Abstract
Flavivirus NS1 is a nonstructural glycoprotein that is expressed on the cell surface and secreted into the extracellular space. Despite its transit through the secretory pathway, NS1 is an essential gene linked to early viral RNA replication. How this occurs has remained a mystery given the disparate localization of NS1 and the viral RNA replication complex, as the latter is present on the cytosolic face of the endoplasmic reticulum (ER). We recently identified an N-terminal di-amino acid motif in NS1 that modulates protein targeting and affected viral replication. Exchange of two amino acids at positions 10 and 11 from dengue virus (DENV) into West Nile virus (WNV) NS1 (RQ10NK) changed its relative surface expression and secretion and attenuated infectivity. However, the phenotype of WNV containing NS1 RQ10NK was unstable, as within two passages heterogeneous plaque variants were observed. Here, using a mutant WNV encoding the NS1 RQ10NK mutation, we identified a suppressor mutation (F86C) in NS4B, a virally encoded transmembrane protein with loops on both the luminal and cytoplasmic sides of the ER membrane. Introduction of NS4B F86C specifically rescued RNA replication of mutant WNV but did not affect the wild-type virus. Mass spectrometry and coimmunoprecipitation studies established a novel physical interaction between NS1 and NS4B, suggesting a mechanism for how luminal NS1 conveys signals to the cytoplasm to regulate RNA replication.
INTRODUCTION
West Nile virus (WNV) is a flavivirus that cycles in nature between birds and Culex mosquitoes and can infect humans and other vertebrate animals. While most human WNV infections are asymptomatic, ∼0.5 to 1% develop a severe neuroinvasive syndrome that manifests as meningitis, acute flaccid paralysis, or encephalitis (39). The Flavivirus genus also contains other human pathogens of global concern, including dengue (DENV), yellow fever (YFV), Saint Louis encephalitis (SLEV), Japanese encephalitis (JEV), and tick-borne encephalitis viruses.
The ∼10.7-kb single-stranded positive-sense flavivirus RNA genome is translated as a single polyprotein, which is cleaved into three structural proteins (C, prM/M, E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) by virus- and host-encoded proteases. Flavivirus RNA replication occurs along the cytosolic face of the endoplasmic reticulum (ER) and requires the enzymatic actions of several NS proteins, including the viral protease (NS3) and RNA-dependent RNA polymerase (NS5). Flavivirus NS1 is a multifunctional 48-kDa glycoprotein that is secreted into the extracellular milieu but is absent from the virion. NS1 also is expressed directly on the plasma membrane of infected cells, although it lacks a canonical transmembrane domain or targeting motif for cellular membranes. The mechanism of cell surface expression of flavivirus NS1 in infected cells remains uncertain, although some fraction may be linked through an atypical glycosyl-phosphatidylinositol anchor (20, 34) or lipid rafts (33). A recent study with WNV and DENV NS1 identified an N-terminal di-amino acid motif (RQ10-11) that modulates targeting to the cell surface or extracellular space (49).
NS1 is synthesized as a monomer, dimerizes after posttranslational modification in the lumen of the ER, and accumulates in extracellular fluid as a hexamer with a lipid core (13, 15, 19, 46, 47). Soluble NS1 also binds back to the plasma membrane of uninfected cells (1) through interactions with selected sulfated glycosaminoglycans (5). NS1 has immune evasive functions in the extracellular space, on the surface of cells, and within cells, as it binds to complement proteins and regulators and antagonizes their functions (3, 4, 9) and disrupts TLR3 signaling pathways (44). Despite NS1's transit through the secretory pathway, NS1 is an essential gene and modulates early viral RNA replication (22, 26, 28). Deletion of NS1 from the viral genome abrogates replication, although an NS1-deleted virus (ΔNS1) can be complemented in trans by ectopic expression of NS1. Prior studies have suggested that the essential intracellular function of NS1 is due to its ability to regulate negative-strand synthesis of viral RNA (26). How this occurs remains uncertain given the disparate localization of NS1 and the viral RNA replication complex, which are positioned on the luminal and cytosolic sides of the ER membrane, respectively. Genetic studies have suggested that YFV NS1 interacts with NS4A, a transmembrane viral protein that spans the ER, which could integrate key NS1 signals into RNA replication occurring in the cytoplasm (25).
Here, we used a mutant WNV encoding the NS1 (RQ10NK) that has enhanced NS1 secretion yet impaired viral replication to identify suppressor mutations. We identified a compensatory mutation in the viral NS4B gene, which rescues replication of mutant WNV but does not independently affect wild-type (WT) WNV replication. Further biochemical and mass spectrometry studies established a physical interaction between NS1 and NS4B, suggesting a mechanism for how luminal NS1 could transmit a signal to the cytoplasmic face of the ER to regulate WNV replication.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney fibroblast (BHK) cells (BHK21-15) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, 10 mM HEPES (pH 7.3), and nonessential amino acids in a 5% humidified CO2 incubator at 37°C. Infection experiments were performed with the New York 1999 strain (strain 385-99) of WNV, which was derived from an infectious cDNA clone (6), or with the Kunjin strain of WNV (strain CH 16532, WNV-KUNV; gift from J. Anderson, New Haven, CT).
Generation of recombinant Sindbis and West Nile viruses.
Recombinant wild-type or RQ10NK NS1 was expressed ectopically using a double subgenomic Sindbis virus (SINV) expression system (p39HK) as described previously (49). To construct recombinant SINV expressing RQ10YK NS1, the RQ10YK mutation was introduced into the wild-type NS1 gene using the QuikChange site-directed mutagenesis kit (Stratagene) in a pSTBlue cloning vector (EMD4 Biosciences) with the following oligonucleotides: NS1 RQ10YK-F, 5′-GCCATAGACATCAGCTACAAAGAGCTGAGATGTGGAAG-3′; RQ10YK-R, 5′-CTTCCACATCTCAGCTCTTTGTAGCTGATGTCTATGGC-3′. NS1-RQ10YK was digested with NotI from the pSTBlue vector and subcloned into the SINV vector that was digested with NotI and treated with alkaline phosphatase. Recombinant SINV were linearized after digestion with XhoI and used as templates for in vitro transcription with SP6 DNA-dependent RNA polymerase (Epicentre).
Recombinant WNV were generated using the two-plasmid (pWN-AB and pWN-CG) infectious cDNA clone (New York 1999, strain 385-99) as described previously (6, 42) with some modifications. To construct recombinant WNV with mutations in NS1, the pSTBlue plasmid encoding NS1 containing the RQ10NK or RQ10YK mutation was digested with DraIII and XbaI and subcloned into the pWN-AB plasmid encoding wild-type or mutant NS1. pWN-AB encoding mutant NS1 was digested with EcoRI and BstEII and ligated overnight at room temperature to pWN-CG that was digested with XbaI and BstEII; a wild-type WNV infectious cDNA template was prepared by digesting pWN-AB and pWN-CG with XbaI, followed by treatment with alkaline phosphatase, NgoMVI digestion, and subsequent ligation overnight at room temperature. Ligated cDNA templates were extracted with phenol-chloroform (1:1) and precipitated with 70% (vol/vol) ethanol. To construct a recombinant WNV with the NS4B F86C mutation, the NS4B portion of plasmid pWN-CG was subcloned into pSTBlue using the KpnI restriction site. The F86C mutation (underlined below) was introduced into NS4B in pSTBlue with the oligonucleotide pair NS4B F86C-F (5′-CACTCGCGCGAGGCTGCCCCTTCGTCGATGT-3′) and NS4B F86C-R (5′-ACATCGACGAAGGGGCAGCCTCGCGCGAGTG-3′) using the QuikChange site-directed mutagenesis kit. The KpnI fragment containing NS4B F86C mutation from pSTBlue was subcloned back into pWN-CG, and the full-length recombinant WNV cDNA template with the F86C NS4B mutation was assembled. WNV with a double mutation in NS1 and NS4B was constructed by ligating the pWN-CG NS4B F86C plasmid with pWN-AB containing RQ10NK or RQ10YK mutations in NS1. After ligation, phenol-chloroform extraction, and ethanol precipitation, DNA-dependent RNA transcription was performed in vitro using the Ampliscribe T7 RNA polymerase transcription kit (Epicentre) with an m7G(5′)ppp(5′)A RNA cap analog (New England BioLabs).
In vitro transcribed SINV or WNV RNA was electroporated to BHK21-15 cells using a GenePulser Xcell electroporator (Bio-Rad) at 850 V, 25 mF, and infinite Ω to generate a P0 virus stock. The titers of recombinant SINV and WNV were determined by plaque assay (49) or focus-forming assay (16), respectively, on BHK21-15 cells. Unless otherwise noted, all experiments were performed with the P0 stock.
Generation of cells propagating Venezuelan equine encephalitis virus (VEEV) replicons expressing NS1.
Wild-type and RQ10NK NS1 were subcloned from a pSTBlue cloning vector into a modified pSC-B W/VEEV shuttle vector. The shuttle vector was constructed by subcloning an XbaI fragment of the VEEV replicon, pVEEVrep/L/eGFP/Pac (36) (gift of I. Frolov, Birmingham, AL) into pSTBlue. The enhanced green fluorescent protein (eGFP) gene was removed from the shuttle vector after digestion with NcoI and BsrGI. Subsequently, the modified shuttle vector was treated with T4 DNA polymerase to blunt the 5′ and 3′ ends and then self-ligated with T4 DNA ligase. The wild-type NS1 gene was subcloned into the shuttle vector using BamHI and EcoRV restriction enzymes. Alternatively, the wild-type NS1 gene was replaced with RQ10NK NS1 from pSTBlue using a NotI restriction enzyme. As a negative control, NS1 with an internal stop codon was constructed: the codon for the eighth amino acid in the signal peptide of NS1 was mutated to a stop codon using the QuikChange site directed mutagenesis kit and the following primers: NS1-stop-F, 5′-CATAGCTCTCACGTAACTCGCAGTTGGAG-3′, and NS1-stop-R, 5′-CTCCAACTGCGAG TTACGTGAGAGCTATG-3′. All wild-type and mutant NS1 genes in the pSC-B W/VEEV shuttle vector were cloned subsequently into a different VEEV replicon (pTC83new/Pac [36]; gift of I. Frolov) for expression using an XbaI site. VEEV replicon plasmids encoding NS1 were linearized with MluI and used as templates for in vitro transcription using SP6 DNA-dependent RNA polymerase. RNA transcripts were electroporated as described above, and cells expressing VEEV replicons were selected with puromycin (5 μg/ml) over 1 week. NS1 expression was confirmed by flow cytometry as described below.
Genome sequencing of WNV.
Full genome sequencing of all revertant viruses containing mutations in NS1 was performed by bidirectional sequencing of amplicons generated by reverse transcription-PCR (RT-PCR) using a series of primers spanning the complete genome (see Table S1 in the supplemental material) and standard molecular biology protocols.
Viral growth kinetics.
BHK21-15 cells (2 × 105) were seeded into 24-well plates and infected with recombinant wild-type or mutant WNV at a multiplicity of infection (MOI) of 1 or 10 depending on the experiment for 1 h at 37°C. Cells were washed three times with phosphate-buffered saline (PBS), and then DMEM supplemented with 10% FBS was added. Supernatants were harvested at indicated time points, aliquoted, and frozen at −80°C prior to titration on BHK21-15 cells using a focus-forming assay (16).
Flow cytometry.
To quantify expression levels of surface and intracellular NS1, flow cytometry was utilized. BHK21-15 cell monolayers were infected with recombinant SINV (MOI of 1) or WNV (MOI of 0.1). At 14 (SINV) or 30 (WNV) hours postinfection, cells were detached with Hanks balanced salt solution (HBSS) supplemented with 5 mM EDTA. For surface staining, cells were washed twice with iced PBS and incubated for 1 h on ice with the NS1-specific monoclonal antibody (MAb) 4NS1 (10). After three washes with PBS, cells were fixed with 2% paraformaldehyde in PBS for 7 min and washed with HBSS. To detect intracellular NS1 or E proteins, after fixation cells were permeabilized with 0.1% (wt/vol) saponin and 0.1% bovine serum albumin (BSA) in HBSS and incubated for 1 h with either mouse-human chimeric 17NS1 (containing a human IgG1 Fc) or humanized E16 antibody (10, 35). After washing, Alexa Fluor 488–goat anti-mouse IgG or Alexa Fluor 647–goat anti-human IgG (Molecular Probes) was added for 30 min on ice. After additional washing, expression levels of WNV NS1 and E were determined by on a FACSArray flow cytometer (Becton, Dickinson) and data were processed with FlowJo software (Tree Star).
Real-time quantitative RT-PCR for WNV RNA.
Wild-type BHK21-15 cells (6 × 105) or BHK21-15 cells propagating VEEV replicons encoding NS1 (wild type or RQ10NK) were infected with WNV (wild type or NS4B F86C) at an MOI of 1. Total cellular RNA was harvested at specified time points using the RNeasy minikit (Qiagen) following the manufacturer's instructions, and viral RNA was quantified using fluorogenic quantitative RT-PCR (qRT-PCR) as described previously (38).
Immunoprecipitation and Western blot analysis.
BHK21-15 cells (6 × 105) were either mock infected or infected with WNV at an MOI of 5. At 28 h postinfection, cells were lysed in 800 μl of a detergent solution (lysis buffer: 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, and 1 mM EDTA) supplemented with a cocktail of protease inhibitors (Sigma) for 10 min on ice. Nuclei were removed after centrifugation (13,800 × g) for 5 min at 4°C. Subsequently, a mixture of 10NS1 and 4NS1 MAbs (10) was added (10 μg per sample), and the samples were incubated for 3 h at 4°C. NS1-MAb complexes were precipitated with protein A-Sepharose beads (Invitrogen) after an additional 1-h incubation at 4°C. Beads were washed three times with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1% sodium deoxycholate, and 1% NP-40), and proteins were eluted by boiling samples at 95°C for 5 min in 4× SDS sample buffer containing 5% (vol/vol) 2-mercaptoethanol. Eluates were separated by 12% SDS-PAGE, and proteins were transferred to nitrocellulose membranes using an iBlot dry blotting system (Invitrogen). The membranes were blocked with 5% milk containing TBST buffer (50 mM Tris, 150 mM NaCl, and 0.05% Tween 20) for 1 h at room temperature and incubated with a 1/500 dilution of a rabbit anti-NS4B polyclonal antibody (gift of W. I. Lipkin, New York, NY) (30) for 3 h at room temperature in 2% reconstituted nonfat dry milk containing TBST buffer. After three serial washes for 15 min each with TBST buffer, the membrane was incubated with a 1/3,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit IgG for 1 h at room temperature. Proteins were visualized using the Amersham ECL plus Western blotting detection system (GE Healthcare) following the manufacturer's instructions, and images were captured using a Fuji phosphorimager.
35S metabolic labeling and immunoprecipitation.
BHK21-15 cells (6 × 105) were infected at an MOI of 5 with recombinant SINV ectopically expressing wild-type NS1, NS1 RQ10NK, or NS1 RQ10YK for 1 h. After the cells were washed three times with PBS, DMEM supplemented with 10% FBS was added. At 15 h after infection, medium was removed, and cells were metabolically starved for 20 min by the addition of DMEM deficient in cysteine and methionine (Invitrogen). Subsequently, cells were pulsed by adding 50 μCi/ml of 35S-labeled methionine and cysteine (EasyTag EXPRES35S protein labeling mix; PerkinElmer Life Science) for 20 min and then chased for specified times by washing and replacing with complete DMEM. Cells were lysed with detergent solution for 10 min on ice, and nuclei were removed by centrifugation (13,800 × g) for 5 min at 4°C. NS1 was immunoprecipitated with 4NS1 and protein A-Sepharose beads as described above. After 12% SDS-PAGE, gels were vacuum dried on filter paper for 1 h at 80°C and proteins were visualized by exposure to Kodak Biomax light film.
Purification of WT and RQ10NK NS1.
WNV-WT and RQ10NK NS1 were produced and isolated from the supernatants of baby hamster kidney fibroblast (BHK) cells that stably propagate VEEV replicons expressing the respective proteins. A two-step purification was performed using immunoaffinity (9NS1 MAb Sepharose) and size exclusion chromatography as described previously (3, 9). Western blotting was performed with 10NS1 MAb.
Affinity purification and mass spectrometry analysis of NS1 interacting proteins.
Immunoaffinity purification of NS1 was performed as described previously (12, 27), with some modifications. Briefly, BHK21-15 cells were mock infected or infected with WNV-KUNV (MOI of 0.1). At 48 h postinfection, cells were washed three times with cold PBS, scraped from the flask, and collected by centrifugation at 300 × g for 5 min. Cell pellets were resuspended in freezing solution (20 mM Na-HEPES, 1.2% [wt/vol] polyvinylpyrrolidone, pH 7.4, phenylmethylsulfonyl fluoride [PMSF], pepstatin A, and protease inhibitor cocktail [Sigma]) dropwise in liquid nitrogen and cryogenically disrupted using a Retsch MM301 mixer mill. Frozen cell powder (0.69 g) was suspended in 3.45 ml of lysis buffer. Lysis buffer conditions of varied stringencies were compared initially for efficiency of NS1 recovery. NS1 isolated using lysis buffers containing 0.3% Triton or 1% Triton and 0.5% sodium deoxycholate was identified by mass spectrometry by peptides covering only 22% or 41% of its sequence, respectively. This sequence coverage further increased to 70% when using the final optimized buffer containing 20 mM HEPES, pH 7.4, 0.1 M potassium acetate, 2 mM MgCl2, 0.1% Tween 20, 1 μM ZnCl2, 1 μM CaCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.3% sodium N-lauroylsarcosine, 0.1 M NaCl, 1/100 protease inhibitor cocktail (Sigma), and 1/100 phosphatase inhibitor cocktail (Sigma). The resulting lysate was homogenized for 15 s with a PT 10-35 Polytron (Kinematica) and centrifuged for 10 min at 1,500 × g at 4°C. To immunoaffinity purify NS1, the supernatant was incubated for 1 h at 4°C with 5 mg M-270 epoxy magnetic beads (Invitrogen Dynal) conjugated with 60 μg of a mixture of anti-NS1 MAbs (4NS1-10NS1-17NS1-23NS1, 1:1:1:1). After incubation, the beads were washed six times with 1 ml of lysis buffer followed by denaturing elution, as described previously (40). The isolated protein complexes were digested in solution with trypsin, using an optimized method of filter-aided sample preparation (23, 40, 48).
Tryptic peptides were analyzed by nanoscale liquid chromatography-tandem mass spectroscopy (nLC-MS/MS) using a Dionex Ultimate 3000 rapid separation liquid chromatography (RSLC) column coupled directly to an LTQ-Orbitrap Velos electron transfer dissociation (ETD) mass spectrometer (ThermoFisher Scientific) as described previously (40). The mass spectrometer was operated in data-dependent acquisition mode, with fragmentation of the top 20 most intense precursor ions by collision-induced dissociation (CID). MS/MS data were analyzed by Proteome Discoverer (version 1.3; ThermoFisher Scientific) and searched by a SEQUEST algorithm (version 1.20) against a protein sequence database compiled of WNV-KUNV protein sequences. Finally, the SEQUEST search results were refined by an X! Tandem algorithm using Scaffold (version Scaffold_3_00_04; Proteome Software). Filters for peptide identification were set to 99% protein confidence, 95% peptide confidence, and a minimum of two unique peptides per protein.
RESULTS
Growth phenotype of WNV NS1 RQ10NK revertants.
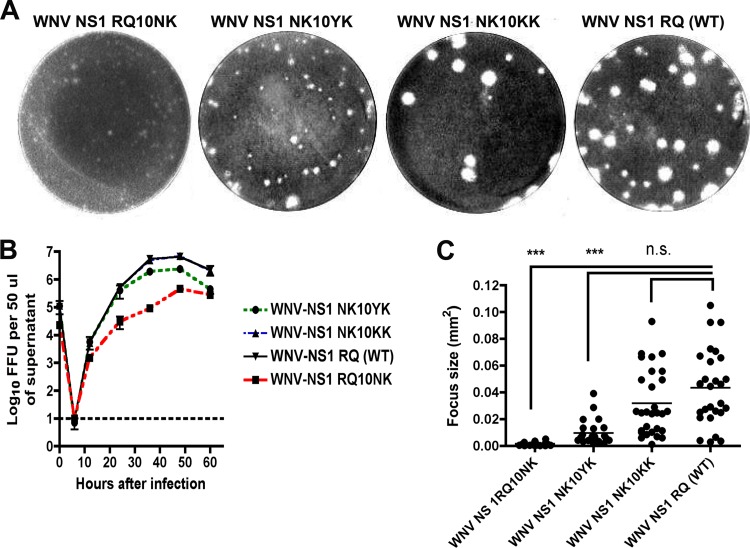
Although NS1 is an essential gene (21, 26), its cellular localization within the lumen of ER has made it difficult to understand how it contributes to negative-strand viral RNA synthesis (26). Previous work in our laboratory demonstrated that an N-terminal di-amino acid motif downstream of the signal sequence at positions 10 and 11 of WNV NS1 modulated its targeting and attenuated WNV replication. While recombinant WNV containing the RQ10NK mutation (RQ, wild type; NK, mutant) in NS1 was attenuated with decreased titers in single-step growth curves and small plaques in BHK21-15 cells (Fig. 1A and B) (49), the phenotype was unstable, as within two passages plaque size variants emerged, and after an additional passage, the growth curve and plaque size defect had reverted (data not shown).
Fig 1.
Comparison of plaque size and growth kinetics of WNV NS1 RQ10NK revertants. (A) WNV NS1 RQ10NK (left) was serially passaged in BHK21-15 cells. Within a few passages emerged a more rapidly growing small-plaque variant (WNV NS1 NK10YK, second left) and a large-plaque variant (WNV NS1 NK10KK, second right), which had a plaque size similar to that of the wild-type virus (WNV NS1 RQ, right). Sequences were generated after plaque purification and were representative of multiple isolates from each group. (B) Single-step growth kinetics of WNV NS1 RQ10NK and revertant viruses. WNV NS1 RQ10NK, WNV NS1 NK10YK, WNV NS1 NK10KK, and WNV NS1 RQ (WT) were added to BHK21-15 cells at an MOI of 1. At each of the indicated time points, virus in the supernatant was titrated by focus-forming assay. The data are the representative results of three independent experiments performed in triplicate. The growth of WNV NS1 NK10YK, WNV NS1 NK10KK, and WNV NS1 RQ (WT) was enhanced compared to that of WNV NS1 RQ10NK at 12, 24, 36, and 48 h (P < 0.05). Note that the growth curves for WNV NS1 NK10KK and WNV NS1 RQ (WT) are quite similar and difficult to distinguish on this graph. (C) Focus size comparison of WNV NS1 RQ10NK, WNV NS1 NK10YK, WNV NS1 NK10KK, and WNV NS1 RQ (WT). The data were obtained from several independent wells and were analyzed using a Biospot counter (Cellular Technology). Asterisks indicate statistical significance (***, P < 0.0001); n.s., not statistically significant. As described in Results, the WNV NS1 NK10YK revertant virus also contained a second site mutation in NS4B (F86C).
To begin to understand the basis for this reversion, 12 plaques of different sizes were selected at passage 3 of the WNV RQ10NK virus, and the NS1 genes were sequenced. These 12 clones were classified into two different groups. In group 1 (10 of 12 clones), the amino acid at position 10 of NS1 was changed back to a basic residue (NK10→KK or NK10→KQ) (NK = mutant; KK or KQ = revertant), as seen in wild-type WNV NS1 (which has an R at position 10). These revertants showed wild-type growth kinetics and large-plaque-size phenotypes, confirming the importance of a positive charge at amino acid 10 for optimal infectivity (Fig. 1A, B, and C). Subsequent full-genome sequencing of viral RNA from these clones revealed no additional mutations in any other genes. In group 2 (2 of 12 clones), the amino acid at position 10 was changed to a polar residue (NK10→YK), which resulted in near-normal growth kinetics. Nonetheless, while the plaque size was slightly larger than that of the parent attenuated WNV-RQ10NK, it was still smaller than that of wild-type WNV (WNV-WT). Full genome sequencing of the NK10YK revertants revealed a single additional mutation in the NS4B gene (F86C) in both of these clones.
RQ10NK and the RQ10→YK display similar NS1 targeting properties.
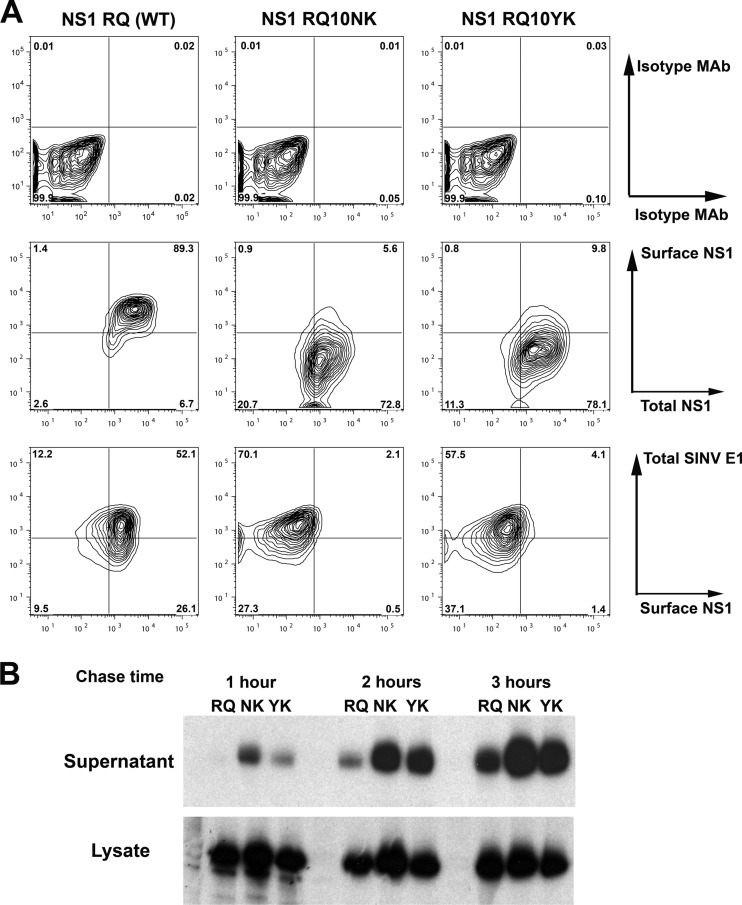
Our previous study with the WNV RQ10NK variant showed changes in NS1 targeting, with accelerated transit from the ER, decreased expression on the cell surface, and increased secretion and accumulation in the extracellular space (49). To define how the NK10→YK revertant mutation affected cellular targeting, we ectopically expressed NS1 using a SINV double genomic expression system, as we did previously with wild-type and RQ10NK NS1 (49), and analyzed surface and total (intracellular and surface) expression by flow cytometry (Fig. 2A). Although total NS1 cellular levels were similar between wild-type protein, RQ10NK, and RQ10YK NS1, surface expression was significantly lower (21-fold; P < 0.001; n = 3) in cells infected with SINV-RQ10NK and SINV-RQ10YK than in those infected with SINV-WT-NS1. As our prior study suggested that surface targeting of WNV NS1 was inversely linked to secretion into the extracellular space (49), we assessed whether this patterns also applied to RQ10YK NS1 using pulse-chase labeling experiments. BHK21-15 cells were infected with recombinant SINV expressing the wild-type or mutant NS1 at equivalent MOIs, and at 15 h postinfection, proteins were labeled with 35S and chased at 1, 2, or 3 h. Subsequently, wild-type and mutant NS1 proteins were immunoprecipitated from cell lysates and supernatants. Notably, NS1 RQ10YK behaved similarly to RQ10NK, and it was secreted into the extracellular space more rapidly and accumulated to higher levels (Fig. 2B).
Fig 2.
Ectopically expressed NS1 RQ10YK showed cellular targeting similar to that of NS1 RQ10NK. (A) BHK21-15 cells were infected with recombinant SINV (MOI of 10) ectopically expressing NS1 RQ (wild type), NS1 RQ10NK, or NS1 RQ10YK. At 14 h after infection, surface NS1 and total NS1 were measured by flow cytometry (middle panels) after sequentially incubating intact and permeabilized cells with different anti-NS1 MAbs (see Materials and Methods). Isotype controls also were performed (top panels). As an infectivity control, total SINV E1 expression level was measured using a polyclonal anti-E1 antibody (bottom panels). Contour plots are shown and are representative of three independent experiments. (B) Biosynthesis of wild-type and mutant NS1. BHK21-15 cells were infected at an MOI of 5 with SINV that ectopically expressed NS1 RQ (wild type), NS1 RQ10NK, or NS1 RQ10YK. At 15 h after infection, cells were placed in cysteine-methionine-free medium for 20 min, pulsed with [35S]cysteine-methionine, and then chased for 1, 2, or 3 h. Cell supernatants or lysates were harvested, immunoprecipitated with 4NS1 MAb and protein A-Sepharose beads, and electrophoresed. The data are the representative results of two independent experiments.
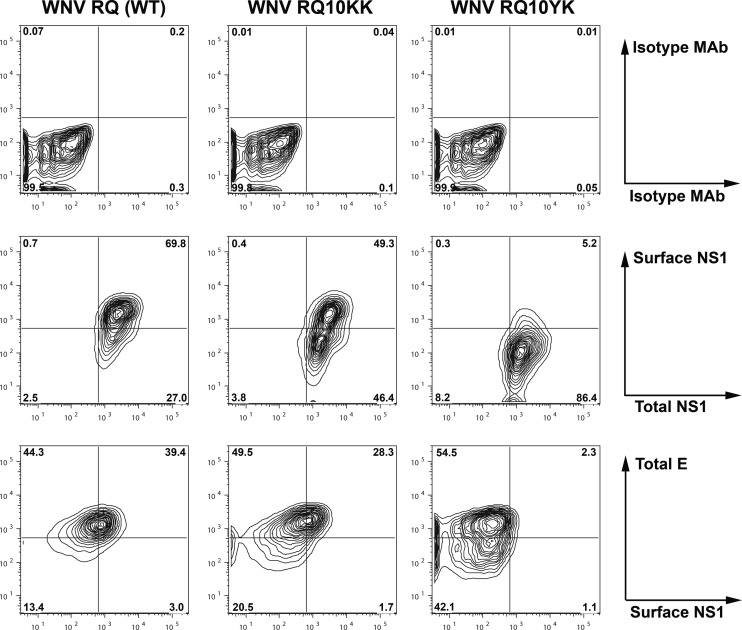
To confirm these findings in the context of WNV, total and cell surface expression levels of NS1 were compared after infection with WNV-WT and two revertant viruses, WNV-NK10→YK and WNV-NK10→KK. Cells were infected independently with all three viruses at an MOI of 0.1, and 30 h later, NS1 expression levels were assessed by flow cytometry. Although total NS1 expression levels were similar in cells infected with the three viruses, smaller amounts of NS1 were detected on the surface of cells infected with WNV-NK10→YK than on those infected with WNV-WT or WNV-NK10→KK (Fig. 3), confirming the results from the SINV expression system. Thus, from a targeting perspective, NS1 NK10→YK behaved similarly to NS1 RQ10NK.
Fig 3.
Cellular targeting of NS1 in the context of infection by revertant WNV. BHK21-15 cells were infected at an MOI of 1 with WNV NS1 RQ (wild type) or the revertants (WNV NS1 RQ10KK or WNV NS1 RQ10YK) generated by passage (Fig. 1), and the cellular targeting of NS1 was analyzed. At 30 h after infection, surface and total NS1 levels were measured by flow cytometry (middle panels) after sequentially incubating intact and permeabilized cells with different anti-NS1 MAbs. Isotype controls also were performed (top panels). As an infectivity control, total WNV E expression level was measured using a specific MAb (WNV E16) (bottom panels). Contour plots are shown and are representative of results from three independent experiments.
An F86C mutation in NS4B is specifically required for growth of WNV with mutant NS1.
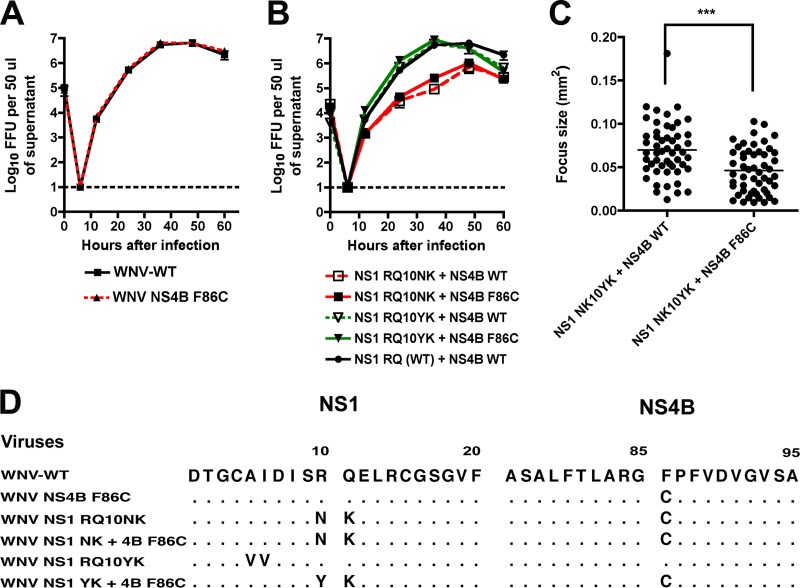
While our sequencing data suggested that the F86C mutation in NS4B might compensate for the loss-of-function NS1 mutation (NK10→YK) in the revertant virus, it remained possible that the mutation was coincidental and unrelated. To address this, we assessed whether the WNV-NS4B F86C mutation conferred a growth advantage for wild-type WNV. Notably, introduction of the NS4B mutation into the WNV-WT did not enhance replication (Fig. 4A), suggesting it was not an adaptive mutation. To establish whether NS4B F86C mutation could compensate for the loss-of-function NS1 variants, we cloned it into the background of NS1 RQ10NK or NS1 RQ10YK. Somewhat surprisingly, the addition of NS4B F86C failed to enhance the replication kinetics of recombinant viruses containing either RQ10NK or RQ10YK in NS1 (Fig. 4B); however, three unusual findings were apparent from these studies: (i) viruses containing NS1-RQ10YK grew to wild-type levels regardless of the introduction of the NS4B mutation, which seemed to conflict with our revertant virus selection data (Fig. 1B); (ii) viruses engineered to have NS1-RQ10YK and NS4B-F86C had smaller foci (0.05 ± 0.003 versus 0.07 ± 0.004, P < 0.0001) than those designed to have NS1-RQ10YK and NS4B-WT (Fig. 4C); and (iii) viruses containing NS1-RQ10NK were attenuated regardless of the introduction of the NS4B mutation (Fig. 4B). Because of these apparent discrepancies, we sequenced viral RNA in all of the P0 stocks. Notably, we found that recombinant viruses designed to have mutant NS1 (RQ10NK or RQ10YK) and NS4B WT had accumulated second site mutations in NS1 or spontaneous mutations in NS4B at position F86C (Fig. 4D). For example, introduction of NS1 RQ10YK by itself was unstable such that the P0 virus stock recovered from the supernatant showed reversion to the wild-type RQ sequence at positions 10 and 11, along with two additional mutations (NS1 AI56VV). Introduction of NS1 RQ10NK by itself resulted in virus that spontaneously acquired the F86C mutation in NS4B; despite several independent attempts and multiple rounds of sequencing, we never recovered a recombinant RQ10NK virus that lacked the F86C mutation, even though we engineered in a WT NS4B gene. Thus, the loss-of-function mutations in NS1 in isolation were unstable in the background of the infectious clone and rapidly were complemented by second site mutations in NS1 or NS4B during the initial virus stock generation. Although viruses containing the RQ10NK change in NS1 were stabilized by the F86C mutation in NS4B, they remained attenuated.
Fig 4.
The F86C mutation in NS4B is specifically required for growth of WNV with mutant NS1. (A) Single-step growth kinetics of WNV NS1 RQ (wild type) and WNV NS1 RQ + NS4B F86C were compared. All viruses were generated from a two-plasmid infectious clone of WNV as detailed in Materials and Methods. BHK21-15 cells were infected at an MOI of 5, and at the indicated time points, virus was harvested from supernatants for titration by focus-forming assay. The data are the representative results of three independent experiments performed in triplicate. (B) Single-step growth kinetics of WNV containing NS1 RQ10NK plus NS4B WT, NS1 RQ10NK plus NS4B F86C, NS1 RQ10YK plus NS4B WT, NS1 RQ10YK plus NS4B F86C, and the parent NS1 RQ (WT) plus NS4B WT. All viruses were generated from an infectious clone of WNV and infected and titrated as described for panel A. The data are representative of results of three independent experiments performed in triplicate, and viruses containing NS1 RQ10NK were statistically different at several time points after infection. (C) Focus size comparison of WNV engineered to have NS1 RQ10YK plus NS4B WT or NS1 RQ10YK plus NS4B F86C. The data are pooled from several independent experiments, and differences were statistically significant (***, P < 0.0001) (D) Amino acid sequence alignment after sequencing of viral RNA harvested from the indicated P0 stocks that were generated after transfection into BHK21-15 cells of in vitro transcribed WNV RNA. Adaptive or stabilizing mutations are explicitly identified. Note that no adventitious mutations were observed in the template cDNA or the in vitro transcribed RNA prior to electroporation.
NS1-RQ10NK has a dominant negative phenotype, which can be overcome by the WNV-NS4B F86C virus.
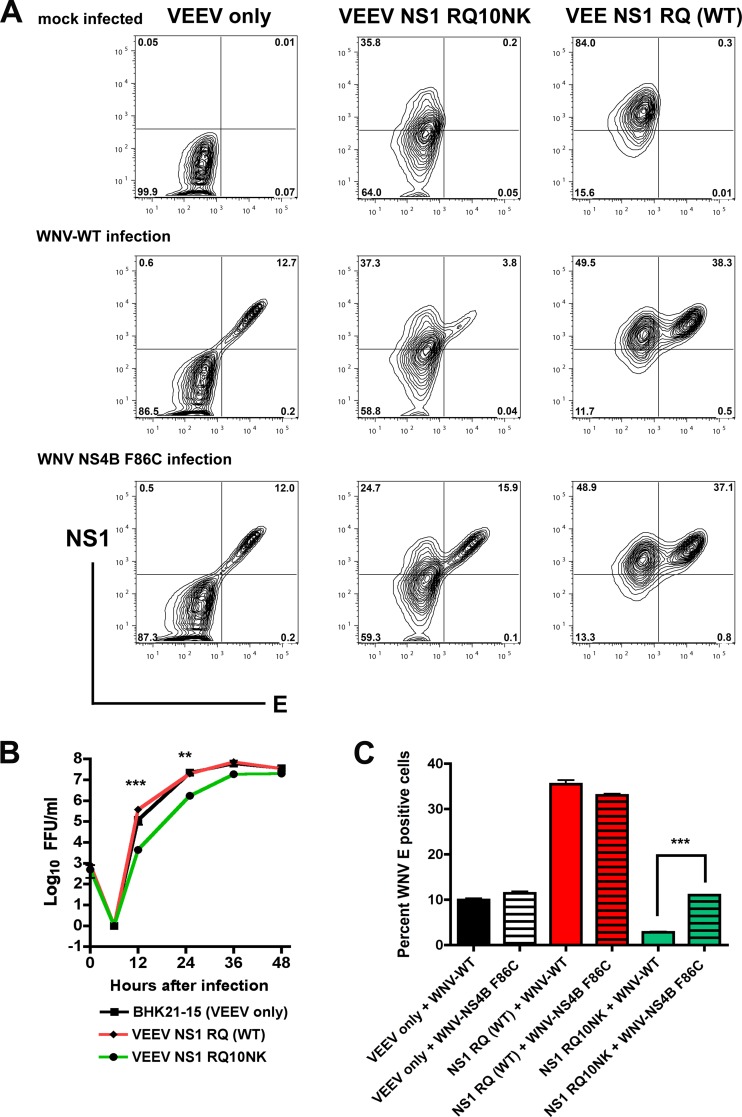
Given that NS1 oligomerizes in the ER, and that WNV-RQ10NK virus was unstable and attenuated in its replication, we evaluated whether expression of this mutant NS1 would alter the infectivity of WNV-WT. BHK21-15 cell lines propagating a VEEV replicon (36) that ectopically expressed wild-type NS1, RQ10NK NS1, or no viral protein (NS1 with an N-terminal stop mutation) were infected with WNV-WT, and viral yield was measured by focus-forming assay on untransfected BHK21-15 cells. WNV-WT propagated in cells expressing RQ10NK NS1 showed a significant growth defect (∼70-fold; P < 0.05) at 12 h compared to cells expressing WT NS1 or the VEEV replicon that did not produce NS1 protein (Fig. 5B). By later times (at 36 h postinfection and beyond), this difference waned, presumably due to accumulation of WT NS1 from WNV-WT. This growth defect was corroborated independently by flow cytometric analysis of the WNV E protein in cells ectopically expressing the WT or RQ10NK form of NS1 (Fig. 5A and C). This dominant negative effect of NS1-RQ10NK, however, was lost in cells infected with WNV NS4B F86C. While cells expressing NS1-RQ10NK and infected with WNV-WT showed less (3.5-fold; P < 0.0001) E protein expression at 24 h by flow cytometry compared to cells expressing the VEEV replicon that did not produce NS1 protein, this difference was abolished (P > 0.3) when WNV-NS4B F86C was used (Fig. 5A and C). Interestingly, ectopic expression of WT NS1 enhanced infection slightly of both WNV-WT and WNV-NS4B F86C at early time points; this manifested as a 2.5-fold increase in viral titer at 12 h and a 3.5-fold increase in E protein expression at 24 h (Fig. 5B and C; P < 0.05). This enhanced infectivity may be related to the effects of soluble NS1, as a prior study showed that preincubation of cells with purified NS1 increased DENV infection in hepatocytes (1).
Fig 5.
Ectopically expressed NS1 RQ10NK acts as a dominant negative for infection of WNV-WT but not WNV containing the NS4B F86C suppressor mutation. (A) BHK21-15 cells propagating VEEV subgenomic replicons encoding NS1 with an N-terminal stop codon (VEEV only), NS1 RQ10NK, or NS1 RQ (wild type) were mock infected (top panels) or superinfected at an MOI of 1 with WNV-WT (middle panels) or WNV containing the NS4B F86C suppressor mutation (WNV NS4B F86C, bottom panels), both of which were generated from the infectious cDNA clone. At 12 h after infection, cells were fixed and permeabilized, and total levels of NS1 and E protein expression were assessed by flow cytometry after immunostaining with 4NS1 or humanized WNV-E16 MAbs. (B) Viral growth curve analysis. BHK21-15 cells (mock infected) or BHK21-15 cells propagating VEEV subgenomic replicons as listed above were mock infected or superinfected at an MOI of 10 with WNV-WT or WNV NS4B F86C. At the indicated time points, virus was harvested from the supernatant and titrated by focus-forming assay. The data are the average of three independent experiments performed in duplicate, and the asterisks indicate statistically significant differences between viruses grown in cells expressing NS1 RQ10NK and NS1 RQ (wild type). (C) Summary of E protein expression by flow cytometry after infection of WNV-WT or WNV-NS4B F86C in cells propagating VEEV replicons with an N-terminal stop codon of NS1 (VEEV only), NS1 RQ10NK, or NS1 RQ (wild type). Results are averaged from three independent experiments, error bars denote standard error of the mean, and asterisks indicate statistically significant differences (***, P < 0.0001).
To confirm the dominant negative effect of NS1-RQ10NK, we measured viral RNA levels by qRT-PCR in cells expressing NS1-WT or NS1-RQ10NK and infected with WNV-WT or WNV NS4B F86C. At 12 h after WNV-WT infection, positive-strand viral RNA levels were lower in cells expressing NS1-RQ10NK than in those expressing NS1-WT or no NS1 protein (Fig. 6A), establishing that NS1-RQ10NK NS1 could trans-dominantly and negatively influence viral RNA accumulation. This negative effect of NS1-RQ10NK, however, was lost in cells infected with WNV NS4B F86C (Fig. 6B). Thus, the RQ10NK mutation of NS1 results in a trans-dominant negative effect on replication of WNV-WT, a phenotype that was complemented by the F86C mutation in NS4B.
Fig 6.

Dominant negative effect of NS1 RQ10NK mutation on WNV RNA replication is rescued by the NS4B F86C suppressor mutation. BHK21-15 cells propagating VEEV replicon expressing NS1 RQ (wild type) or NS1 RQ10NK NS1 were superinfected with WNV-WT (A) or WNV NS4B F86C (B) at an MOI of 10. At 3, 6, and 12 h after infection, total RNA was extracted from cells and WNV RNA was measured by real-time quantitative RT-PCR and compared to 18S rRNA. A standard curve of 18S rRNA per BHK21-15 cell was used for normalization. The data are the average of results from three independent experiments performed in duplicate, and asterisks indicate statistically significant differences (**, P < 0.01; ***, P < 0.0001).
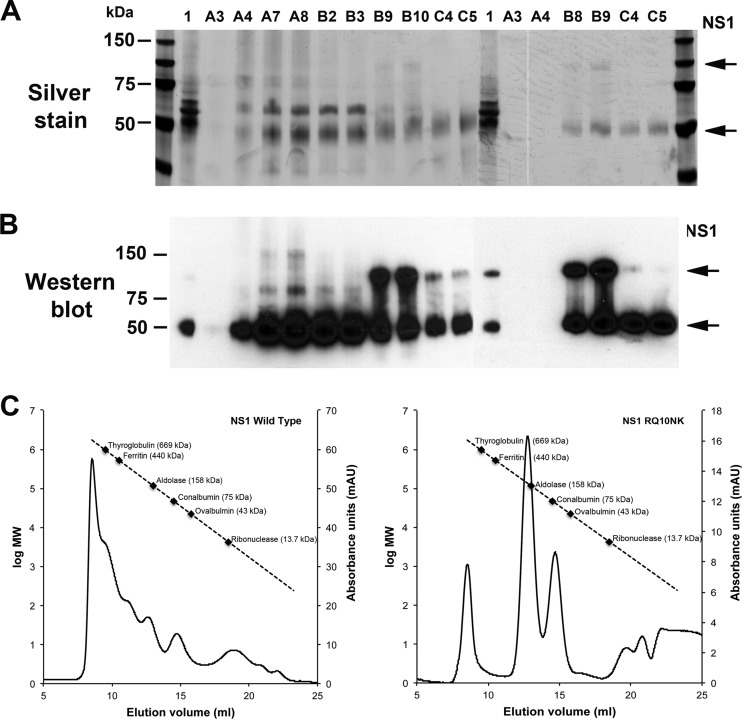
A prior study showed that preincubation of hepatoma cells with soluble hexameric DENV NS1 increased the production of infectious DENV (1). These authors suggested that accumulation of soluble NS1 in the late endosomes could potentiate DENV infection through an uncharacterized mechanism. Because NS1-RQ10NK trans-dominantly and negatively affected WNV replication yet was associated with enhanced levels of NS1 secretion (Fig. 2) (49), we evaluated the effect of this mutation on the oligomeric forms of secreted NS1. We purified WT and RQ10NK NS1 by antibody affinity and size exclusion chromatography from supernatants of BHK21-15 cell lines propagating a VEEV replicon that ectopically expressed the WT and variant NS1 proteins. While both proteins were readily purified and antigenically intact (Fig. 7A and B), the distribution of oligomers was distinct: WT WNV NS1 was observed as a dodecamer, hexamer, tetramer, and dimer as described previously (3, 9), whereas RQ10NK NS1 was present primarily as a tetramer and dimer, with little of the higher order forms (Fig. 7C). Thus, the RQ10NK mutation appears to affect the oligomeric assembly of NS1 in supernatant, which could contribute to effects on viral replication.
Fig 7.
Oligomeric forms of soluble WT and RQ10NK NS1 after purification. WT and RQ10NK NS1 were purified by sequential antibody affinity (9NS1 Sepharose) and size exclusion (Superdex-200) chromatography from supernatants of BHK21-15 cell lines propagating a VEEV replicon that ectopically expressed the WT and variant NS1 proteins. Fractions corresponding to the peaks of the size exclusion chromatography were subjected to SDS-PAGE and silver staining (A) or Western blotting (B) with 10NS1 MAb. Fractions A7 and A8, higher-order NS1 oligomers (WT only); B2 and B3, hexamer (WT only); B8 to B10, tetrameric form; and C4 and C5, dimer. (C) Based on a calibration curve (standards ranging from 13.7 to 669 kDa), the approximate molecular sizes were determined and are indicated in the chromatography tracings.
WNV NS1 interacts with NS4B.
While the above results suggested a genetic interaction between NS1 and NS4B, they did not provide insight as to how the RQ10NK mutation altered viral replication. We hypothesized that a direct physical interaction between NS1 and the transmembrane protein NS4B could link processes in the lumen and the cytosolic face of the ER, the latter the site of viral replication (17, 28, 29). To begin to assess this, BHK21-15 cells were mock infected or infected with WNV-WT at an MOI of 10. At 28 h after infection, cells were lysed and NS1 was immunoprecipitated with a monoclonal antibody against NS1. Western blotting demonstrated the coisolation of NS1 with WNV NS4B. The ∼27-kDa band that was recognized by the anti-NS4B polyclonal antibody was detected only in anti-NS1 Sepharose bead immunoprecipitates from WNV-infected but not mock-infected cells (Fig. 8A). As an independent confirmation, we performed mass spectrometry on proteins that were immunoaffinity purified by anti-NS1 MAb-conjugated magnetic beads from cells infected with WNV-KUNV or mock infected (Fig. 8B); this strain was used because of its lower biosafety level status, which enabled the biochemical analysis to be performed. To obtain an effective isolation of NS1 and focus on strong interactions, we selected a stringent lysis buffer containing 1% Triton X-100, 0.5% sodium deoxycholate, and 0.3% sodium N-lauroylsarcosine. Based on mass spectrometry of NS1 immunopurifications, we identified peptides for prM/M, E, NS2A, NS2B, NS3, NS4B, and NS5 proteins (Fig. 8C). To determine whether these viral proteins are associated with NS1 as mature forms, and not as part of the polyprotein, we searched for the presence of unique semitryptic peptides at either the N or C terminus that represent polyprotein cleavage sites. We confirmed the presence of prM/M, NS3, and NS4B as mature proteins. As illustrated in Fig. 8D, NS4B was identified with high confidence by collision-induced dissociation (CID) MS/MS from anti-NS1 immunoprecipitates from WNV-KUNV but not mock-infected cells. Thus, using two independent approaches, we show that WNV NS1 and NS4B can interact physically within infected cells.
Fig 8.
Physical interaction between NS1 and NS4B. (A) Coimmunoprecipitation and Western blotting. BHK21-15 cells were mock infected or infected with WNV-WT at an MOI of 10. At 28 h after infection, cells were lysed and NS1 proteins were immunoprecipitated with anti-NS1 MAb and protein A-Sepharose. After extensive washing, beads were boiled in SDS sample buffer, and the eluate was electrophoresed and subjected to Western blotting with a polyclonal anti-NS4B antibody. (B to D) Mass spectrometry (MS) identifies an interaction between WNV NS1 and NS4B. (B) Workflow for the immunoaffinity isolation of NS1 protein complexes. (C) Summary of viral proteins identified by mass spectrometry (nLC-MS/MS using an LTQ Orbitrap Velos) as coisolated with NS1 in WNV-KUNV-infected cells. The number of unique peptides and identifications of boundary peptides that indicate mature forms are shown. Three viral proteins coisolated with NS1 and validated as mature proteins are shown in bold. M, molecular mass. (D) Representative collision-induced dissociation (CID) MS/MS spectrum of a tryptic peptide of NS4B demonstrating its association with WNV NS1.
DISCUSSION
Flavivirus NS1 is a glycosylated nonstructural protein that oligomerizes in the ER lumen, is expressed on the plasma membrane of infected cells, and is secreted into the extracellular space. NS1 also binds back to uninfected cells and antagonizes complement function in solution and on the plasma membrane surface. Additionally, even though NS1 is localized to the lumen of the ER and viral replication occurs on the cytosolic face of the ER, NS1 is an essential gene that regulates early viral RNA replication (21, 26) through mechanisms that remain to be defined. Previously, we identified a short N-terminal di-amino acid motif that modulated cellular targeting of NS1 and affected replication (49). In the current study, we passaged an attenuated WNV containing NS1 RQ10NK in culture and identified revertant viruses containing a mutation (F86C) in the viral NS4B transmembrane protein, which spans the luminal and cytoplasmic faces of the ER. Reverse genetic analysis confirmed that the F86C change in NS4B was not an adaptive mutation per se, as it failed to enhance replication when introduced into the backbone of the wild-type WNV. However, the F86C substitution in NS4B was required for efficient replication of WNV containing loss-of-function mutations in NS1 lacking a key basic residue at position 10. This became most apparent in studies with recombinant WNV, in which introduction of the loss-of-function mutations in NS1 alone was unstable and rapidly complemented by second site mutations in either NS1 or NS4B. Mechanistic experiments revealed that RQ10NK mutation of NS1 had a trans-dominant negative effect on viral replication and RNA accumulation, and this phenotype was reversed by the F86C mutation in NS4B. As immunoprecipitation and mass spectrometry studies established a physical linkage between NS1 and NS4B, our data suggest that NS1 within the ER lumen may regulate viral RNA replication by associating with the transmembrane protein NS4B, which has direct proximity to the replication complex on the cytosolic face of the ER membrane (31) and can modulate its activity (18, 37).
Our prior study suggested that the rate of NS1 transit from the ER to the extracellular space negatively correlated with viral replication (49). Because mutations that caused more rapid egress from the ER had lower infectivity, we postulated that ER retention of NS1 was a critical factor regulating flavivirus replication. However, experiments with the NK→YK revertant dissociate the ER retention of the NS1 phenotype from viral replication. The NK→YK virus showed an enhanced secretion rate of NS1 but near-wild-type multistep growth kinetics, although a small-sized-plaque phenotype was observed. Thus, ER retention of NS1 per se does not likely affect the RNA replication; more likely, NS1 within the ER must interact with other proteins to transmit key signals that regulate flavivirus genome amplification.
Ectopic expression of RQ10NK NS1 in BHK21-15 cells using a VEEV replicon had a trans-dominant negative effect on replication of wild-type WNV. This observation was interesting from a functional perspective because it suggests that the interaction in the ER lumen that facilitates flavivirus RNA replication might utilize an oligomeric (e.g., dimer or hexamer) form of NS1. The RQ10NK variant oligomer may be inherently less efficient at this function and thus, by the formation of mixed oligomers (wild type plus RQ10NK), the efficiency of replication, and the dominant negative phenotype are observed. Indeed, we observed fewer higher-order oligomer species, including hexamers, in RQ10NK compared to WT NS1. The viral growth kinetics supported this model, as the replication defect was observed at early time points (e.g., 12 h) when the amount of RQ10NK NS1 exceeded that of WT NS1, but at later time points (e.g., beyond 24 h) when the amount of WT NS1 exceeds that of RQ10NK NS1, the relative fraction of homogenous wild-type NS1 oligomers with greater functional activity should increase. Soluble hexameric NS1 has been proposed to augment DENV infection in hepatoma cells through an undefined mechanism (1). Consistent with this hypothesis, a point mutation in NS1 of WNV-KUNV and Murray Valley encephalitis virus that reduced the efficiency of dimerization also resulted in attenuated viral growth kinetics in vitro and in vivo (11). Alternatively, at early time points when it is present in greater abundance in the replicon cells, the RQ10NK homo-oligomer could compete with the WT NS1 homo-oligomer for a limiting host or viral factor, which results in impaired replication.
The topology of flavivirus NS4B remains uncertain, although protease protection studies suggest that after the 2K-signal sequence, the N-terminal region lies within the lumen of the ER and is followed by three transmembrane domains with alternating peptide loops on the cytoplasmic and ER faces (31). This model places the F86 amino acid residue within the ER lumen, in a location such that it could interact with newly synthesized NS1 prior to its transit through the secretory pathway. NS4B also has been reported to have antagonist activity for the type I interferon (IFN) signaling cascade (2, 14, 32). Whereas the F86C mutation was not adaptive in BHK21-15 cells when it was introduced into the WNV-WT infectious clone, we observed attenuated growth kinetics in wild-type mouse embryonic fibroblasts, and this defect was rescued in cells with a genetic deficiency of the type I IFN receptor (S. Youn and M. Diamond, unpublished results); thus, while mutation at F86 of NS4B can rescue viruses with mutant NS1 in vitro, it may be at the expense of another function (e.g., IFN antagonism) which is not evident in BHK21-15 cells lacking intact cell-intrinsic antiviral immune responses. As such, proteomic studies with NS4B-WT and NS4B-F86C are planned in cells sufficient and deficient for type I IFN signaling to identify binding partners that may define the mechanism of IFN antagonism. While ectopic expression of NS1 has not been reported to directly antagonize IFN functions, it appears to inhibit signaling through TLR3 (44).
Although our study establishes a genetic and physical interaction between NS1 and NS4B, it remains uncertain whether the linkage is direct or indirect. NS1 has been reported to interact with NS4A (25) and the viral structural proteins (7, 45) and to colocalize by immunofluorescence with NS2B and NS3 (43). Thus, it is plausible that other viral or host proteins may mediate the physical interaction between NS1 and NS4B; indeed four other viral nonstructural proteins (NS2A, NS2B, NS3, and NS5) were detected by mass spectrometry analysis in NS1 immunoprecipitates, one other of which was confirmed (NS3) as being a mature protein. Prior studies with YFV suggested that an interaction between NS1 and NS4A, another transmembrane viral protein that spans the ER membrane, could integrate NS1 signals for RNA replication occurring in the cytoplasm (25). While this study provided compelling evidence for a genetic interaction based on the identification of a suppressor mutation in NS4A for an attenuated YFV strain, no direct interaction between NS1 and NS4A was reported, although the two viral proteins cosediment with membrane fractions enriched in RNA-dependent RNA polymerase activity (8). While it is conceivable that the two viral ER transmembrane proteins NS4A and NS4B function together to transmit a signal from NS1 to the cytoplasm, our mass spectrometry experiments after NS1 immunoprecipitation identified peptides corresponding to NS4B but not NS4A. While direct binding assays (e.g., enzyme-linked immunosorbent assay [ELISA] or surface plasmon resonance) could in theory address this question, it has proven difficult to produce a recombinant NS4A or NS4B protein with the correct topology and folding.
So how does a sequence change in NS4B complement the attenuating mutation in NS1? This remains uncertain, although we speculate that an amino acid change in NS4B affects direct binding to NS1 or binding to a partner protein that interacts with NS1. We believe this interaction most likely occurs in the lumen of ER given the location of nascently synthesized NS1 (24). While it is theoretically possible that some “misfolded” NS1 could enter the cytoplasm via the retrotranslocation machinery (41), preliminary experiments have failed to detect NS1 (ubiquitinated or nonubiquitinated) in cytoplasmic fractions by Western blotting with MAbs recognizing linear epitopes or observe effects on WNV replication after silencing of genes associated with ER-associated degradation (Youn and Diamond, unpublished). More detailed mass spectrometry experiments are planned with WNV expressing wild-type and RQ10NK NS1 and/or wild-type and F86C NS4B to define how these sequence changes impact the interactome with other viral and host proteins. In summary, our studies establish a novel genetic and physical interaction between NS1 and NS4B, which suggests a mechanism for how luminal NS1 can transmit a signal to the cytoplasmic face of the ER to regulate WNV replication.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Klimstra for the SINV expression constructs, I. Frolov for the VEEV replicons, W. I. Lipkin for the anti-NS4B antibody, and S. Johnson for the mouse-human chimeric 17NS1.
This work was supported by the Pediatric Dengue Vaccine Initiative and by a grant from the Midwest Regional Centers of Excellence for Biodefense and Emerging Infectious Disease Research (U54-AI057160) to M.S.D. and by NIH NIDA grant DP1DA026192 and HFSPO award RGY0079/2009-C to I.M.C.
Footnotes
Published ahead of print 2 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alcon-LePoder S, et al. 2005. The secreted form of dengue virus nonstructural protein NS1 is endocytosed by hepatocytes and accumulates in late endosomes: implications for viral infectivity. J. Virol. 79:11403–11411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 85:2723–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Avirutnan P, et al. 2010. Antagonism of the complement component C4 by flavivirus non-structural protein NS1. J. Exp. Med. 207:793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avirutnan P, et al. 2011. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J. Immunol. 187:424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avirutnan P, et al. 2007. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 3:e183 doi:10.1371/journal.ppat.0030183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beasley DW, et al. 2005. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J. Virol. 79:8339–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blitvich BJ, Mackenzie JS, Coelen RJ, Howard MJ, Hall RA. 1995. A novel complex formed between the flavivirus E and NS1 proteins: analysis of its structure and function. Arch. Virol. 140:145–156 [DOI] [PubMed] [Google Scholar]
- 8. Chu PW, Westaway EG. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177–191 [DOI] [PubMed] [Google Scholar]
- 9. Chung KM, et al. 2006. West Nile virus non-structural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc. Natl. Acad. Sci. U. S. A. 103:19111–19116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung KM, et al. 2006. Antibodies against West Nile virus non-structural (NS)-1 protein prevent lethal infection through Fc gamma receptor-dependent and independent mechanisms. J. Virol. 80:1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clark DC, et al. 2007. In situ reactions of monoclonal antibodies with a viable mutant of Murray Valley encephalitis virus reveal an absence of dimeric NS1 protein. J. Gen. Virol. 88:1175–1183 [DOI] [PubMed] [Google Scholar]
- 12. Cristea IM, et al. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 281:30269–30278 [DOI] [PubMed] [Google Scholar]
- 13. Crooks AJ, Lee JM, Easterbrook LM, Timofeev AV, Stephenson JR. 1994. The NS1 protein of tick-borne encephalitis virus forms multimeric species upon secretion from the host cell. J. Gen. Virol. 75(Pt 12):3453–3460 [DOI] [PubMed] [Google Scholar]
- 14. Evans JD, Seeger C. 2007. Differential effects of mutations in NS4B on West Nile virus replication and inhibition of interferon signaling. J. Virol. 81:11809–11816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flamand M, et al. 1999. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J. Virol. 73:6104–6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fuchs A, Pinto AK, Schwaeble WJ, Diamond MS. 2011. The lectin pathway of complement activation contributes to protection from West Nile virus infection. Virology 412:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 84:10438–10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grant D, et al. 2011. A single amino acid in nonstructural protein NS4B confers virulence to dengue virus in AG129 mice through enhancement of viral RNA synthesis. J. Virol. 85:7775–7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gutsche I, et al. 2011. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. U. S. A. 108:8003–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacobs MG, Robinson PJ, Bletchly C, Mackenzie JM, Young PR. 2000. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction. FASEB J. 14:1603–1610 [DOI] [PubMed] [Google Scholar]
- 21. Khromykh AA, Sedlak PL, Westaway EG. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khromykh AA, Sedlak PL, Westaway EG. 1999. trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J. Virol. 73:9247–9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kramer T, Greco TM, Enquist LW, Cristea IM. 2011. Proteomic characterization of pseudorabies virus extracellular virions. J. Virol. 85:6427–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindenbach BD, Rice CM. 2001. Flaviviridae: the viruses and their replication, p 991–1041 In Knipe DM, Howley PM. (ed), Fields virology, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Lindenbach BD, Rice CM. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 73:4611–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindenbach BD, Rice CM. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo Y, Li T, Yu F, Kramer T, Cristea IM. 2010. Resolving the composition of protein complexes using a MALDI LTQ Orbitrap. J. Am. Soc. Mass Spectrom. 21:34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mackenzie JM, Jones MK, Young PR. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232–240 [DOI] [PubMed] [Google Scholar]
- 29. Mackenzie JM, Kenney MT, Westaway EG. 2007. West Nile virus strain Kunjin NS5 polymerase is a phosphoprotein localized at the cytoplasmic site of viral RNA synthesis. J. Gen. Virol. 88:1163–1168 [DOI] [PubMed] [Google Scholar]
- 30. Medigeshi GR, et al. 2007. West Nile virus infection activates the unfolded protein response leading to CHOP induction and apoptosis. J. Virol. 81:10849–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller S, Sparacio S, Bartenschlager R. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854–8863 [DOI] [PubMed] [Google Scholar]
- 32. Munoz-Jordan JL, et al. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Noisakran S, et al. 2008. Association of dengue virus NS1 protein with lipid rafts. J. Gen. Virol. 89:2492–2500 [DOI] [PubMed] [Google Scholar]
- 34. Noisakran S, et al. 2007. Characterization of dengue virus NS1 stably expressed in 293T cell lines. J. Virol. Methods 142:67–80 [DOI] [PubMed] [Google Scholar]
- 35. Oliphant T, et al. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petrakova O, et al. 2005. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in mammalian cells. J. Virol. 79:7597–7608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Puig-Basagoiti F, et al. 2007. A mouse cell-adapted NS4B mutation attenuates West Nile virus RNA synthesis. Virology 361:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Samuel MA, et al. 2006. PKR and RNAse L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sejvar JJ, et al. 2003. Neurologic manifestations and outcome of West Nile virus infection. JAMA 290:511–515 [DOI] [PubMed] [Google Scholar]
- 40. Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. 2012. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol. Cell. Proteomics 11:M111.015156. doi:10.1074/mcp.M111.015156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vembar SS, Brodsky JL. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vogt MR, et al. 2009. Human monoclonal antibodies induced by natural infection against West Nile virus neutralize at a postattachment step. J. Virol. 83:6494–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilson JR, de Sessions PF, Leon MA, Scholle F. 2008. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 82:8262–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Winkelmann ER, Widman DG, Suzuki R, Mason PW. 2011. Analyses of mutations selected by passaging a chimeric flavivirus identify mutations that alter infectivity and reveal an interaction between the structural proteins and the nonstructural glycoprotein NS1. Virology 421:96–104 [DOI] [PubMed] [Google Scholar]
- 46. Winkler G, Maxwell SE, Ruemmler C, Stollar V. 1989. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology 171:302–305 [DOI] [PubMed] [Google Scholar]
- 47. Winkler G, Randolph VB, Cleaves GR, Ryan TE, Stollar V. 1988. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology 162:187–196 [DOI] [PubMed] [Google Scholar]
- 48. Wisniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat. Methods 6:359–362 [DOI] [PubMed] [Google Scholar]
- 49. Youn S, Cho H, Fremont DH, Diamond MS. 2010. A short N-terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition. J. Virol. 84:9516–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.