Abstract
Specific major histocompatibility complex (MHC) class I alleles are associated with an increased frequency of spontaneous control of human and simian immunodeficiency viruses (HIV and SIV). The mechanism of control is thought to involve MHC class I-restricted CD8+ T cells, but it is not clear whether particular CD8+ T cell responses or a broad repertoire of epitope-specific CD8+ T cell populations (termed T cell breadth) are principally responsible for mediating immunologic control. To test the hypothesis that heterozygous macaques control SIV replication as a function of superior T cell breadth, we infected MHC-homozygous and MHC-heterozygous cynomolgus macaques with the pathogenic virus SIVmac239. As measured by a gamma interferon enzyme-linked immunosorbent spot assay (IFN-γ ELISPOT) using blood, T cell breadth did not differ significantly between homozygotes and heterozygotes. Surprisingly, macaques that controlled SIV replication, regardless of their MHC zygosity, shared durable T cell responses against similar regions of Nef. While the limited genetic variability in these animals prevents us from making generalizations about the importance of Nef-specific T cell responses in controlling HIV, these results suggest that the T cell-mediated control of virus replication that we observed is more likely the consequence of targeting specificity rather than T cell breadth.
INTRODUCTION
Studies of human immunodeficiency virus (HIV) “elite controllers” implicate major histocompatibility complex (MHC) class I genes, and by extension the CD8+ T cell responses they restrict, in immunologic containment of viral replication (25). Identifying the specific attributes of CD8+ T cell responses that are fundamental to HIV control could clarify whether immunologic containment is possible for the majority of infected individuals who do not become elite controllers and could facilitate the design of prophylactic vaccines that elicit cellular immune responses capable of mediating viral control.
A T cell response directed against a large number of distinct viral epitopes, i.e., high “T cell breadth,” has frequently been proposed to be a favorable immunologic attribute (21, 30). This breadth hypothesis is attractive because it is conceptually simple: if immunologic control fails because too few epitope-specific CD8+ T cell responses are elicited, interventions designed to broaden the CD8+ T cell response should be beneficial. The data in support of the breadth hypothesis are mixed. Perhaps the most striking supporting evidence is the observation that individuals who are MHC homozygous experience faster disease progression than fully heterozygous individuals (8, 29). Because MHC-heterozygous individuals have twice as many different MHC class I molecules available to bind and present HIV-derived peptides, the theoretical breadth of their CD8+ T cell response at any moment is double that of their MHC-homozygous counterparts. Several studies have suggested that broadly directed CD8+ T cell responses are associated with low virus loads and/or high CD4+ T cell counts (9, 11, 12). However, other studies of HIV-infected humans have yielded conflicting results (1, 16, 19). Furthermore, Indian rhesus macaques that express Mamu-B*17, an MHC class I allele associated with simian immunodeficiency virus (SIV) elite control, do not segregate into controllers and progressors based on the breadth of their CD8+ T cell responses (13, 20). All of these studies are confounded by complex host MHC genetics, which contributes to the persisting controversy.
Unlike humans and other macaques, Mauritian cynomolgus macaques (MCM) are geographically isolated and descend from a small founder population that arrived on Mauritius within the last 500 years (17). Consequently, MCM have extremely limited MHC genetic diversity, with only seven major haplotypes (6, 22), termed M1 through M7. The MHC alleles on each of these seven haplotypes are known.
In an earlier study, we showed that MHC-heterozygous MCM maintained lower chronic-phase viremia than MHC-homozygous animals, although this was in a cohort enriched for the M3 haplotype (24). Here, we expand this study to include 27 SIVmac239-infected MCM representing all seven MHC haplotypes. The previous observation that M3 homozygous macaques have higher viral loads than do M1/M3 heterozygous macaques was confirmed. Overall, however, we found no significant correlation between MHC zygosity and viral load. Control of SIV replication, defined as a plasma viral load below 1,000 RNA copies/ml, was observed only in MCM possessing the M1 or M7 haplotypes, regardless of MHC zygosity. Moreover, animals that successfully controlled viral replication consistently mounted durable, immunodominant responses against a region of the Nef protein. These responses were subdominant or absent in animals that failed to control SIV replication.
MATERIALS AND METHODS
Animals and infections.
This study was based on an initial prospective evaluation of 18 MCM (6 M1/M1, 6 M1/M3, and 6 M3/M3). Following our initial study, we included 10 additional MCM of various haplotypes (Table 1). Animals were infected intrarectally with molecularly cloned SIVmac239 nef-open virus (50% tissue culture infective dose [TCID50], from 100 to 50,000), accession number M33262 (27). All animals were cared for by the Wisconsin National Primate Research Center (University of Wisconsin, Madison, WI) according to experimental protocols approved by the University of Wisconsin Graduate School Animal Care and Use Committee. A panel of microsatellite markers spanning the MHC region was used to determine individual haplotypes as described previously (31). Genotypes were confirmed using Roche/454 pyrosequencing (6).
Table 1.
Animals used in study, ordered by plasma viral load
| Animal name | Haplotype | Infection dose (TCID50) | Data from 52 weeks postinfection |
||
|---|---|---|---|---|---|
| Log viral load | No. of SFCs per 106 PBMC | No. of T cell responses | |||
| cy0321 | M1/M1 | 50,000 | 1 | 5,780 | 6 |
| cy0322 | M1/M1 | 50,000 | 1 | ||
| cy0326 | M1/M3 | 50,000 | 1 | 2,250 | 6 |
| cy0323 | M1/M1 | 50,000 | 1.7 | 1,410 | 5 |
| cy0330 | M1/M3 | 50,000 | 1.7 | 180 | 1 |
| cy0342 | M1/M2 | 50,000 | 1.7 | 1,700 | 9 |
| cy0389 | M4/M7 | 1,000 | 1.7 | 405 | 4 |
| cy0390 | M7/M7 | 500 | 1.7 | 495 | 3 |
| cy0327 | M1/M3 | 50,000 | 2 | 2,700 | 5 |
| cy0332 | M3/M3 | 50,000 | 4 | 6,450 | 7 |
| cy0334 | M3/M3 | 50,000 | 4 | 6,620 | 5 |
| cy0335 | M3/M3 | 50,000 | 4 | 2,140 | 2 |
| cy0331 | M1/M3 | 50,000 | 4.1 | 7,280 | 6 |
| cy0325 | M1/M1 | 50,000 | 4.3 | 2,880 | 4 |
| cy0354 | M3/M5 | 1,000 | 4.4 | 805 | 4 |
| cy0320 | M1/M1 | 50,000 | 4.6 | 1,510 | 3 |
| cy0324 | M1/M1 | 50,000 | 4.7 | 730 | 3 |
| cy0351 | M2/M4 | 100 | 4.8 | 1,510 | 8 |
| cy0337 | M3/M3 | 50,000 | 4.9 | 400 | 4 |
| cy0328 | M1/M3 | 50,000 | 5.1 | 2,930 | 3 |
| cy0350 | M2/M2 | 500 | 5.4 | 715 | 4 |
| cy0355 | M1/M5 | 500 | 5.4 | 1,480 | 7 |
| cy0353 | M3/M5 | 100 | 5.6 | 3,700 | 9 |
| cy0356 | M1/M6 | 1,000 | 5.7 | 1,130 | 3 |
| cy0329 | M1/M3 | 50,000 | 5.9 | 9,160 | 7 |
| cy0333 | M3/M3 | 50,000 | 6 | 3,710 | 2 |
| cy0352 | M2/M4 | 100 | 6.1 | 50 | 1 |
| cy0336 | M3/M3 | 50,000 | 6.3 | 540 | 2 |
Plasma viral load analysis.
Virion-associated SIV RNA in plasma was measured using a real-time (RT) quantitative reverse transcription-PCR assay, essentially as described previously (10).
Peptides.
The NIH AIDS Research and Reference Reagent Program (Germantown, MD) provided 15-mer peptides with an 11-residue overlap spanning the full SIVmac239 proteome. Additional peptides used for mapping Nef 196-203 HW8 were synthesized by ProImmune Inc. (Sarasota, MN). All peptide sequences were derived from the SIVmac239 genome.
IFN-γ ELISPOTs.
Gamma interferon enzyme-linked immunosorbent spot assays (IFN-γ ELISPOTs) were conducted according to the manufacturer's protocol as described previously (7). Briefly, peripheral blood mononuclear cells (PBMC) were isolated using Ficoll-Paque Plus (GE Healthcare Bioscience, Uppsala, Sweden) density centrifugation from whole blood containing EDTA at 24 and 52 weeks postinfection. Cells were suspended in RPMI 1640 (HyClone, Logan, UT) supplemented with 10% fetal calf serum, 1% antibiotic/antimycotic (HyClone), and 1% l-glutamine (HyClone) (R-10 medium) and then added at 105 cells per well in 100 μl of R-10 to a precoated monkey IFN-γ ELISPOT Plus plate (Mabtech Inc., Mariemont, OH) with the peptides of interest. Traditional, full proteome peptides contained pools of 10 peptides at a concentration of 10 μM or 1 μM each. To identify single peptides that elicited IFN-γ release, we performed a second ELISPOT in tandem using matrix pools containing a randomized assortment of 10 peptides at a concentration of 10 μM each. Concanavalin A (10 μM) was used as a positive control in duplicate wells. A minimum of six wells (three samples in duplicate) did not receive any stimulant and served as negative controls. All samples were repeated in duplicate or triplicate. Wells were imaged using an AID ELISPOT reader, and spots were counted using an automated program with fixed parameters. For the assay to be considered successful, a minimum of 100 spots was required in each positive-control well. Experimental responses exceeding the arithmetic mean of the negative-control wells plus 2 standard deviations were considered positive. The limit of detection was set at 70 spot-forming cells (SFCs) per 106 PBMC. To account for the difference in SFCs between animals, responses were normalized using the following equation: 100 × (SFCs from a positive peptide)/(SFCs from all positive peptides).
Viral sequencing.
Viral RNA was isolated from cell-free plasma using the QIAmp MinElute Virus Spin kit (Qiagen, Valencia, CA). Four overlapping PCR amplicons spanning the entire SIVmac239 genome were then amplified using the SuperScript III One-Step RT-PCR system with Platinum Taq High Fidelity (Invitrogen, Carlsbad, CA) as described previously (2). Products were gel purified and fragmented with the Nextera DNA Sample Prep kit (Epicentre, Madison, WI), and then multiplex identifier (MID) tags and adapter sequences for Roche/454 pyrosequencing were added. Purified fragments were pyrosequenced using a Roche/454 GS Junior instrument. Variants present in >1% of reads were considered authentic, a threshold that we had previously determined empirically (2). Amino acid alignments were derived from the sequence read data using a custom data analysis pipeline in Galaxy, an open-source system for processing next-generation sequence data previously described (3, 4, 14, 23).
ICS.
Peptide-specific cytokine release by CD8+ T cell lines was detected by flow cytometry as described previously (5). Briefly, peptide-pulsed B-lymphoblastoid cell line (BLCL) or MHC class I transferents were incubated with CD8+ T cell lines specific for Nef 196-203 HW8 in the presence of brefeldin A (10 μg/ml; Sigma-Aldrich, St. Louis, MO). After 5 h, the cells were surface stained with the directly conjugated monoclonal antibodies (MAbs) αCD3-Alexa Fluor 700 (clone SP34-2; BD Biosciences, San Jose, CA) and αCD8-Pacific Blue (clone RPA-T8; BD Biosciences) and then washed, fixed, and permeabilized prior to intracellular staining with α-IFN-γ-fluorescein isothiocyanate (αIFN-γ-FITC) (clone 4S.B3; BD Biosciences) and tumor necrosis alpha-alpha-phycoerythrin (αTNF-α-PE) (clone MAb11; BD Biosciences). Stained cells were fixed in 2% paraformaldehyde and acquired using an LSRII flow cytometer (BD Biosciences). Data were analyzed using FlowJo software version 8.8.2 (TreeStar, Ashland, OR). All intracellular cytokine staining (ICS) results were confirmed in at least two individual experiments.
Tetramer staining.
Soluble tetrameric peptide-MHC class I complexes were generated as described previously (26). Cryopreserved PBMC were resuspended in 150 μl of R-10 with 5 μg/ml of phycoerythrin (PE)-conjugated tetramer for 20 min at 37°C and washed once with fluorescence-activated cell sorter (FACS) buffer. Subsequently, cells were surface stained with αCD3-Alexa Fluor 700, αCD14 ECD (clone RMO52; Beckman Coulter), αCD19 ECD (clone J4.119, Beckman Coulter), and αCD8-Pacific Blue and then washed and fixed with 2% paraformaldehyde. Stained cells were acquired using an LSRII flow cytometer (BD Biosciences) and data analyzed using FlowJo software version 8.8.2.
Statistical analysis.
Replicates were graphed using GraphPad Prism (GraphPad Software, La Jolla, CA). A two-way or one-way analysis of variance (ANOVA) with Tukey's multiple-comparison posttest was used to compare the columns and determine significance. Correlations were also assessed in GraphPad Prism assuming Gaussian populations. Bars on graphs represent the standard errors of the means.
RESULTS
MCM with low viral loads do not have increased T cell breadth.
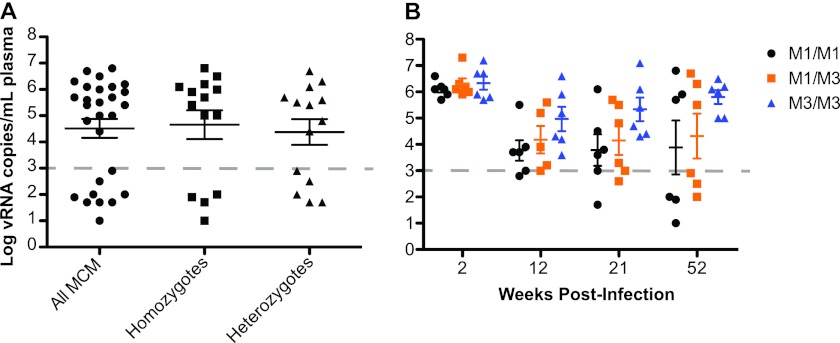
We analyzed a cohort of 27 MCM infected with SIVmac239; each of the major MHC haplotypes, M1 through M7, was expressed in at least one of these animals (Table 1). The cohort included six M1/M1 homozygous macaques, six M1/M3 heterozygous macaques, and six M3/M3 homozygous macaques. Nine of the 27 MCM, including three M1/M1 homozygotes, four M1 heterozygotes, one M7/M7 homozygote, and one M7 heterozygote, consistently maintained low plasma viral loads (<103 copies of viral RNA per ml plasma) in the chronic phase of infection through 52 weeks postchallenge (Fig. 1A). In agreement with our previous study (24), M3/M3 homozygous MCM fared poorly, with none of these animals exhibiting chronic-phase viral load control (Fig. 1B). Low viral loads in four homozygous MCM (three M1/M1 and one M7/M7) reduced the mean viral load of the entire homozygous group, and as a result, there was no significant difference in mean viral loads between heterozygous and homozygous MCM at 52 weeks postinfection.
Fig 1.
Effects of heterozygosity on chronic-phase viral loads. (A) Mean viral loads (number of viral RNA copies per ml plasma) for all 27 MCM at 52 weeks postinfection. (B) Mean viral loads for a subgroup of 18 MHC-matched (six M1/M1, six M1/M3, and six M3/M3) MCM at 2, 12, 21, and 52 weeks postinfection with SIVmac239. The horizontal dashed line represents the threshold of viral control as defined for this cohort (1,000 copies/ml). There were no statistically significant differences between the different haplotypes.
SIV control in four MHC-homozygous macaques was somewhat unexpected but fortuitous, as it allowed us to examine the attributes of T cell responses in the context of both MHC-homozygous and MHC-heterozygous viral control. We first examined T cell breadth at 24 and 52 weeks postinfection to test the hypothesis that controllers mount T cell responses targeting a greater number of epitopes than animals that fail to control SIVmac239. We stimulated freshly isolated PBMC with peptide pools spanning the entire viral proteome and measured IFN-γ production using ELISPOTs. Cells from one controller, cy0322, could not be analyzed by IFN-γ ELISPOT due to a high level of nonspecific cytokine secretion.
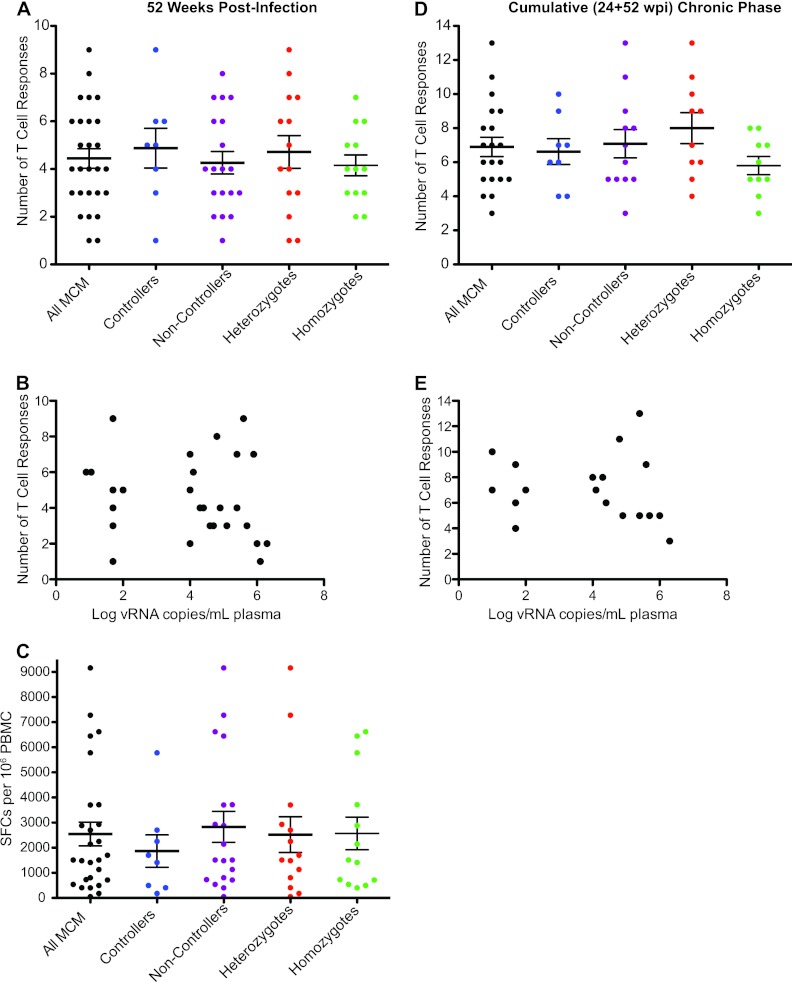
To examine T cell breadth, we tabulated the number of T cell responses targeting distinct peptide pools at 52 weeks postinfection. Each animal made between one and nine distinct T cell responses. Neither MHC zygosity nor SIV controller status correlated with the breadth of the measured T cell response (Fig. 2A and B). We also assessed the strength of the total SIV-specific T cell response by summing the magnitudes of individual T cell responses observed at 52 weeks postinfection. There was no correlation between the total response magnitude, as measured by SFCs per million PBMC, and viral load at 52 weeks postinfection (Fig. 2C).
Fig 2.
The number and strength of T cell responses detected by IFN-γ ELISPOT do not correspond to viral load or MHC haplotype. (A) Number of positive T cell responses in 26 MCM at 52 weeks postinfection separated into controller, noncontroller, homozygote, and heterozygote populations. (B) Comparison of the number of distinct T cell responses and viral loads at 52 weeks postinfection. (C) Sum of all SIV peptide-specific SFCs per million PBMC separated into controller, noncontroller, homozygote, and heterozygote populations at 52 weeks postinfection. (D) Cumulative (24 + 52 weeks postinfection) chronic-phase T cell responses in 20 MCM separated into controller, noncontroller, homozygote, and heterozygote populations. (E) Comparison of the number of cumulative T cell responses and viral loads at 52 weeks postinfection.
Some responses were detected at 24 weeks postinfection but not at 52 weeks, and vice versa. Therefore, to approximate the total T cell breadth in each animal over time, we determined the total number of distinct T cell responses detected at either or both of these time points (Fig. 2D). For technical reasons, data are not available from six animals (two M1/M1, two M1/M3, and two M3/M3). When considering both time points in aggregate, the measured breadth increased slightly, up to 13 T cell responses per animal. Heterozygous MCM had slightly more chronic phase T cell responses than homozygous MCM in this analysis, but the difference was not significant. Again, the number of cumulative chronic-phase T cell responses did not appear to correlate with SIV control (Fig. 2E).
MCM with low viral loads share similar immunodominant CD8+ T cell responses.
CD8+ T cell responses against particular epitopes in individual HIV and SIV proteins have been associated with viral control. Indeed, the benefits of CD8+ T cell responses specific for Gag, Pol, Vif, Vpr, Vpu, Tat, Rev, Env, and/or Nef have been championed by various studies and in various contexts (15, 16, 18, 28). We therefore analyzed the ELISPOT data described above to examine CD8+ T cell responses by protein target.
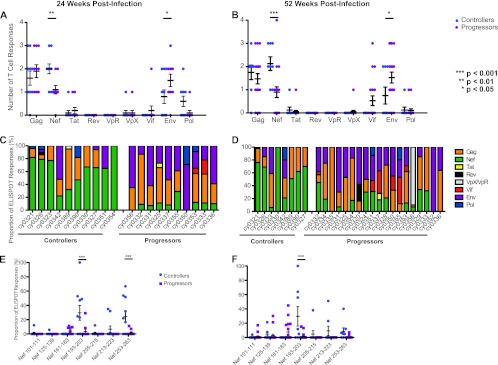
First, we compared T cell breadth at the protein level in controllers and progressors, determining the number of distinct peptides derived from each viral protein targeted at both 24 and 52 weeks postinfection (Fig. 3A and B, respectively). Notably, a similar level of T cell breadth was observed over the course of time for each protein. The relative number of Nef-specific T cell responses was significantly greater in MCM with low viral loads (P < 0.01 and P < 0.001 at 24 and 52 weeks postinfection, respectively). In contrast, the relative number of Env-specific T cell responses was significantly (P < 0.05 at both time points) greater in MCM with high viral loads.
Fig 3.
The number and magnitude of T cell responses to Nef peptides during chronic infection differ between controllers and progressors. (A and B) Number of T cell responses per SIV protein in controllers and progressors at 24 (A) and 52 (B) weeks postinfection. Controllers (animals with viral loads of <103 RNA copies/ml plasma) target significantly more Nef peptides than do progressors (P < 0.01), while progressors target significantly more Env peptides than do controllers (P < 0.05). (C and D) Proportion of the total T cell response directed against each SIV protein in each animal at 24 (C) and 52 (D) weeks postinfection. (E and F) Individual T cell responses within the Nef protein at 24 (E) and 52 (F) weeks postinfection. MCM with low viral loads mounted significantly larger responses against Nef 193-203 at both 24 and 52 weeks postinfection (P < 0.001 in each case).
Next, to identify which proteins immunodominant T cell populations targeted, we examined the percent contribution of each T cell response to the total SIV-specific response. Within each animal, the dominant T cell responses were mainly directed against one of three proteins: Gag, Env, or Nef. Interestingly, a majority of MCM with low viral loads (8/10 and 6/8 at 24 and 52 weeks postinfection, respectively) mounted dominant T cell responses against epitopes in the Nef protein. In contrast, Nef was not the dominant target in any progressors at 24 weeks postinfection, and only one progressor made a dominant Nef-specific T cell response at 52 weeks postinfection (Fig. 3C and D).
To extend these findings, we determined the regions within Nef that were recognized by these T cell responses. In total, seven Nef-specific T cell epitopes were identified. At 24 weeks postinfection, T cell responses against two epitopes, Nef 193-203 and Nef 253-263, were detected more frequently and accounted for a greater proportion of the total virus-specific response in controllers than in progressors (Fig. 3E). By 52 weeks postinfection, only the T cell response against Nef 193-203 accounted for a significantly higher proportion of the total SIV-specific response in controllers versus progressors (Fig. 3F). The two M7 controllers each mounted a T cell response against Nef 213-223, but due to the small number of animals with the M7 haplotype, the significance of this observation could not be determined. Interestingly, all three putatively beneficial responses clustered in a narrow region near the carboxy terminus of Nef, and two of them, Nef 193-203 and Nef 213-223, were restricted by MHC class I A alleles (Fig. 4).
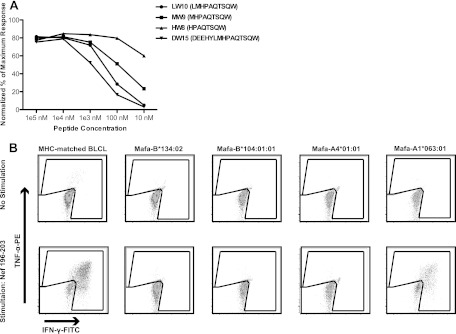
Fig 4.
Identification of the Nef 196-203 HW8 epitope. (A) MHC-matched BLCLs were pulsed with serial dilutions of prospective optimal peptides and cocultured with a Nef 196-203 HW8-specific T cell line. Activation of CD8+ T cells was measured by ICS using αIFN-γ-FITC and αTNF-α-PE. When responses were normalized to the percentage of the maximum response, Nef 196-203 HW8 was determined to be the optimal epitope. (B) MHC class I transferent K562 antigen-presenting cells were pulsed with cognate peptide and cocultured with a Nef 196-203 HW8-specific T cell line. Activation of CD8+ T cells was measured by ICS as described for panel A. Nef 196-203 HW8 was found to be restricted by Mafa-A1*063. Results are representative of at least two separate experiments.
High viral load is associated with rapid accumulation of substitutions in Nef 196-203.
The observation that a subset of the M1/M1 homozygous and M1/M3 heterozygous MCM controlled SIVmac239 and mounted strong responses against Nef 196-203 is paradoxical. Why did the other animals with identical MHC genetics not recognize this epitope? To explore this issue, we first confirmed the MHC restriction of Nef 196-203, the minimal optimal epitope. We constructed transferents expressing Mafa-A1*063:01, -A4*01:01, -B*104:01, or -B*134:02 and used them in an intracellular cytokine staining assay (Fig. 4). A CD8+ T cell line specific for Nef 196-203 HW8 released cytokines when stimulated by peptide-pulsed, MHC-matched BLCLs as well as transferents expressing only Mafa-A1*063.
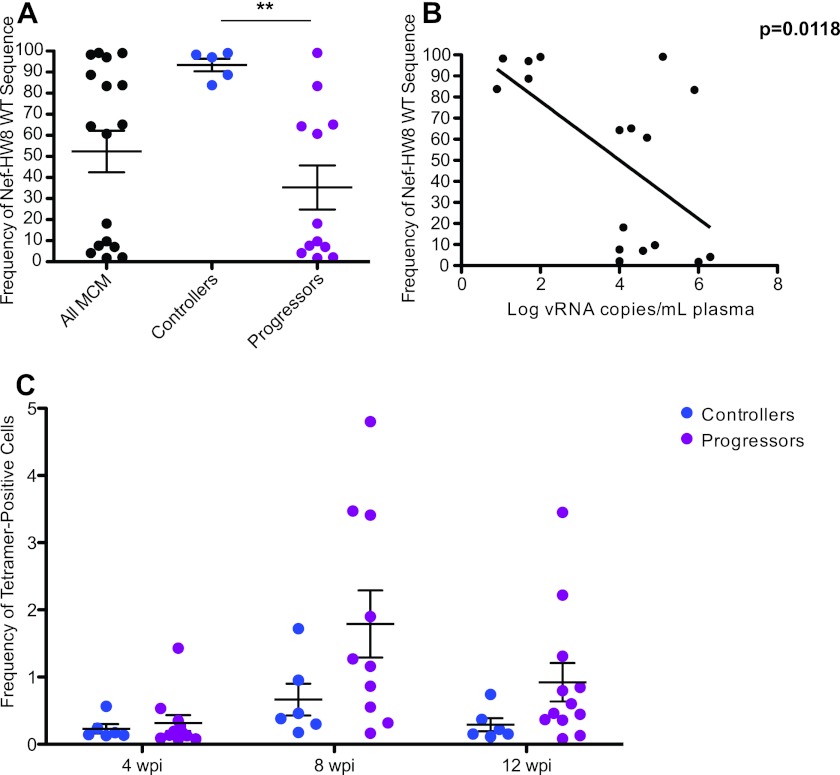
One potential explanation for the apparent absence of responses against Nef 196-203 HW8 in some chronically infected animals expressing the M1, M2, or M3 haplotypes is that SIV effectively escapes from CD8+ T cells recognizing this epitope early during infection in some, but not all, animals. Accordingly, we examined whether viruses isolated from the subgroup of six M1/M1 homozygous, six M1/M3 heterozygous, and six M3/M3 homozygous MCM accumulated amino acid substitutions in this region at 12 weeks postinfection. One M1/M1 homozygous MCM that maintained low viral loads could not be analyzed. Interestingly, viruses from all five controllers almost exclusively displayed wild-type sequence across Nef 196-203 HW8, while only 2/12 progressors harbored viral populations with dominant wild-type sequence at this epitope (Fig. 5A). The frequency of wild-type Nef 196-203 HW8 correlated significantly with chronic-phase viral loads at 52 weeks postinfection (Fig. 5B).
Fig 5.
High viral loads are associated with the rapid accumulation of substitutions in Nef 196-203. (A) The wild-type (WT) frequency of Nef 196-203 HW8 was examined by 454 pyrosequencing at 12 weeks postinfection. There was a significant difference (P < 0.01) in the frequency of wild-type Nef 196-203 HW8 sequences between controllers and progressors. (B) Comparison of wild-type Nef 196-203 HW8 sequence frequency at 12 weeks postinfection and viral loads at 52 weeks postinfection. (C) Nef 196-203 HW8-specific CD8+ T cell populations quantified by tetramer staining. Although most progressors mounted substantial responses to this epitope by 8 weeks postinfection, there were no significant differences between controllers and progressors at any time point.
To confirm that differential CD8+ T cell pressure was driving viral escape at Nef 196-203 HW8 in progressors prior to 12 weeks postinfection, we examined response kinetics using the corresponding peptide-MHC class I tetramer. Although not significantly different, progressors tended to have higher frequencies of Nef 196-203 HW8 tetramer-positive cells at 8 weeks postinfection than controllers (Fig. 5C). Collectively, these observations suggest that early events in viral infection, specifically the timing of CD8+ T cell responses and viral escape, may have a significant impact on whether or not individuals are able to maintain low viral loads during chronic infection.
DISCUSSION
Full heterozygosity in MHC class I alleles is associated with delayed progression to AIDS (8). However, the complex MHC genetics of humans and Indian rhesus macaques have frustrated efforts to discern the underlying mechanism of this association. Two possibilities have received considerable attention. One is that fully heterozygous individuals simply have a greater likelihood of possessing one or more protective MHC class I alleles (16), imbuing these individuals with the ability to mount unusually efficacious CD8+ T cell responses. A second explanation is that at any moment, cells from MHC-heterozygous individuals present a broader array of virus-derived epitopes than do homozygous ones, allowing a larger number of distinct CD8+ T cell specificities to target infected cells. A broader T cell repertoire may simply be more robust, i.e., better able to maintain control over a diversity of viral variants, than a narrow one. These mechanisms are not mutually exclusive. Our data suggest that in the model studied here, SIV-infected MCM, MHC heterozygosity maximizes the likelihood of having an allele that restricts one or more durable CD8+ T cell responses that correlate with viral control. To our knowledge, this is the first comprehensive demonstration that directly examined both of these possibilities in the same study, leading to the conclusion that in this model favorable CD8+ T cell responses and not epitopic breadth per se influence the heterozygote advantage.
Care must be taken when assessing T cell breadth using ex vivo immunologic assays. We measured T cell breadth by IFN-γ ELISPOT in blood samples, but it is possible that the breadth of the CD8+ T cell response in other anatomic compartments may be different. In addition, the ELISPOT method measures only a single functional attribute of T cell responses, i.e., IFN-γ secretion, and may therefore underestimate those T cell populations that primarily elaborate other effector molecules. It seems unlikely, however, that these caveats would generate a systematic bias leading to the detection of greater T cell breadth only in heterozygous individuals. Furthermore, all ex vivo assays suffer to a greater or lesser extent from a lack of sensitivity. It is possible that bona fide responses below the limit of ELISPOT detection exist, and we cannot exclude the possibility that a difference in CD8+ T cell breadth exists among such minor responses.
It is important to note that while we examined T cell responses at two different time points, we could not rigorously evaluate cumulative T cell breadth in this study. We stopped the study at 52 weeks postinfection, before the development of AIDS. It is possible that, given enough time, the heterozygous animals would suppress virus replication for a longer time than their homozygous counterparts, perhaps because some CD8+ T cell specificities can be engaged in heterozygotes only after the failure of frontline responses detected in the first year. The importance of cumulative T cell breadth may therefore become apparent only on time scales longer than that of the present study, and importantly, cumulative T cell breadth could give heterozygous macaques longer disease-free survivorship without affecting the level of SIV replication. It is also possible that T cell responses would be affected if different SIV strains, instead of a single clonal virus, were used.
Previously, we demonstrated that MHC-heterozygous MCM maintained lower chronic-phase viremia than MHC-homozygous animals in a cohort heavily enriched for the M3 haplotype. In the present study, we again observed that M3 homozygous macaques had higher viral loads than did M1/M3 heterozygous macaques (24). Surprisingly, however, we found no significant correlation between MHC zygosity and viral load when examining MCM of all seven haplotypes. This does not indicate that the heterozygous advantage does not exist in HIV-seropositive cohorts. Instead, considering the limited genetic diversity of this population, we suspect that our results are strongly influenced by the presence of two protective haplotypes that associate with durable viral control regardless of zygosity. Despite this limitation, the MCM system will allow us to dissect the differences between “favorable” and “unfavorable” CD8+ T cell responses that are and are not associated with virologic control in future experiments. Further, due to the limited genetic diversity of both MCM and the clonal pathogenic virus SIVmac239, we will be able to more accurately assess how stochastic immunological and viral events influence durable control.
Additional studies will be required to understand why MHC-identical animals exhibited different patterns of CD8+ T cell immunodominance, particularly with regard to M1 macaques in which a dominant Nef-specific response is correlated with viral control. The simplest explanation is that vigorous T cell responses against this epitope were mounted in all animals, with immune escape variants arising rapidly only in a subset of the animals, effectively “knocking out” a favorable response. If this is true, then the ability of an individual to contain viral replication durably may be at the mercy of stochastic events that are impossible to predict a priori. Recently, we demonstrated “conditional” T cell escape during the first weeks of SIVmac239 infection, in which the selection of escape variants within an immunodominant epitope was influenced by the selection of escape variants elsewhere in the genome (23). Such conditional escape could be resolved only by using a clonal virus, studying the initial responses that select immune escape variants during acute infection, and examining MHC-defined animals. Similar interepitope relationships may pertain to chronic infection and are amenable to identification with substantial data sets that could be generated relatively easily, using deep-sequencing methodologies such as those employed here.
Claims that specific domains of the HIV or SIV proteome represent magic bullets as universally effective CD8+ T cell targets should be viewed skeptically. Nonetheless, it is possible that some epitopes may make more effective targets than others. Indeed, the benefit of CD8+ T cell responses specific for Gag has been championed by various studies and in various contexts (15, 16, 18, 28). One potential mechanism for this effect is that Gag is subject to strong evolutionary constraints and escape mutations in Gag frequently exact a cost to viral fitness. In our study, viral control correlated with a single response directed against Nef 196-203 HW8. There may be isolated examples of Nef-specific responses that are similarly protective in HIV-seropositive individuals but whose effect is masked in population studies with heterogeneous HLA representation. Certain regions of Nef may also be under significant constraint. Conversely, there may be Gag-specific CD8+ T cell responses in other macaque populations that can control SIV viral loads but are restricted by MHC class I alleles not represented in MCM.
In conclusion, our data indicate that in SIV-infected MCM there is no relationship between viral control and either the number or the magnitude of CD8+ T cell responses at a given moment. Instead, controllers shared stable CD8+ T cell recognition of epitopes located in a small region of the SIV Nef protein. The presence of the MHC class I allele capable of restricting these beneficial epitopes is not, by itself, enough to confer control of plasma viremia. Rather, durable control may depend on both definable parameters, such as the presence of a protective MHC class I allele, and stochastic ones, such as the direction and rate of viral evolution under CD8+ T cell pressure. It seems likely that both T cell breadth and durable effective targeting of selected, evolutionarily constrained epitopes can contribute to control of AIDS virus replication by CD8+ T cells; the critical goal of outflanking the ability of the virus to mutationally escape T cell responses can be achieved by multiple mechanisms. However, in the SIV-infected MCM studied here, stable CD8+ T cell recognition of epitopes located in a small region of the SIV Nef protein appears to be the key factor associated with control of viral replication.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grants R01 AI077376, awarded to David H. O'Connor, and R01 AI08747, awarded to Thomas C. Friedrich and David H. O'Connor. This research was also supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E (Michael Piatak, Jr., and Jeffrey D. Lifson). This work was conducted at facilities supported in part by grant number P51 RR000167 from the National Center for Research Resources, a component of the National Institutes of Health. Additionally, work for the manuscript was conducted in a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01.
We acknowledge the staff of the Wisconsin National Primate Research Center's Animal Services and Scientific Protocol Implementation Units for their assistance with this study.
Footnotes
Published ahead of print 9 May 2012
REFERENCES
- 1. Addo MM, et al. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bimber BN, et al. 2010. Whole-genome characterization of human and simian immunodeficiency virus intrahost diversity by ultradeep pyrosequencing. J. Virol. 84:12087–12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blankenberg D, et al. 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26:1783–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blankenberg D, et al. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19:Unit 19.10.1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Budde ML, et al. 2011. Transcriptionally abundant major histocompatibility complex class I alleles are fundamental to nonhuman primate simian immunodeficiency virus-specific CD8+ T cell responses. J. Virol. 85:3250–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Budde ML, et al. 2010. Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 62:773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burwitz BJ, et al. 2009. Mauritian cynomolgus macaques share two exceptionally common major histocompatibility complex class I alleles that restrict simian immunodeficiency virus-specific CD8+ T cells. J. Virol. 83:6011–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carrington M, et al. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752 [DOI] [PubMed] [Google Scholar]
- 9. Chouquet C, et al. 2002. Correlation between breadth of memory HIV-specific cytotoxic T cells, viral load and disease progression in HIV infection. AIDS 16:2399–2407 [DOI] [PubMed] [Google Scholar]
- 10. Cline AN, Bess JW, Piatak MJ, Lifson JD. 2005. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34:303–312 [DOI] [PubMed] [Google Scholar]
- 11. Dalod M, et al. 1999. Broad, intense anti-human immunodeficiency virus (HIV) ex vivo CD8(+) responses in HIV type 1-infected patients: comparison with anti-Epstein-Barr virus responses and changes during antiretroviral therapy. J. Virol. 73:7108–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frahm N, et al. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedrich TC, et al. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 81:3465–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86 doi:10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Honeyborne I, et al. 2007. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J. Virol. 81:3667–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kiepiela P, et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53 [DOI] [PubMed] [Google Scholar]
- 17. Lawler SH, Sussman RW, Taylor LL. 1995. Mitochondrial DNA of the Mauritian macaques (Macaca fascicularis): an example of the founder effect. Am. J. Phys. Anthropol. 96:133–141 [DOI] [PubMed] [Google Scholar]
- 18. Loffredo JT, et al. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 82:1723–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo M, et al. 2012. For protection from HIV-1 infection, more might not be better: a systematic analysis of HIV Gag epitopes of two alleles associated with different outcomes of HIV-1 infection. J. Virol. 86:1155–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maness NJ, et al. 2008. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive simian immunodeficiency virus-infected rhesus macaques. J. Virol. 82:5245–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McElrath MJ, et al. 2008. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 372:1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mee ET, et al. 2009. MHC haplotype frequencies in a UK breeding colony of Mauritian cynomolgus macaques mirror those found in a distinct population from the same geographic origin. J. Med. Primatol. 38:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Connor SL, et al. 2012. Conditional CD8+ T cell escape during acute simian immunodeficiency virus infection. J. Virol. 86:605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Connor SL, et al. 2010. MHC heterozygote advantage in simian immunodeficiency virus-infected mauritian cynomolgus macaques. Sci. Transl. Med. 2:22ra18 doi:10.1126/scitranslmed.3000524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pereyra F, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price DA, et al. 2005. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 202:1349–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Regier DA, Desrosiers RC. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 6:1221–1231 [DOI] [PubMed] [Google Scholar]
- 28. Rolland M, et al. 2008. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One 3:e1424 doi:10.1371/journal.pone.0001424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang J, et al. 1999. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses 15:317–324 [DOI] [PubMed] [Google Scholar]
- 30. Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. 2008. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 14:617–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wiseman RW, et al. 2007. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J. Virol. 81:349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]