Abstract
Broadly neutralizing antibodies to the CD4 binding site (CD4bs) of gp120 are generated by some HIV-1-infected individuals, but little is known about the prevalence and evolution of this antibody response during the course of HIV-1 infection. We analyzed the sera of 113 HIV-1 seroconverters from three cohorts for binding to a panel of gp120 core proteins and their corresponding CD4bs knockout mutants. Among sera collected between 99 and 258 weeks post-HIV-1 infection, 88% contained antibodies to the CD4bs and 47% contained antibodies to resurfaced stabilized core (RSC) probes that react preferentially with broadly neutralizing CD4bs antibodies (BNCD4), such as monoclonal antibodies (MAbs) VRC01 and VRC-CH31. Analysis of longitudinal serum samples from a subset of 18 subjects revealed that CD4bs antibodies to gp120 arose within the first 4 to 16 weeks of infection, while the development of RSC-reactive antibodies was more varied, occurring between 10 and 152 weeks post-HIV-1 infection. Despite the presence of these antibodies, serum neutralization mediated by RSC-reactive antibodies was detected in sera from only a few donors infected for more than 3 years. Thus, CD4bs antibodies that bind a VRC01-like epitope are often induced during HIV-1 infection, but the level and potency required to mediate serum neutralization may take years to develop. An improved understanding of the immunological factors associated with the development and maturation of neutralizing CD4bs antibodies during HIV-1 infection may provide insights into the requirements for eliciting this response by vaccination.
INTRODUCTION
A major goal of HIV-1 vaccination is to elicit antibodies that inactivate or neutralize the majority of circulating HIV-1 strains. These potentially protective antibodies target the HIV-1 spike, which is composed of a trimer of noncovalently linked surface glycoprotein (gp120) and transmembrane protein (gp41) molecules. While most HIV-1 envelope glycoprotein (Env)-containing candidate vaccines generate robust levels of Env-specific antibodies, these antibodies neutralize only a minority of HIV-1 strains. In contrast, the sera of some HIV-1-infected subjects contain potent and broadly reactive neutralizing antibodies. Detailed analysis of such sera has provided information about the viral epitopes targeted by neutralizing antibodies (2, 14, 16, 17, 28, 32, 37). In addition, the recent isolation of numerous broadly reactive neutralizing monoclonal antibodies (MAbs) and their liganded structures has helped to define the relatively conserved regions of Env that are vulnerable to antibody neutralization. These epitopes include the membrane-proximal region (MPER) of gp41 (6, 23, 24) and several regions on gp120, including glycan-containing regions at the base of variable regions 1 and 2 (V1V2) (3, 19, 36) and the base of V3 (25, 35) and an epitope within the CD4 binding site (CD4bs) of gp120 (9, 29, 41, 44).
Until recently, only one known CD4bs-directed neutralizing MAb was able to neutralize primary HIV-1 strains. Monoclonal antibody b12 was first described in 1994 (5), and its crystal structure bound to the core of gp120 was resolved in 2007 (45). We recently isolated MAb VRC01, which is capable of neutralizing about 90% of HIV-1 strains (39), and in the last 2 years, additional broadly neutralizing CD4bs (BNCD4) MAbs have been isolated from a few HIV-1-infected donors whose sera were known to contain highly potent neutralizing activity (29, 41). The genetic and structural analysis of these antibodies has identified several common characteristics: the heavy chains of these CD4bs MAbs use variable gene VH1-2 or the closely related VH1-46, and both the heavy and light chains contain high levels of somatic mutations that are required for effective neutralization.
The existence of these BNCD4 MAbs provides proof of concept that the immune system can produce potent antibodies to this conserved region of Env, but all MAbs isolated to date were from a few highly selected donors who had been HIV-1 infected for at least several years. In addition, broadly reactive serum neutralization is rarely detected before 2 to 3 years of HIV-1 infection, and it is not clear if BNCD4 antibodies would be present in some donors prior to this time but not detected in current neutralization assays. We therefore used a set of well-characterized recombinant core gp120 probes to analyze the prevalence and the time course of BNCD4-related antibodies in three cohorts of HIV-1 seroconverters. We previously described versions of gp120 core constructs (containing deletions of V1, V2, and V3 and truncations at the N and C termini) that react with most known CD4bs MAbs (15, 16), as well as the design of a resurfaced stabilized core (RSC3) that was used to isolate MAb VRC01 (39). Here, we used these gp120 core and RSC3 proteins to detect antibodies antigenically related to MAb VRC01 in the sera of HIV-1-infected subjects. We analyzed a total of 113 sera from the Center for HIV/AIDS Vaccine Immunology (CHAVI), the Center for the AIDS Programme of Research in South Africa (CAPRISA), and Amsterdam cohorts to determine the prevalence of CD4bs antibodies and further studied the evolution of this response in a subset of 18 subjects.
MATERIALS AND METHODS
Human subjects.
The sera and plasmas used in this study were from three previously described HIV-1 seroconverter cohorts. The 45 subjects from the CAPRISA 002 acute-infection cohort were women from South Africa (13); of the 30 subjects from the CHAVI 001 cohort (21 male and 8 female), 14 were from sites in Malawi, 11 from South Africa, and 5 from the United States (12, 18, 33), and 38 male subjects were in the Amsterdam Cohort (10). In the CAPRISA cohort, acute infection was diagnosed following a positive HIV-1 RNA PCR in the absence of HIV-1 antibodies or detection of HIV-1 antibodies within 3 months of a previously negative antibody test. The time of infection was defined as 14 days prior to a positive RNA PCR assay or as the midpoint between the last HIV-1-seronegative test and the first seropositive test. In the Amsterdam cohort, subjects were tested approximately every 3 months, and infection was defined as the midpoint between the last HIV-1-seronegative time point and the first positive enzyme-linked immunosorbent assay (ELISA). In the CHAVI 001 cohort, high-risk subjects were screened for HIV-1 infection by ELISA, Western blotting, and plasma RNA and categorized using the Fiebig staging classification of acute infection, which allows an estimation of the time of initial HIV-1 infection (11, 12, 20, 33). Subjects were included in this analysis if their Fiebig stage was calculated to be between 1 and 5 at screening, except for CH018, who was Fiebig 6 (11). The samples used for initial screening assays depended on the maturity of the seroconverter cohort, and the median time points were 99, 147, and 258 weeks postinfection in the CHAVI, CAPRISA, and Amsterdam cohorts, respectively. All donor sera analyzed were from time points prior to the initiation of antiretroviral therapy.
Probes for detection of CD4bs-specific antibodies.
To detect antibodies to the CD4bs region of gp120, we used several well-characterized recombinant gp120 core glycoproteins and their respective CD4bs knockout mutants. The YU2-based gp120 core, which lacks variable regions V1, V2, and V3 and is truncated at the N- and C-terminal regions, and its D368R mutant have been described previously (15–17). The replacement of aspartic acid with an arginine at position 368 within the CD4 binding pocket of gp120 has been shown to reduce or knock out binding of most CD4bs MAbs (31, 42). The resurfaced stabilized gp120 core (RSC3), along with its RSC3 Δ371I mutant (39), has been described previously. The RSC3 protein is a resurfaced version of the stabilized core of gp120 (HXB2 Ds12F123) (8, 45). Removal of the isoleucine at position 371 of RSC3 was shown to reduce binding by b12 and VRC01, and this probe pair was used to isolate the VRC01 MAb (39). The RSC3/G367R probe was designed, based on modeling of available crystal structures, to further improve the specificity of RSC3 for VRC01-like MAbs by abrogating or substantially reducing binding to MAbs b12 and b13 while maintaining VRC01 binding. RSC3 Δ371I/P363N was designed as an additional CD4bs knockout mutant of RSC3. Of note, all the RSC3 probes were designed to maintain binding to MAb 2G12, which reacts with outer-domain glycans. Thus, both the RSC3 probes and their CD4bs knockout mutants reacted similarly with 2G12, and the MAb served as a positive control in ELISAs.
HIV-1 core protein binding assays.
ELISAs were performed as previously described (41). Briefly, 96-well ELISA plates were coated with 2 μg/ml of the specified recombinant protein in phosphate-buffered saline (PBS) overnight at 4°C. The following day, the plates were blocked with B3T buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 3.3% fetal bovine serum, 2% bovine albumin, 0.07% Tween 20) and incubated with 4-fold serial dilutions of heat-inactivated sera starting at a dilution of 1:100, followed by peroxidase-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch). All incubations were for 1 h at 37°C, and all volumes were 100 μl, except for blocking, which was 200 μl. The plates were washed between incubations with 0.1% Tween 20 in PBS, detected using SureBlue TMB substrate (Kirkegaard & Perry Laboratories), and subsequently read at 450 nm. Endpoint titers were calculated based on the final reciprocal serum dilution with a background-corrected optical density (OD) greater than or equal to 0.1. Sera were considered reactive when the reciprocal endpoint titer was greater than 200. Anti-gp120 MAb 2G12 was purchased from Polymun Scientific Inc. (Vienna, Austria) (34), and CD4-induced (CD4i) MAb 17b was provided by James Robinson (Tulane University) (30). Anti-CD4bs MAbs were provided as follows: F91 was provided by James Robinson (Tulane University, New Orleans, LA) (22), MAb F105 was provided by Marshall Posner (Dana Farber Cancer Institute, Boston, MA) (27), MAb b6 was provided by Dennis Burton (Scripps Research Institute, La Jolla, CA) (1), MAb m18 IgG was provided by Dimiter Dimitrov (National Cancer Institute, Frederick, MD) (43), and HJ16 was provided by Davide Corte (Institute for Research in Biomedicine, Bellinzona, Switzerland) (7). The heavy- and light-chain genes of MAbs b12 (5), b13 (1), VRC01 (39), VRC-PG04, VRC-CH31 (41), NIH45-46, 3BNC60, 3BNC117, 12A12, and 12A21 (29) were synthesized and cloned into the expression vector VRC-1012 containing a modified cytomegalovirus immediate-early 1 gene promoter (CMV/R promoter) and the coding sequence for the constant region of IgG1. Full-length IgGs were expressed from transient transfection of 293F cells and purified by affinity chromatography using HiTrap Protein A HP Columns (GE Healthcare).
HIV-1 neutralization assays.
HIV-1 neutralization was measured using a single round of infection by Env pseudoviruses and Tzm-bl target cells as previously described (38, 40). CD4bs-directed neutralization was assessed in a neutralization competition assay format using specific protein probes to block antibody-mediated neutralization, as previously described (39). Briefly, 25 μg/ml of RSC3 or 25 μg/ml of the mutant RSC3 Δ371I/P363N or PBS was incubated with sera serially diluted 4-fold from 1:20 for 15 min, after which 40 μl of HXBc2 pseudovirus (multiplicity of infection [MOI], approximately 0.1) was added for a 30-min incubation. Tzm-bl cells were added at a concentration of 104 per well, and the single-round infection proceeded for 48 h. All incubations were at 37°C, and each infection was performed in duplicate wells of a 96-well flat-bottom culture plate. Neutralization curves were fitted by nonlinear regression using a four-parameter hill slope equation programmed into JMP statistical software (JMP 5.1; SAS Institute Inc., Cary, NC). The 50% inhibitory dilutions (ID50s) were reported as the reciprocal serum dilutions required to inhibit infection by 50%. CD4bs-directed activity was calculated as a ≥30% reduction in the ID50 values of the sera in the presence of RSC3 compared to RSC3 Δ371I/P363N.
RESULTS
Probes designed to preferentially react with broadly neutralizing CD4bs antibodies.
The known CD4bs MAbs bind strongly to recombinant gp120, but only a subset of these MAbs are able to access the CD4bs of the native viral spike and potently neutralize primary HIV-1 isolates. These neutralizing CD4bs antibodies include b12, HJ16, VRC01, VRC-PG04, and VRC-CH31 (5, 7, 41), as well as MAbs from several additional donors that were recently described (29). The prototypical MAbs F105, m18, b6, b13, and F91 are examples of CD4bs MAbs with neutralization limited to a subset of tier 1 HIV-1 strains. Here, we evaluated the abilities of various protein probes to distinguish between these specificities. The gp120 core has been previously shown to bind CD4bs-directed antibodies (8, 16), and we confirmed this pattern of reactivity. All CD4bs MAbs tested reacted strongly with gp120 core by ELISA and, with the exception of MAbs b6 and HJ16, bound less efficiently to the D368R mutant (Table 1). In contrast, the RSC3 protein did not bind to the weakly neutralizing CD4bs MAbs F105, F91, and b6 but maintained strong reactivity to the broader and more potently neutralizing MAbs, such as b12 and VRC01. Since RSC3 also retained reactivity to MAbs b13 and m18, which are MAbs with limited neutralization breadth, we designed additional mutant versions of RSC3. One such mutant, RSC3/G367R, was designed to create a steric clash for MAb b12 and b13 binding while causing little interference with VRC01 binding. ELISA data verified that the RSC3/G367R probe displayed reduced binding to b12, b13, and m18 while maintaining strong ELISA reactivity to VRC01. The RSC3/G367R probe also maintained modestly strong binding to the other BNCD4 MAbs of VH1-2 origin (VRC-PG04, VRC-CH31, NIH45-46, 3BNC60, 3BNC117, and 12A12), with the exception of MAb 12A21 (Table 1). Thus, the RSC3/G367R mutant was a useful adjunct to RSC3 in antigenically distinguishing most BNCD4 MAbs from the weaker neutralizing group. We, therefore used the gp120 core, as well as these resurfaced core protein probes, to detect CD4bs antibodies in sera and to further discriminate antibody specificities related to VRC01 and BNCD4 MAbs.
Table 1.
Antigenic profile of proteins used to characterize CD4bs antibody specificity
| Antibodya | Bindingd |
||||||
|---|---|---|---|---|---|---|---|
| YU2 gp120-based proteinsb | Resurfaced core proteinsc | ||||||
| Type | Name | gp120 core | gp120 core D368R | RSC3 | RSC3/G367R | RSC3 Δ371I | RSC3 Δ371I/ P363N |
| CD4bs | F105 | ++++ | − | − | − | − | − |
| F91 | ++++ | − | − | − | − | − | |
| m18 | ++++ | − | ++++ | − | − | − | |
| b6 | ++++ | ++++ | − | − | − | − | |
| b13 | ++++ | − | ++++ | + | − | − | |
| b12 | ++++ | − | ++++ | + | − | − | |
| VRC01 | ++++ | ++ | ++++ | +++ | ++ | + | |
| VRC-PG04 | ++++ | ++ | ++++ | +++ | +++ | ++ | |
| VRC-CH31 | +++ | ++ | +++ | ++ | + | − | |
| NIH45-46 | ++++ | − | ++++ | ++ | − | − | |
| 3BNC60 | ++++ | +++ | ++++ | ++ | − | − | |
| 3BNC117 | ++++ | +++ | ++++ | ++ | − | − | |
| 12A12 | ++++ | +++ | ++++ | ++ | + | − | |
| 12A21 | ++++ | +++ | ++++ | + | − | − | |
| HJ16 | ++ | ++ | ++ | +++ | ++ | + | |
| CD4i | 17b | − | − | − | − | − | − |
| Glycan | 2G12 | + | − | +++ | +++ | +++ | ++ |
Antibodies are grouped by category: CD4bs, CD4i, and glycan reactive (Glycan). Of the CD4bs MAbs, VRC01, VRC-PG04, VRC-CH31, NIH45-46, 3BNC60, 3BNC117, 12A12, and 12A21 are all broadly neutralizing antibodies derived from the VH1-2*02 germ line allele.
Mutant residue numbers are based on the HXBc2 sequence, and gp120 D368R is the CD4bs knockout mutation.
Mutant residue numbers are based on the HXBc2 sequence. RSC3 Δ371I has an isoleucine deletion at position 371, and RSC3 Δ371I/P363N contains an additional P363N mutation. Both were designed as CD4bs knockout versions of RSC3.
Binding was categorized based on ELISA OD at 450 nm (OD450) values at the highest concentration of antibody tested (5 μg/ml) and the 50% effective concentration (EC50) values as follows: ++++, OD450 ≥ 3.0 and EC50 ≥ 0.10; +++, OD450 ≥ 3.0 and EC50 > 0.10; ++, 1.0 ≤ OD450 < 3.0; +, 0.2 ≤ OD450 < 1.0; and −, OD450 < 0.2.
CD4bs antibodies are commonly elicited in HIV-1 infection.
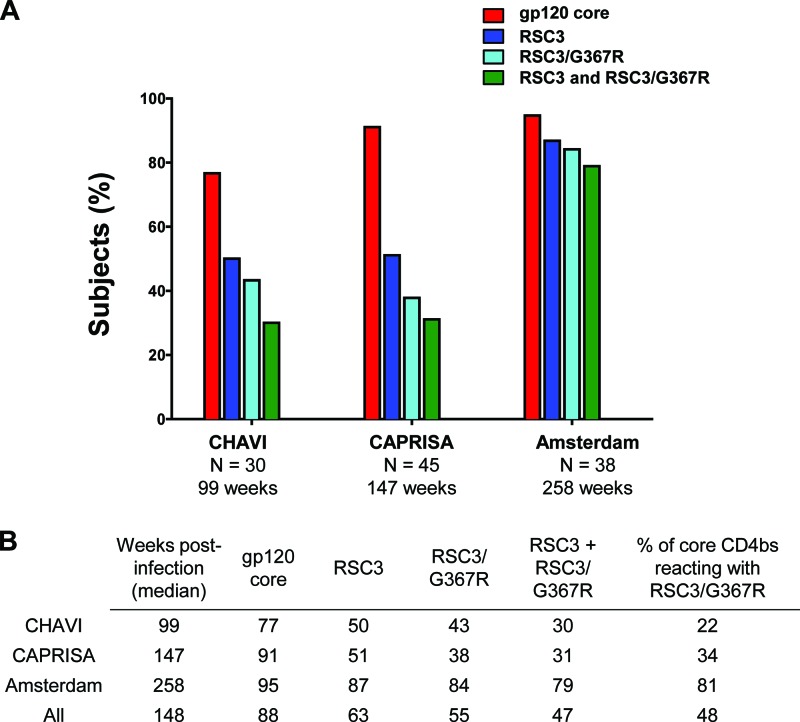
The time period of longitudinal follow-up for the three HIV-1 cohorts varied from approximately 99 weeks (CHAVI) to greater than 250 weeks (Amsterdam). Since we were interested in the evolution of the CD4bs antibody response, we first screened sera obtained from a late time point in each cohort. Thus, the 30 CHAVI sera were obtained at ∼99 weeks postinfection, the 45 CAPRISA sera were from ∼147 weeks, and the 38 Amsterdam sera were from ∼258 weeks postinfection. All 113 sera were screened by ELISA for reactivity to the gp120 core and RSC3 protein probes described above. A serum was considered reactive to a specific probe if its endpoint titer was greater than 1:200 and there was a greater than 2.5-fold loss of activity on the cognate CD4bs mutant. A majority of sera in each cohort contained CD4bs antibodies to the gp120 core, indicating the overall immunogenicity of the CD4bs during infection (Fig. 1A). In each cohort, the prevalence of serum reactivity to the more specific RSC3 probes was lower than the overall prevalence of core CD4bs antibodies. Since most BNCD4 MAbs reacted to both RSC3 and RSC3/G367R (Table 1), we classified sera with reactivity to both of these proteins as containing antibodies similar to this group of broadly neutralizing VRC01-like antibodies that derive from the VH1-2 heavy-chain germ line gene. Based on this criterion, sera with VRC01-like binding activity were found in 30%, 31%, and 79% of the CHAVI, CAPRISA, and Amsterdam cohorts, respectively. It is notable that the Amsterdam sera were obtained approximately 5 years postinfection compared to less than 3 years for CAPRISA and less than 2 years for the CHAVI cohort. Among those subjects that made any CD4bs antibody response (i.e., gp120 core reactive), the percentages that also made RSC3-reactive antibodies were 22%, 34%, and 81% in the CHAVI, CAPRISA, and Amsterdam cohorts, respectively (Fig. 1B). This observation suggests that a longer time of infection may foster the evolution of CD4bs antibodies that attain reactivity to RSC3.
Fig 1.
Prevalence of CD4 binding site antibodies among donors in three HIV-1 seroconverter cohorts. (A) The percentages of donors reacting by ELISA with the indicated protein probes are shown by the colored bars. A 2.5-fold or greater difference in serum binding to the wild type compared to the corresponding CD4bs knockout mutant probe was considered positive. For each cohort, the number of subjects analyzed (N) and the median time post-HIV-1 infection are indicated. (B) Summary of percent positive serum ELISA data for each core protein probe or the combination of both RSC3 and RSC3/G367R. Samples reactive with both RSC3 and RSC3/G367R have binding profiles similar to those of BNCD4 MAbs. The last column indicates the percentages of sera reactive with gp120 core that also reacted with both RSC3 and RSC3/G367R.
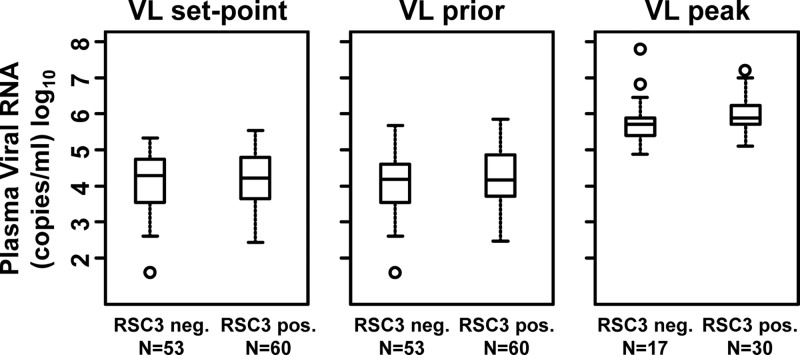
Since the development of serum neutralizing antibody breadth has been associated with the level of plasma viremia (13, 26, 28), we analyzed the association between the plasma viral load and the development of RSC3-reactive CD4bs antibodies. Among the 113 antiretroviral-naïve subjects, there was no association between the viral-load set point (the mean viral load between 24 weeks postinfection and the time of serum screening) or the mean viral load 1 year prior to serum screening and the presence of RSC3 binding antibodies in sera (Fig. 2), nor was there a correlation between the peak viral load (analyzed in a subset of 47 subjects) and the development of RSC3-reactive antibodies. An analysis of peripheral CD4 T cell levels also found no association, though the large majority of subjects had CD4 T cell levels greater than 300/μl (data not shown). Thus, although the level of viremia may play a role in the development of neutralizing antibodies, and perhaps even BNCD4-like antibodies, there was no association between the viral load and the presence of RSC3 binding antibodies in these three cohorts.
Fig 2.
Lack of association between plasma viral load and RSC3-reactive CD4bs antibodies. RSC3-positive sera were those reactive with both RSC3 and RSC3/G367R. The viral load (VL) was calculated as the geometric mean of VL values between 24 weeks postinfection and the time of screening (set point), VL values 52 weeks prior to screening (prior), or the peak viral load in a subset of donors with early sampling time points (peak). The box plot shows the median values and the 25% and 75% quartiles. The range and outliers are shown by the bars and circles, respectively. neg., negative; pos., positive.
Evolution of the CD4bs antibody response.
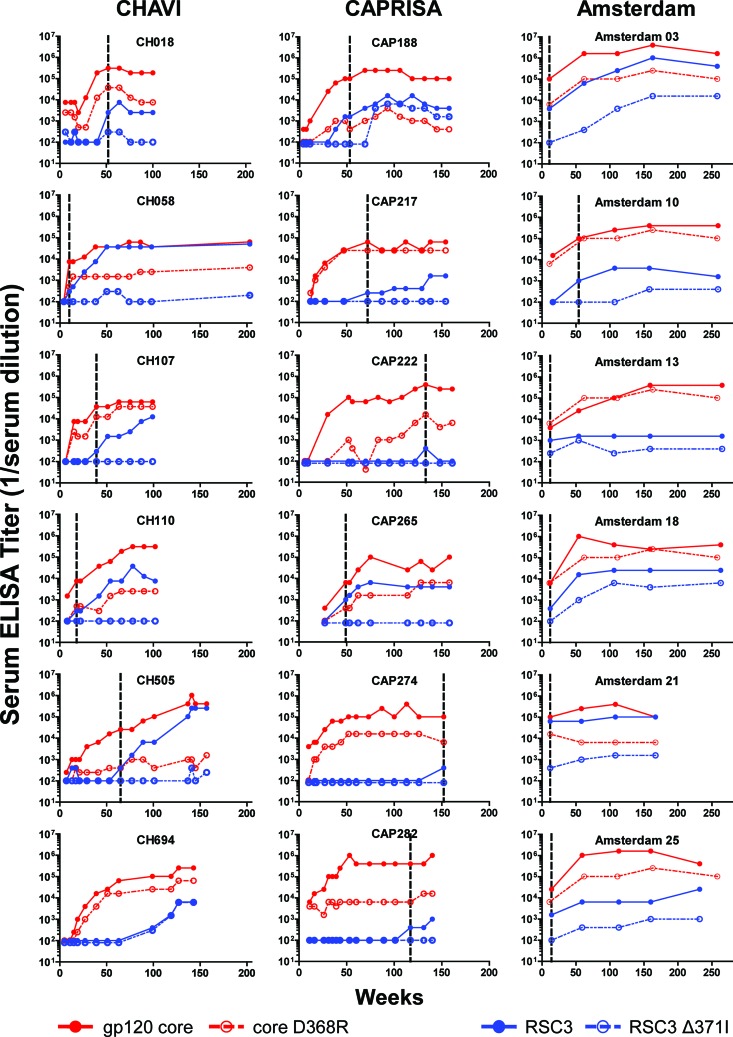
To assess the kinetics of CD4bs antibody development, we analyzed longitudinal serum samples for six subjects from each cohort whose sera contained RSC3-reactive antibodies at a later time point (Table 2). CD4bs antibodies that bind to gp120 core were detectable within 16 weeks postinfection in 15 of 18 subjects (Fig. 3). Among the six CHAVI subjects for whom we had the earliest time points postinfection, serum CD4bs antibodies arose between 7 and 15 weeks after infection, in agreement with previously published work suggesting the early, but not immediate, development of CD4bs antibodies (33). CD4bs antibodies that bound to the RSC3 probes generally developed after these core antibodies, ranging from as early as 10 weeks (CH058) to as late as 152 weeks (CAP274) postinfection. Two subjects from the CHAVI cohort, CH058 and CH505, demonstrated a common pattern of antibody development in which CD4bs core antibodies were detected first, followed by RSC3-reactive CD4bs antibodies; however, other patterns were observed (Fig. 3). Overall, these data suggest that CD4bs antibodies occur in most subjects and develop early, while antibodies that bind to the more precise neutralizing epitope defined by VRC01 and related BNCD4 MAbs develop later, although usually within the first 2 years of infection.
Table 2.
Serum endpoint ELISA titers against protein probes used to characterize CD4bs reactivity
| Subject | Wka | Titer |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| YU2 core | D368R | Ratiob | RSC3 | RSC3 Δ371I | Ratiob | RSC3/G367R | RSC3 Δ371I/P363N | Ratiob | ||
| CH018 | 100 | 187,500 | 7,500 | 25 | 2,500 | 100 | 25 | 300 | 100 | 3.0 |
| CH058 | 98 | 37,500 | 2,500 | 15 | 37,500 | 100 | 375 | 2,500 | 100 | 25 |
| CH107 | 99 | 62,500 | 37,500 | 1.7 | 12,500 | 100 | 125 | 300 | 100 | 3.0 |
| CH110 | 102 | 312,500 | 2,500 | 125 | 7,500 | 100 | 75 | 100 | 100 | NC |
| CH505 | 101 | 102,400 | 400 | 256 | 6,400 | 100 | 64 | 100 | 100 | NC |
| CH694 | 99 | 102,400 | 25,600 | 4.0 | 400 | 400 | 1.0 | 1,600 | 250 | 6.4 |
| CAP188 | 145 | 102,400 | 400 | 256 | 4,000 | 1,600 | 2.5 | 6,400 | 250 | 26 |
| CAP217 | 154 | 64,000 | 25,600 | 2.5 | 1,600 | 100 | 16 | 400 | 100 | 4.0 |
| CAP222 | 148 | 256,000 | 4,000 | 64 | 100 | 100 | n/c | 100 | 100 | NC |
| CAP265 | 158 | 102,400 | 6,400 | 16 | 4,000 | 100 | 40 | 400 | 100 | 4.0 |
| CAP274 | 152 | 102,400 | 6,400 | 16 | 400 | 100 | 4.0 | 100 | 100 | NC |
| CAP282 | 156 | 1024,000 | 16,000 | 64 | 1,000 | 100 | 10 | 100 | 100 | NC |
| Amsterdam 03 | 258 | 1638,400 | 102,400 | 16 | 409,600 | 16,000 | 25.6 | 64,000 | 250 | 256 |
| Amsterdam 10 | 259 | 409,600 | 102,400 | 4.0 | 1,600 | 400 | 4.0 | 1,000 | 400 | 2.5 |
| Amsterdam 13 | 265 | 409,600 | 25,600 | 16 | 1,600 | 400 | 4.0 | 1,600 | 100 | 16 |
| Amsterdam 18 | 263 | 409,600 | 25,600 | 16 | 25,600 | 6,400 | 4.0 | 4,000 | 1,600 | 2.5 |
| Amsterdam 21 | 167 | 102,400 | 6,400 | 16 | 102,400 | 1,600 | 64 | 6,400 | 1,000 | 6.4 |
| Amsterdam 25 | 232 | 409,600 | 64,000 | 6.4 | 25,600 | 1,000 | 25.6 | 1,000 | 400 | 2.5 |
Number of weeks post HIV-1 infection, as defined in Materials and Methods.
Ratio of previous two endpoint titers. Ratio was not calculated (NC) when reciprocal serum endpoint titers were both <200.
Fig 3.
Kinetics of the CD4 binding site antibody responses in 18 selected subjects. Serial longitudinal serum samples from six subjects from each cohort were analyzed. The ELISA endpoint titers (y axis) are graphed for multiple weeks after HIV-1 infection (x axis). The line colors correspond to the protein probes shown in Fig. 1. The solid lines show reactivity to the probe, while the dashed lines indicate the corresponding CD4bs knockout mutant. The first time point reactive with RSC3 is indicated by the vertical dotted lines.
Serum neutralizing activity.
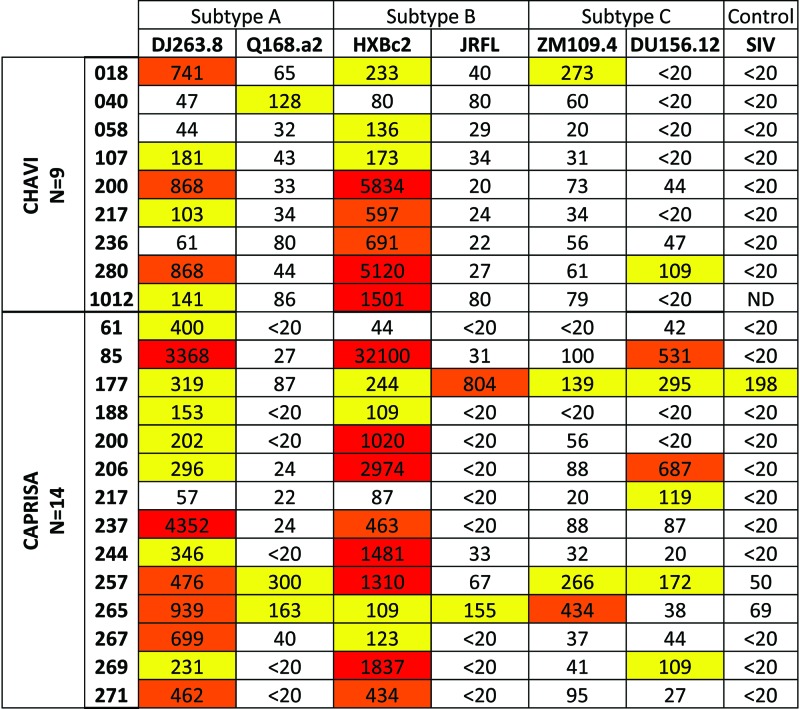
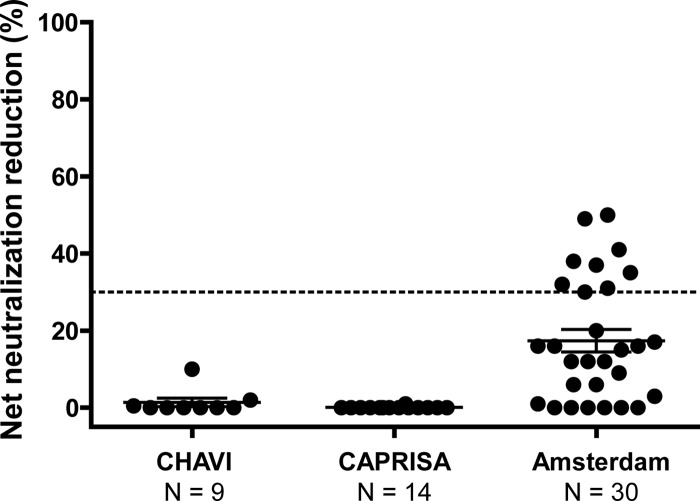
Subjects from the CHAVI and CAPRISA cohorts whose sera exhibited the antigenic profile of BNCD4 antibodies (reactivity to the RSC3 and RSC3/G367R probes) (Fig. 1A) were screened against a panel of 6 heterologous Env pseudoviruses (Fig. 4). The 23 sera analyzed were from the same time point shown in Fig. 1 (medians, 99 weeks postinfection for the CHAVI subjects and 147 weeks postinfection for the CAPRISA subjects). These sera displayed relatively modest neutralization potency and breadth. This finding is consistent with prior published data demonstrating that neutralization breadth generally develops after 2 to 3 years of HIV-1 infection (13, 21, 28). The lack of potent neutralization in these sera, which contain RSC3-reactive antibodies, suggests that this antibody specificity has not yet achieved either the affinity or serum concentration necessary to mediate broad virus neutralization. Unfortunately, the volume of sera required for these neutralization assays was not available from the Amsterdam cohort. Since most of the sera neutralized the sensitive HXBc2 viral strain, we performed competition neutralization assays on all 53 RSC3-reactive sera to determine if the virus neutralization was mediated by CD4bs antibodies. Protein competition assays were performed using the RSC3 and RSC3 Δ371I/P363N proteins, which do not bind CD4 and therefore do not block viral entry (39, 41). A 30% net reduction in the serum ID50 value after incubation with RSC3 (compared to RSC3 Δ371I/P363N) was considered evidence of CD4bs-directed neutralization. None of the CHAVI or CAPRISA sera contained detectable CD4bs-directed neutralization activity, but 9 sera from the Amsterdam cohort had detectable CD4bs neutralizing antibodies that were blocked by RSC3 (Fig. 5).
Fig 4.
Neutralization breadth of RSC3-reactive serum binding. Selected sera from the CHAVI and CAPRISA cohorts (rows) that were ELISA reactive with both RSC3 and RSC3/G367R were screened for neutralization against six Env pseudoviruses (columns) representing three genetic subtypes. All viruses are tier 2 primary Envs, except for the tier 1 HXBc2, which was used to allow subsequent neutralization competition assays. Reciprocal serum ID50 values of >100 are highlighted in yellow, >400 in orange, and >1,000 in red. SIV, simian immunodeficiency virus.
Fig 5.
Analysis of serum CD4bs-directed neutralization. Sera that were ELISA reactive with both RSC3 and RSC3/G367R (BNCD4-like probes) were assayed for inhibition of neutralization, using both RSC3 and RSC3/Δ371I/P363N as inhibitors. The net percent reduction in neutralization of HXBc2 attributed to RSC3 compared to RSC3/Δ371I/P363N is plotted (y axis), with each dot representing one subject. The number of subjects analyzed in each cohort is indicated on the x axis. A greater than 30% reduction is considered positive. The mean and standard error of the mean (SEM) for each cohort are indicated by the long and short horizontal lines.
DISCUSSION
The CD4bs of HIV-1 gp120 is a desirable target for vaccine design because it is a highly conserved region of Env that interacts with the host CD4 receptor and because MAbs to the CD4bs can potently neutralize the large majority of HIV-1 strains. Thus, a major aim of HIV-1 vaccination is to elicit BNCD4 antibodies similar to those generated during HIV-1 infection. However, not all HIV-1-infected individuals develop these neutralizing antibodies, and the existing BNCD4 MAbs have been isolated from highly selected individuals who were infected for a minimum of several years. In this study, we sought to determine the prevalence of HIV-1-infected subjects with serum antibodies similar to the known BNCD4 MAbs. Since potent primary isolate neutralization may take years to reach detectable serum levels, we used a set of gp120 probes to detect serum CD4bs antibodies with antigenic profiles similar to those of VRC01 and related BNCD4 MAbs. In addition, we studied the time course of development of general CD4bs- and BNCD4-related antibodies and show that the latter antibody specificity occurs in a subset of those with CD4bs antibodies and develops weeks to months later.
We used a set of well-characterized gp120 core and RSC3 protein probes, and their cognate CD4bs knockout mutants, to detect serum antibodies. The antigenic profiles of these proteins were confirmed with a panel of known MAbs, and preferential reactivity to the RSC3 and RSC3/G367R probes was shown to distinguish most of the BNCD4 MAbs (e.g., VRC01, VRC-PG04, and VRC-CH31) from the remainder of the CD4bs MAbs, which demonstrated weak or no binding. However, it remains possible that the RSC3/G367R probe reacts strongly with some as yet unidentified non-broadly neutralizing CD4bs antibodies. As such, the use of these probes is clearly an indirect measure of the presence of neutralizing antibodies, such as VRC01. We have therefore attempted to be cautious in our conclusions, referring to serum reactivity patterns as evidence of antibody specificities that are similar to those of known broadly neutralizing antibodies. The fact that RSC3-reactive antibodies were detected in sera with weak HIV-1 neutralization activity suggests that either the level of RSC3-reactive antibodies was too low to mediate neutralization or, as noted above, that these antibodies were not sufficiently affinity matured to mediate virus neutralization. Of note, we were able to confirm the presence of CD4bs-directed neutralizing antibodies in some subjects via serum RSC3 competition neutralization assays; however, the assay detects CD4bs neutralizing activity only when it constitutes at least 30% of serum neutralization.
Overall, our data show that the CD4bs of gp120 is highly immunogenic. More than 80% of the subjects studied had evidence of such antibodies at time points 99 or more weeks postinfection, and longitudinal studies indicated that CD4bs antibodies were present within 16 weeks postinfection. However, a more detailed analysis suggests that the precise region of the CD4bs bound by MAbs such as VRC01 is less immunogenic. At approximately 2 to 3 years postinfection, only 30% and 31% of the CHAVI and CAPRISA subjects, respectively, had CD4bs antibodies with specificities similar to those of BNCD4 MAbs. This percentage may increase with the time of infection, because 79% of the Amsterdam cohort (infected for ∼5 years) had the antibodies. The longitudinal sampling in a subset of 18 seroconverters confirms the delayed appearance of BNCD4-like antibodies, which often arose months after the first appearance of core-directed CD4bs antibodies. It is possible that the different demographics and infecting viral clades among the cohorts account for these differences, but since the CD4bs is highly conserved, this seems unlikely. Of note, 21 of the 38 sera from the Amsterdam cohort were included in our study because there was some prior evidence of moderate neutralizing activity and the remaining 17 sera did not show such activity (10). The percentages of sera with RSC3 binding activity did not differ among these two sets of sera, and we do not think this selection accounts for the higher RSC3 reactivity in the cohort.
The virological or immunological characteristics that are associated with the development of neutralizing antibodies to the CD4bs remain to be elucidated. In our cohorts of antiretroviral-naive subjects, there was no association between the peak or set point plasma viral load and the presence of RSC3-reactive antibodies. Thus, it is not yet clear why only a subset of HIV-1-infected individuals make antibodies that bind epitopes similar to those of BNCD4 MAbs, and it is possible that virologic or host genetic factors play a role. Some subjects that can make BNCD4 can also make broadly neutralizing V2V3-directed antibodies (4, 32), suggesting that common factors may affect the ability to make broadly neutralizing antibodies.
Among subjects in this study who generated BNCD4-like antibody responses, the longitudinal data suggested that weeks to months were required for the induction of these antibodies. This finding may correspond to the high level of affinity maturation that is characteristic of CD4bs neutralizing antibodies with heavy-chain nucleotide substitution rates of 24% to 31% compared to germ line antibody gene sequence (29, 39). Thus, it is possible that the delayed induction of BNCD4-like antibodies is due in part to the level of affinity maturation required for the antibody to acquire potent neutralization activity. To test this hypothesis, MAbs from these subjects could be isolated and analyzed. Once the gene family derivation of specific neutralizing MAbs is known, it should be possible to use deep-sequencing technologies to track the genetic footprint of the antibody lineage from early in infection through the time point of potent neutralization. Such studies could provide insights into the pathway for the development of neutralizing antibodies to the CD4bs and could be applied similarly to understand the evolution of antibodies to other neutralization epitopes. This highlights the value of HIV-1 seroconverter cohorts that acquire samples early after infection and that follow subjects over time to the development of broadly neutralizing antibodies.
In summary, these data from three HIV-1 seroconverter cohorts provide the first prospective estimates of the prevalence of neutralizing antibodies to the CD4bs of gp120. While this region of the HIV-1 Env is highly immunogenic overall, the precise region bound by neutralizing antibodies, such as VRC01, VRC-PG04, and VRC-CH31, is less immunogenic, and induction of such antibodies is delayed compared to other CD4bs antibody specificities. Future studies to understand immunologic factors and antibody maturation pathways associated with the successful development of BNCD4 antibodies may provide insights for the design of improved immunogens and immunization strategies.
ACKNOWLEDGMENTS
This research was supported by CHAVI grant AI067854 from the Division of AIDS, NIAID, NIH, and by the intramural research program of the Vaccine Research Center, NIAID, NIH. We thank the U.S. National Institutes of Health Comprehensive International Program of Research on AIDS (CIPRA) (grant AI51794) and the Columbia University-Southern African Fogarty AIDS International Training and Research Programme (AITRP) (grant D43TW00231) for the research infrastructure and training that made the CAPRISA 002 Acute Infection study possible. The Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, the University Medical Center Utrecht, and the Jan van Goyen Clinic, are part of the Netherlands HIV Monitoring Foundation and are financially supported by the Center for Infectious Disease Control of the Netherlands National Institute for Public Health and the Environment. This study was financially supported by the Netherlands Organization for Scientific research (grant 918.66.628), the European Community's Six Framework Program European HIV Enterprise (EUROPRISE; FP6/2007-2012) under grant agreement 037611, and the European Community's Seventh Framework Program Next Generation HIV-1 Immunogens Inducing Broadly Reactive Neutralizing Antibodies (NGIN) (FP7/2007-2013) under grant agreement 201433.
We thank the study participants and the clinical and laboratory staffs of all cohorts and at each clinical site for providing specimens. We thank the CHAVI 001 clinical team (T. Taha, N. Kumwenda, G. Kamanga, F. Martinson, I. Hoffman, H. Rees, V. Edward, P. Thumbi, J. Eron, C. Gay, J. Kuruc, and G. Churchyard) and Nicole Casale for providing technical assistance with serum ELISAs.
Footnotes
Published ahead of print 9 May 2012
REFERENCES
- 1. Barbas CF, III, et al. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. U. S. A. 89:9339–9343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Binley JM, et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonsignori M, et al. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85:9998–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonsignori M, et al. 2012. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J. Virol. 86:4688–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burton DR, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 6. Cardoso RM, et al. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22:163–173 [DOI] [PubMed] [Google Scholar]
- 7. Corti D, et al. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805 doi:10.1371/journal.pone.0008805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dey B, et al. 2009. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 5:e1000445 doi:10.1371/journal.ppat.1000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diskin R, et al. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Euler Z, et al. 2010. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J. Infect. Dis. 201:1045–1053 [DOI] [PubMed] [Google Scholar]
- 11. Fiebig EW, et al. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879 [DOI] [PubMed] [Google Scholar]
- 12. Goonetilleke N, et al. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gray ES, et al. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gray ES, et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kwong PD, et al. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329–1339 [DOI] [PubMed] [Google Scholar]
- 16. Li Y, et al. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, et al. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu P, et al. 2011. Dynamic antibody specificities and virion concentrations in circulating immune complexes in acute to chronic HIV-1 infection. J. Virol. 85:11196–11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McLellan JS, et al. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10:11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mikell I, et al. 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7:e1001251 doi:10.1371/journal.ppat.1001251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moore JP, Ho DD. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67:863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morris L, et al. 2011. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One 6:e23532 doi:10.1371/journal.pone.0023532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ofek G, et al. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pejchal R, et al. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piantadosi A, et al. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J. Virol. 83:10269–10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. 6:7–14 [PubMed] [Google Scholar]
- 28. Sather DN, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scheid JF, et al. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thali M, et al. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thali M, et al. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomaras GD, et al. 2011. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J. Virol. 85:11502–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tomaras GD, et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trkola A, et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walker LM, et al. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walker LM, et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walker LM, et al. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6:e1001028 doi:10.1371/journal.ppat.1001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wei X, et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu X, et al. 2009. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J. Virol. 83:10892–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu X, et al. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wyatt R, et al. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711 [DOI] [PubMed] [Google Scholar]
- 43. Zhang MY, et al. 2003. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods 283:17–25 [DOI] [PubMed] [Google Scholar]
- 44. Zhou T, et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou T, et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]