1 responses, and most (28/30, 93%) had detectable IgA against each of the three vaccine proteins (Table 1). In all cases, IgG1 was found to have the highest serum concentration among the IgG subclasses to the three vaccine components (mean concentration of serum antibody to gp120 was 5 μg/ml; to Nef, 146 μg/ml; to Tat 67, μg/ml). IgG2 vaccine-induced antibodies were detected in a minority (8/30, 27%) of subjects (Table 1). When IgG3 and IgG4 vaccine-induced antibodies were detected, they were found in concentrations less than IgG2 vaccine-induced antibodies (Fig. 1A to C). Mean concentrations of antibodies induced to Tat and Nef were higher than those specific for gp120, with anti-gp120 IgG1 antibody concentrations 7.5% and 3.4% of antibody to Tat or Nef, respectively.
Table 1.
Binding antibody response to vaccine components
| Ab isotype | gp120 |
Nef |
Tat |
|||
|---|---|---|---|---|---|---|
| Isotype response ratea | Day 182 concn (μg/ml)b | Isotype response rate | Day 182 concn (μg/ml) | Isotype response rate | Day 182 concn (μg/ml) | |
| IgG1 | 30/30 | 5 | 30/30 | 146 | 30/30 | 67 |
| IgG2 | 8/30 | 0.2 | 24/30 | 1.5 | 23/30 | 1.4 |
| IgG3 | 20/30 | 0.014 | 30/30 | 2.0 | 30/30 | 1.1 |
| IgG4 | 18/30 | 0.01 | 27/30 | 0.02 | 29/30 | 0.04 |
| IgA | 30/30 | NDc | 28/30 | ND | 30/30 | ND |
Antibody response in postvaccination serum samples of 30 vaccine recipients. Number shown is proportion of subjects with serum antibodies against specific vaccine components out of the total number tested.
Mean concentration of antigen-specific antibody detected in day 182 serum samples in responsive vaccine recipients.
ND, not determined.
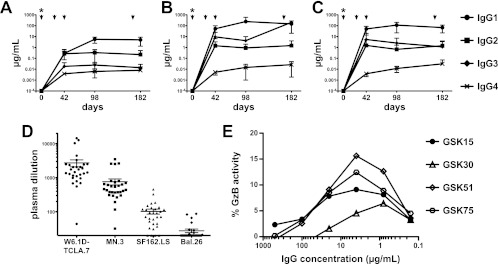
Fig 1.
Plasma antibody activity induced by GSK PRO HIV-002. (A to C) Vaccine-induced reactivity by isotype. Serial serum samples from vaccine recipients were analyzed for vaccine-specific antibody. Binding antibody multiplex assay data are plotted as estimated antibody concentrations for IgG subclasses; asterisks indicate that antibodies at day 0 were below the limit of detection. IgG subclass responses to HIV-1 gp120 appeared at day 42, with IgG1 responses peaking at day 98 (A). Antibody responses to Nef (B) and Tat (C) also appeared at day 42. The overall concentrations of HIV-1-specific antibody responses mirror the relative concentrations present in blood. (D) Neutralization activity of d182 serum. Serum samples were tested for activity against a panel of Env-pseudotyped viruses in the TZM-bl assay. No neutralizing activity was detected at a 1:20 dilution of preimmunization samples. In contrast, serum samples from day 182 showed neutralization of autologous W6.1D-TCLA.7 Env-pseudotyped virus (geometric mean titer, 1:1645) and neutralization against HIV-1 MN.3 (geometric mean titer, 1:448) in all vaccine recipients. Most vaccine recipients (26/30, or 87%) developed neutralizing activity against HIV-1 SF162.LS (geometric mean titer, 1:75) while a minority developed activity against HIV-1 BaL.26 (9/30, or 30% of subjects with neutralizing activity). (E) ADCC activity of serum IgG. Serum IgG from the four subjects (identified by GSK numbers) from whom rMAbs were recovered was tested for ADCC activity as described. No activity was detected in samples prior to immunization. In contrast, ADCC activity was detected in all samples from day 182 with a peak in granzyme B (GzB) activity comparable to activity levels of the recovered rMAbs (Fig. 4).
Specificities of vaccine-induced serum antibody.
Preimmune and day 182 (2 weeks after final boosting) serum samples from 20 subjects were analyzed for HIV-1 neutralizing activity using the TZM-bl pseudovirus neutralization assay (33). No neutralizing activity was detected at a 1:20 dilution of preimmunization samples. Following the vaccination regimen (24), neutralization of autologous W6.1D-TCLA.7 Env pseudovirus was detected in all vaccine recipients (geometric mean titer, 1:1645) as was neutralization against the tier 1 HIV-1 isolate MN.3 (geometric mean titer, 1:448) (Fig. 1D). Most (26/30, 87%) vaccine recipients developed neutralizing activity against the tier 1 HIV-1 isolate SF162.LS (geometric mean titer, 1:75) while a minority (9/30, 30%) developed activity against tier 1 HIV-1 BaL.26 (Fig. 1D). Taken together, and in agreement with a previous report (24), the serum antibody response confirms the immunogenicity of this vaccine and its capacity to elicit a robust neutralizing antibody response against autologous and heterologous tier 1 viruses.
The Env immunogen used in this trial formulated with a different adjuvant system (AS02A) was previously shown to induce antibody-dependent cellular cytotoxicity (ADCC) activity (12). Purified IgG isolated from the serum of four vaccine recipients in this study from preimmunization (day 0) samples and from samples after the fourth immunization (day 182) was tested. ADCC activity was found in 4/4 subjects only in postimmunization samples, with maximal granzyme B activity ranging from 6% to 15% (Fig. 1E), consistent with prior observations of the vaccine response (12).
gp120-specific memory B cells circulate 2 weeks after vaccination.
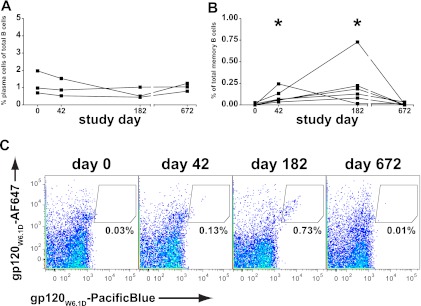
Previous studies of influenza immunization have shown that vaccine-specific plasma cells circulate with an increased frequency in blood 7 days after vaccination while vaccine-specific memory B cells are most numerous in blood ∼14 days after vaccination (60). We studied PBMC from three subjects for the presence of plasma cells (CD3/14/16/235a− CD19+ CD20−/lo CD27hi CD38hi) at four time points: preimmunization (day 0), 2 weeks after the second immunization (day 42), 2 weeks after the fourth immunization (day 182), and 18 months following the last immunization (day 672). Circulating plasma cells were calculated as a percentage of the total B cell population (CD3/14/16/235a− CD19+) and were similarly low at all time points for the three subjects tested (Fig. 2A) (overall mean, 0.97%; P = 0.44, Kruskal-Wallis test). Thus, consistent with what has been observed for other vaccines (60), we did not detect significant changes in total plasmacytosis at 14 days after administration of this gp120 vaccine.
Fig 2.
B cell populations circulating after vaccination. (A) Plasma cells were quantitated as a fraction of total B cells in three subjects at four available time points. No difference between preimmunization and postimmunization time points was observed (values are mean ± standard error; day 0, 1.2% ± 0.4%, day 42, 1.0% ± 0.3%; day 182, 0.7% ± 0.3%; day 672, 1.0% ± 0.1%). (B) Antigen-specific memory B cells quantitated as a fraction of memory B cells in six subjects. Preimmunization and convalescent samples showed few circulating antigen-specific memory B cells (day 0, 0.007% ± 0.004%; day 672, 0.010% ± 0.005%) while samples taken 2 weeks after immunization had a significantly larger fraction of circulating antigen-specific memory B cells (day 42, 0.10% ± 0.03%; day 182, 0.23% ± 0.10%; P = 0.001 (asterisks); Kruskal-Wallis statistic, 16.3). (C) Quantitation of antigen-specific memory B cells by flow cytometry. B cells stained with fluorescently labeled gp120W6.1D were gated for memory B cells. Antigen-specific B cells appear as a diagonal in the shown gate; low frequencies were observed at early and late time points. Elevated numbers of circulating antigen-specific B cells were found 2 weeks after the second and fourth immunizations.
We conjugated one component of the vaccine immunogen, gp120W6.1D (12), with two different fluorochromes to identify antigen-specific B cells. PBMC from six vaccine recipients were studied at the same four time points as above. Memory B cells were gated (CD3/14/16/235a− CD19+ surface IgD−), and antigen-specific B cells were identified within the memory population (positive for gp120W6.1D in both colors) (Fig. 2C) using a strategy similar to that previously described (14, 36). Calculated as a fraction of memory B cells, gp120W6.1D-reactive cells were absent prior to vaccination, were found to circulate at an elevated frequency 2 weeks after vaccination (day 42, 0.10% ± 0.03%; day 182, 0.23% ± 0.10%), and returned to baseline by 18 months after the last vaccination (Fig. 2B) (P = 0.001, Kruskal-Wallis test).
Isolation of gp120-reactive antibodies from plasmacytes and memory B cells.
Using the same flow cytometry protocol, we sorted single plasma cells and antigen-specific memory B cells from vaccine recipients for recovery of immunoglobulin (Ig) genes and production of rMAbs (14, 26). We recovered 453 rMAbs from four subjects; 402 were isolated from plasma cells, and 51 were isolated from antigen-specific memory B cells (Fig. 3). We isolated gp120W6.1D-reactive rMAbs from each of the postvaccination time points (days 42 and 182); no MAbs isolated from prevaccination samples or at the 18-month follow-up sample (day 672) were specific for gp120W6.1D. Only 5/402 (1%) of rMAbs derived from plasma cells were reactive with HIV-1 Env while 29/51 (57%) of rMAbs derived from cells sorted with antigen-specific labels were Env reactive (Fig. 3) (χ2 = 193, P < 0.0001). Thus, vaccination with gp120W6.1D induced circulation of Env-reactive B cells 2 weeks after vaccination, and these antigen-specific B cells could be efficiently isolated by flow cytometry for the production of human rMAbs to permit characterization of the vaccine-induced Ab repertoire.
Fig 3.
Recovery of Env-specific antibodies from vaccine recipients. (A) Plasma cells yielded 402 rMAbs distributed across all four time points sorted. Env-reactive rMAbs were isolated only from postvaccination time points; 5/402 (1.2%) of rMAbs reacted with gp120 (orange wedge). (B) Antigen-specific memory B cells yielded 51 rMAbs across three postimmunization time points; no antibodies were isolated from the day 0 samples. Env-reactive rMAbs predominated; 29/51 (57%) of isolated rMAbs reacted with gp120. Antigen-specific memory B cell sorting was more efficient in isolating Env-reactive antibodies (P < 0.0001; χ2 = 193).
IgG1 predominance of isolated rMAbs.
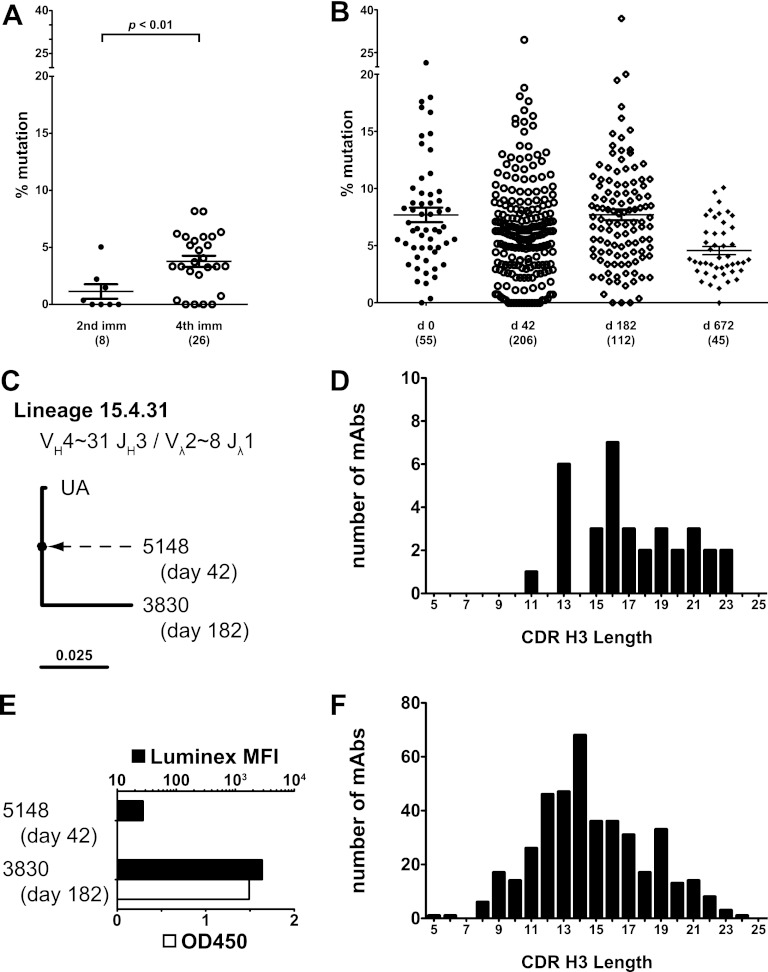
HIV-1 Env-specific rMAbs were mostly (33/34, 97%) (see Table S1 in the supplemental material) IgG1, consistent with the measurements of serum antibody (Table 1 and Fig. 1D). Only one antibody of isotype IgG3 was isolated (1/34, 3%), and no MAbs of isotype IgG2 or IgG4 were recovered. The IgG3 rMAb was isolated from the day 42 time point, while all rMAbs isolated at day 182 were IgG1 (see Table S1). Env-reactive rMAbs used both kappa and lambda chains (kappa, 17/34 or 50%; there was no preferential usage of either light chain at any time point) (see Table S2). A similar usage of variable gene segments for both heavy and light chains was found in HIV-1-specific rMAbs recovered from the day 42 and day 182 time points (see Tables S3 and S4). The most commonly isolated gene segments of the HIV-1-specific rMAbs were VH3∼11 and Vλ2∼14 (see Table S5) (used by 8/34 rMAbs each, or 24%). Complementarity-determining region 3 of heavy chain (CDR H3) for HIV-1-specific rMAbs ranged from 11 to 23 amino acids (median, 16.5 amino acids) (Fig. 4D). For the group of rMAbs not reactive with HIV-1 antigens, CDR H3 length was from 5 to 24 amino acids (median, 14 amino acids) (Fig. 4F). Kolmogorov-Smirnov comparison of the two distributions showed that they were different (test statistic = 0.33, P = 0.002).
Fig 4.
Increased maturation of recovered antibodies following repeat immunization. (A) HC mutation frequencies of Env-reactive antibodies after two immunizations (day 42) was 1.8% ± 0.6%; antibodies recovered after four immunizations (day 182 and 672) had a mutation frequency of 3.8% ± 0.5% (P = 0.0095; Mann-Whitney U = 40). (B) HC mutation frequencies of antibodies not reactive with HIV-1 gp120 or gp140 in any assay are shown. The number of antibodies shown appears below each column. Mean mutation frequency ± standard error for each time point was as follows: day 0, 7.7% ± 0.6%; day 42, 6.4% ± 0.3%; day 182, 7.7% ± 0.5%; day 672, 4.6% ± 0.4%. Antibodies recovered at day 672 had a lower mean mutation frequency than those recovered at the other three time points (P < 0.001; Kruskal-Wallis statistic, 20.5). (C) Clonal lineage 15.4.31 consisted of two rMAbs, one isolated at day 42 and one at day 182. Antibody 5148 was nearly unmutated (see Fig. S1 in the supplemental material). (D) CDR H3 length of Env-reactive rMAbs shown as a histogram. Median CDR H3 length was 16.5 amino acids; the mean was 17.2 amino acids. (E) Reactivity of clonal lineage 15.4.31 in binding antibody multiplex (Luminex) assay and ELISA, tested at 1 μg/ml. Antibody 3830 had a 4.7% HC mutation frequency and reacted strongly to gp120W6.1D in both assays. Antibody 5148 had a 0% HC mutation frequency, reacted weakly in the multiplex binding assay, and did not react in ELISA. MFI, mean fluorescence intensity. (F) CDR H3 lengths of all recovered rMAbs, regardless of reactivity, shown as a histogram. Median CDR H3 length was 14 amino acids; the mean was 14.7.
Neutralization profile of isolated rMAbs.
We further characterized 5/34 isolated HIV-1 Env-specific rMAbs for their ability to recapitulate serum neutralization activity. These five rMAbs were tested in the TZM-bl neutralization assay against a panel of 22 tier 1 and tier 2 Env-pseudotyped viruses (Table 2). Two of the tested rMAbs demonstrated potent autologous neutralizing activity against the W6.1D-TCLA.7 Env pseudovirus, and each of these rMAbs also more weakly neutralized the tier 1 SF162.LS strain (Table 2). One rMAb (3489) potently neutralized the tier 1 MN.3 strain while the other (3491) did not; in contrast rMAb 3491 neutralized tier 1 HIV-1 BaL.26 while rMAb 3489 did not (Table 2). Thus, the activity of these rMAbs recapitulated that of the subject serum (Fig. 1D).
Table 2.
Neutralization of Env-pseudotyped viruses by rMAbs
| Virusa | IC50 in TZM-bl cells for the indicated MAb (μg/ml) |
||||
|---|---|---|---|---|---|
| 3487 | 3489 | 3491 | 3493 | 3506 | |
| MN.3 | >50 | 0.17 | >50 | >50 | >50 |
| SF162.LS | >50 | 5.4 | 4.1 | >50 | >50 |
| BaL.26 | >50 | >50 | 15 | >50 | >50 |
| W6.1D-TCLA.7 | >50 | 0.07 | 0.05 | >50 | >50 |
| 6535.3 | >50 | 42 | >50 | >50 | >50 |
Isolates shown are subtype B and tier 1 except for 6535.3, which is tier 2. All other viruses tested (17 isolates) were not neutralized; they consist of the following: clade B tier 2, QH0692.42, WEAU-d15.410.787, and BB1006-11.C3.1601; clade A tier 1, 92RW020.2; clade A tier 2, Q23.17, Q842.d12, Q461.e2, and Q769.d22; clade C tier 1, TV1.21; clade C tier 2, Du422.1, ZM197 M.PB7, Ce2010_F5, and 704809221_1B3; clade CRF01_AE tier 1, TH023.6, and NP03.13; clade CRF01_AE tier 2, CM244.ec1 and C3347.c11.
ADCC activity of isolated rMAbs.
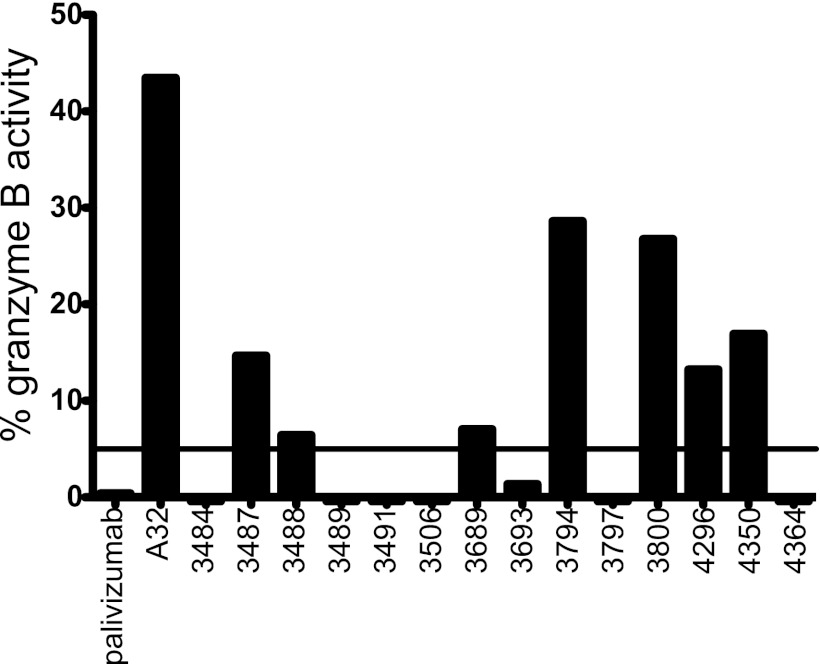
A panel of rMAbs was next tested for ability to mediate antibody-dependent cell mediated cytotoxicity (ADCC) against HIV-1-infected target cells. The two rMAbs found to mediate neutralization (3489 and 3491) did not mediate ADCC in this assay; however, 7 of 14 total rMAbs tested were able to mediate ADCC against A1953.B-infected CEM.NKRCCR5 cells (Fig. 5). Thus, similar to the finding for neutralization activity, rMAbs isolated from vaccinated subjects recapitulated the ADCC activity observed in serum (Fig. 1E).
Fig 5.
ADCC activity of selected rMAbs. A subset of 14 antibodies was tested for ADCC activity; the granzyme B activity is shown for each rMAb. The cutoff of reactivity is 5%, shown by a horizontal line. Seven rMAbs mediated ADCC, recapitulating the serum activity (Fig. 1E).
Recovery of rMAbs reflective of affinity maturation associated with vaccination.
Recovered Ig genes were analyzed for mutation away from inferred germ line or unmutated ancestor (UA) sequences (25, 56). HIV-1 Env-reactive rMAbs recovered after two immunizations had an average mutation frequency of 1.8% ± 0.6% while those recovered after four immunizations had an increased mutation frequency of 3.8% ± 0.5% (P = 0.0095; Mann-Whitney U of 40) (Fig. 4A). The distribution of mutation frequencies after four immunizations included four rMAbs with no mutations, suggesting that repeated immunization might have continued to recruit unmutated B cells in addition to promoting somatic hypermutation of previously recruited memory B cells. The increase in mutation frequency was not due to an overall shift in mutation frequency among all B cells as rMAbs recovered from the same time points that did not react with Env had consistent and higher overall mutation frequencies than Env-specific rMAbs (Fig. 4B).
Clonal lineages of HIV-1-reactive rMAbs.
An important question for HIV-1 vaccine design is whether repeated immunization with Env promotes affinity maturation of previously stimulated memory B cells or if sequential recruitment of unmutated B cells predominates. To address this question, we analyzed all recovered rMAbs for evidence of clonal relatedness and found three clonal lineages that had Env-specific rMAbs. One of these, lineage 15.4.31, was notable as it consisted of one rMAb isolated from day 42 and another from day 182 (Fig. 4C; see also Fig. S1 in the supplemental material). The rMAb isolated from day 42 (5148) had no mutations in the heavy chain and only one mutation in the lambda chain compared with the UA (see Fig. S1); this rMAb bound weakly to gp120W6.1D (Fig. 4E). In contrast, rMAb 3830 from day 182 had 18 mutations in the heavy chain and 5 mutations in the lambda chain and bound strongly to gp120W6.1D, consistent with antigen drive in this clonal lineage. Taken together, these rMAbs were consistent with clonal persistence of this lineage during the immunization series.
The two other clonal lineages found did not cross multiple time points; both were recovered from day 182 samples. Lineage 13.1.2 was a two-member linage of modestly mutated rMAbs (3.7 to 5.9% heavy chain [HC] mutation) (see Fig. S2 in the supplemental material). Lineage 16.3.11 was a four-member lineage of rMAbs with low mutation frequency (0 to 2.6% HC mutation) (see Fig. S3), a mutation pattern consistent with the recruitment of an unmutated naïve or memory B cell that expanded following the fourth immunization. These nearly unmutated Env-reactive antibodies isolated from the fourth immunization, combined with data from lineage 15.4.31 that spanned time points in this study, are consistent with both vaccine-driven somatic mutation and ongoing recruitment of new unmutated B cells during sequential immunization.
Changing specificity of rMAbs during affinity maturation.
Using inferred germ line antibody sequences derived by computational methods (25, 39, 56), we constructed UAs of three rMAbs. UA binding was compared to that of mature rMAbs using intact HIV-1 Env proteins and overlapping peptides spanning gp120 (Table 3). Antibodies 3506 and 3506_UA both reacted with Env protein, although neither antibody bound to linear peptides (Table 3). In contrast, antibody 3491_UA bound to a linear peptide in the C2 region of gp120 (Table 3). Interestingly, the recovered rMAb 3491 did not bind this linear sequence, suggesting that the binding specificity may have become more conformationally dependent following somatic hypermutation. Furthermore, the recovered rMAb 3491 mediated neutralization (Table 2) but 3491_UA did not, consistent with antigen-driven maturation of binding leading to neutralizing activity in the mutated antibody that was absent in the UA. Thus, this vaccine was capable of recruiting unmutated naïve B cells to undergo affinity maturation leading to HIV-1 neutralizing activity.
Table 3.
Epitope mapping of rMAbs
| rMAb | Peptide sequence bounda | gp120 sequence locationb | Neutralization or ADCC activityc | Env bindingd | HEp-2 reactivityd |
|---|---|---|---|---|---|
| 3487 | − | − | ADCC | + | + |
| 3489 | − | − | Neutralization | + | ± |
| 3489_UA | − | − | Neg | − | + |
| 3491 | − | − | Neutralization | + | + |
| 3491_UA | PAGFAILKCNDKKFS | 220-234 | Neg | − | + |
| 3506 | − | − | Neg | + | + |
| 3506_UA | − | − | Neg | + | + |
| 4364 | EDIISLWDQSL | 106-116 | Neg | + | + |
−, no binding/not applicable.
Env sequences are numbered relative to HIV-1HXB2CG using the system described by Korber et al. (23) and represent amino acid positions.
Neutralization data are from Table 2; ADCC data are from Fig. 3. Neg, not active in either neutralization or ADCC assays.
+, reactive; ±, equivocal reactivity; −, negative. HEp-2 reactivity data are from Fig. 6.
Isolation of polyreactive rMAbs reactive with gp120W6.1D.
Acute and chronic HIV-1 infection has been shown to induce polyreactive antibodies that react with HIV-1 proteins (25, 37). To address whether HIV-1 Env can similarly induce polyreactive antibodies in the setting of vaccination, we tested 22 Env-specific rMAbs for their ability to bind to an array of autoantigens and HEp-2 epithelial cells; 3/22 (14%) rMAbs reactive with Env were also reactive with HEp-2 cells (Fig. 6A to C). Interestingly, 1/2 rMAbs that mediated virus neutralization (Table 2, 3491) was also reactive with HEp-2 cells (Fig. 6A); the UA of that rMAb was also HEp-2-reactive (Fig. 6G, 3491_UA). When the two rMAbs that mediated neutralization were tested against a panel of autoantigens, both rMAbs (3489 and 3491) showed reactivity to multiple antigens including ribonucleoproteins (RNP) (Table 4).
Fig 6.
HEp-2 cell reactivities of recovered rMAbs. All images were taken with a 40× objective; all antibodies were tested at 25 μg/ml, except 13D5, which was tested at 50 μg/ml. The exposure was 10 s unless otherwise noted. Scale bar, 25 μm. Images show results with the following antibodies: 3491, 12-s exposure, diffuse staining and dividing cells (A); 3797, 8-s exposure, dividing cells (B); 4364, 7-s exposure, nuclear and cytoplasmic fibrillar pattern (C); 2F5 (positive control), diffuse (D); 17b (negative control), no staining (E); 13D5 (murine MAb), 15s exposure, fibrillar pattern (F); 3491_UA, diffuse (G); 4364, 7-s exposure, magnified to show fibrillar staining; 13D5, 15-s exposure, magnified to show fibrillar staining (I).
Table 4.
Autoreactivity of rMAbs
| rMAba | Athena Luminex result with the indicated autoantigenb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| SS-A | SS-B | Sm | RNP | Scl-70 | Jo-1 | dsDNA | Centromere B | Histone | |
| 4E10 | + | − | − | − | − | − | − | − | − |
| 3487 | − | − | − | + | − | − | +++ | − | − |
| 3488 | − | − | + | ++ | + | − | ++ | + | ++ |
| 3489 | − | − | − | − | − | − | + | + | − |
| 3491 | ++ | + | + | +++ | − | − | ++ | +++ | ++ |
| 3794 | − | − | − | − | − | − | +++ | ++ | + |
| 3800 | − | − | − | + | − | − | ++ | +++ | ++ |
| 4296 | − | − | − | − | − | − | − | − | − |
| 4350 | − | − | − | − | − | − | − | − | − |
| 4364 | +++ | + | + | + | − | +++ | − | + | + |
Antibodies were screened at 50μg/ml.
Positive for this assay when used in a clinical setting for human serum is 120. −, 0 to 119; +, 120 to 239; ++, 240 to 360; +++, >360. dsDNA, double-stranded DNA; RNP, ribonucleoprotein.
One of the HEp-2 reactive rMAbs (4364) bound to two linear Env peptides within the C1 region of gp120 with the core sequence EDIISLWDQSL, an epitope overlapping that of anti-Env mouse MAb 13D5 (11) (Table 3). Both mouse MAb 13D5 and the human gp120-induced rMAb 4364 bound to HEp-2 cells by indirect immunofluorescence with similar patterns, suggesting that these antibodies directed to the C1 region of gp120 may cross-react with a nuclear self-antigen (Fig. 6C, F, H, and I). Thus, HIV-1 Env in the setting of vaccination can recruit B cells from the normal B cell pool with polyreactive characteristics, as occurs in acute (25) and chronic (37) HIV-1 infection.
DISCUSSION
Advances in molecular techniques have allowed the efficient recovery of immunoglobulin genes from human B cells in a variety of conditions including influenza vaccination (34, 60), influenza virus infection (34, 59, 64), and acute (25) and chronic (37, 43, 61) HIV-1 infection. Sorting methods using fluorescently labeled antigen hooks have been used to isolate broadly neutralizing antibodies (43, 61); however, the use of fluorescently labeled Env proteins to study the vaccine-induced repertoire resulting from candidate HIV-1 vaccination has not been reported. Using Env-specific sorting, we were able to interrogate HIV-1 vaccine-induced antibody maturation (Fig. 3). The activities of the antibodies recovered in this study recapitulated both the neutralization activity and ADCC activity observed in the serum of vaccine recipients although we cannot exclude the possibility that this vaccine induced rare antibodies with greater breadth or activity. In-depth characterization of binding activity, structure, and mutational pathways of antibodies isolated from recipients of candidate HIV-1 vaccines, like that reported here, will permit iterative rational design of future vaccine candidates.
Polyreactivity is a trait of the normal human B cell repertoire, with ∼40% of immature peripheral B cells and ∼20% of memory B cells showing polyreactivity using the same methods used in this study (48, 58). Natural autoreactive antibodies have been associated with protection from infectious diseases (5, 28, 40). Many broadly neutralizing antibodies against HIV-1 isolated to date have evidence of polyreactivity; long, hydrophobic CDR H3 regions; and/or high numbers of somatic mutations (55). These characteristics suggest that HIV-1 Env may preferentially trigger the normal human polyreactive B cell pool. Thus, it was interesting that this vaccine elicited HIV-1-reactive antibodies that also had evidence of polyreactivity (Table 4 and Fig. 6). If polyreactive B cell precursors are required as the starting points for the development of broad activity, then the ability of this vaccine to elicit polyreactive antibodies suggests that at least one hurdle for vaccine development can at least be partially cleared. The Env-reactive antibodies recovered from this study had slightly longer CDR H3 regions than the antibodies that did not bind Env (Fig. 4D and F); this difference may have resulted from the smaller sample size of the Env-reactive MAbs. However, we did not find exceptionally long CDR H3 regions (Fig. 4D), a high degree of somatic mutation (Fig. 4A), or broadly neutralizing activity (Table 2), suggesting that additional or new vaccine designs will be required to elicit antibodies with those characteristics. It was recently suggested that polyreactive antibodies would be more effective at neutralizing HIV-1 due to polyreactivity-mediated heterotypic binding to virions (37, 38). Whether the induction of polyreactive antibodies by vaccination, such as observed in the present study, will augment the risk of autoimmune disease is not corroborated by other observations reporting vaccine-induced polyreactive and autoreactive antibodies in 10 to 17% of those receiving hepatitis A (18), hepatitis B (6, 29), and influenza vaccines (19, 53), while no long-term adverse effects were reported (6, 54). Using techniques similar to those employed in this study, we found that ∼20% of isolated rMAbs from subjects immunized with trivalent inactivated influenza vaccine were polyreactive (B. F. Haynes, unpublished data). Vaccinations have been rarely reported to reactivate autoimmune disease (35); however, most evidence supports the safety of killed or recombinant vaccines in autoimmune disease patients (1, 7, 32, 52). In cases where it is observed, autoantibody increases following vaccination have been reported to have no clinical consequences (1, 52).
Given the high mutation frequency of many broadly neutralizing antibodies (43, 55, 57), a fundamental question for HIV-1 vaccine development is whether repeated immunization recruits naïve B cells in successive waves or if previously mutated memory B cells are driven to mutate further. The results of this study are consistent with both mechanisms; we found evidence of both unmutated B cells stimulated at late immunization time points (Fig. 4A; see also Fig. S3 in the supplemental material) and recall of previously circulating B cells (Fig. 4C and 4E; see Fig. S1). Whether a vaccine-adjuvant combination can be created that can preferentially drive memory B cells to higher levels of mutation and whether such a strategy will result in the development of breadth of activity remain to be determined.
Using a combination of computational and molecular techniques, we created inferred unmutated ancestor antibodies to further characterize the effect of mutation of HIV-1 specificity. Two antibodies, 3489 and 3491, mediated pseudovirus neutralization; neither UA was able to neutralize, suggesting that neutralization was an activity acquired during somatic hypermutation. Interestingly, 3491_UA bound to a linear peptide in the gp120 C2 region while the recovered rMAb 3491 did not, suggesting that this antibody specificity became more conformationally dependent during affinity maturation. Antibody 3491 was moderately mutated (5.2% HC mutation frequency), and both 3491 and its UA reacted with HEp-2 cells (Fig. 6A and G). This suggested that polyreactivity and Env-epitope reactivity were able to coexist early during antigen-driven somatic hypermutation although loss and regain of these specificities cannot be excluded.
In summary, we have demonstrated that four immunizations with gp120 Env in human volunteers induced both sequential recruitment of unmutated B cells and stimulation of previously recruited B cells. The degree of somatic mutation induced by gp120 Env after four immunizations did not reach the level of mutation reported for broadly neutralizing anti-HIV-1 antibodies. Repetitive boosting with modified Env immunogens may induce higher degrees of somatic mutation in vaccine-elicited antibodies. The RV144 trial (42) showed nondurable efficacy in the absence of broadly neutralizing antibodies, suggesting that anti-HIV-1 antibodies with narrow neutralization profiles or other activities such as ADCC may be protective in vaccine recipients. Thus, study of the affinity maturation of vaccine-induced antibodies with a variety of anti-HIV-1 activities can guide vaccine development. Future designs that can elicit antibodies that inhibit virus replication through ADCC or other mechanisms (50) may block infection if the antibodies are present at the time of HIV-1 exposure.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following individuals for expert technical assistance in this body of work: Kenesha D. Luney, Michele Donathan, and R. Glenn Overman.
Support for this work was provided by a Collaboration for AIDS Vaccine Discovery grant to B.F. Haynes from the Bill and Melinda Gates Foundation and by the Center For HIV/AIDS Vaccine Immunology (grant U19 AI067854).
G.V. is an employee of GSK Biologicals.
Footnotes
Published ahead of print 2 May 2012
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1. Abu-Shakra M, Press J, Buskila D, Sukenik S. 2007. Influenza vaccination of patients with systemic lupus erythematosus: safety and immunogenicity issues. Autoimmun. Rev. 6:543–546 [DOI] [PubMed] [Google Scholar]
- 2. Alam SM, et al. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alam SM, et al. 2008. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J. Virol. 82:115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batista FD, Neuberger MS. 1998. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity 8:751–759 [DOI] [PubMed] [Google Scholar]
- 5. Baumgarth N, et al. 2000. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. J. Exp. Med. 192:271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belloni C, et al. 2002. No evidence of autoimmunity in 6-year-old children immunized at birth with recombinant hepatitis B vaccine. Pediatrics 110:e4 doi:10.1542/peds.110.1.e4 [DOI] [PubMed] [Google Scholar]
- 7. Conti F, Rezai S, Valesini G. 2008. Vaccination and autoimmune rheumatic diseases. Autoimmun. Rev. 8:124–128 [DOI] [PubMed] [Google Scholar]
- 8. Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 9. Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 10. Felsenstein J. 2005. PHYLIP (phylogeny inference package), 3.6 ed Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 11. Gao F, et al. 2009. Cross-reactive monoclonal antibodies to multiple HIV-1 subtype and SIVcpz envelope glycoproteins. Virology 394:91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goepfert PA, et al. 2007. Durable HIV-1 antibody and T-cell responses elicited by an adjuvanted multi-protein recombinant vaccine in uninfected human volunteers. Vaccine 25:510–518 [DOI] [PubMed] [Google Scholar]
- 13. Gray D, MacLennan IC, Lane PJ. 1986. Virgin B cell recruitment and the lifespan of memory clones during antibody responses to 2,4-dinitrophenyl-hemocyanin. Eur. J. Immunol. 16:641–648 [DOI] [PubMed] [Google Scholar]
- 14. Gray ES, et al. 2011. Isolation of a monoclonal antibody that targets the alpha-2 helix of gp120 and represents the initial autologous neutralizing-antibody response in an HIV-1 subtype C-infected individual. J. Virol. 85:7719–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gray ES, et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haynes BF, et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906–1908 [DOI] [PubMed] [Google Scholar]
- 17. Haynes BF, Liao HX, Tomaras GD. 2010. Is developing an HIV-1 vaccine possible? Curr. Opin. HIV AIDS 5:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a. Haynes BF, Kelsoe G, Harrison SC, Kepler TB. 2012. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 30:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karali Z, Basaranoglu ST, Karali Y, Oral B, Kilic SS. 2011. Autoimmunity and hepatitis A vaccine in children. J. Investig. Allergol. Clin. Immunol. 21:389–393 [PubMed] [Google Scholar]
- 19. Katerinis I, et al. 2011. De novo anti-HLA antibody after pandemic H1N1 and seasonal influenza immunization in kidney transplant recipients. Am. J. Transplant. 11:1727–1733 [DOI] [PubMed] [Google Scholar]
- 20. Kepler TB, Perelson AS. 1993. Somatic hypermutation in B cells: an optimal control treatment. J. Theor. Biol. 164:37–64 [DOI] [PubMed] [Google Scholar]
- 21. Kepler TB, et al. 2010. Chiropteran types I and II interferon genes inferred from genome sequencing traces by a statistical gene-family assembler. BMC Genomics 11:444 doi:10.1186/1471-2164-11-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim JH, Rerks-Ngarm S, Excler JL, Michael NL. 2010. HIV vaccines: lessons learned and the way forward. Curr. Opin. HIV AIDS 5:428–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korber B, et al. 1998. Numbering Positions in HIV Relative to HXB2CG. Los Alamos National Laboratory, Los Alamos, NM: http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/HXB2.html [Google Scholar]
- 24. Leroux-Roels I, et al. 2010. Strong and persistent CD4+ T-cell response in healthy adults immunized with a candidate HIV-1 vaccine containing gp120, Nef and Tat antigens formulated in three adjuvant systems. Vaccine 28:7016–7024 [DOI] [PubMed] [Google Scholar]
- 25. Liao HX, et al. 2011. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J. Exp. Med. 208:2237–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liao HX, et al. 2009. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods 158:171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liao HX, et al. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353:268–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin F, Kearney JF. 2000. B-cell subsets and the mature preimmune repertoire. Marginal zone and B1 B cells as part of a “natural immune memory.” Immunol. Rev. 175:70–79 [PubMed] [Google Scholar]
- 29. Martinuc Porobic J, et al. 2005. Anti-phospholipid antibodies following vaccination with recombinant hepatitis B vaccine. Clin. Exp. Immunol. 142:377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mietzner B, et al. 2008. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc. Natl. Acad. Sci. U. S. A. 105:9727–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mikell I, et al. 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7:e1001251 doi:10.1371/journal.ppat.1001251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Millet A, Decaux O, Perlat A, Grosbois B, Jego P. 2009. Systemic lupus erythematosus and vaccination. Eur. J. Intern. Med. 20:236–241 [DOI] [PubMed] [Google Scholar]
- 33. Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12, unit 12.11. doi:10.1002/0471142735.im1211s64 [DOI] [PubMed] [Google Scholar]
- 34. Moody MA, et al. 2011. H3N2 Influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One 6:e25797 doi:10.1371/journal.pone.0025797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morihara K, Arakawa Y, Takenaka H, Morihara T, Katoh N. 2011. Systemic lupus erythematosus following vaccination against 2009 influenza A (H1N1). Lupus 20:775–776 [DOI] [PubMed] [Google Scholar]
- 36. Morris L, et al. 2011. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One 6:e23532 doi:10.1371/journal.pone.0023532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mouquet H, et al. 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mouquet H, Warncke M, Scheid JF, Seaman MS, Nussenzweig MC. 2012. Enhanced HIV-1 neutralization by antibody heteroligation. Proc. Natl. Acad. Sci. U. S. A. 109:875–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Munshaw S, Kepler TB. 2010. SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics 26:867–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ochsenbein AF, et al. 1999. Control of early viral and bacterial distribution and disease by natural antibodies. Science 286:2156–2159 [DOI] [PubMed] [Google Scholar]
- 41. Pollara J, et al. 2011. High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A 79:603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rerks-Ngarm S, et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 43. Scheid JF, et al. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shlomchik M, et al. 1990. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J. Exp. Med. 171:265–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith K, et al. 2009. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4:372–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith TF, Waterman MS. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195–197 [DOI] [PubMed] [Google Scholar]
- 47. Stamatatos L, Morris L, Burton DR, Mascola JR. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15:866–870 [DOI] [PubMed] [Google Scholar]
- 48. Tiller T, et al. 2007. Autoreactivity in human IgG+ memory B cells. Immunity 26:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tomaras GD, et al. 2011. Polyclonal B Cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J. Virol. 85:11502–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tomaras GD, Haynes BF. 2010. Strategies for eliciting HIV-1 inhibitory antibodies. Curr. Opin. HIV AIDS 5:421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tomaras GD, et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Toplak N, Avcin T. 2009. Influenza and autoimmunity. Ann. N. Y. Acad. Sci. 1173:619–626 [DOI] [PubMed] [Google Scholar]
- 53. Toplak N, et al. 2008. Autoimmune response following annual influenza vaccination in 92 apparently healthy adults. Autoimmun. Rev. 8:134–138 [DOI] [PubMed] [Google Scholar]
- 54. Van Damme P, et al. 2001. Long-term persistence of antibodies induced by vaccination and safety follow-up, with the first combined vaccine against hepatitis A and B in children and adults. J. Med. Virol. 65:6–13 [PubMed] [Google Scholar]
- 55. Verkoczy L, Kelsoe G, Moody MA, Haynes BF. 2011. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr. Opin. Immunol. 23:383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Volpe JM, Cowell LG, Kepler TB. 2006. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics 22:438–444 [DOI] [PubMed] [Google Scholar]
- 57. Walker LM, et al. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wardemann H, et al. 2003. Predominant autoantibody production by early human B cell precursors. Science 301:1374–1377 [DOI] [PubMed] [Google Scholar]
- 59. Wrammert J, et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208:181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wrammert J, et al. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453:667–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xiao X, et al. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390:404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yates NL, et al. 2011. Multiple HIV-1-specific IgG3 responses decline during acute HIV-1: implications for detection of incident HIV infection. AIDS 25:2089–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu X, et al. 2008. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang J, Shakhnovich EI. 2010. Optimality of mutation and selection in germinal centers. PLoS Comput. Biol. 6:e1000800 doi:10.1371/journal.pcbi.1000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.